Abstract
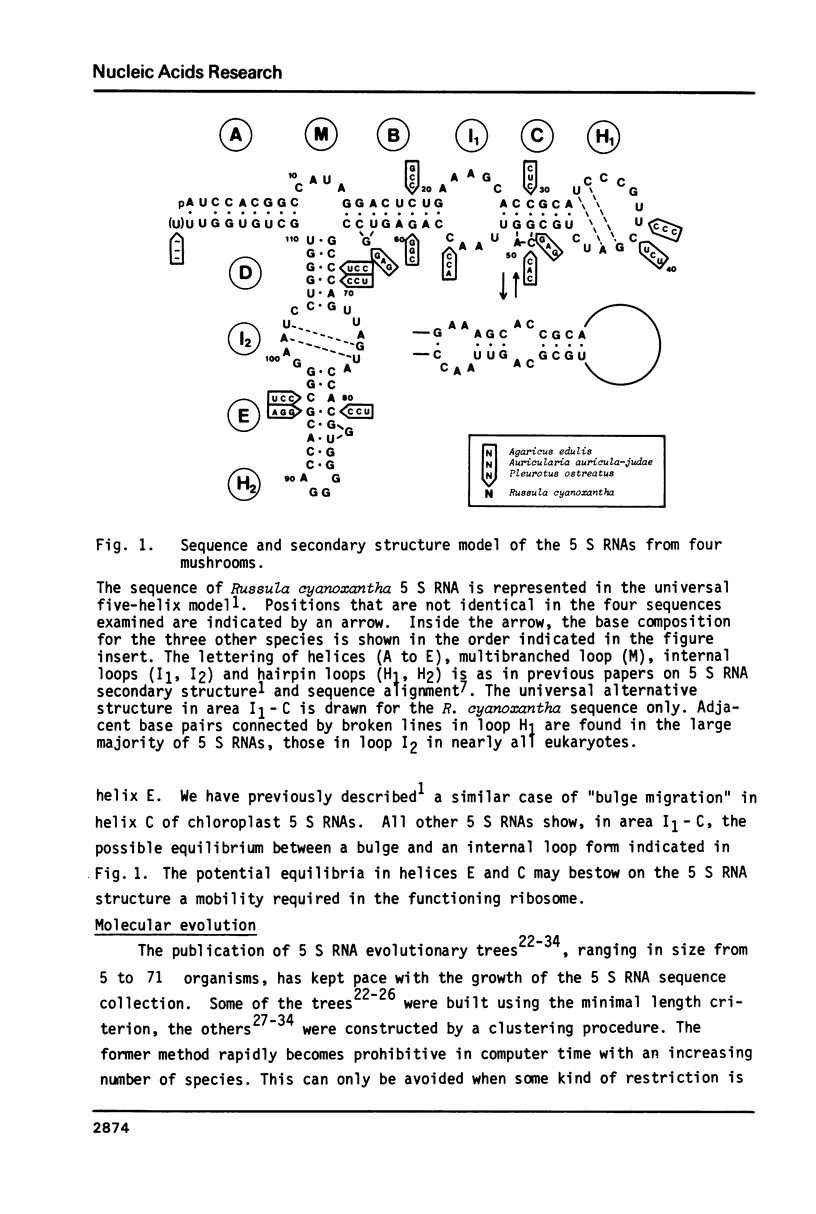
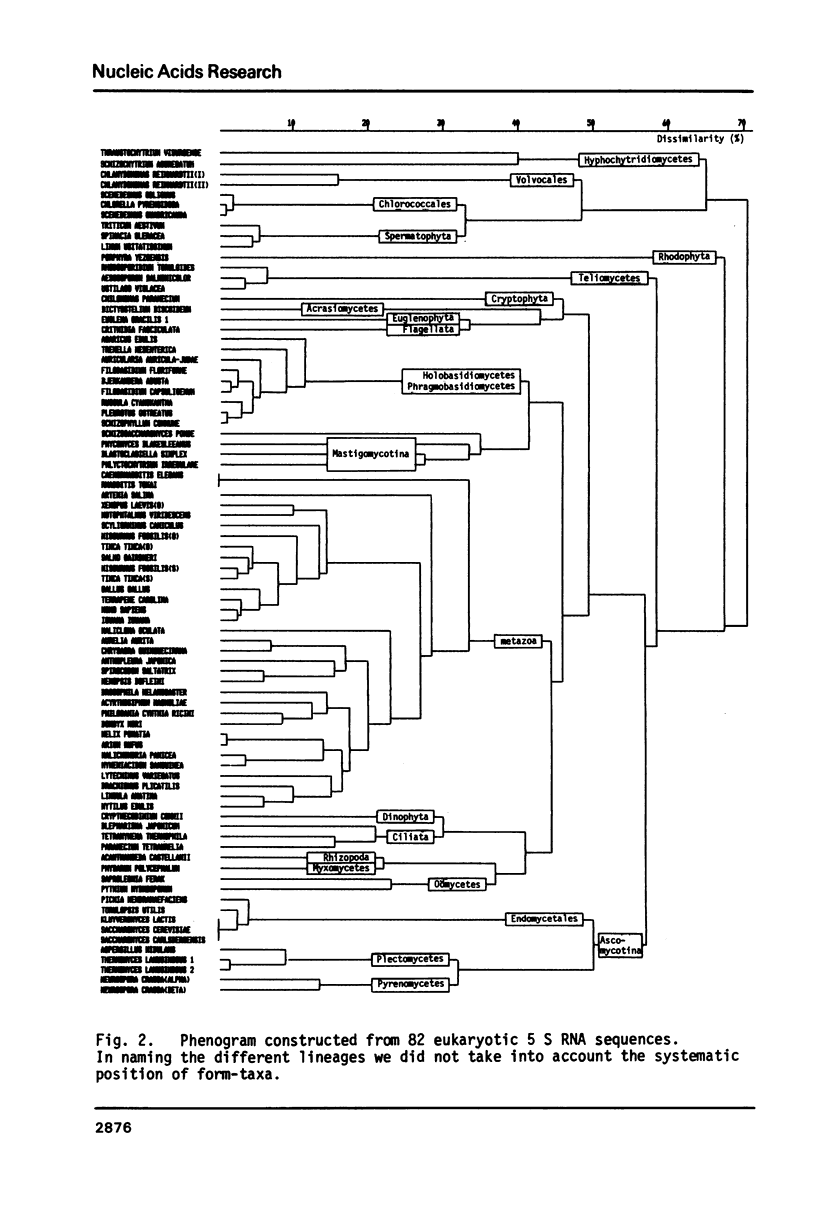
The nucleotide sequences of the 5 S ribosomal RNAs of the mushrooms Russula cyanoxantha, Pleurotus ostreatus, Agaricus edulis, and Auricularia auricula-judae were determined. The sequences fit in a universal five-helix secondary structure model for 5 S RNA. As in most other 5 S RNAs, some helical areas contain non-standard base pairs. A clustering method was used to reconstruct an evolutionary tree from 82 eukaryotic 5 S RNA sequences. It allows to make a choice between alternative systematic classifications for basidiomycetes and reveals that the fungal kingdom is highly polyphyletic.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dams E., Vandenberghe A., De Wachter R. Sequences of the 5S rRNAs of Azotobacter vinelandii, Pseudomonas aeruginosa and Pseudomonas fluorescens with some notes on 5S RNA secondary structure. Nucleic Acids Res. 1983 Mar 11;11(5):1245–1252. doi: 10.1093/nar/11.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wachter R., Chen M. W., Vandenberghe A. Conservation of secondary structure in 5 S ribosomal RNA: a uniform model for eukaryotic, eubacterial, archaebacterial and organelle sequences is energetically favourable. Biochimie. 1982 May;64(5):311–329. doi: 10.1016/s0300-9084(82)80436-7. [DOI] [PubMed] [Google Scholar]
- Denis H., Wegnez M. Biochemical research on oogenesis. Oocytes and liver cells of the teleost fish Tinca tinca contain different kinds of 5S RNA. Dev Biol. 1977 Sep;59(2):228–236. doi: 10.1016/0012-1606(77)90256-1. [DOI] [PubMed] [Google Scholar]
- Denis H., Wegnez M. Evolution of the 5 S RNA genes in vertebrates. J Mol Evol. 1978 Oct 27;12(1):11–15. doi: 10.1007/BF01732542. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A. Collection of published 5S and 5.8S RNA sequences and their precursors. Nucleic Acids Res. 1982 Jan 22;10(2):r93–115. doi: 10.1093/nar/10.2.762-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A., Huysmans E., Vandenberghe A., De Wachter R. Collection of published 5S and 5.8S ribosomal RNA sequences. Nucleic Acids Res. 1983 Jan 11;11(1):r105–r133. [PMC free article] [PubMed] [Google Scholar]
- Fang B. L., De Baere R., Vandenberghe A., De Wachter R. Sequences of three molluscan 5 S ribosomal RNAs confirm the validity of a dynamic secondary structure model. Nucleic Acids Res. 1982 Aug 11;10(15):4679–4685. doi: 10.1093/nar/10.15.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsbrough P. B., Ellis T. H., Lomonossoff G. P. Sequence variation and methylation of the flax 5S RNA genes. Nucleic Acids Res. 1982 Aug 11;10(15):4501–4514. doi: 10.1093/nar/10.15.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Klotz L. C., Blanken R. L., Loeblich A. R., 3rd An evaluation of the phylogenetic position of the dinoflagellate Crypthecodinium cohnii based on 5S rRNA characterization. J Mol Evol. 1981;17(6):334–337. doi: 10.1007/BF01734355. [DOI] [PubMed] [Google Scholar]
- Hori H. Evolution of 5sRNA. J Mol Evol. 1975 Dec 31;7(1):75–86. doi: 10.1007/BF01732181. [DOI] [PubMed] [Google Scholar]
- Hori H. Molecular evolution of 5S RNA. Mol Gen Genet. 1976 May 7;145(2):119–123. doi: 10.1007/BF00269583. [DOI] [PubMed] [Google Scholar]
- Hori H., Ohama T., Kumazaki T., Osawa S. Nucleotide sequences of 5S rRNAs from four jellyfishes. Nucleic Acids Res. 1982 Nov 25;10(22):7405–7408. doi: 10.1093/nar/10.22.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Ota T. On the stochastic model for estimation of mutational distance between homologous proteins. J Mol Evol. 1972 Dec 29;2(1):87–90. doi: 10.1007/BF01653945. [DOI] [PubMed] [Google Scholar]
- Kumazaki T., Hori H., Osawa S., Ishii N., Suzuki K. The nucleotide sequences of 5S rRNAs from a rotifer, Brachionus plicatilis, and two nematodes, Rhabditis tokai and Caenorhabditis elegans. Nucleic Acids Res. 1982 Nov 11;10(21):7001–7004. doi: 10.1093/nar/10.21.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazaki T., Hori H., Osawa S. The nucleotide sequence of 5 S ribosomal RNA from a protozoan species Chilomonas paramecium belonging to the class Phytomastigophorea. FEBS Lett. 1982 Nov 29;149(2):281–284. doi: 10.1016/0014-5793(82)81117-4. [DOI] [PubMed] [Google Scholar]
- Kumazaki T., Hori H., Osawa S. The nucleotide sequence of 5 S ribosomal RNA from a sea anemone, Anthopleura japonica. FEBS Lett. 1982 Sep 20;146(2):307–310. doi: 10.1016/0014-5793(82)80940-x. [DOI] [PubMed] [Google Scholar]
- Küntzel H., Heidrich M., Piechulla B. Phylogenetic tree derived from bacterial, cytosol and organelle 5S rRNA sequences. Nucleic Acids Res. 1981 Mar 25;9(6):1451–1461. doi: 10.1093/nar/9.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küntzel H., Piechulla B., Hahn U. Consensus structure and evolution of 5S rRNA. Nucleic Acids Res. 1983 Feb 11;11(3):893–900. doi: 10.1093/nar/11.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay R. M., Doolittle W. F. Two thraustochytrid 5S ribosomal RNAs. Nucleic Acids Res. 1982 Dec 20;10(24):8307–8310. doi: 10.1093/nar/10.24.8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J. Prediction of pairing schemes in RNA molecules-loop contributions and energy of wobble and non-wobble pairs. Biochimie. 1979;61(10):1133–1150. doi: 10.1016/s0300-9084(80)80227-6. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankoff D., Cedergren R. J., McKay W. A strategy for sequence phylogeny research. Nucleic Acids Res. 1982 Jan 11;10(1):421–431. doi: 10.1093/nar/10.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankoff D., Morel C., Cedergren R. J. Evolution of 5S RNA and the non-randomness of base replacement. Nat New Biol. 1973 Oct 24;245(147):232–234. doi: 10.1038/newbio245232a0. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Van der Walt J. P. The perfect and imperfect states of Sporobolomyces salmonicolor. Antonie Van Leeuwenhoek. 1970;36(1):49–55. doi: 10.1007/BF02069007. [DOI] [PubMed] [Google Scholar]
- Walker W. F., Doolittle W. F. Redividing the basidiomycetes on the basis of 5S rRNA sequences. Nature. 1982 Oct 21;299(5885):723–724. doi: 10.1038/299723a0. [DOI] [PubMed] [Google Scholar]
- Wegnez M., Denis H., Mazabraud A., Clérot J. C. Biochemical research on oogenesis. RNA accumulation during oogenesis of the dogfish Scyliorhinus caniculus. Dev Biol. 1978 Jan;62(1):99–111. doi: 10.1016/0012-1606(78)90095-7. [DOI] [PubMed] [Google Scholar]
- Wildeman A. G., Nazar R. N. Structural studies of 5 S ribosomal RNAs from a thermophilic fungus, Thermomyces lanuginosus. A comparison of generalized models for eukaryotic 5 S RNAs. J Biol Chem. 1982 Oct 10;257(19):11395–11404. [PubMed] [Google Scholar]



