Abstract
Motivation: Metal ions are essential for the folding of RNA molecules into stable tertiary structures and are often involved in the catalytic activity of ribozymes. However, the positions of metal ions in RNA 3D structures are difficult to determine experimentally. This motivated us to develop a computational predictor of metal ion sites for RNA structures.
Results: We developed a statistical potential for predicting positions of metal ions (magnesium, sodium and potassium), based on the analysis of binding sites in experimentally solved RNA structures. The MetalionRNA program is available as a web server that predicts metal ions for RNA structures submitted by the user.
Availability: The MetalionRNA web server is accessible at http://metalionrna.genesilico.pl/.
Contact: iamb@genesilico.pl
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
RNA plays a key role in many biological processes. It takes part in almost every aspect of processing genetic information, including decoding codon triplets, alternative splicing, peptide bond formation and the regulation of these mechanisms (Pyle, 2002). The function of many RNA molecules is dependent on their 3D structure (Holbrook, 2008). The RNA backbone is negatively charged. The neutralization of the electrostatic repulsion by the binding of cations is essential for the formation of compact tertiary structures. It has been shown in folding studies that tRNA stability increases remarkably in the presence of monovalent (especially Na+ and K+) and divalent (Mg2+) cations (Urbanke et al., 1975). However, divalent Mg2+ cations are more effective in stabilizing the native structure of RNA (Romer and Hach, 1975; Stein and Crothers, 1976). The higher the charge density of the RNA, the higher the concentration of cations near the surface and the greater the entropic advantage in using divalent ones, because fewer cations are confined near the RNA. Thus, a small number of ‘strong’ Mg2+ binding sites maybe responsible for the effective stabilization of RNA tertiary structure [reviewed by (Draper, 2004, 2008; Serra et al., 2002)].
Metal ions also serve as essential cofactors in many reactions catalyzed by ribozymes. The hammerhead ribozyme, group I and group II introns, as well as ribonuclease P (RNaseP) ribozymes are examples of catalytic RNA that need divalent cations to perform their functions [reviewed by (Schnabl and Sigel, 2010)]. For example, the cleavage of a phosphodiester bond by the hammerhead ribozyme depends on the presence of metal ions that are required for both folding and activity (Sigurdsson and Eckstein, 1995).
The formation of RNA–metal ion complexes occurs in an aqueous environment. The energy of electrostatic interactions of a cation with water molecules depends on its charge and radius. Mg2+ has a small radius (~0.72Å) and can tightly organize six water molecules in an octahedral arrangement, followed by organization of further layers of water. Theoretical calculations combined with experimental analyses suggest a total hydration-free energy for Mg2+ of −455 kcal/mol (Markham et al., 2002). K+ is larger (radius ~1.38Å), has a smaller charge and it organizes eight or nine water molecules in a less ordered manner, with the hydration energy of −80 kcal/mol (Draper et al., 2005).
Three different binding modes of magnesium ions can be distinguished [reviewed by (Draper, 2004; Draper et al., 2005)]. First, partially dehydrated cations can interact with RNA directly, chelated by electronegative atoms, such as phosphate oxygens, creating very strong interactions. Second, fully solvated cations can be stably bound to RNA via one or two layers of water molecules. Third, cations may contribute to RNA stability without occupying discrete sites, in a diffuse manner, where they interact with the RNA only by electrostatic interactions, without making direct contacts or perturbing their hydration layers.
Despite the growing number of experimentally solved RNA structures, the positions of cations in these structures still cannot be easily determined. Mg2+, Na+ and H2O have 10 electrons each and can be distinguished only in high-resolution crystal structures. Hence, many bound cations can be easily mistaken for water molecules or may be missing from crystal structures. The positions of metal ions are also difficult to determine by nuclear magnetic resonance (NMR). This situation motivated us to develop a computational predictor which only uses information about the RNA structure to identify the most likely metal ion-binding sites in this structure.
The statistical approach has been applied with great success in prediction of protein and RNA structures and in prediction of metal ion-binding sites in protein structures, and is based on a solid probabilistic framework (Hamelryck, 2009). The basic assumption of this method is that the free energy associated with a given molecular interaction is strictly correlated with the relative frequency by which this interaction occurs among known structures. Here, we present the MetalionRNA tool that employs an anisotropic knowledge-based potential to predict metal ion-binding sites in 3D structures of RNA.
2 METHODS
2.1 Preparation of input structures
To generate a knowledge-based potential and test MetalionRNA, a 5-fold cross-validation test was performed using RNA-metal ion complexes. We used a representative set of 113 crystallographically determined structures containing RNA and metal ions (including structures of e.g. protein–RNA complexes), available from the Protein Data Bank (PDB). Since the resolution of crystallographic structures is a key factor for an accurate determination of the identity and position of cations, we only used structures with a resolution <2.0Å for Mg2+ and <3.0Å for K+ and Na+ as only the higher resolution limit allowed us to collect a sufficient number of structures (Supplementary Table S1). For groups of RNAs with a sequence identity >90%, we used only one structure with the highest resolution. For residues with more than one alternative conformation, we used the first variant. We intended to take into account only cations interacting exclusively with RNA atoms, whose binding is not caused by other molecules. Therefore, we excluded metal ions closer than 9Å to any atom other than RNA, water or another cation.
For additional tests of MetalionRNA, we used a set of 116 crystallographically determined structures containing DNA and Mg2+ cations, with a resolution <3.0Å (for PDB codes, see Supplementary Table S1). Like with RNA, for groups of DNA molecules with sequence identity >90% we only used one structure with the highest resolution. For residues with more than one alternative conformation, we used the first variant.
2.2 Compilation of an anisotropic statistical potential
Klebe et al. developed an isotropic statistical potential for the prediction of protein–ligand interactions (Gohlke et al., 2000). We applied this approach to create a distance and angle-dependent anisotropic potential describing interactions between metal ions and RNA atom pairs. An n-particle correlation function g(n)(d1, α1;…. dn, αn) is translated into a knowledge-based potential W(n)(d1, α1;…. dn, αn) via the following equation:
where g(n) indicates the observed frequency of contacts of a cation c with all adjacent atom pairs [a, b] [d is the distance between cation and atom b; α is the angle (a, b, c)] and W(n) indicates the potential for a given position. We derived the function g(n)(d1, α1;…. dn, αn) from crystal structures by sampling the frequencies of RNA atom pair and metal ion contacts.
The maximum radius of interaction between an RNA atom pair and a metal ion, to be considered for the statistical potential, was limited to 9Å. This radius directly influences the specificity of the potential. A short distance emphasizes specific interactions between cations and the atoms of its binding site. On the other hand, a generous threshold allows to take indirect long-distance interactions into account, e.g. those mediated by solvent molecules. For example, the Klebe group developed a potential for short distances of up to 6Å (Gohlke et al., 2000), and another group applied a larger threshold of 12Å in their studies of protein–ligand interactions (Muegge and Martin, 1999). We chose a medium threshold value, since we wanted to cover highly specific direct RNA–cation contacts, as well as water-mediated interactions.
2.3 Anisotropic contact statistics based on atom pairs
We based our predictor of RNA-metal ion interactions on contacts formed by cations with oxygen and nitrogen atoms that are known to make the strongest contribution to metal binding. First, we defined a list of atom pairs [a, b] in nucleotides, of which b is an O or N atom that may directly interact with a cation, and a is covalently bound to b (Table 1).
Table 1.
RNA atom pairs used to derive RNA-ion contacts
| Ribose/phosphate backbone | Adenine side chain | Guanine side chain | Cytosine side chain | Uracil side chain |
|---|---|---|---|---|
| P, OP1 | C2, N1 | C2, N1 | C2, N3 | C2, N3 |
| P, OP2 | C2, N3 | C2, N2 | C2, O2 | C2, O2 |
| P, O5′ | C4, N3 | C2, N3 | C4, N3 | C4, N3 |
| C1′ , O4′ | C5, N7 | C4, N3 | C4, N4 | C4, O4 |
| C2′ , O2′ | C6, N1 | C5, N7 | ||
| C3′ , O3′ | C6, N6 | C6, N1 | ||
| C4′ , O4′ | C8, N7 | C6, O6 | ||
| C5′ , O5′ | C8, N7 |
For post-transcriptionally modified nucleotides identified by our in-house program ModeRNA (Rother et al., 2011), we took into account only the pairs [a, b] that were chemically identical to those in the unmodified ‘parent’ nucleotides [see the MODOMICS database (Dunin-Horkawicz et al., 2006) for details of RNA modification pathways]. Second, to derive contact statistics, all RNA structures were scanned for the presence of metal ions within a 9Å sphere of O or N atoms (all possible atoms b). For each identified cation c, its distance d to the respective atom b, and the angle α (a, b, c) were calculated. Thus, the relative position of a cation to a pair [a, b] can be described by a distance d and an angle α. To generate statistics from a set of measured values for d and α, they were discretized by statistical binning, using steps of 0.25Å and 5○ and thus creating a radial grid R. Figure 1 illustrates the principle of deriving the statistics for cations around an RNA atom pair [P, OP2]. Next, the counts per bin were normalized, since the spatial units defined by discrete steps of d and α had different sizes (the bin volume is dependent on the distance and angle). Accordingly, we divided the count of cations obtained from each d, α pair by the corresponding volume V of the radial grid R bin. To avoid overrating the contribution of atom pair couples [a, b] in which the same atom b is present twice (endocyclic N atoms of nucleobases and O4′ and O5′ atoms in the backbone e.g. [C2, N3], [C4, N3]), their relative weights were assigned to 0.5, compared to pairs with a unique atom b.
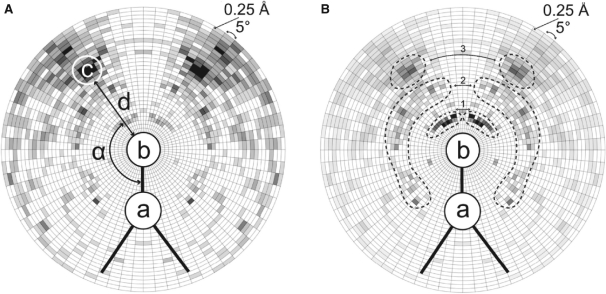
Fig. 1.
Schematic views of the radial grid R used for deriving contact statistics for RNA atom pairs [a, b] in contact with cation c. The grid used for counting uses radial steps of 0.25Å and 5○ around atom b (an O or N atom) and a (covalently bound to b). (A) Statistics of cation presence. For each cation, its distance d to the respective atom b, and the angle α (a, b, c) are calculated. The contact statistics derived for the RNA atom pair [P, OP2] and Mg2+ ions are shown in a gray scale (the more Mg2+ in a given bin, the darker the area). (B) The diagram shows the distribution of values for a normalized potential derived from contact statistics (A) for the RNA atom pair [P, OP2] and Mg2+ ions (the darker the area, the more negative value of the potential for the given bin). The three possible states of magnesium binding to RNA (Draper, 2004) are represented by the three peak tuples of the darkest areas. The first peak tuple (i) corresponds to Mg2+ chelated and partially dehydrated by phosphate groups of RNA. The second peak tuple (ii) corresponds to the water-mediated state. The third peak tuple (iii) represents the situation where the Mg2+ ion remains hydrated and interacts with the RNA via a layer of water molecules.
3 IMPLEMENTATION
3.1 Algorithm for prediction of metal ion positions
We implemented a grid-based function to calculate the potential for predicting metal ions in a target structure. The most important advantage of using a grid is that the discretization of space obviates the need to solve the potential function analytically, and allows mapping of the statistical data into well-defined portions of space. A grid-based approach has been successfully applied in small molecule docking, for instance in the AutoDock program (Goodsell et al., 1996). In MetalionRNA, the search space was divided into a cubic grid C with a grid width of 0.25Å (or of 0.5Å). We chose these values on the basis of the minimal distance of chelated cations in contact with RNA, such that at least a few grid cells are between an RNA atom and a cation. For a larger grid width of the cubic grid C, the statistics would be biased by the cell boundaries, negatively affecting the prediction quality. On the other hand, the calculation time and memory usage grow with the third power to the inverse grid width, without much influence on the prediction quality (See Section 4.3). Grid widths 0.25 and 0.5Å are less than the shortest bond, but big enough to run MetalionRNA calculations in a reasonable time. To avoid unnecessary calculations, the value of the potential in the target structure is calculated for cells of grid C around the previously defined RNA atom pairs only.
For each RNA atom pair [a, b] (of which b is an O or N atom and a is covalently bound to b; see Table 1), the program computes the value in all cells of grid C within the radius of 9Å around the atom b. Since cations cannot overlap with RNA atoms, all cells of grid C that are ‘occupied’ by RNA atoms, i.e. are within the van der Waals radius of an RNA atom, are excluded from the computation. Subsequently, the anisotropic potential value W(n) is calculated for all ‘unoccupied’ cells. The potential W(n) is additive for cells of grid C in a distance of 9Å from more than one RNA atom pair. Finally, all cells of grid C are sorted according to their W(n) value. For the top-scoring cells of grid C, all cells within a radius corresponding to half of the minimal distance between two cations of the same type (the default value was derived from known RNA structures, see Supplementary Table S1 for PDB codes) are examined. The radius of the new candidate cation cannot overlap with the radius of a previously proposed cation with a better score. If this condition is fulfilled, MetalionRNA places a cation in the center of the top-scoring cell, calculates the sum of W(n) of the cells and removes the cells covered by the new cation from further consideration.
Figure 2 illustrates the idea of calculating the potential for grid C, resulting from the presence of two RNA atom pairs [P, OP2] and identifying the most likely positions of cations. This procedure is repeated until a default or user-defined number of preferred cation positions is determined. The default value depends on the number of residues in the target structure. We calculated an average number of metal ions per nucleotide from the representative set of 113 crystallographically determined RNA-metal ion complexes (Supplementary Table S1), and in our calculations we generate by default the average number of metal ions plus one.
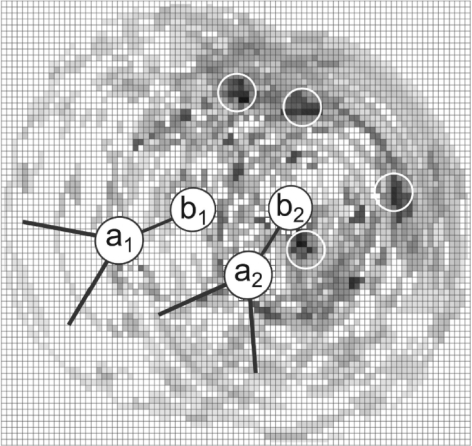
Fig. 2.
A schematic view of the cubic grid C used for deriving the potential for two [P, OP2] pairs. Only one layer of grid cells is represented and for simplicity we consider that all atoms are within this single layer. The potential W(n) is additive for cells <9Å from more than one OP2 atom. (the darker the area, the more negative value of the potential for the given grid cell). MetalionRNA places the center of a predicted cation in the grid cell with the lowest value, calculates the sum of W(n) of cells covered by the cation introduced and removes these cells from further consideration.
3.2 Cross-validation
We employed a cross-validation procedure for all three sets of PDB structures (containing Mg2+, Na+ and K+) (see Supplementary Table S1 for PDB codes). We randomly split the PDB structures into five subsets and carried out a 5-fold cross-validation, using one of the subsets for testing and the other four for training the potential. This approach ensures that the same cation-binding sites are not used for training and testing. The results of the complete cross-validation test for each of the structure subsets were summed up to estimate the prediction accuracy. We calculated the true positive rate (TPR) and the false positive rate (FPR) defined as:
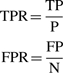 |
where TP (true positives) is the number of predicted cation positions within a cut-off distance d from the true position in a structure from the test set (i.e. predicted cations that are close to the experimentally observed ones), P (positives) is the total number of cations observed in the crystal structures, FP (false positives) is the number of predicted cations that are far (beyond the distance cut-off d) from the cations in the crystal structures and N (negatives) is the maximum number of cations that can be predicted for a given structure in the space within 9Å from any O or N RNA atom considered as an ‘ion-binder,’ minus P. We analyzed the accuracy of the predictor for a series of distance cut-offs and illustrated the results in the form of receiver operating characteristics (ROC) plots (Fawcett, 2006). The area under the ROC curve (AUC) was calculated to assess the accuracy of MetalionRNA.
4 RESULTS
We developed MetalionRNA, a computational method for the prediction of metal ion-binding sites in RNA 3D structures, using a statistical potential and a grid-based calculation approach. The potential is based on the analysis of known metal ion-binding sites present in 113 RNA structures. As an input, MetalionRNA takes an RNA 3D structure in the PDB format, and returns PDB files with the calculated RNA potential surface and the coordinates of cations predicted for the target RNAstructure.
4.1 Web server
To make our method easily available to the research community, we developed a web server available at http://metalionrna.genesilico.pl (server mirror is available at http://metalionrna.amu.edu.pl). The submission form accepts an RNA structure only in the PDB format. Every other file format is rejected and the server displays an adequate error message. One can specify the cation type, the number of cation positions expected to bind to the query structure, the minimal distance between predicted cations, width of the cubic grid, the ionic radius of the cation or use default values. The default cation is Mg2+. The default number of predicted ions is calculated on the basis of the number of residues in the target structure; the minimal default distance between predicted cations is the one observed in known structures (Supplementary Table S1), and the default width of the cubic grid C is 0.5Å. The results returned by the server are available as a separate web page, including a file with the predicted cation positions in text and PDB formats, a script to display the predicted cations in the PyMOL viewer, and a PDB file containing the target structure with the calculated potential surface. The page with the output files is kept on the server for 1 week.
The time required for MetalionRNA to return predictions depends mainly on the size of the molecule. Currently, we use a simple queuing system that allows running one prediction at a time. For a tRNA molecule (PDB id: 1EHZ) 76 nt long, with the default number of 7 Mg2+ hits, it takes ~5 min to obtain the results. The server was implemented in Python using the Django web framework.
One of the weaknesses of the statistical approach is the relative paucity of high-resolution crystal structures of RNA molecules with accurately determined cation-binding sites. The MetalionRNA web server once per week (every Saturday at 12 p.m. Central European Time) downloads structures released in the PDB that have resolution better than 2Å. RNA structures containing Mg2+, Na+ or K+ cations that fulfill the conditions described in Section 2.1 are added to the original training set and the statistical potential is recalculated. In time, the structures with the resolution worse between 2Å and 3Å will be outnumbered by those with the resolution better than 2Å, hopefully leading to a constant improvement of the potential. The MetalionRNA web site allows the user to select whether to perform predictions with the original potential described in this article or with the updated one.
4.2 RNA-metal ion statistical preferences
To calculate the anisotropic statistical potential for RNA-ion contact prediction, we derived statistics for the most common cations from 50 RNA structures containing Mg2+ (182 binding sites), 25 RNA structures containing Na+ (88 binding sites) and 38 RNA structures containing K+ (123 binding sites). The graph showing the statistical potential in Figure 1B depicts the preferred interaction geometries for direct contacts and solvated Mg2+ ions. The distribution function for the RNA atom pair [P, OP2] and Mg2+ has three peak tuples (the darkest areas). The peak tuples correspond to the three possible states of magnesium binding to RNA (Draper, 2004). The first peak tuple is present at a distance of ~2Å, with an acute angle of 15–60○. It corresponds to magnesium ions chelated and partially dehydrated by phosphate groups of RNA. In this state, the Mg2+ ion interacts with RNA atoms directly. The second peak tuple is at a distance of 4–5Å with a bimodal angle distribution. Acute angles (15–90○) correspond to the water-mediated state, in which the cation retains one layer of hydrating water molecules that in turn interact with the RNA atoms. Obtuse angles (120○ and higher) correspond to cations chelated with the OP2 atom, and the same Mg2+ ions appear as the first peak (in the distance of about 2–2.5Å) for the [P, OP1] pair (data not shown). Finally, the third peak tuple corresponds to a distance of 6–7Å and represents the situation where the Mg2+ ion remains hydrated and interacts with the RNA via a layer of water molecules. For these distances, angles of 30–60○ are predominant.
4.3 MetalionRNA predicts metal ion with high accuracy
In order to assess the accuracy of MetalionRNA, a 5-fold cross-validation test was performed using RNA–metal ion complexes (Supplementary Table S1). We used a cubic grid C with edge width of 0.25 and 0.5Å, a Mg2+ ionic radius of 0.75Å and a minimal distance between predicted cations of 1.5Å. The results for the cubic grid of 0.5Å edge width are illustrated in the form of ROC plots (RNA-Mg2+ in Fig. 3A, RNA-Na+ in Fig. 3B and RNA−K+ in Fig. 3C).
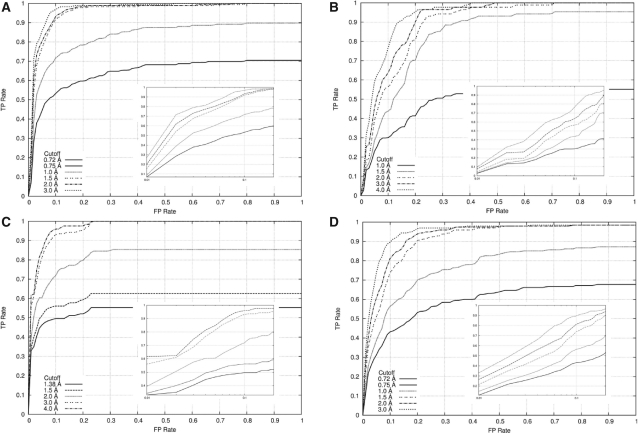
Fig. 3.
ROC curves to assess the classification performance of MetalionRNA with the width of 0.5Å for the cubic grid C using (A) the RNA-Mg2+ dataset, (B) the RNA-Na+ dataset, (C) the RNA−K+ dataset, (D) the DNA-Mg2+ dataset and various cut-off distance values (the maximum distance between a predicted and a real metal ion in which the prediction is marked as correct). In the big picture overall graph is shown, in the small picture only a range between 0 and 0.2Å is illustrated on a logarithmic scale.
The AUC values that describe the degree of successful predictions for the Mg2+ ions were calculated for the following cut-off distances (the maximum distances between a predicted and a real metal ion, for which the prediction was regarded as correct): 0.72, 0.75, 1.0, 1.5, 2.0 and 3.0Å. The ionic radius of Mg2+ is 0.72Å, the other values are multiples of grid width of C. Using these values, the AUC values for the Mg2+ ions were 50, 56, 81, 93, 95, 96% for the grid C of 0.25Å and 62, 62, 81, 95, 96, 97% for the grid C of 0.5Å. The solid line in Figure 3A illustrates predictions that lie within the ionic radius of Mg2+ (0.72Å) and hence are within the space occupied by the cation in the crystallographic model.
For Na+ ions, AUC values were calculated to be 43, 82, 87, 91, 93% (0.25Å grid) and 47, 78, 85, 88, 91% (0.5Å grid) for the cut-off distances 1.0, 1.5, 2.0, 3.0 and 4.0Å, respectively. The ionic radius of Na+ is 1.0Å. Predictions for Na+ are slightly less accurate than those for Mg2+, most likely because of the smaller number of cations in the training dataset. The solid line in Figure 3B illustrates predictions within the ionic radius of Na+ (1.0Å). For K+ ions, AUC values were 54, 61, 84, 96, 97% (0.25Å grid) and 54, 61, 81, 97, 98% (0.5Å grid) for the cut-off distances 1.38, 1.5, 2.0, 3.0 and 4.0Å, respectively. The ionic radius of K+ is 1.38. Figure 3C shows predictions for K+.
We also conducted the predictions and ROC analysis for a set of DNA–Mg2+ complexes (Fig. 3D) using the statistical potential derived from RNA–Mg2+ PDB complexes. Interestingly, our method works for DNA structures that were not considered in the training of the potential: the AUC curve corresponding to the cut-off distances of 0.72, 0.75, 1.0, 1.5, 2.0 and 3.0Å was calculated to be 44, 49, 74, 90, 91, 93% (for the grid C of 0.25Å) and 56, 56, 72, 88, 91, 93% (for the grid C of 0.5Å) respectively. These results are only slightly worse than those for RNA and indicate that our approach captured a general aspect of the metal ion binding by nucleic acids.
FEATURE is another method for predicting metal ions in RNA structures (Banatao et al., 2003). It applies supervised learning on a training set consisting of positive and negative examples of Mg2+ ion binding sites to create a statistical model that describes the micro-environments surrounding site-bound and diffusely bound cations. To create a statistical model, 126 physicochemical and structural properties that influence or take part in RNA–Mg2+ ion interactions were used, and the method was tested on a 58 nt fragment of Bacillus stearothermophilus 23S rRNA (PDB code 1HC8). To compare the performance of MetalionRNA with that of WebFEATURE, we made predictions for this structure using our default settings, as well as after retraining our potential on the FEATURE training set.
Table 2 and Figure 4 summarize predictions for seven Mg2+ ions present in the 1HC8 structure. MetalionRNA calculated it for the molecule of that size, with six Mg2+ ions expected to be observed in a crystal structure solved under ‘average’ conditions; hence, the six top-scoring predictions are considered as strong bets, and further positions in the ranking correspond to alternative, low-confidence sites, potentially occupied e.g. at higher Mg2+ concentrations. The six predictions reported with top scores by MetalionRNA with the default potential included four out of the seven Mg2+ ions, identified with accuracy of 0.6–1.9Å. The remaining ions were predicted with ranks 10, 13 and 29. Using a potential calculated from the FEATURE training set, MetalionRNA predicted only two of the seven ions at the first six positions of the ranking, with accuracy of 0.8 and 0.6Å, respectively. The remaining five ions were ranked at positions 8, 9, 21, 29 and 33. FEATURE correctly identified only two site-bound Mg2+ ion positions within its seven top-scored predictions with accuracy of 1.5 and 3.6Å, respectively. The diffuse ions were all scored relatively poorly by FEATURE, all outside the top positions of its ranking.
Table 2.
A list of Mg2+ ions in the 23s rRNA structure (PDB ID: 1HC8) for which predictions using MetalionRNA and FEATURE were done
| Mg2+ (atom no.) | MetalionRNA |
FEATURE deviation (Å), rank out of 224 | |
|---|---|---|---|
| MetalionRNA training set: deviation (Å), rank (out of 224) | FEATURE training set deviation (Å) rank (out of 224) | ||
| 1159 | 0.8 (1) | 0.8 (1) | 1.6 (15), 0.4 (58) |
| 1160 | 1.9 (6) | 1.7 (33) | 0.6 (17), 2.0 (8) |
| 1161 | 2.9 (29) | 3.7 (29) | 1.5 (102) |
| *1163 | 0.6 (3) | 0.6 (4) | 1.5 (5a) |
| 1164 | 1.4 (2) | 1.1 (21) | 0.7 (181), 1.6 (88) |
| *1167 | 3.8 (10) | 1.1 (8) | 3.6 (2a) |
| 1172 | 3.2 (13) | 3.4 (9) | 2.5 (202) |
The first column from the left lists real Mg2+ ions' identifiers as labeled in the PDB file 1HC8. The site-bound Mg2+ ions are labeled with an asterisk. Column 2 describes the predictions made by MetalionRNA using the Mg2+ training set (Supplementary Table S1). Column 3 describes the MetalionRNA predictions made using statistical potential derived from the FEATURE training set. In both columns, the first value is a prediction distance to the respective Mg2+ ion, the value in the brackets is the prediction rank by score with respect to the total number of all generated binding sites. Column 4 contains the predictions made by FEATURE. The first value is a prediction distance to the respective Mg2+ ion, the value in the brackets is the prediction rank by score with respect to the total number of all hits above the cut-off score [for details see Banatao et al. (2003)].
a Rank out of 9.
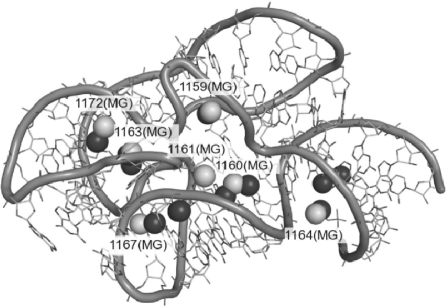
Fig. 4.
Structure of the 23S rRNA fragment (PDB ID: 1HC8) with the experimentally determined positions of Mg2+ cations indicated by white labeled balls. Top-scoring Mg2+ cations predicted by MetalionRNA are shown as black balls. For detailed comparison of predicted and experimentally observed ions, see Table 2.
MetalionRNA with both variants of the potential were able to identify four out of five diffuse Mg2+ ions much better than FEATURE. The only exception was Mg2+ ion 1160, for which FEATURE found a more accurate match, but only at the 17th position of the ranking, while MetalionRNA reported a reasonable prediction at the sixth position of its ranking (i.e. above the default threshold). Predictions for two of the diffuse ions (1161 and 1172) were reported with relatively low scores by both methods. MetalionRNA also predicted one of the two site-bound Mg2+ ions (*1163) with very high accuracy and high position in the ranking (using our training set: 0.6Å, Rank 3, using the FEATURE training set: 0.6Å, Rank 4). For this cation, FEATURE performed only slightly worse (accuracy 1.5Å, Rank 5 in a separate prediction for site-bound ions alone). The second site (*1167) was predicted by MetalionRNA with accuracy of 3.8Å (Rank 10) and 1.1Å (Rank 8) for the two training sets, while FEATURE reported it with accuracy of 3.6Å, Rank 2 (again, in a separate prediction for site-bound ions). Hence, both methods performed similarly well for site-bound ions. Summarizing, MetalionRNA was able to identify four out of seven true Mg2+ sites in the 1HC8 structure with just two false positives, while FEATURE identified these ions with a much higher number of false positives.
Interestingly, the top-scoring Mg2+ binding site predicted by FEATURE corresponds to a K+ binding site in the 1HC8 structure (*1162, accuracy 1.4Å). MetalionRNA predicted this site at the fourth position of the ranking specific for K+ cations (accuracy 2.2Å), with the three alternative predictions coinciding with the Mg2+ binding sites observed in the experimentally determined structure. Among the Mg2+ binding sites predicted by MetalionRNA, this K+ binding site is found at the 18th position in our ranking (accuracy 1.7Å). This partial overlap of predicted Mg2+ and K+ binding sites suggests that cations compete with each other for binding to the RNA molecule. MetalionRNA does not yet support simultaneous prediction of different ions and does not take the ion concentration into account. Such features will be implemented when the number of high-resolution RNA structures determined at a range of different ion concentrations (and with confidently assigned ions) reaches the level required for statistical significance of training and testing the knowledge-based potential.
5 DISCUSSION
MetalionRNA is a novel tool for predicting metal ion-binding sites in RNA structures. It uses an anisotropic statistical potential trained on a database of known structures. The current implementation is capable of making predictions for Mg2+, Na+ and K+ cations, and further ions will be added as the database of RNA structures is expected to grow. The 5-fold cross-validation test proved that ion positions are predicted by MetalionRNA with useful accuracy, as the method successfully reproduces the crystallographically determined positions of Mg2+, Na+ and K+ cations in dozens of different RNA molecules. A similar accuracy was achieved by the prediction of Mg2+ in DNA structures, which were not used for training, revealing that the general mechanism of ion binding by both types of the nucleic acids is sufficiently similar to be captured by a coarse-grained method such as ours. Comparison with another fully automated method FEATURE demonstrated that MetalionRNA can identify true Mg2+ sites in RNA structure with a relatively low rate of false positives, suggesting that it may be a practically useful tool.
There are alternative approaches for predicting metal ion-binding sites in RNA structures with high accuracy. Hermann and Westhof (1998) applied Brownian dynamics (BD) simulations of cations diffusing under the influence of random Brownian motion within the electrostatic field to predict metal ion-binding sites. Misra and Draper (2000) presented an analytical model based on the non-linear Poisson–Boltzmann (PB) equation that describes the energetic and stoichiometric linkage between the Mg2+ binding and RNA folding. Tan and Chen (2005, 2010) developed a statistical mechanical model based on the PB theory, which considers and ensemble of discrete ion distributions; it models electrostatics and steric interactions for tightly bound ions and uses the mean-field fluid model to describe the diffuse ions. The advantage of these methods is that they model the physicochemistry of the system and therefore can be used to infer dynamic and thermodynamic parameters of the systems under study and its individual elements. An important feature of these and similar methods is the examination of the system under physical conditions defined by the user, such as temperature, concentration of different ions, possible presence of other molecules, etc. These methods are, however, computationally very costly, and require specialized expertise to set up and run the simulations, and to interpret their results. The simulation methods are not available as ‘black box’ packages that can take an RNA structure as an input and generate defined positions of ions as an output. For these reasons, they cannot be used to make predictions for a large series of test structures. They serve different purpose than the automated predictive methods such as FEATURE or MetalionRNA and these two types of tools cannot be directly compared. The advantage of MetalionRNA is that it is relatively fast, can be accessed by a user friendly web interface and does not require special skills to interpret the results. Ion sites predicted by MetalionRNA are ranked according to their score, which can be used to infer the relative order and strength of binding consecutive metal ions by the given RNA molecule e.g. with increasing ion concentration.
MetalionRNA requires the 3D structure of a nucleic acid as an input. However, predictions of ion-binding sites in nucleic acid structures may be validated experimentally with methods that do not require the experimental determination of nucleic acid structure. In particular, Fenton chemistry makes use of the ability of Fe2+ to replace Mg2+ and to generate highly reactive hydroxyl radicals that can cleave nucleic acid backbones in spatial proximity of the ion-binding site; the sites of cleavage can be then mapped with standard biochemical methods (Berens et al., 1998). This and other methods of experimental determination of ion-binding sites can be used in conjunction with MetalionRNA to model RNA structures in the more physically and biologically realistic ion-bound state. The next steps in the development of MetalionRNA will be to assess its ability to predict ion-binding sites in low-accuracy structures and to explore the possibilities of integrating the modeling of metal ions with software for automated RNA 3D structure modeling by comparative (Rother et al., 2011) or de novo assembly approaches (Das and Baker, 2007). We also intend to explore the possibility to include the ion concentration as a parameter of the prediction, and to enable predictions for mixed solutions with different cations present simultaneously and potentially competing for similar binding sites.
6 CONCLUSIONS
We developed MetalionRNA, a novel bioinformatics tool for prediction of metal ion-binding sites in RNA. The anisotropic potential in MetalionRNA outperforms the previously published FEATURE method based on a statistical approach. Our method can be used to assist crystal structure determination e.g. by identifying tentative metal ion sites to be further validated by comparison with experimental data or to propose metal positions for structural models that lack coordinates of cations, e.g. RNA structures determined by NMR spectroscopy (Shen et al., 1995) or theoretical models. MetalionRNA is freely available as a web server, at http://metalionrna.genesilico.pl/ and has a mirror at http://metalionrna.amu.edu.pl.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Russ Altman, Mike Wong and Rey Banatao for their help with WebFEATURE, and Magdalena Rother for help in using ModeRNA. We also thank Irina Tuszynska and Marcin Pawlowski for critical reading of the manuscript.
Funding: Foundation for Polish Science (grant TEAM/2009-4/2 and ‘Ideas for Poland’ fellowship to J.M.B.); Polish Ministry of Science and Higher Education (for the development of software in the Bujnicki Laboratory; POIG.02.03.00-00-003/09); EU Seventh Framework Programme (Health-Prot, contract number 229676). German Academic Exchange Service (D/09/42768 to K.R.); European Research Council (RNA+P = 123D to J.M.B.).
Conflict of Interest: none declared.
REFERENCES
- Banatao D.R., et al. Microenvironment analysis and identification of magnesium binding sites in RNA. Nucleic Acids Res. 2003;31:4450–4460. doi: 10.1093/nar/gkg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens C., et al. Visualizing metal-ion-binding sites in group I introns by iron(II)-mediated Fenton reactions. Chem. Biol. 1998;5:163–175. doi: 10.1016/s1074-5521(98)90061-8. [DOI] [PubMed] [Google Scholar]
- Das R., Baker D. Automated de novo prediction of native-like RNA tertiary structures. Proc. Natl Acad. Sci. USA. 2007;104:14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D.E. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D.E. RNA folding: thermodynamic and molecular descriptions of the roles of ions. Biophys. J. 2008;95:5489–5495. doi: 10.1529/biophysj.108.131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D.E., et al. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- Dunin-Horkawicz S., et al. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2006;34:D145–D149. doi: 10.1093/nar/gkj084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006;27:861–874. [Google Scholar]
- Gohlke H., et al. Knowledge-based scoring function to predict protein-ligand interactions. J. Mol. Biol. 2000;295:337–356. doi: 10.1006/jmbi.1999.3371. [DOI] [PubMed] [Google Scholar]
- Goodsell D.S., et al. Automated docking of flexible ligands: applications of AutoDock. J. Mol. Recognit. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Hamelryck T. Probabilistic models and machine learning in structural bioinformatics. Stat. Methods Med. Res. 2009;18:505–526. doi: 10.1177/0962280208099492. [DOI] [PubMed] [Google Scholar]
- Hermann T., Westhof E. Exploration of metal ion binding sites in RNA folds by Brownian-dynamics simulations. Structure. 1998;6:1303–1314. doi: 10.1016/s0969-2126(98)00130-0. [DOI] [PubMed] [Google Scholar]
- Holbrook S.R. Structural principles from large RNAs. Annu. Rev. Biophys. 2008;37:445–464. doi: 10.1146/annurev.biophys.36.040306.132755. [DOI] [PubMed] [Google Scholar]
- Markham G.D., et al. The arrangement of first- and second-sphere water molecules in divalent magnesium complexes: results from molecular orbital and density functional theory and from structural crystallography. J. Phys. Chem. B. 2002;106:5118–5134. [Google Scholar]
- Misra V.K., Draper D.E. Mg(2+) binding to tRNA revisited: the nonlinear Poisson-Boltzmann model. J. Mol. Biol. 2000;299:813–825. doi: 10.1006/jmbi.2000.3769. [DOI] [PubMed] [Google Scholar]
- Muegge I., Martin Y.C. A general and fast scoring function for protein-ligand interactions: a simplified potential approach. J. Med. Chem. 1999;42:791–804. doi: 10.1021/jm980536j. [DOI] [PubMed] [Google Scholar]
- Pyle A.M. Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- Romer R., Hach R. tRNA conformation and magnesium binding. A study of a yeast phenylalanine-specific tRNA by a fluorescent indicator and differential melting curves. Eur. J. Biochem. 1975;55:271–284. doi: 10.1111/j.1432-1033.1975.tb02160.x. [DOI] [PubMed] [Google Scholar]
- Rother M., et al. ModeRNA: a tool for comparative modeling of RNA 3D structure. Nucleic Acids Res. 2011;39:4007–4022. doi: 10.1093/nar/gkq1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabl J., Sigel R.K. Controlling ribozyme activity by metal ions. Curr. Opin. Chem. Biol. 2010;14:269–275. doi: 10.1016/j.cbpa.2009.11.024. [DOI] [PubMed] [Google Scholar]
- Serra M.J., et al. Effects of magnesium ions on the stabilization of RNA oligomers of defined structures. RNA. 2002;8:307–323. doi: 10.1017/s1355838202024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L.X., et al. RNA structure at high resolution. F. ASEB J. 1995;9:1023–1033. doi: 10.1096/fasebj.9.11.7544309. [DOI] [PubMed] [Google Scholar]
- Sigurdsson S.T., Eckstein F. Structure-function relationships of hammerhead ribozymes: from understanding to applications. Trends Biotechnol. 1995;13:286–289. doi: 10.1016/S0167-7799(00)88966-0. [DOI] [PubMed] [Google Scholar]
- Stein A., Crothers D.M. Equilibrium binding of magnesium(II) by Escherichia coli tRNAfMet. Biochemistry. 1976;15:157–160. doi: 10.1021/bi00646a024. [DOI] [PubMed] [Google Scholar]
- Tan Z.J., Chen S.J. Electrostatic correlations and fluctuations for ion binding to a finite length polyelectrolyte. J. Chem. Phys. 2005;122:044903–1-044903-16. doi: 10.1063/1.1842059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z.J., Chen S.J. Predicting ion binding properties for RNA tertiary structures. Biophys. J. 2010;99:1565–1576. doi: 10.1016/j.bpj.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanke C., et al. Tertiary structure of tRNAPhe (yeast): kinetics and electrostatic repulsion. Eur. J. Biochem. 1975;55:439–444. doi: 10.1111/j.1432-1033.1975.tb02180.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.