Summary
Histone H2B ubiquitylation is a transcription-dependent modification that not only regulates nucleosome dynamics but also controls the trimethylation of histone H3 on lysine 4 by promoting ubiquitylation of Swd2, a component of both the histone methyltransferase COMPASS complex and the Cleavage and Polyadenylation Factor(CPF). We show that preventing either H2B ubiquitylation or H2B-dependent modification of Swd2 results in nuclear accumulation of poly(A) RNA due to a defect in the integrity and stability of APT, a subcomplex of the CPF. Ubiquitin-regulated APT complex dynamics is required for the correct recruitment of the mRNA export receptor, Mex67 to nuclear mRNPs. While H2B ubiquitylation controls the recruitment of the different Mex67 adaptors to mRNPs, the effect of Swd2 ubiquitylation is restricted to Yra1 and Nab2 which, in turn, controls poly(A) tail length. Modification of H2B thus participates in the crosstalk between co-transcriptional events and assembly of mRNPs linking nuclear processing and mRNA export.
Introduction
Transcripts generated by RNA polymerase II (RNAPII) undergo a precisely orchestrated cascade of processing steps involving 5′-end capping, splicing, 3′-end cleavage and polyadenylation, before being transported to the cytoplasm as export-competent mRNA ribonucleoprotein particles (mRNPs). These biochemically distinct reactions occur co-transcriptionally and are tightly coupled, impacting the efficiency of one another (Moore and Proudfoot, 2009; Perales and Bentley, 2009). This functional coordination results from physical interactions between the different mRNA biogenesis machineries, from histones to the mRNA export apparatus, which synchronize or cross-stimulate connected processes (Luna et al., 2008; Pandit et al., 2008). In this context, the RNAPII C-terminal domain (CTD) serves as a major recruitment platform for distinct mRNA processing machineries that depend on the phosphorylation state of the CTD (Buratowski, 2009; Egloff and Murphy, 2008). Furthermore, several ‘reverse-coupling’ scenarios have been described, where processing events impact the subsequent round of transcription (Damgaard et al., 2008; Furger et al., 2002; Mapendano et al.).
In addition to CTD phosphorylation, other factors and modifications coordinate mRNA biogenesis. In particular, ubiquitin conjugation influences the dynamics of molecular scaffolds responsible for functional coupling events (Fleming et al., 2008; Gwizdek et al., 2006). Consistent with this view, we reported that the Bre1 ubiquitin ligase as well as ubiquitylation of histone H2B on lysine 123 control the monoubiquitylation of Swd2, a sub-unit of the S. cerevisiae SET1 H3K4 methyltransferase complex, through modification of two distinct sites, lysine residues K25 and K68 or K69. We proposed that transcription and H2B-dependent recruitment of Rad6/Bre1 occur in the vicinity of Swd2 and lead to ubiquitylation of Swd2 (Vitaliano-Prunier et al., 2008). We further showed that the K68,69 modification represents a key event in the crosstalk between H2B ubiquitylation and H3K4 trimethylation on transcribing genes (Vitaliano-Prunier et al., 2008). Swd2 also interacts with six other proteins (Pta1; Pti1; Ref2; Syc1; and two phosphatases Ssu72 and Glc7) to form the APT (Associated with Pta1) complex which co-purifies via Pta1 with the Cleavage and Polyadenylation Factors (CPF) (Nedea et al., 2003; Roguev et al., 2001). While the CPF complex is involved in mRNA 3′-end formation and transcription termination, the APT complex also contributes to the Nrd1/exosome-dependent 3′-end formation of snoRNAs (Cheng et al., 2004; Dheur et al., 2003; Dichtl et al., 2004; Kim et al., 2006; Nedea et al., 2003; Nedea et al., 2008). In addition, the APT complex has been implicated in the formation of a gene loop between promoter and terminator, proposed to assist the recycling of the transcription machinery for the next round of transcription and to confer transcriptional memory (Ansari and Hampsey, 2005). These studies raise the possibility that ubiquitylation of H2B could directly participate in the formation of export-competent mRNPs through control of the ubiquitylation of Swd2 and subsequent regulation of Swd2-containing complexes.
Results and Discussion
Ubiquitylation of H2B and Swd2 contributes to efficient mRNA nuclear export
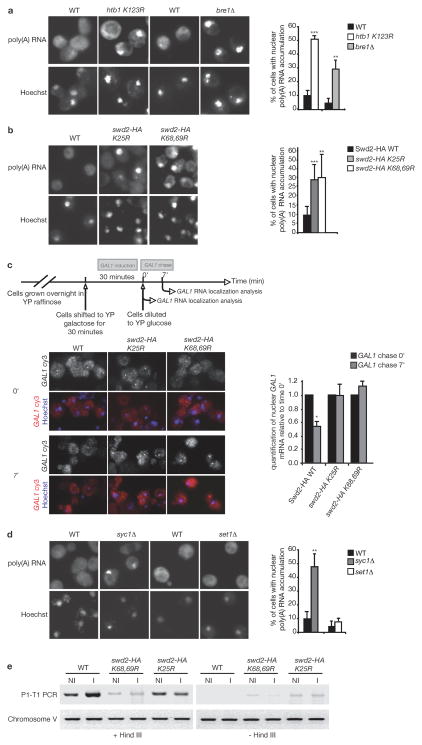
To test whether ubiquitylation of H2B modulates mRNPs formation, we examined nuclear export, the ultimate step of nuclear mRNP biogenesis. Accumulation of mRNPs in the nucleus reflects either defects in the mRNA export process per se or deficiencies in upstream processes such as mRNA 3′-end formation (Apponi et al., 2007; Hammell et al., 2002; Jensen et al., 2001; Qu et al., 2009). No significant export defect was observed in an H2B ubiquitylation mutant (htb1-K123R) at 30°C as detected by in situ hybridization with an oligo(dT) probe. Increasing temperature has been reported to amplify mRNA export defects of ubiquitylation mutants (Gwizdek et al., 2006; Iglesias et al., 2010). Following a 3h shift to 39°C, 50% of H2B ubiquitylation mutant cells indeed accumulated poly(A)RNA in the nucleus. Consistently, deletion of the ubiquitin ligase BRE1, induced an export defect under the same experimental conditions (Figure 1a). In response to heat-shock at 42°C, the mRNA export pathway is altered to allow efficient transport of heat-shock mRNAs while other mRNAs are retained in the nucleus (Saavedra et al., 1997). However, mRNA expression levels determined by genome-wide expression profiling were highly similar at 30°C and after a 3h shift at 39°C. In particular, heat-shock genes such as SSA1 are no longer induced after a 3h shift at 39°C, confirming that the transient heat-shock response is over in the experimental conditions employed for this experiment (Suppl. Figure S1a). Together these results indicate that preventing H2B ubiquitylation impacts mRNA nuclear export.
Figure 1. Ubiquitylation of H2B controls mRNA export through Swd2 ubiquitylation in the APT complex.
(a,b,d) Subcellular localization of poly(A) RNA was analyzed by FISH using Cy3-labelled oligo dT probe in (a) Htb1 (WT), htb1-K123R and bre1Δ, (b) Swd2–HA wt, swd2-HA K25R or swd2-HA K68,69R shuffle strains or in (d) WT, set1Δ and syc1Δ strains grown overnight at 30°C and shifted to 39°C for 3 h. The percentage of cells with nuclear accumulation of poly(A) RNA was examined for at least 300 DAPI-stained cells from each condition (mean ± standard deviation (SD)) (see also Figure S1). (c) GAL1 mRNA localization was analyzed in a chase experiment by FISH using a specific probe as outlined in the scheme. For each cell analyzed, the intensity of the transcription site was measured and normalized to the intensity of a constant area in the cytoplasm. The signal at 7 min of chase is expressed relative to the signal at t=0 for each cell type. An average of 30 cells was analyzed for each time point (mean ± SD) (e) 3C analysis. Chromatin was isolated from the indicated cells grown in raffinose (non induced, NI) or in galactose (induced, I). 3C analysis of gene looping (P1T1) was performed on the GAL10 gene (Ansari and Hampsey, 2005), on chromatin digested (+HindIII) or not (−HindIII). Chromosome V represents a control to ensure that equal amounts of template DNA were present in all reactions.
To determine whether the impact of ubiquitylated H2B (ubH2B) on mRNA export was mediated by ubiquitylation of Swd2, poly(A)RNA localization was analyzed in cells mutant for either the K25 or K68, 69 ubiquitylation site in Swd2. This analysis revealed an export defect in 30% of these mutant cells following a 3hr shift to 39°C (Figure 1b). Notably, the K68,69R mutation did not influence the mRNA expression pattern (Suppl. Figure S1 b, c). Although no significant nuclear accumulation of poly(A)RNA was detected at 30°C, a change in the kinetics of export can not be excluded as previously reported (Gwizdek et al., 2006). We thus examined the subcellular localization of a specific inducible transcript, GAL1 mRNA, with a “transcriptional pulse-chase” experiment in which GAL1 mRNA transcription was induced for 30 min in galactose at 30°C and then inhibited by addition of glucose. Newly transcribed GAL1 mRNA mainly accumulated within a nuclear dot at or close to the site of transcription (Figure 1c). After transcription shut off, disappearance of the nuclear dots, reflecting GAL1 mRNA export to the cytoplasm, was quantified and compared to the cytoplasmic content. Whereas 50% of GAL1 transcripts were transported from these dots to the cytoplasm within 7 minutes in wild-type cells, no mRNA export could be detected in Swd2 ubiquitylation mutants. (Figure 1c). Longer time points were necessary to detect “nuclear dot” extinction in the mutant cells indicating that this delayed transport probably results from a delayed release from the transcription site.
To determine whether the transport defect observed in Swd2 ubiquitylation mutants cells was due to APT or SET1 complexes function, poly(A) RNA localization was analyzed in APT complex mutant syc1Δ cells, which revealed nuclear accumulation of poly(A) RNA in 47% of cells at 39°C (Figure 1d). In contrast, no export defect was observed in set1Δ cells. Together, these results indicate that ubH2B is required for efficient poly(A) RNA export at 39°C and that this function is mediated by ubiquitylation of Swd2 and the role of Swd2 in the APT/CPF complex. In contrast, Set1 function, and presumably H3K4 trimethylation, are not involved in poly(A) RNA nuclear export.
The APT complex has been implicated in transcriptional crosstalk with the 5′ end through facilitating the formation of a gene loop between gene promoter and terminator (Ansari and Hampsey, 2005). Swd2 is recruited to both the 5′ and 3′-end of transcribing genes and in its ubiquitylated form, plays a major role in controlling 5′ (H3K4 trimethylation) and 3′ events. We investigated the possible involvement of Swd2 ubiquitylation in loop interactions of the GAL10 gene through Chromosome Conformation Capture (Singh et al., 2009). Using targeted 3C primers (Suppl. Table S2), a transcription-dependent gene loop was detected in GAL10 whose formation was significantly impaired in ubiquitylation mutants of Swd2 (Figure 1e), reinforcing the hypothesis that ubH2B-controlled Swd2 ubiquitylation in APT plays a general role in coordinating co-transcriptional events with mRNA export from the nucleus.
Ubiquitylation of Swd2 controls the integrity and stability of the APT complex as well as its interaction with the mRNA nuclear export machinery
To assess the composition of the APT complex with respect to Swd2 ubiquitylation, SYC1 was genomically ProtA-tagged in cells expressing wild-type Swd2 (wt) or ubiquitylation mutants of Swd2 (Nedea et al., 2008). Surprisingly, combining Syc1 tagging with the K68,69R mutation of Swd2 (but not with swd2-HA K25R) caused a strong growth defect at 30°C and lethality at 39°C whereas Ref2-ProtA had no effect (Figure 2a). Western blot analysis in swd2-HA K68,69R/Syc1-ProtA cells at 30°C showed a significant decrease in both Swd2 and Syc1-ProtA levels compared to wt (Figure 2b) that probably caused dissociation of Glc7 and Ref2 as observed upon deletion of SWD2 (Nedea et al., 2008). The composition of the APT complex was thus severely affected in swd2-HA K68,69R/Syc1-ProtA cells thus impairing their viability. Noteworthy, growth defect of both swd2-HA K68,69R and syc1Δ cells at 39°C likely reflects a partial destabilization of the APT when either mutant is submitted to increased temperature (Figure 2a, not shown).
Figure 2. Ubiquitylation of Swd2 controls the integrity of the APT complex and the interaction with the export machinery.
(a) Serial dilutions of the indicated strains were incubated at the indicated temperature. (b) Steady-state levels of indicated proteins was analyzed by western blot in the indicated strains grown at 30°C. (c) The stability of the wt or mutant Swd2-HA protein was analyzed upon treatment with cycloheximide (CX) for the indicated times (min) at 39°C. The graph represents the percentage of Swd2-HA remaining relative to time 0 (mean ± SD) (d) Lysates (Input) of Swd2-HA tagged or nontagged (−) control strains were immunoprecipitated (IP) using an anti-HA antibody and analyzed by Western Blot using the indicated antibodies. (f) left panel, Two-hybrid analysis of the Lex-Mex67 baits versus Gal4-Swd2 derived preys. Right panel, Chromatin fraction from cross-linked and sonicated extracts were immunoprecipitated (IP) using anti-HA antibody and analyzed by Western blotting using anti-HA and anti-Mex67 antibodies.
Intriguingly, the growth defect resulting from the Swd2 K68,69R mutation was reversed when Swd2 K25 was replaced by an arginine residue (Figure 2b). Consistently, the K25R mutation of Swd2 restored expression level of both Swd2 and Syc1-ProtA observed upon mutation of K68,69 to R (Figure 2b). These data suggest that K25 ubiquitylation could participate in the degradation of Swd2. Indeed, cycloheximide chase experiments revealed that the half-life of swd2-HA K25R is twice as long (28 min) as that of wt Swd2-HA or swd2-HA K68,69R (12 min) (Figure 2c). Accordingly, a fraction of Swd2 is poly-ubiquitylated by K48-linked ubiquitin chains which targets Swd2 to the 26S proteasome (Vitaliano-Prunier et al., 2008). Similarly to combined mutations of K25 and K68, 69, deletion of the ubiquitin ligase Bre1, which controls ubiquitylation of Swd2 on both sites (Vitaliano-Prunier et al., 2008), did not induce any steady-state change in Swd2 or Syc1-ProtA expression and increased Swd2 half-life (Figure 2b and not shown). Finally, preventing ubiquitylation of Swd2 on both K25 and K68, 69 decreased the co-transcriptional recruitment of Syc1-ProtA, indicating that Swd2 ubiquitylation is required for the recruitment of a whole APT complex to actively transcribed genes (Suppl. Figure S1e). Together these results indicate that the separate ubiquitylation sites within Swd2 play distinct functions within the APT complex with K68,69 being involved in maintaining the integrity of the complex on transcribing genes and K25 controlling the degradation of Swd2 and the consequent APT complex stability.
Fully mature and correctly packaged S. cerevisiae mRNPs are released from the transcription site and transported to the cytoplasm by the heterodimeric export receptor Mex67/Mtr2, which promotes translocation through the nuclear pore complexes (NPC) by direct interactions with phenylalanine/glycine (FG)-nucleoporins (Gruter et al., 1998; Segref et al., 1997). Mex67 requires mRNA-binding adaptors that associate with transcripts during the transcription process. Current models predict the existence of at least two different but partially redundant mRNA export pathways defined by the SR-like protein, Npl3 (Gilbert and Guthrie, 2004), and the Nab2/Yra1 complex (Iglesias et al., 2010). Interestingly Npl3, Nab2 and Mex67 co-immuno-precipitated with Swd2 (Figure 2d). Interaction between Mex67 and Swd2 is mediated by the UBiquitin-Associated (UBA) domain of Mex67 and affected by the Swd2 K68,69R mutation suggesting that ubiquitylation of Swd2 couples the APT complex to the nuclear export machinery (Figure 2e).
Ubiquitylation of H2B and Swd2 are required for the correct recruitment of nuclear export machinery to mRNA
To further dissect how H2B ubiquitylation and consequently Swd2 ubiquitylation control mRNA transport, nuclear mRNPs were purified from strains expressing wt or ubiquitylation-defective mutant H2B using a genomically TAP-tagged version of the nuclear cap binding protein Cbp20 (Iglesias et al., 2010). As expected, Mex67 and its adaptors specifically co-purified with mRNPs from wt cells (Figure 3a). Although Swd2 was not detected in association with mRNPs, a transient interaction cannot be formally excluded. Preventing ubiquitylation of H2B severely affected the recruitment of both the Nab2/Yra1 complex and Npl3 to nuclear mRNPs (Figure 3a). Consequently, association of Mex67 with mRNPs was significantly decreased in H2B mutant cells resulting in nuclear export defect. The amount of Cbp20-bound nuclear mRNAs was decreased by only 20% upon H2B-K123R mutation indicating that the transcriptional defect conferred by this mutation cannot explain the drastic effect on the mRNP composition (not shown). To confirm a role of ubH2B in the recruitment of export machinery to mRNPs, recruitment of Mex67 to chromatin-associated mRNAs corresponding to precise transcribed genes was analyzed by RT-qPCR (RNA-ChIP). The expression of the constitutively expressed PMA1 transcript in htb1 K123R cells corresponded to 80% of the wt level. In contrast, the recruitment of Mex67 to PMA1 mRNA (relative to total PMA1 mRNA) in mutant cells corresponded to only 40% of the recruitment in wt cells along the length of the transcript (Figure 3b). These data indicate that ubH2B tightly controls the recruitment of the nuclear export machinery to transcribing mRNA.
Figure 3. Ubiquitylation of H2B and Swd2 are required for the correct recruitment of the mRNA nuclear export machinery.
(a, c) Nuclear mRNPs were purified from indicated Cbp20-TAP tagged strains using IgG-Sepharose beads. Co-purifying proteins were detected by Western Blot using anti-Nab2, anti-Yra1, anti-Npl3 or anti-Mex67 antibodies. The ratio of mRNPs-associated export factors relative to the wt cells and to the immuno-purified Cbp20-TAP was determined from 3 independent experiments (mean ± SD). (b, d) RNA ChIP on the PMA1 mRNA was performed with cross-linked chromatin from indicated strains using the Mex67 antibody. (b, left panel) mRNA expression level of PMA1 was normalized by polIII-transcribed scR1 RNA expression and is represented as a percentage of the wt strain (mean ± SD). (b, middle panel; see also Figure S2) Distribution of Mex67 along the PMA1 mRNA in wt cells (b, right panel and d) Recruitment of Mex67 to PMA1 mRNA in wt and mutant cells is represented as a percentage of the recruitment detected in the control wt strain (mean ± SD). Any DNA contamination of the RNA-ChIP was controlled by omitting the reverse transcription step prior to qPCR analysis (No RT). (e) Bulk poly(A) tail length was analyzed in indicated strains at 30°C or 39°C. The amount of poly(A) longer than 70–80 nt was quantified by measuring the intensity of the signal above 70–80 nt normalized by the intensity of the signal around 30 nt and by the background of the blot (mean ± SD) (see also Figure S2). (f) Localization of poly(A) RNA was analyzed by FISH using Cy3-labelled oligo dT probe in Swd2–HA wt, swd2-HA K25R or swd2-HA K68,69R strains overexpressing Nab2 (pRS426-Nab2) or not (Empty vector) after a 3 hr shift to 39°C. The ratio of cells with nuclear accumulation of poly(A) RNA relative to wt control cells was determined by examining at least 300 DAPI-stained cells from three independent FISH experiments (mean ± SD). (g) Serial dilutions of the corresponding strains were spotted onto YPD plates and grown at the indicated temperature (right panel).
In agreement with these results, Swd2 mutation of K68,69R and to a lesser extent K25R, caused a substantial decrease in the amount of Nab2, Yra1 and Mex67 (Figure 3c) that co-purified with Cbp20-containing mRNPs. Similar defects in Mex67 recruitment were observed when analyzed by RNA-ChIP assay on PMA1 mRNA (Figure 3d and Suppl. Figure 3). In contrast to Nab2 and Yra1, recruitment of the Npl3 adaptor was not significantly affected in ubiquitylation mutants of Swd2 (Figure 3c). We also observed a severe growth defect when a thermosensitive allele of NPL3 (npl3-1) was combined with swd2-HA K25R at permissive temperature (not shown). Together these results indicate that the Npl3-dependent export pathway is not affected in ubiquitylation mutants of Swd2 and provide an explanation for why inhibition of Swd2 ubiquitylation leads to a less severe mRNA export defect than blocking H2B ubiquitylation (Figure 1a, b).
Nab2 is a shuttling hnRNP protein that is not only required for nuclear export of mRNA but also restricts the poly(A) tail length (Anderson et al., 1993; Green et al., 2002; Hector et al., 2002). In agreement with the decrease of Nab2 in nuclear mRNPs purified from swd2-HA K68,69R mutant cells, the poly(A) tail length distribution of the global poly(A) RNA pool was ~15 nucleotides longer in swd2-HA K68,69R cells compared to wt or swd2-HA K25R cells following a 3 h shift at 39°C (Figure 3e). In contrast to thermosensitive mutants (ts) or unstable mutants of Swd2 (Cheng et al., 2004; Dichtl et al., 2004; Nedea et al., 2003; Nedea et al., 2008), Swd2 ubiquitylation did not affect RNAPII transcription termination at either 30°C or 39°C, or polyadenylation of snoRNA prior to their maturation by the exosome (Suppl. Figure S2). Thus preventing Swd2 ubiquitylation does not globally affect the CPF function but modulates poly(A) tail length. Proper ubiquitylation of H2B leading to subsequent ubiquitylation of Swd2 thus facilitates the recruitment of Yra1/Nab2 to mRNPs allowing proper control of poly(A) tail length and efficient nuclear export. This model is supported by the finding that overexpression of Nab2 restored wild-type mRNA export in ubiquitylation mutants of Swd2 (Figure 3f), as well as a wild type growth of swd2-HA K68,69R/Syc1-ProtA cells at both 30°C and 39°C (Figure 3g).
In this study, we show that H2B ubiquitylation controls the formation of export-competent mRNPs. This function is in part, mediated by the ability of ubH2B to facilitate ubiquitylation of Swd2 on both K25 and K68, 69 residues, a process that occurs at the 5′ end of transcribing genes (Vitaliano-Prunier et al., 2008) and that respectively regulates the stability and the integrity of the APT complex at the 3′ end. This tight control is required for the correct recruitment of the mRNA export receptor, Mex67, to mature nuclear mRNPs as well as the Yra1/Nab2 adaptor complex which, in turn, controls poly(A) tail length and facilitates efficient nuclear export of mRNPs (Figure 4). Such a mechanism accounts for the well described functional coupling between 3′-end maturation, and particularly poly(A) tail length, and nuclear export of mRNA and suggests that longer poly(A) tails do not result from nuclear export defects but rather are a consequence of a Nab2/Yra1 recruitment defect. Interestingly, ubH2B also controls the recruitment of Npl3 to nuclear mRNPs independent of Swd2 modification, which is consistent with the synthetic lethality observed between NPL3 and BRE1 (Pan et al., 2006). Another APT component, the Glc7 phosphatase, promotes the dephosphorylation of Npl3, a molecular event required for proper mRNA export (Gilbert and Guthrie, 2004). Finally, Swd2 ubiquitylation but also Ssu72 and Pta1, two members of the APT complex participate in the formation of a gene loop between the 5′ and 3′ ends of transcribing genes (Ansari and Hampsey, 2005), further supporting the view that the APT complex is at the intersection of the crosstalk between 5′ and 3′ co-transcriptional events and in particular, histone modification, mRNA 3′-end formation and nuclear export (Figure 4). Consistent with the results of this study implicating ubiquitin modification in coupling post-transcriptional events with mRNA export, ubiquitylation of Yra1 by the Tom1 ligase controls dissociation of Yra1 from the Nab2-bound mRNP complex facilitating the export of mature and properly assembled mRNPs (Iglesias et al., 2010). In addition, ubiquitylation of Hpr1, a component of the THO complex associated with the transcription machinery, facilitates the co-transcriptional recruitment of Mex67 through its UBA domain. This interaction also coordinates recruitment of the mRNA export machinery with transcription and early mRNP assembly (Gwizdek et al., 2005; Gwizdek et al., 2006; Hobeika et al., 2007). Taken together this evidence suggests that ubiquitin modification tightly controls the timing of the inter-connected molecular events that culminate in formation of export-competent mRNPs.
Figure 4. Model recapitulating the role of ubH2B in the crosstalk between 3′ end processing and nuclear export.

UbH2B facilitates subsequent ubiquitylation of the Swd2 protein on both K25 and K68, 69 residues, a process that precisely regulates both the stability and the integrity of the APT complex at the 3′ end of transcribing genes. Ubiquitylated Swd2 is required for the correct recruitment of the mRNA export receptor Mex67 and its Yra1/Nab2 adaptor complex to mature mRNPs. Independent of Swd2, ubH2B also controls the recruitment of the Mex67 adaptor Npl3 to mRNPs. Preventing H2B ubiquitylation leads to mRNPs devoid of mRNA export machinery that are consequently retained in the nucleus.
Experimental Procedures
Strains and Plasmids
The strains and plasmids used in this study are described in Supplementary table S1.
RNA-ChIP assay
RNA-ChIP experiments were performed as previously described (Gwizdek et al 2006) with the following modifications. Cells were grown in YPD at 30°C and shifted to 39°C for 3h to an OD600 = 0.8–1.0. Cells were then crosslinked with 1% formaldehyde for 10 min at 39°C. Sonicated extracts were centrifuged for 5 min at 2500g prior to overnight immunoprecipitation using anti-Mex67-coated Protein G–Sepharose beads. After reversing crosslinking, total and immunoprecipitated RNAs were purified using the Nucleospin RNA II kit from Macherey Nalgen. After retro-transcription with random oligonucleotides (Roche) using the SuperScript™ II reverse Transcriptase (Invitrogen), real time qPCR was performed using the Syber Green mix (Roche) and the Light Cycler 480 system (Roche) with gene specific primers corresponding to a 150 bp fragments described in Suppl Table S2.
Statistical analysis
Histograms depicts the mean and standard deviations of at least three independent experiments. Significance of the differences observed between wild-type and mutant cells was evaluated using Student t test. No asterisk corresponds to p value above 0,05, one asterisk corresponds to p values between 0,01 and 0,05, two asterisks corresponds to p value between 0,001 and 0,01 and three asterisks corresponds to p value below 0,001.
Supplementary Material
Highlights.
H2B ubiquitylation controls mRNA export via ubiquitylation of the CPF component Swd2.
Monoubiquitylation of Swd2 controls the integrity and stability of a CPF subcomplex.
The CPF subcomplex APT coordinates polyadenylation and export via Nab2 recruitment.
H2B-dependent modification of Swd2 facilitates gene looping.
Acknowledgments
We would like to thank C. Moore, L. Minvielle-Sebastia, C. Guthrie for reagents and V. Oréal for his help. We are most grateful to B. Palancade, F. Stutz, J. Weitzman and V. Géli for critical reading of the manuscript. This study was funded by grants from the Agence Nationale pour la Recherche (BLAN1227-01 to CD) and the Ligue contre le Cancer (CD’s team is “Equipe labellisee”). LH is supported by the University Paris V, AVP by the Agence Nationale pour la Recherche and AB by the Association de Recherche contre le Cancer.
Footnotes
Additional procedures are detailed in the supplementary information.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson JT, Wilson SM, Datar KV, Swanson MS. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Molecular and cellular biology. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A, Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes & development. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apponi LH, Kelly SM, Harreman MT, Lehner AN, Corbett AH, Valentini SR. An interaction between two RNA binding proteins, Nab2 and Pub1, links mRNA processing/export and mRNA stability. Molecular and cellular biology. 2007;27:6569–6579. doi: 10.1128/MCB.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, He X, Moore C. The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Molecular and cellular biology. 2004;24:2932–2943. doi: 10.1128/MCB.24.7.2932-2943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell. 2008;29:271–278. doi: 10.1016/j.molcel.2007.11.035. [DOI] [PubMed] [Google Scholar]
- Dheur S, Vo le TA, Voisinet-Hakil F, Minet M, Schmitter JM, Lacroute F, Wyers F, Minvielle-Sebastia L. Pti1p and Ref2p found in association with the mRNA 3′ end formation complex direct snoRNA maturation. Embo J. 2003;22:2831–2840. doi: 10.1093/emboj/cdg253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Aasland R, Keller W. Functions for S. cerevisiae Swd2p in 3′ end formation of specific mRNAs and snoRNAs and global histone 3 lysine 4 methylation. RNA (New York, NY) 2004;10:965–977. doi: 10.1261/rna.7090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Furger A, O’Sullivan JM, Binnie A, Lee BA, Proudfoot NJ. Promoter proximal splice sites enhance transcription. Genes & development. 2002;16:2792–2799. doi: 10.1101/gad.983602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell. 2004;13:201–212. doi: 10.1016/s1097-2765(04)00030-9. [DOI] [PubMed] [Google Scholar]
- Green DM, Marfatia KA, Crafton EB, Zhang X, Cheng X, Corbett AH. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J Biol Chem. 2002;277:7752–7760. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber BK, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- Gwizdek C, Hobeika M, Kus B, Ossareh-Nazari B, Dargemont C, Rodriguez MS. The mRNA nuclear export factor Hpr1 is regulated by Rsp5-mediated ubiquitylation. J Biol Chem. 2005;280:13401–13405. doi: 10.1074/jbc.C500040200. [DOI] [PubMed] [Google Scholar]
- Gwizdek C, Iglesias N, Rodriguez MS, Ossareh-Nazari B, Hobeika M, Divita G, Stutz F, Dargemont C. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Natl Acad Sci U S A. 2006;103:16376–16381. doi: 10.1073/pnas.0607941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Gross S, Zenklusen D, Heath CV, Stutz F, Moore C, Cole CN. Coupling of termination, 3′ processing, and mRNA export. Molecular and cellular biology. 2002;22:6441–6457. doi: 10.1128/MCB.22.18.6441-6457.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector RE, Nykamp KR, Dheur S, Anderson JT, Non PJ, Urbinati CR, Wilson SM, Minvielle-Sebastia L, Swanson MS. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. Embo J. 2002;21:1800–1810. doi: 10.1093/emboj/21.7.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobeika M, Brockmann C, Iglesias N, Gwizdek C, Neuhaus D, Stutz F, Stewart M, Divita G, Dargemont C. Coordination of Hpr1 and ubiquitin binding by the UBA domain of the mRNA export factor Mex67. Mol Biol Cell. 2007;18:2561–2568. doi: 10.1091/mbc.E07-02-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Von Dach E, Corbett AH, Dargemont C, Stutz F. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes & development. 2010;24:1927–1938. doi: 10.1101/gad.583310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Patricio K, McCarthy T, Rosbash M. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol Cell. 2001;7:887–898. doi: 10.1016/s1097-2765(01)00232-5. [DOI] [PubMed] [Google Scholar]
- Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S. Distinct pathways for snoRNA and mRNA termination. Mol Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Luna R, Gaillard H, Gonzalez-Aguilera C, Aguilera A. Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma. 2008;117:319–331. doi: 10.1007/s00412-008-0158-4. [DOI] [PubMed] [Google Scholar]
- Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell. 2010;40:410–422. doi: 10.1016/j.molcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, Greenblatt J. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem. 2003;278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- Nedea E, Nalbant D, Xia D, Theoharis NT, Suter B, Richardson CJ, Tatchell K, Kislinger T, Greenblatt JF, Nagy PL. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol Cell. 2008;29:577–587. doi: 10.1016/j.molcel.2007.12.031. [DOI] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Lykke-Andersen S, Nasser T, Saguez C, Bertrand E, Jensen TH, Moore C. Assembly of an export-competent mRNP is needed for efficient release of the 3′-end processing complex after polyadenylation. Molecular and cellular biology. 2009;29:5327–5338. doi: 10.1128/MCB.00468-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. Embo J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra CA, Hammell CM, Heath CV, Cole CN. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes & development. 1997;11:2845–2856. doi: 10.1101/gad.11.21.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. Embo J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BN, Ansari A, Hampsey M. Detection of gene loops by 3C in yeast. Methods (San Diego, Calif) 2009;48:361–367. doi: 10.1016/j.ymeth.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaliano-Prunier A, Menant A, Hobeika M, Geli V, Gwizdek C, Dargemont C. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol. 2008;10:1365–1371. doi: 10.1038/ncb1796. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.