SUMMARY
Pol II(G) is a distinct form of RNA polymerase II that contains the tightly associated Gdown1 polypeptide (encoded by POLR2M). Unlike Pol II, Pol II(G) is highly dependent upon Mediator for robust activator-dependent transcription in a biochemically defined in vitro system. Here, in vitro studies show that Gdown1 competes with TFIIF for binding to the RPB1 and RPB5 subunits of Pol II, thereby inhibiting an essential function of TFIIF in preinitiation complex assembly, but also that Mediator can actually facilitate Pol II(G) binding to the promoter prior to subsequent Mediator functions. Complementary ChIP and RNAi analyses reveal that Pol II(G) is recruited to promoter regions of subsets of actively transcribed genes, where it appears to restrict transcription. These and other results suggest that Pol II(G) may act to modulate some genes while simultaneously, as a poised (non-initiated) polymerase, setting the stage for Mediator-dependent enhancement of their activity.
INTRODUCTION
In the eukaryotic genome, Pol II is the central transcription machine that is responsible for transcription of protein-coding genes. Gene transcription is regulated at multiple steps that involve initiation, elongation and termination. Upon specific signals, a constellation of transcription factors converges on Pol II to achieve transcriptional activation of specific genes. Among the factors that act in concert with Pol II at core promoters, the general transcription factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (GTFs) are essential core components that are involved in formation of a functional preinitiation complex (PIC) (Thomas and Chiang, 2006). PIC formation is one of the critical steps in the transcription cycle that may directly regulate transcription. However, recent genome-wide analyses have revealed that Pol II may be enriched at promoter-proximal regions of both transcriptionally active and inactive genes, indicating that regulation at these genes might take place at a post-recruitment step (Guenther et al., 2007, Muse et al., 2007, Zeitlinger et al., 2007). Another factor important for transcriptional activation is the Mediator, an evolutionarily conserved 30-subunit complex that facilitates transcription activation of numerous genes and acts at least in part as a bridge between Pol II and DNA-binding transcriptional factors (Malik and Roeder, 2010).
From yeast to human, Pol II is composed of 12 highly conserved subunits, designated RPB1 to RPB12. However, recent studies of Hu et al. (2006) have revealed a distinct form of Pol II that contains an additional, tightly associated polypeptide called Gdown1 (encoded by POL2RM). Because of the high affinity of Gdown1 for Pol II, this form of Pol II was named Pol II(G). All mammalian species have orthologs of Gdown1; and genomic sequence comparisons have further identified an ortholog in Drosophila melanogaster, but not in lower organisms such as Caenorhabditis elegans. In an in vitro assay system reconstituted with pure factors, Pol II(G), unlike the 12-subunit Pol II (designated Pol II), fails to activate transcription in the absence of Mediator (Hu et al., 2006), indicating that Pol II(G) has unique properties relative to Pol II. The fact that Mediator reverses the repressed transcriptional capacity of Pol II(G) indicates that Pol II(G) depends on Mediator to activate transcription. However, the mechanisms underlying the Gdown1-mediated repression of Pol II and the reversal of this repression by Mediator have not been described.
Here we show that Gdown1 competes with TFIIF for binding to Pol II in vitro, resulting in an inhibition of a critical step in PIC assembly that is controlled by TFIIF. Despite this competition, Pol II(G) can be recruited to the promoter in an activator- and Mediator-dependent manner in vitro, with Mediator playing a critical role in this recruitment. Complementary ChIP and RNAi studies further reveal that Pol II(G) is recruited to the promoter regions of a subset of actively transcribed genes and that a mechanism entailing modulation by Gdown1 is at work at these loci. Our findings identify a distinct role for Pol II(G), relative to Pol II, in transcriptional regulation through a mechanism that involves inhibitory effects of Gdown1 on TFIIF functions and establishment of a poised (non-initiated) polymerase for subsequent Mediator-dependent activation.
RESULTS
Gdown1 represses transcription by inhibiting TFIIF function in pre-initiation complex assembly
In extending our previous studies (Hu et al., 2006), we first determined whether transcription activation by the tumor suppressor p53 is also subject to regulation by Pol II(G). In an in vitro system reconstituted with purified factors (GTFs and the cofactor PC4) and a model template containing a GADD45-derived p53 responsive element upstream of the adenovirus major late (AdML) core promoter, we observed that Pol II(G) was markedly less efficient than Pol II in mediating p53-dependent transcription (Figure 1A, lane 2 versus 6). However, in the presence of Mediator, Pol II and Pol II(G) exhibited equivalent levels of activity comparable to that observed with Pol II in the absence of Mediator (lanes 4 and 8 versus lane 6), indicating that Gdown1 represses p53-dependent transcription by Pol II and that Mediator effectively reverses this repression (lane 4). These results are similar to those observed with HNF4 (Hu et al., 2006), and further establish the potential for Gdown1-elicited Mediator function with different activators. Notably, in these and the following in vitro experiments, p53 has been used as a model activator, along with the model promoter, to establish basic mechanistic principles. However, because of this reductionist approach, and in view of subsequent Gdown ChIP-seq analyses (described below), Gdown1 function may be restricted to a limited set of p53 target genes.
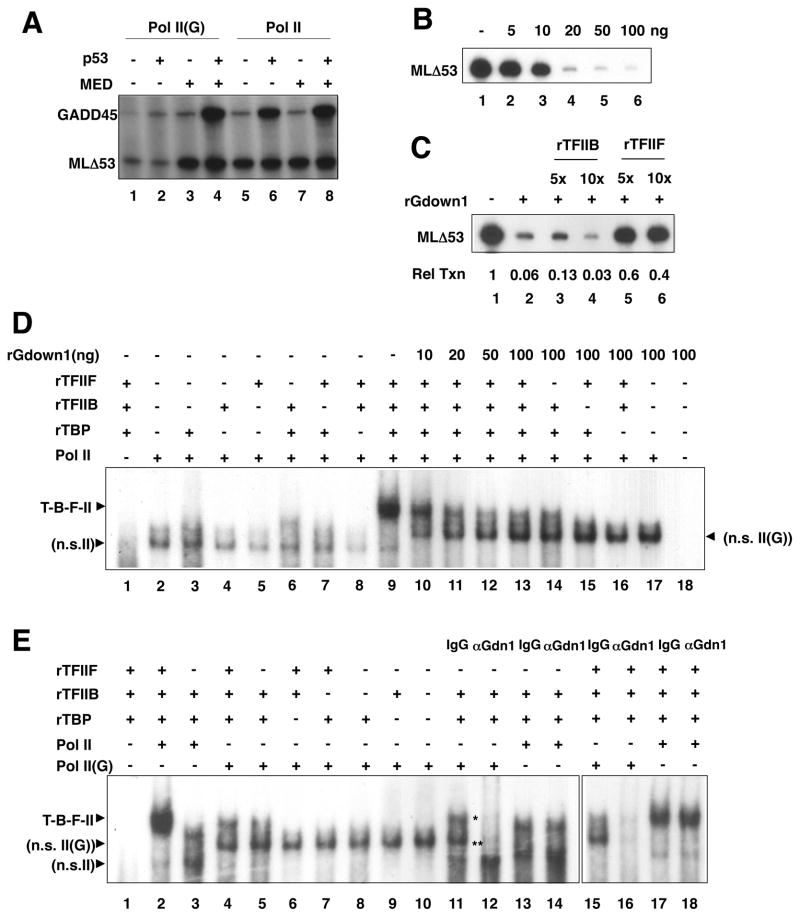
Figure 1. Gdown1 competes with TFIIF for binding to Pol II in vitro.
(A) p53-dependent and basal transcription with Pol II and Pol II(G). Transcription was assayed in a completely defined system containing GTFs, PC4 and plasmids containing the adenovirus major late (AdML) core promoter and either p53-binding sites (GADD45) (An et al., 2004) or no activator binding sites (MLΔ53). p53 and Mediator were added as indicated. (B) Inhibitory effect of recombinant Gdown1 on basal transcription. In vitro transcription reactions were reconstituted with 10 ng of TBP, 10 ng of TFIIB, 25 ng of TFIIF and 50 ng of Pol II. The indicated amounts of rGdown1 were added to reaction mixtures. Reaction mixtures also contained 50 ng of pMLΔ53. (C) Partial recovery of basal transcription by addition of excess TFIIF. In vitro transcription reaction mixtures were as described in (B). Reaction mixtures contained 5- or 10-fold excesses of rTFIIB or rTFIIF and 20 ng of rGdown1 as indicated. (D) Inhibition of PIC assembly by Gdown1. An end-labeled Ad ML oligonucleotide (−40 to +20) probe was incubated with the indicated combinations of factors: TFIIB (10 ng, lane 2–5), TFIIF (50 ng, lanes 2–5), and bovine Pol II (50 ng). All reactions contained PC4 (65 ng) and TBP (10 ng). Reactions were incubated at 30°C for 40 min and purified RNA products resolved by native PAGE. (E) Pol II(G)-promoter complex formation in the absence of TFIIF. EMSA was performed as described in D. Indicated combinations of factors were incubated with the probe. All the reactions contained TFIIB (10 ng), PC4 (65 ng), and TBP (10 ng). One μg of a purified IgG (lane 6) or Gdown1 antibody (lane 7) was added to the reactions. A single asterisk denotes the upper band and two asterisks denote the middle band. See also Figure S1.
Importantly, we also observed that under these conditions Mediator could reverse not only the Gdown1-mediated repression of activator-dependent transcription but also the Gdown1-mediated repression of activator-independent (basal) transcription (lane 3) from the MLΔ53 control template. This suggested that factor interactions common to basal and activated transcription are affected by an interplay between Gdown1 and Mediator. Therefore, and in order to elucidate the mechanism of inhibition by Pol II(G) in the absence of Mediator, we investigated the effect of Gdown1 on activator-independent transcription. To this end, we employed a reconstituted in vitro transcription assay containing Pol II, TBP, TFIIB, and TFIIF, which are sufficient (without TFIIE and TFIIH) for basal transcription from supercoiled TATA-containing DNA templates (Tyree et al., 1993). The resulting basal activity was dramatically reduced in a dose-dependent manner by recombinant Gdown1 (rGdown1), with near complete inhibition at a level of 20 ng (Figure 1B, lane 4). We reasoned that Gdown1 impedes the function of one or more of the general transcription factors and that addition of an excess amount of that factor(s) would reverse the inhibitory effect by Gdown1. To test this possibility, 5- to 10-fold molar excesses of rTFIIB and rTFIIF were added independently to basal transcription reactions in the presence of 20 ng of Gdown1 and the basal activities were measured. Whereas the addition of excess rTFIIB showed no effect (Figure 1C, lane 2 versus lane 4), a 5-fold molar excess of rTFIIF resulted in recovery of up to 60% of the basal activity observed in the absence of Gdown1 (lane 1 versus lane 5). Consistent with its ability to reverse Gdown1-mediated inhibition of basal transcription in the complete assay system (Figure 1A) and Gdown1 effects on TFIIF, Mediator also countered Gdown1-mediated inhibition in the minimal assay with TBP, TFIIB and TFIIF (data not shown).
Since TFIIF is critical for PIC formation and directly associates with Pol II (Thomas and Chiang, 2006), the observed inhibitory effect of Gdown1 on transcription might reflect an inhibition of PIC assembly. To test this possibility, an electrophoretic mobility-shift assay (EMSA) was employed. An oligonucleotide containing AdML core promoter sequences from nucleotide positions −40 to +20 relative to the transcription start site was used as a probe to test effects of Gdown1 on formation of promoter sub-complexes (minimal PICs) by TBP, TFIIB, TFIIF, and Pol II in the presence of PC4 (Malik et al., 1998) (Figure 1D). The efficient formation of the expected higher order (TBP-TFIIB-TFIIF-Pol II) promoter complex was observed only if all three factors (TBP, TFIIB and TFIIF) were added with Pol II (lane 9), while no comparable level of a higher order promoter complex was seen when Pol II was incubated with any subset of these GTFs (lanes 3 to 8). Under our conditions, neither TBP-TFIIB nor TBP-TFIIB-TFIIF complex formation was observed (lane 1). Some non-specific (GTF-independent) binding of Pol II to the probe was observed; however, this level of non-specific complex formation was much lower than that of the bona fide TBP-TFIIB-TFIIF-Pol II complex (compare lanes 2 and 9). When rGdown1 was included in the binding reactions, formation of this TBP-TFIIB-TFIIF-Pol II promoter complex was inhibited in a dose-dependent manner (lanes 10 to 13), consistent with the transcription inhibition data (Figure 1B, lanes 4–6). Nonetheless, in contrast to the results of the transcription data, some higher order promoter complex formation was detectable even in the presence of 50 ng or 100 ng of rGdown1 (Fig. 1D, lanes 12 and 13 versus Figure 1B, lanes 5 and 6). Thus, concomitant with the partial loss of the slower-migrating TBP-TFIIB-TFIIF-Pol II promoter complex, a weaker faster-migrating band was seen upon addition of Gdown1 to these reactions. Further analyses revealed, surprisingly, that the residual slower-migrating Pol II complex is likely a TBP-TFIIB-Pol II-Gdown1 complex since it was formed in the absence of TFIIF (lane 14) but not when either TBP (lane 16) or TFIIB (lane 15) was omitted. By contrast, and because Gdown1 does not directly bind to the probe (lane 18), the new faster-migrating band formed in the absence of all these GTFs (lanes 14–17) likely reflects a non-specific Pol II(G) complex. Overall, these EMSA data show that Gdown1 inhibits authentic PIC assembly (with Pol II) while allowing formation of an alternate, but considerably weaker, higher order promoter complex (with Pol II(G)).
In the next series of experiments, instead of adding ectopic Gdown1 to the reactions, we analyzed promoter complex formation by a standard (12-subunit) Pol II preparation and by a Pol II(G) preparation that contains near-stoichiometric Gdown1 (Hu et al., 2006). Once again, Pol II formed a higher order promoter complex in the presence of TBP, TFIIB, and TFIIF (Figure 1E, lane 2). Notably, this analysis reaffirmed the TFIIF requirement for efficient PIC assembly by Pol II since its omission resulted in formation, primarily, of a non-specific Pol II complex (lane 3). By contrast, Pol II(G) was significantly less efficient than Pol II in forming a comparable higher order promoter complex in the presence of TBP, TFIIB and TFIIF (lane 4 versus lane 2). Interestingly, however, some residual complexes were still formed with Pol II(G) (lane 4). These included both a complex that migrated similarly to the authentic Pol II promoter complex (upper band) and a faster-migrating species (lower band). The upper band was observed even in the absence of TFIIF (lane 5), but did require both TBP and TFIIB (lanes 6 to 10). On the other hand, the lower band was observed independently of TBP and TFIIB and thus, as in the EMSA of Fig. 1D, likely represents a non-specific Pol II(G) complex (note that Pol II(G) has a stronger tendency than Pol II to form non-specific complexes). These data suggest that Pol II(G) can form a higher order promoter complex that is independent of TFIIF but dependent upon TBP and TFIIB. However, this complex is formed less efficiently than the Pol II-TBP-TFIIB-TFIIF complex, at least in the absence of other components such as Mediator, and, based on the functional assays, is less capable of productive transcription.
To further ascertain the composition of the TFIIF-independent Pol II(G) complexes, we included anti-Gdown1 antibodies in the EMSA reactions. Formation of complexes with Pol II (both upper and lower bands) was not inhibited by addition of either an anti-Gdown1 antibody (lane 14) or control IgG (lane 13). However, formation of Pol II(G) complexes, including both the TBP- and TFIIB-dependent higher order complex (upper band, single asterisk) and the non-specific Pol II(G) complex (middle band, double asterisk), was specifically inhibited by addition of an anti-Gdown1 antibody (lane 12) and not by control IgG (lane 11). This inhibition was seen regardless of whether or not TFIIF was included in the reactions (lane 15 and 16 versus lane 17 and 18). Hence, the higher order TBP- and TFIIB-dependent promoter complex formed by Pol II(G) indeed contains Gdown1.
Overall, we conclude that Gdown1 can exclude TFIIF from the higher order promoter complex, which prevents efficient PIC formation. This, in part, could explain the lower levels of promoter-dependent transcription by Pol II(G), relative to Pol II, that are seen in the absence of Mediator.
Gdown1 and TFIIF show mutually exclusive interactions with Pol II
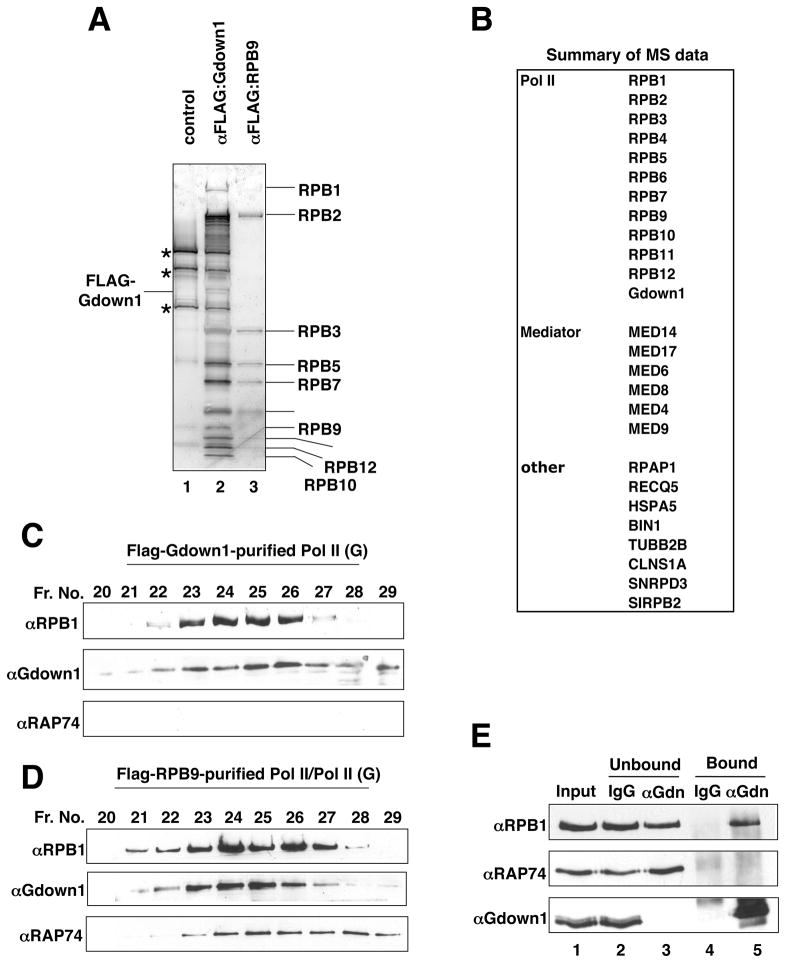
Since TFIIF directly interacts with Pol II, the above results predicted that Gdown1 might prevent TFIIF from binding to Pol II. To test this possibility and to identify other factors that might interact with Gdown1, we established a HeLa cell line (f:Gdown1) that stably expresses FLAG-tagged Gdown1. Derived nuclear extract was subjected to immuno-affinity purification on anti-FLAG antibody (M2 agarose) beads. In parallel, nuclear extracts from a HeLa cell line (f:RPB9) that stably expresses FLAG-tagged RPB9 and from control HeLa cells (“empty vector”) were processed as positive and negative controls, respectively. As shown in Figure 2A, the f:Gdown1 purified material contained polypeptides (lane 2) that co-migrated with Pol II subunits in the f:RPB9 purified preparation (lane 3), but not in the mock-purified preparation (lane1). Mass spectrometric analysis of the f:Gdown1 fraction identified 25 proteins, including 11 Pol II subunits and 6 Mediator subunits, that were not evident in the mock-purified fraction (Figure 2B). Of the remaining 8 proteins, RPAP1 (Jeronimo, et al., 2004) and RECQ5 (Aygun et al., 2008) were reported as Pol II-associating proteins. No TFIIF or other GTFs were observed.
Figure 2. Analysis of Pol II complexes.
(A) Purification of Gdown1 complex. Nuclear extracts from HeLa cells (lane 1) or from cell lines that stably express FLAG-tagged Gdown1 (lane 2) or FLAG-tagged RPB9 (lane 3) were directly purified on M2 agarose in Buffer C containing 0.18 M KCl and 0.1% NP40 and bound proteins were eluted and analyzed by SDS-PAGE with silver staining. (B) Mass spectrometric analysis of the Gdown1 complex. Proteins identified by MALDI mass spectrometry are indicated. (C and D) Gel filtration (Superose 6) analyses of FLAG-Gdown1 (C) and FLAG-RPB9 (D) complexes. Fractions were analyzed by immunoblot. (E) Pol II(G) does not interact with TFIIF. Fractions 24 and 25 in (C) were immunoprecipitated with rabbit IgG or anti-Gdown1 antibodies. The antibody-bound or unbound fractions were analyzed by immunoblot with indicated antibodies.
The affinity-purified f:Gdown1 and f:RPB9 preparations were further analyzed and compared by gel filtration on a Superose 6 column. Immunoblots of the Superose 6 fractions (Figure 2C and D) showed that, under these conditions, Pol II fractions purified from f:RPB9 nuclear extracts contained detectable levels of both TFIIF and Gdown1 that are presumably present in substoichiometric amounts (Figure 2D). By contrast, TFIIF was not detected in the Pol II fractions purified from f:Gdown1 nuclear extracts (Figure 2C). The Pol II fractions (Figure 2D, fractions 23–26) then were subjected to immunoprecipitation with anti-Gdown1 antibody to separate Pol II(G). As shown in Figure 2E, TFIIF was not detected in the Pol II(G)-containing immunoprecipitate (lane 3 versus lane 5), but was present with Pol II in the Gdown1-free unbound fraction.
Altogether, these data suggest that Pol II(G) and Pol II-TFIIF represent distinct enzyme forms and that, in cells, Gdown1 and TFIIF associate with Pol II in a mutually exclusive manner, leading to the hypothesis that Gdown1 competes with TFIIF for binding to Pol II.
Gdown1 competes with TFIIF by directly interacting with RPB5 and RPB1
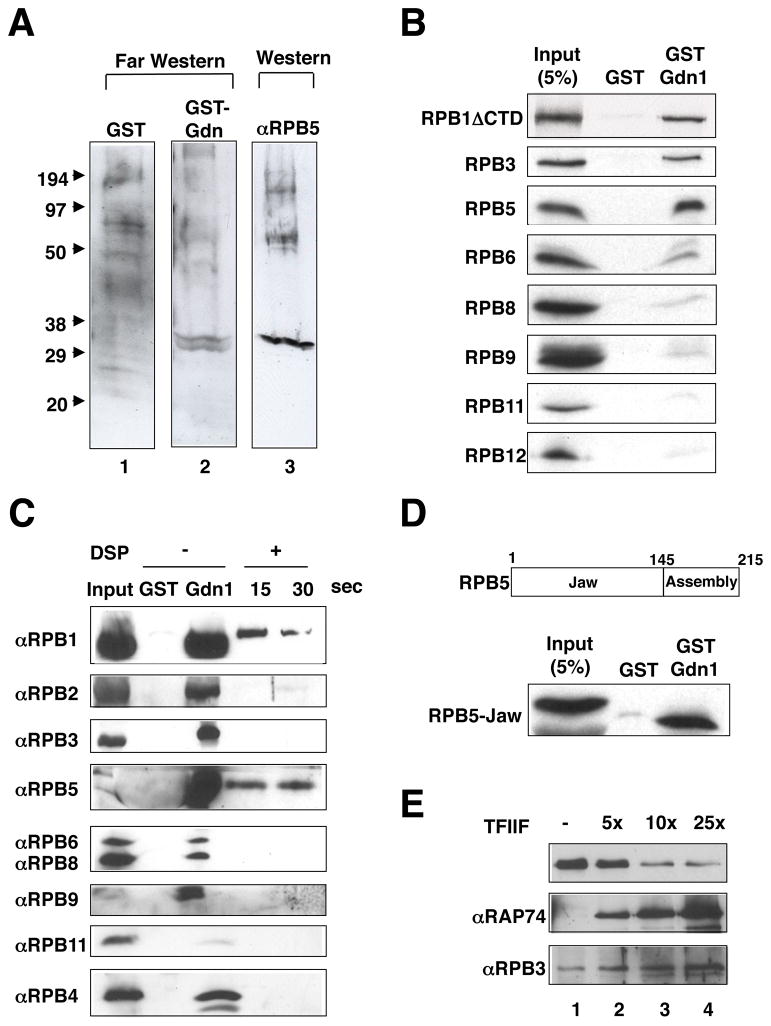
To further confirm our hypothesis, we determined the Pol II subunits that interact directly with Gdown1. To this end, we first employed a Far Western blot analysis. An affinity-purified Pol II (f:RPB9) preparation was separated by SDS-PAGE and processed for Far Western blotting with GST or GST-fused Gdown1. A prominent signal was observed around 29 kDa, which coincides with the molecular mass of the RPB5 subunit. By re-probing the blot membrane with anti-RPB5 antibody, the signal was confirmed to be RPB5 (Figure 3A). This result was further confirmed by a GST pull-down assay, with several in vitro translated Pol II subunits, which revealed a strong interaction of GST-fused Gdown1 not only with RPB5 but also with RPB1ΔCTD and RPB3 (Figure 3B).
Figure 3. Gdown1 interacts with RPB5 and RPB1.
(A) Far Western blot assay. Purified Pol II was resolved by SDS-PAGE and blotted to a membrane that then was incubated with GST (lane 1) or GST-Gdown1 (lane 2) protein as a probe. Membrane bound proteins were detected with HRP-labeled anti-GST antibody. The membrane that was probed with GST-Gdown1 was stripped and immunoblotted with an anti- RPB5 antibody (lane 3). (B) GST pulldown assay. Selected subunits of Pol II were expressed as 35S-labeled proteins in a rabbit reticulocyte coupled transcription and translation system and incubated with immobilized GST or GST-Gdown1. Bound proteins were analyzed by SDS-PAGE and autoradiography. (C) Cross-linking assay. Immobilized GST or GST-Gdown1 was incubated with purified Pol II. Proteins bound to the beads were incubated with/without DSP for the indicated times. After washing with 8 M urea buffer, the crosslinks were reversed and bound proteins were analyzed by immunoblot. (D) GST pulldown assay. Immobilized GST or GST-Gdown1 was incubated with 35S-labeled RPB5-jaw domain and bound proteins were analyzed by SDS-PAGE and autoradiography. (E) Dissociation of Gdown1 from Pol II by excess TFIIF. Pol II(G) was purified as described in Experimental Procedures. A 5-, 10-, or 25-fold excess of rTFIIF was added to the Pol II(G) immobilized beads in Buffer C containing 0.3 M KCl and 0.1% NP40. After washing the beads with Buffer C containing 0.1 M KCl and 0.1% NP40, the proteins that remained on the beads were analyzed by immunoblot.
To assess direct Gdown1-Pol II subunit interactions in the natural context of the intact Pol II(G) complex, we employed a protein-protein cross-linking analysis. Immobilized GST or GST-fused Gdown1 was incubated with HeLa nuclear extract and subjected to cross-linking with dithiobis (succinimidyl proprionate) (DSP) as detailed in the figure legend. Of the nine Pol II subunits tested, RPB1 and RPB5 showed strong cross-linking signals that are indicative of direct Gdown1 interactions whereas a suggested interaction with RPB6 (above) was not confirmed (Figure 3C). These results are consistent with the co-localization of RPB1 (“foot” and “cleft” regions) and the associated RPB5 (N-terminal “lower jaw” and C-terminal “assembly” domains) to the “shelf” module of Pol II (Cramer et al, 2001). Consistent with the reported function of RPB5 as a protein-protein interaction surface (Miyao and Woychik, 1998), an in vitro synthesized fragment containing the RPB5 jaw domain bound to GST-fused Gdown1 but not to GST (Figure 2D). These results show that Gdown1 directly interacts with RPB5 and RPB1 in the context of the intact Pol II and, further, that the RPB5 interaction minimally involves its jaw domain.
To directly test our hypothesis that Gdown1 may compete with TFIIF, we employed an in vitro competition assay. Pol II(G), which was purified from f:Gdown1 nuclear extracts, was immobilized on protein A-Sepharose beads that were saturated with an anti-CTD monoclonal antibody (8W16) and incubated with increasing amounts of TFIIF. The result (Figure 3E) shows that a 10-fold molar excess of TFIIF over Gdown1 significantly decreased the Gdown1 signal (lane 3), indicating that TFIIF is able to remove Gdown1 from Pol II, at least under the conditions (0.3 M KCl) employed. Although the competition was not observed at moderate salt (0.1 M KCl), the result clearly shows that Gdown1 can compete with TFIIF for binding to Pol II.
Taken together, these results suggest that Gdown1 competes with TFIIF for association with Pol II by directly interacting with RPB1 and RPB5 subunits of Pol II.
Pol II(G) is recruited to the promoter in an activator-enhanced manner and Mediator is essential for this recruitment
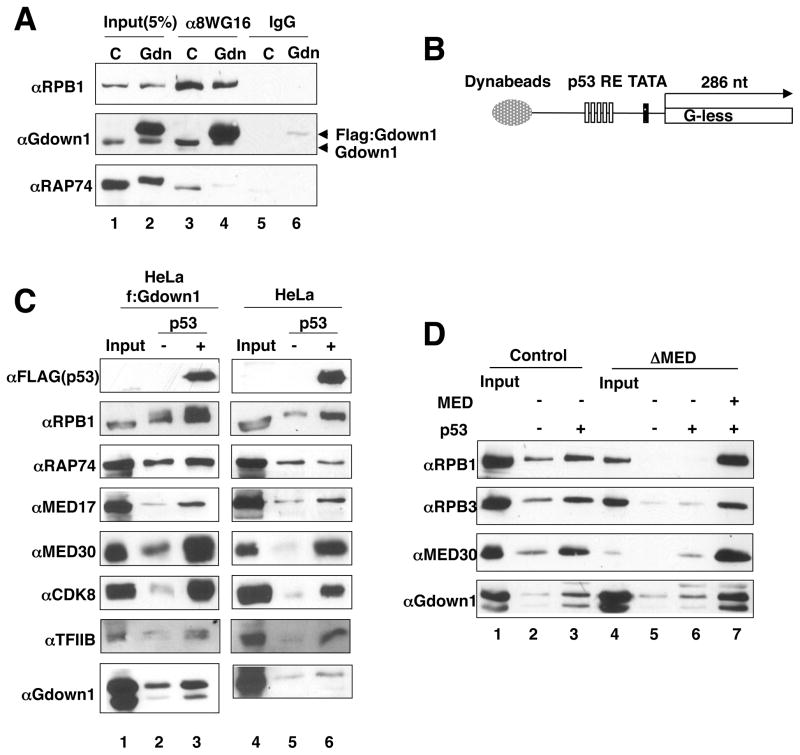
In view of the above results indicating that Pol II(G) is compromised in its ability to form a functional PIC, we next investigated the basis for our previous observation that Mediator could restore full activity to Pol II(G) (Hu et al., 2006; Fig. 1A). An immobilized template assay was employed in conjunction with nuclear extract from f:Gdown1-expressing cells. In these cells, ectopic Gdown1 was over-expressed relative to endogenous Gdown1 (Figure 4A lane 2). Indeed, as shown by immunoprecipitation with an anti-CTD monoclonal antibody (8WG16), these extracts contain proportionately more Pol II(G) (lane 4). To assess a possible activator-dependent Pol II(G) binding, a DNA fragment containing five p53 binding sites upstream of the AdML core promoter (Figure 4B) was immobilized on paramagnetic beads. The DNA template-immobilized beads were incubated with either f:Gdown1 or control HeLa nuclear extract in the absence or presence of p53. After a 1.5 h incubation, beads were washed to remove unbound proteins and proteins retained on the beads were analyzed by immunoblot. As shown in Figure 4C, Pol II (RPB1), Mediator (MED17, MED30 and CDK8), and TFIIB were recruited to the DNA template in a p53-enhanced manner in both f:Gdown1 (lane 2 versus lane 3) and control (lane 5 versus lane 6) HeLa extracts, confirming the reliability of the assay for analyzing the recruitment of other factors. Whereas a comparably high p53-dependent recruitment of TFIIF was not evident, likely because of the non-specific binding of TFIIF to DNA (Baek et al., 2006), p53-enhanced binding of Gdown1 was observed in both the f:Gdown1 HeLa extract (lane 2 versus lane 3; upper f:Gdown1 band and lower endogenous Gdown1) and the control HeLa extract (lane 5 versus lane 6; endogenous Gdown1 only). Since most of the endogenous Gdown1 is associated with Pol II, this result indicates that Pol II(G) is recruited in an activator-enhanced manner.
Figure 4. Pol II(G) is recruited to the promoter in an activator- and Mediator-dependent fashion.
(A) Comparison of the levels of Pol II(G) in HeLa or HeLa f:Gdown1 nuclear extracts. Pol II in HeLa (C) or HeLa f:Gdown1 (Gdn) nuclear extracts was immunoprecipitated with mouse IgG (lane 5 and 6) or 8WG16 (lane 3 and 4) antibodies. Immunoprecipitates were analyzed by immunoblot. (B) Schematic of the immobilized template used. (C) Pol II(G) is recruited to the promoter in an activator- and Mediator-dependent fashion. M280-streptavidin Dynabeads carrying a biotinylated DNA fragment were incubated with HeLa f:Gdown1 or HeLa nuclear extract with p53 (lane 2) or without p53 (lane 1). After 1.5 h incubation, the beads were washed and bound proteins were analyzed by immunoblot. (D) Mediator-dependent Pol II(G) recruitment to DNA template. Mediator was immuno-depleted from HeLa f:Gdown1 nuclear extract as described in Experimental Procedures. Factor recruitment was monitored as described in (C). See also Figure S2.
To determine whether the activator-dependent recruitment of Pol II(G) is also Mediator-dependent, the template was incubated with f:Gdown1 nuclear extract from which Mediator had been immunodepleted (Malik and Roeder, 2003). Importantly, Pol II(G) recruitment was significantly decreased in the absence of Mediator (Figure 4D, lane 6 versus lane 3), and addition of purified Mediator restored the recruitment (lane 7 versus lane 6). Consistent with previous results indicating that Mediator reverses the inactive state of Pol II(G) in a reconstituted transcription assay (Figure 1A) and associates with Pol II(G) (Figure 2B), the present results indicate that recruitment of Pol II(G) to the promoter is enhanced by the Mediator. Although Gdown1 and TFIIF show competitive binding to Pol II, these results suggest that Gdown1 can be recruited to the promoter in an activator-enhanced manner and that Mediator is required for this recruitment. This activator- and Mediator-enhanced recruitment of Pol II(G) may facilitate its above-described (Figure 1D,E) intrinsic ability to form a partial (TFIIF-deficient) pre-initiation complex with TBP and TFIIB.
Pol II(G) is recruited to promoter regions in vivo
To assess Pol II(G) recruitment to promoters in vivo, we employed ChIP-seq analysis to determine which genes are occupied by Pol II(G) in primary human lung fibroblast cells (IMR90). For this purpose, antibodies against Gdown1 and Pol II, along with a purified rabbit IgG as the negative control, were used for ChIP. IMR90 is an extensively studied cell line, and a recent genome-wide analysis of global run-on sequencing (GRO-seq) has mapped all transcriptionally engaged RNA polymerases using this cell line (Core et al., 2008). Thus, the results of GRO-seq enabled us to map Pol II(G) in conjunction with the corresponding data on nascent RNA products. In interpreting these results we have assumed, based on the extreme stability of Pol II(G) (even in 2M urea) and no apparent presence of Gdown1 within other complexes, that the presence of Gdown1 on a gene reflects Pol II(G).
The MACS program (Zhang et al., 2008) was used to identify regions with enriched ChIP-seq signals (i.e., binding sites for Pol II(G) and Pol II). Of 22,594 genes, 8625 promoters were bound by Pol II and, at least 60 promoters were bound by Pol II(G) (i.e., reacted with Gdown1 antibody). Although this latter number is smaller than expected based on the apparently high content of Pol II(G) in liver and calf thymus (Hu et al., 2006), it is based on very stringent criteria for scoring genes as Gdown1 positive (see the technical note in Supplementary Information) – such that the number of actual Gdown1 target genes could be considerably greater. The 60 high-confidence genes identified here were further classified on the basis of their pausing P value (Table S1), which is defined as the comparison of the density of reads in the sense strand promoter proximal peak to the density of reads in the body of the gene as compared to a uniform distribution of all these reads based on the number of mappable bases (Core et al., 2008). Among the 60 identified genes, 35 belong to Class II genes, which contain a paused Pol II and are actively transcribed. Twenty five genes belong to Class I genes, which are active but display no paused Pol II. None of the 60 identified genes were found to belong to either Class III (inactive, paused Pol II) or Class IV (inactive, no paused Pol II). Thus, and most importantly, these results indicate that Gdown1 is associated with actively transcribed genes.
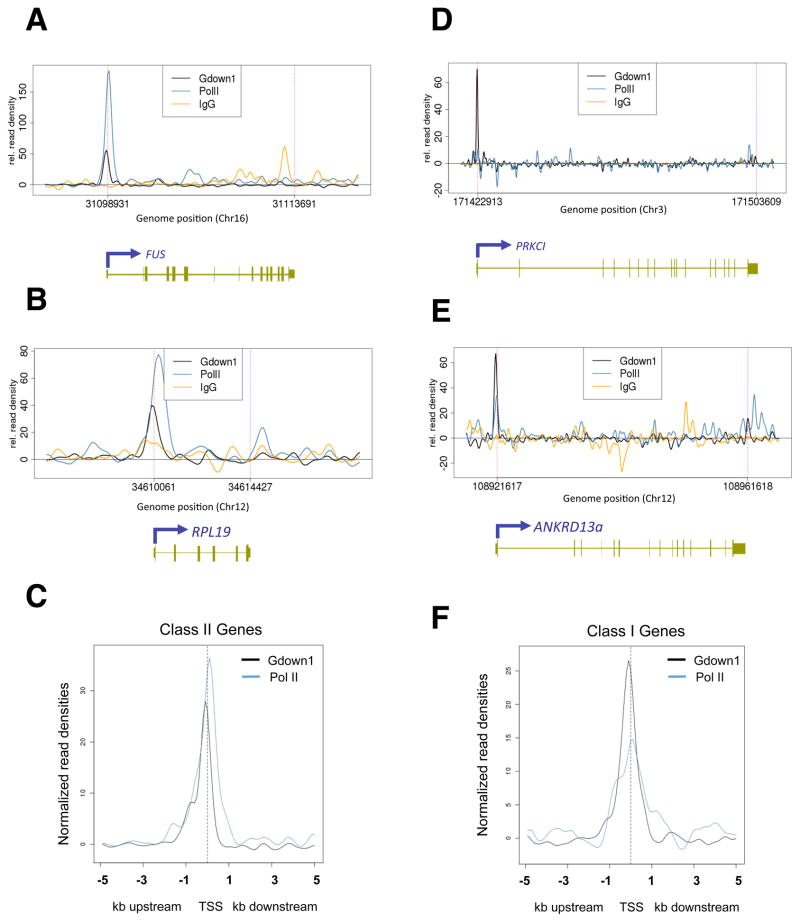
Importantly, Pol II(G) was found mostly in the promoter regions of the identified genes (Figure 5A,B,D,E). In Class II genes, the Pol II peak of the read density is mostly located at the promoter proximal region due to the pausing, which is clearly observed in FUS and RPL19 (Figure 5A and B). Interestingly, the Pol II(G) peaks that are located at the promoter regions (from −1 to −200 relative to the TSS) of these genes do not coincide with the Pol II peaks (Figure 5A and B). Thus, the averaged Pol II peak from the 20 identified genes in this group maps ~50 bp downstream of the transcriptional start sites, whereas the averaged Pol II(G) peak maps ~50 bp upstream of the transcriptional start sites (Figure 5C). Similarly, for Class I genes, which do not contain a paused Pol II (Figure 5D and E), the averaged Pol II peak from 13 identified genes does not coincide with, and lies downstream of, the averaged Pol II(G) peak (Figure 5F). These results indicate that Pol II(G) is associated with actively transcribed genes, primarily at a region(s) just upstream of the transcription start site, and that Gdown1 does not necessarily cause promoter-proximal pausing of Pol II as had been anticipated. Although interpretation of these results is potentially complicated by divergent transcription events, an analysis of the IMR90 GRO-seq data for these cells (Core et al., 2008) has indicated that some of the highlighted Gdown1 target genes do not have divergent transcription -- indicating that Pol II(G) is not necessarily involved in divergent transcription. These and other results have implications for a proposed model (see Discussion) involving poised Pol II(G).
Figure 5. Pol II(G) is enriched at promoter regions of a subset of genes.
ChIP-seq data for Gdown1, Pol II and IgG using antibodies that recognize Gdown1 (black), the RPB1 subunit (blue) and IgG (orange) are shown for four genes, plotted as relative read density versus base pair units. The start site and direction of transcription are shown by arrows, with boxes depicting exons and lines representing introns. (A) FUS (B) RPL19 (D) PRKCI (E) ANKRD13a. (C, F) Averaged read densities of Pol II (blue) and Pol II(G) (black) with Class II (C) or Class I (F) genes identified by ChIP-seq are shown. See also Table S1.
Pol II(G) regulates transcription at its target genes
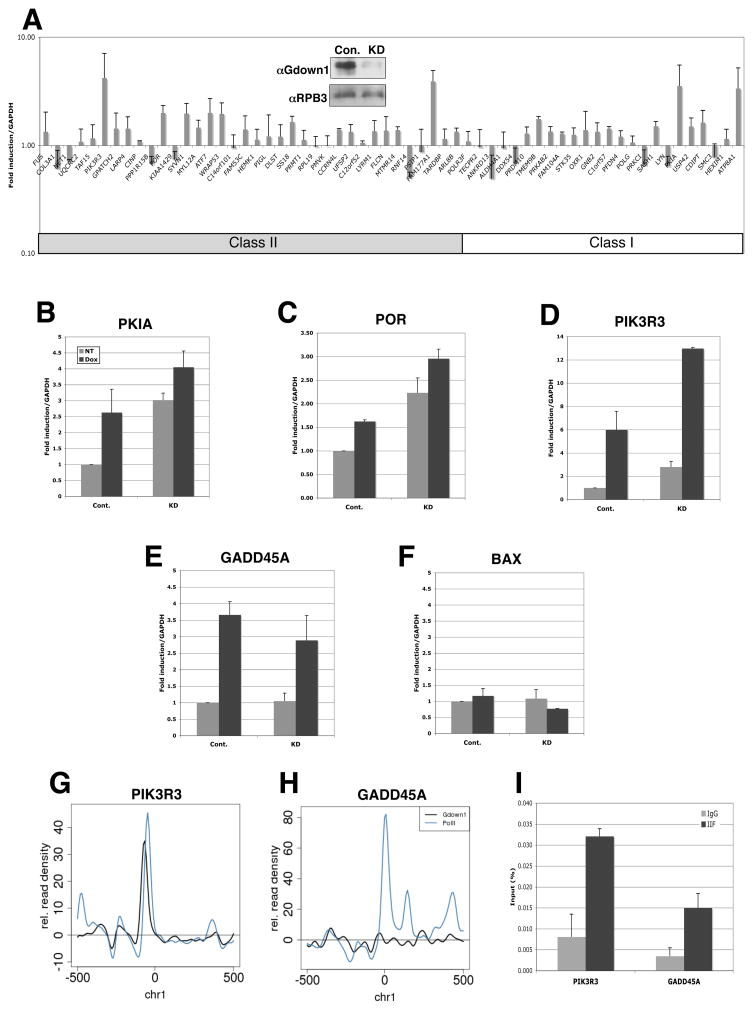
Our in vitro results have demonstrated that Gdown1 has the potential to negatively regulate PIC formation. To ascertain whether Gdown1 also negatively regulates transcription in vivo, IMR90 cells were infected with lentivirus vectors expressing Gdown1-targeted shRNA and effects on expression of Gdown1 and its target genes identified by the ChIP-seq analysis were examined. This treatment reduced the Gdown1 level RNA by more than 90% (data not shown) and, as well, significantly reduced the level of the Gdown1 protein (Figure 6A, inset). Quantitation of RNA levels of the target genes indicated that most were up-regulated 2- to 4-fold upon Gdown1 knockdown, whereas at least two genes (ANKRD13a and RNF14) were significantly down-regulated (Figure 6A). Expression of genes that showed no enrichment of Gdown1, such as GAPDH and histone genes, was not affected by Gdown1 knockdown (data not shown). These results suggest that Pol II(G) negatively regulates expression of a majority of the target genes identified here.
Figure 6. Effects of Gdown1 knockdown on gene expression.
(A) Gene expression of Pol II(G) target genes in Gdown1 knockdown cells. Error bars correspond to standard deviation from five independent experiments. (B) Gdown1 knockdown in IMR90 cells. Inset, whole cell lysates were prepared from control or knockdown cells and analyzed by immunoblot. (B)–(F) Doxorubicin effects on expression of the Pol II(G) target genes (B to D) or apoptotic genes (E, F). Error bars correspond to standard deviation from three independent experiments. (G and H) ChIP-seq data for PIK3R3 (G) and GADD45A (H). (I) ChIP analysis on the promoter regions of PIK3R3 and GADD45A genes. ChIP analysis was performed with indicated antibodies. Error bars correspond to standard deviation from three independent experiments. See also Figures S3 and S4.
We have demonstrated that Gdown1 is recruited to promoters in an activator/Mediator-dependent manner in vitro. To further explore the role of Pol II(G) as a negative regulator in vivo, we investigated the impact of Gdown1 knockdown on activation of several target genes that were found to be up-regulated by doxorubicin treatment in IMR90 cells. Doxorubicin induces a DNA damage stress response that results in activation of p53-target genes. We found that among Gdown1 target genes, the PKIA and PIK3R3 genes, in particular, were strongly induced by doxorubicin treatment (Figure 6B,C).
Importantly, doxorubicin treatment also induced a marked decrease in cell viability of Gdown1 knockdown cells (30% survival) relative to control cells (Figure S3), which suggests that Pol II(G) may be involved in the transcriptional regulation of stress-inducible genes. Consistent with this possibility, the expression of several well-documented p53-target genes appears to be repressed by Gdown1 (Figure S4), although our initial ChIP-seq analyses failed to detect significant enrichment of Gdown1 on these genes. In relation to other Gdown1 target genes, PKIA and POR were induced significantly by doxorubicin treatment in control cells. These levels approached those observed upon Gdown1 knockdown in control (untreated) cells (Figure 6B,C). Expression of PKIA was further increased in the knockdown cells by doxorubicin treatment, albeit only by 30–35% (Figure 6B), suggesting that the expression of PKIA is normally down-regulated by Pol II(G). In the case of PIK3R3, doxorubicin treatment of control cells induced the expression by 6-fold. This level of induction is greater than what was seen either by Gdown1 knockdown (3-fold) or by subsequent doxorubicin treatment of Gdown1 knockdown cells (ca. 4-fold relative to untreated knockdown cells). However, the absolute levels in the latter case exceeded the doxorubicin-treated levels of control cells by 2-fold (Figure 6D). This suggests that PIK3R3 transcription is also negatively regulated by Pol II(G). Expression of GADD45A and BAX was not affected by Gdown1 knockdown (Figure 6E, F), consistent with the failure to find Gdown1 on these genes by ChIP-seq.
Although Gdown1 competes with TFIIF in vitro, immobilized template assays demonstrated that Gdown1 is recruited to promoters in a Mediator-dependent fashion in the presence of TFIIF. To confirm the in vitro results, ChIP assays were employed to assess TFIIF occupancy on promoter regions of PIK3R3 (Gdown1 target gene, as shown in Figure 6G) and GADD45A (non-target gene, as shown in Figure 6H). As shown in Figure 6I, significant occupancies of TFIIF were detected on both genes, which confirms that TFIIF is indeed recruited to the promoter regions of Gdown1 target genes in vivo.
Taken together, these results suggest that Pol II(G) negatively regulates transcription at specific target genes, presumably by inhibiting TFIIF function.
DISCUSSION
In following up our earlier demonstration (Hu et al., 2006) of Mediator-dependent transcription by Pol II(G), we now have demonstrated (i) that Gdown1 can inhibit basal as well as activator-dependent transcription, indicative of functions through the general transcriptional machinery, (ii) that Gdown1 competes with TFIIF for binding to Pol II in vitro, which can result in an inhibition of PIC formation, (iii) that Gdown1, like TFIIF, directly interacts with the RPB5 and RPB1 subunits of Pol II, (iv) that Pol II(G) is recruited to promoter regions of subsets of active genes in vivo, but does not co-localize with paused polymerases on these genes, and (v) that Pol II(G) modulates transcription at these target genes. These results suggest additional mechanisms for regulation of transcription initiation on specific genes, by a poised but non-initiated Pol II, as well as an additional mechanism of action of the Mediator complex.
Mechanism of transcription inhibition by Gdown1
Given that Gdown1 and TFIIF interact competitively with Pol II (this study) and that TFIIF plays critical roles in PIC assembly (Luse et al., 2011, Thomas and Chiang, 2006) Gdown1 functions likely relate mechanistically to the regulation of TFIIF functions. In this regard, studies of yeast TFIIF, whose heterotrimeric subunit composition is somewhat different from the metazoan heterodimeric TFIIF, have suggested dynamic interactions between its various structured domains and discrete regions across Pol II -- as well as the likelihood that additional unstructured TFIIF segments might interact with Pol II in a context-determined manner. Thus, an electron microscopy analysis of the yeast Pol II-TFIIF complex suggested that, among other domains, TFIIF density extends broadly along the Pol II cleft to span the RPB5-containing jaw (Chung et al., 2003). By contrast, more recent cross-linking analyses (Eichner et al., 2010; Chen et al., 2010) localized the bulk of the yeast TFIIF interactions to the RPB2 lobe on one side of the cleft, while leaving open the possibility that certain TFIIF segments (Tfg1/RAP74 WH domain) might hover in the vicinity of the Pol II jaw domain. Earlier studies with individual polypeptides additionally suggested an interaction of human RAP30 with the RPB5 subunit of Pol II (Wei et al., 2001). We have demonstrated here that Gdown1 directly binds to the jaw domain of RPB5, although we cannot exclude additional interactions of Gdown1 with the assembly domain. Consistent both with this observation and with the close proximity of the C-terminal domain of RPB5 to RPB1 (Cramer et al., 2001), our analyses have shown that Gdown1 also interacts with RPB1. We further have found that Gdown1 forms a dimer in vitro (unpublished result), which may facilitate the extended interactions spanning RPB5 and RPB1. The significance of a joint interaction of Gdown1 with RPB1 and RPB5 is underscored by the fact that whereas RPB5 is a common subunit of Pols I, II and III, Gdown1 interacts only with Pol II. Thus, the Gdown1 specificity rests on its joint interaction with RPB1 and RPB5. Notably, the competition between TFIIF and Gdown1 that we describe here can be explained either by their binding to the same (or overlapping) domains on Pol II or through mutual steric effects.
The idea of competition between TFIIF and Gdown1 for Pol II binding is further supported by our observation that Pol II(G) purified from various nuclear extracts is not associated with TFIIF and, reciprocally, that an extract-derived Pol II-TFIIF complex contains no associated Gdown1. Functionally, this competition can result in an inhibition of PIC assembly that leads to repression of transcription. Indeed, gel shift assays have demonstrated that whereas TBP, TFIIB, TFIIF, and Pol II form a minimal functional promoter complex, the presence of Gdown1 results in the formation of an analogous promoter complex that lacks TFIIF. Although the formation of a minimal promoter complex with TBP, TFIIB and Pol II(G) is inefficient compared to complex formation with TBP, TFIIB, TFIIF and Pol II, it appears that Gdown1 can form a significant level of a minimal promoter complex in the absence of TFIIF. This raises the possibility that under appropriate conditions (e.g., in the presence of Mediator, see below, and possibly other Mediator/Pol II(G)-associated factors described herein) Gdown1 may partly substitute for TFIIF in facilitating recruitment of the Pol II complex to the promoter. Indeed, TFIIF-independent Pol II(G) promoter complex formation may provide another possible mechanism wherein Pol II might occupy a promoter, as a “poised” polymerase, but not transition to an active PIC because of Gdown1-mediated constraints to TFIIF interactions with Pol II.
Intriguingly, the C-terminus of Gdown1 shows significant sequence similarity to a RAP30 C-terminal region (residues 162 to 249) that possesses cryptic DNA-binding activity (Tan et al., 1994) and belongs to the eukaryotic “winged” HTH family of DNA binding domains (Groft et al., 1998). In particular, the upstream region of α-helix H1 of RAP30 is strikingly similar to the C-terminal region of Gdown1 (Figure S1), with six of the 17 potential DNA contact sites in RAP30 being conserved in Gdown1. Thus, this C-terminal region of Gdown1 may contribute to the binding of Pol II(G) to target genes.
A role for Mediator in Pol II(G)-mediated transcription
Consistent with the competitive binding of Gdown1 and TFIIF to Pol II, excess TFIIF can counter the Gdown1-mediated repression of basal transcription in vitro. However, whereas the recovery by excess TFIIF is partial (60%), Mediator can completely reverse the inhibitory effect of Gdown1 in a reconstituted system (Figure 1A; Hu et al., 2006). Thus, it is conceivable that Mediator might compensate for an otherwise inefficient formation of a Pol II(G)-containing promoter complex by stabilizing such a complex during PIC formation, with consequent full recovery of transcription in the reconstituted system. The normal association of Gdown1 with Pol II is very strong, as evidenced by the fact that, in the absence of other factors, Gdown1 does not dissociate from Pol II even under denaturing conditions (Hu et al., 2006). Despite such a tight association, and surprisingly, a 25-fold molar excess of TFIIF can partially remove Gdown1 from Pol II under high salt conditions. However, since these conditions are far from physiological, it is likely that Mediator may facilitate TFIIF interactions with Pol II during formation of a functional PIC involving Pol II(G). While the nature of such a mechanism remains unknown, it is interesting to note that Gdown1 is phosphorylated both in vivo (Dephoure et al., 2008) and during transcription in vitro (Figure S2). Since protein phosphorylation events often play critical roles in regulating protein-protein interactions, it is tempting to speculate that phosphorylation of Gdown1 might contribute to its dissociation from Pol II. Moreover, a mass spectrometry analysis (Ishihama et al., 2008) has revealed that the Gdown1 phosphorylation sites are evolutionarily conserved from flies to human, suggesting that the phosphorylation of Gdown1 is important for Gdown1 function.
A role for Gdown1 in the regulation of gene expression in vivo
Our global ChIP-seq analysis identified at least 60 genes in human fibroblasts that had peaks of Gdown1 within 200 bp of the transcription start site. This result suggests that Pol II(G) regulates transcription in a gene-specific manner rather than globally. This notion is also supported by the Gdown1 knockdown analysis in IMR90 cells, which showed no apparent defects for maintaining normal cell growth. A potential caveat is that low residual levels of Gdown1 in the knockdown analysis or incomplete crosslinking in the ChIP analysis might have precluded detection of a broader group of Gdown1-regulated genes, although this would not detract from the significance of the present results indicating a select group of target genes that may be more dependent on normal (high) levels of Gdown1. More importantly, our results provide in vivo evidence showing that Pol II(G) is indeed recruited to promoter regions of genes that are actively transcribed. This is consistent with our demonstration (discussed above) that Pol II(G) can form a minimal preinitiation complex in vitro.
In conjunction with GRO-seq results (Core et al., 2008), the global ChIP-seq analysis has revealed that the identified Gdown1 target genes belong to either Class I or Class II, but not to Class III or Class IV, genes. This clearly indicates that Pol II(G) associates with actively transcribed genes. Although Gdown1 has the potential to negatively regulate transcription under selective conditions in vitro, the primary function in vivo seems, paradoxically, not to completely inactivate gene transcription per se but rather to serve a modulatory role and/or to poise Pol II for promoter activation. Among 16,882 active genes in IMR90 cells, 30% contain a paused Pol II at promoter-proximal regions (Core et al., 2008), which has led to the suggestion that the regulation of transcription at a post-recruitment step(s) also plays an important role in gene transcription. However, our results indicate that Pol II(G) resides in part on Class I (unpaused) promoters, which suggests that Pol II(G) does not directly or necessarily cause pausing. Furthermore, the Pol II(G) peak, which is localized to a promoter region just upstream of the transcriptional start site, does not coincide with the Pol II peak in Class II (paused) genes (Figure 5C), which is centered just downstream of the transcriptional start site. Although it remains unclear whether Gdown1 fully dissociates from Pol II during initiation, these observations reinforce our hypothesis that Gdown1 acts upstream of transcription initiation and promoter-proximal pausing events, serving rather to reversibly constrain a poised (promoter-associated) Pol II from transitioning to an active PIC.
Expression of most of the currently identified Gdown1 target genes in IMR90 cells was up-regulated by Gdown1 knockdown, suggesting that Gdown1 might negatively regulate transcription. Interestingly, the increased expression of several Gdown1 target genes in Gdown1 knockdown cells was comparable to the induced expression by doxorubicin in control cells. Although it is not yet known whether these doxorubicin-inducible Gdown1 target genes are direct p53 target genes, these observations suggest that Pol II(G) may be involved in the transcriptional regulation of stress-inducible genes. Given that transcription by Pol II(G) is critically dependent on Mediator, we further hypothesize that under stress-free conditions, in which Mediator is not stably recruited to the genes (or is otherwise not fully functional), transcription by Pol II(G) that is detected at the genes remains relatively inefficient. At the same time, transient occupancy of Pol II(G) at promoter regions might preclude the recruitment of functional Pol II-TFIIF. However, once Mediator is stably recruited (or “activated”) by activators upon a stress response, Pol II(G) can execute transcription as efficiently as normal Pol II. Therefore, Gdown1 may play a role in maintaining diminished (but not fully inhibited) expression of these stress-inducible genes. This regulatory mechanism would be particularly suitable for these genes as it would enable cells to immediately or coordinately respond to the stress signals. Since only a fraction of the target genes are regarded as stress-inducible (Table S1), similar considerations would apply to the other Gdown1 target genes in other signaling pathways.
In relation to the Pol II(G)–Mediator connection established here and in previous studies (Hu et al, 2006), our current finding of a limited number of intracellular Gdown1 target genes may seem inconsistent with the broad involvement of Mediator in transcription of protein-coding genes (Malik and Roeder, 2010) and, as well, with the potentially high fractional content of Pol II(G) in the total cellular Pol II pool (Hu et al, 2008) and the lack of a transcription factor-specific inhibition by Gdown1 in our in vitro assays. However, several points are relevant to these issues. First, beyond Gdown1, which elicits a Mediator requirement in our in vitro assays, other natural constraints to transcription that are not present in these assays (Malik et al., 2007) may also elicit Mediator requirements in vivo. Second, and as noted earlier, technical matters (detailed in Experimental Procedures) may have precluded our identification of a much broader group of Gdown1 target genes and led us to focus here only on high-confidence Gdown1-associated genes. Thus, there may well be a much larger number of Gdown1 target genes and functions other than those indicated here. In this regard, our demonstration of antagonistic Gdown1-TFIIF functions, along with previous indications of TFIIF functions in transcription elongation (Saunders et al., 2006) and post-initiation Mediator functions (Malik and Roeder, 2010), raise the interesting possibility of Gdown1 functions in transcription elongation. Consistent with this possibility, we have shown an ability of Gdown1 to affect transcription elongation in vitro (data not shown).
In summary, we propose that Pol II(G) plays a distinct role in regulating the expression of selected genes by a mechanism that modulates the inhibition of TFIIF function and that this in turn elicits a role for Mediator in counteracting these inhibitory functions under appropriate inducing conditions.
EXPERIMENTAL PROCEDURES
Generation of a Gdown1 epitope-tagged stable cell line and purification of Pol II(G)
Human Gdown1 was cloned into the pIRES1neo-derived vector VP5, which allows expression of target proteins as fusions with FLAG and HA. HeLa S cells were transfected with the resulting plasmid and stable G418-resistant clones expressing Gdown1 were selected as described (Malik and Roeder, 2003). For identification of Gdown1-associated proteins, nuclear extract from the FLAG-tagged Gdown1 stable cell line was directly applied to M2 agarose and bound proteins eluted with FLAG peptide. For purification of Pol II(G), nuclear extracts were first fractionated on a DE52 column; the 0.3 M KCl fraction was then purified over M2 agarose. The eluted protein complex was further purified on 8WG16-protein A-Sepharose for some experiments, as indicated.
Antibodies and immunodepletion of nuclear extracts
Antibodies against RAP74, RPB5, RPB6, MED17, and MED30 were raised in rabbits and have been described elsewhere. Antibodies against RPB1, CDK8 and p53 were from Santa Cruz. Antibodies against RPB2, RPB3, RPB8, RPB9, RPB11 and RPB4 were a gift of Koji Hisatake (University of Tsukuba). Two different antibodies against Gdown1 were generated: one using full-length human Gdown1 that was expressed in bacteria and used to immunize rabbits (Covance) and one by genomic antibody technology (Strategic Diagnostics). For immunodepletion, affinity-purified antibodies were cross-linked to protein A-Sepharose and incubated with nuclear extracts as described previously (Malik and Roeder, 2003). For Mediator depletion, nuclear extracts were incubated sequentially with immobilized anti-MED6 anti-MED30 antibodies.
In vitro transcription and electrophoretic mobility-shift assays
Pol II, GTFs, PC4 and Mediator were purified as described previously (Malik and Roeder, 2003). Recombinant Gdown1 and bovine Pol II and Pol II(G) were purified as described previously (Hu et al., 2006). In vitro transcription with purified components was performed as described (Malik and Roeder, 2003). Electrophoretic mobility-shift assays were performed as described (Malik et al., 1998).
ChIP-seq
Detailed methods are presented in Supplemental Information. However, on an important technical matter related to the relatively small number of reported Gdown1 peaks, we note that both input and IgG control samples showed many peaks, especially around promoter regions. Therefore, we chose a conservative strategy by focusing on those Gdown1 peaks that did not overlap with input and/or IgG peaks, thereby reducing the number of Gdown1 peaks that also overlapped with Pol II. Thus, the results that are presented reflect only high-confidence peaks, and do not exclude the possibility of a much broader group of Gdown1 target genes on which Gdown1 functions at some of the other binding sites.
For further Experimental Procedures, see the Supplemental Information available online.
Supplementary Material
Highlights.
Gdown1 down-regulates transcription by competing with TFIIF for binding to Pol II
Mediator is required for PIC assembly entailing Pol II(G)
Pol II(G) is recruited to promoter regions in vivo to regulate gene transcription
Pol II(G) is a poised but not initiated Pol II
Acknowledgments
We are grateful to Leighton Core and John Lis for providing unpublished data, Marc Vigneron and Richard Ebright for RPB1 expression vectors, Wei-Yi Chen for discussion of the project, Debabrata Biswas for supporting data, Satoshi Iida for critical reading of the manuscript, and Zheng-Yuan Fu and Adam Nock for technical assistance. This work was supported by NIH grants CA129325 (R.G.R.), DK071900 (R.G.R.), GM64474 (A.G.), and RR00862 (B.T.C.); by the Natural Science Foundation of China (30800169)(X.H.); and by the Ludwig Institute for Cancer Research and the California Institute of Regenerative Medicine (RN2-00905) (B.R.).
Footnotes
ACESSION NUMBERS
Data analyzed herein have been deposited in GEO with accession number GSE33128. The data has not yet been made public, however for the time being the following link will give reviewer access to the data: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=fhevlgykcwgwkhq&acc=GSE33128
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aygun O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci U S A. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek HJ, Kang YK, Roeder RG. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J Biol Chem. 2006;281:15172–15181. doi: 10.1074/jbc.M601983200. [DOI] [PubMed] [Google Scholar]
- Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, et al. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. Embo Journal. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WH, Craighead JL, Chang WH, Ezeokonkwo C, Bareket-Samish A, Kornberg RD, Asturias FJ. RNA polymerase II/TFIIF structure and conserved organization of the initiation complex. Molecular Cell. 2003;12:1003–1013. doi: 10.1016/s1097-2765(03)00387-3. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichner J, Chen HT, Warfield L, Hahn S. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. Embo Journal. 2010;29:706–716. doi: 10.1038/emboj.2009.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groft CM, Uljon SN, Wang R, Werner MH. Structural homology between the Rap30 DNA-binding domain and linker histone H5: implications for preinitiation complex assembly. Proc Natl Acad Sci U S A. 1998;95:9117–9122. doi: 10.1073/pnas.95.16.9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Malik S, Negroiu CC, Hubbard K, Velalar CN, Hampton B, Grosu D, Catalano J, Roeder RG, Gnatt A. A Mediator-responsive form of metazoan RNA polymerase II. Proc Natl Acad Sci U S A. 2006;103:9506–9511. doi: 10.1073/pnas.0603702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, Kyono Y, Sugiyama N, Imami K, Tomita M. Successive and selective release of phosphorylated peptides captured by hydroxy acid-modified metal oxide chromatography. Journal of Proteome Research. 2008;7:4585–4593. doi: 10.1021/pr800305y. [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Langelier MF, Zeghouf M, Cojocaru M, Bergeron D, Baali D, Forget D, Mnaimneh S, Davierwala AP, Pootoolal J, et al. RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol Cell Biol. 2004;24:7043–7058. doi: 10.1128/MCB.24.16.7043-7058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luse DS, Cabart P, Ujvari A, Pal M. Transcription factor TFIIF is not required for initiation by RNA polymerase II, but it is essential to stabilize transcription factor TFIIB in early elongation complexes. P Natl Acad Sci USA. 2011;108:15786–15791. doi: 10.1073/pnas.1104591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Barrero MJ, Jones T. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. P Natl Acad Sci USA. 2007;104:6182–6187. doi: 10.1073/pnas.0608717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Guermah M, Roeder RG. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proc Natl Acad Sci U S A. 1998;95:2192–2197. doi: 10.1073/pnas.95.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Isolation and functional characterization of the TRAP/mediator complex. Methods Enzymol. 2003;364:257–284. doi: 10.1016/s0076-6879(03)64015-2. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao T, Woychik NA. RNA polymerase subunit RPB5 plays a role in transcriptional activation. Proc Natl Acad Sci U S A. 1998;95:15281–15286. doi: 10.1073/pnas.95.26.15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Tan S, Garrett KP, Conaway RC, Conaway JW. Cryptic DNA-binding domain in the C terminus of RNA polymerase II general transcription factor RAP30. Proc Natl Acad Sci U S A. 1994;91:9808–9812. doi: 10.1073/pnas.91.21.9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Tyree CM, George CP, Lira-DeVito LM, Wampler SL, Dahmus ME, Zawel L, Kadonaga JT. Identification of a minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II. Genes Dev. 1993;7:1254–1265. doi: 10.1101/gad.7.7a.1254. [DOI] [PubMed] [Google Scholar]
- Wei W, Dorjsuren D, Lin Y, Qin W, Nomura T, Hayashi N, Murakami S. Direct interaction between the subunit RAP30 of transcription factor IIF (TFIIF) and RNA polymerase subunit 5, which contributes to the association between TFIIF and RNA polymerase II. J Biol Chem. 2001;276:12266–12273. doi: 10.1074/jbc.M009634200. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.