Summary
MRG15 is a member of the mortality family of transcription factors that targets a wide variety of multi-protein complexes involved in transcription regulation, DNA repair, and alternative splicing to chromatin. The structure of the apo-MRG15 MRG domain implicated in interactions with diverse proteins has been described, but not in complex with any of its targets. Here we structurally and functionally characterize the interaction between MRG15 and Pf1, two constitutively-associated subunits of the histone deacetylase-associated Rpd3S/Sin3S corepressor complex. The MRG domain adopts a structure reminiscent of the apo-state whereas the Pf1 MRG-binding domain engages two discrete hydrophobic surfaces on the MRG domain via a bipartite motif comprising an α-helix and a segment in an extended conformation, both of which are critical for high-affinity interactions. Multiple MRG15 interactors share an FxLP motif in the extended segment but equivalent sequence/helical motifs are not readily evident, implying potential diversity in MRG-recognition mechanisms.
Introduction
The chromatin promotes a variety of processes critical for cell growth and survival including transcription, replication, recombination, repair, and splicing. Emerging evidence suggests that besides serving as a substrate for many of these processes, chromatin takes an active role in their regulation by harboring signals in the form of diverse post-translational modifications of histones. These signals are better-characterized in transcription biology, providing the basis for the histone code/language hypothesis (Gardner et al., 2011; Jenuwein and Allis, 2001), and are interpreted at the molecular level by specific chromatin-binding modules embedded within multi-protein coregulator complexes to yield specific transcriptional outcomes. Although a broad range of transcription factors have been implicated in the recognition of these signals, the MRG15 protein, a chromodomain containing protein and a member of the so-called mortality family of transcription factors (Chen et al., 2010), appears to transcend its role in transcription regulation by also playing crucial roles in recombinational repair and alternative splicing.
The genes encoding MRG15 and two other members of the family, MORF4 and MRGX, were originally identified in a screen for genes involved in cellular senescence (Bertram et al., 1999). Subsequent studies revealed that only MORF4 is involved in replicative senescence whereas MRG15 and MRGX promote cell cycle progression and cell proliferation (Chen et al., 2011; Chen et al., 2009; Tominaga et al., 2005). MRG15 is a subunit of a number of multi-protein coregulator complexes involved in both transcriptional activation and repression including the Rb-associated MAF1 complex and at least three disparate complexes containing chromatin-modifying activities including the histone acetyltransferase (HAT)-associated MAF2 and NuA4/Tip60 complexes and the histone deacetylase (HDAC)-associated Rpd3S/Sin3S complex (Carrozza et al., 2005b; Doyon et al., 2004; Jelinic et al., 2011; Pardo et al., 2002; Yochum and Ayer, 2002). More recent studies have revealed that the MRG15 also associates with the BRCA complex involved in DNA damage repair by homologous recombination and a co-transcriptional splicing complex involved in alternative splicing (Hayakawa et al., 2010; Luco et al., 2010; Sy et al., 2009). Consistent with its varied roles, MRG15 knockouts result in embryonically lethal phenotype with significant defects in cell proliferation, differentiation and organ development as well as defects in DNA repair (Garcia et al., 2007; Tominaga et al., 2005).
MRG15 is unique in that it possesses an N-terminal chromodomain that is absent in its paralogs. This atypical chromodomain appears to bind specifically, albeit with low affinity, to histones enriched in H3 K36(me2/3) found in the intragenic regions of actively transcribed genes (Carrozza et al., 2005b; Joshi and Struhl, 2005; Keogh et al., 2005; Sun et al., 2008; Xu et al., 2008). The C-terminal MRG domain on the other hand is directly involved in protein-protein interactions with diverse proteins in the aforementioned complexes. Crystal structures of the MRG15 MRG domain have been described, but only in the apo-state and not in complex with any of its targets (Bowman et al., 2006; Zhang et al., 2006). The sequence and structural requirements for effective interactions with MRG domains were not defined, although a hydrophobic groove on the surface of the domain was implicated by genetic and biochemical studies as being crucial for function (Bowman et al., 2006; Zhang et al., 2006).
We recently showed that the MRG15 MRG domain competed rather than collaborated with the PAH2 domain of Sin3 for a segment of Pf1, a subunit of the 0.6 megadalton mammalian Rpd3S/Sin3S complex and a molecular adaptor that links MRG15 on the one hand through multivalent interactions with Sin3 and the rest of the complex on the other (Kumar et al., 2011). The evolutionarily-conserved Rpd3S/Sin3S chromatin-modifying complex comprises at least five subunits, of which three subunits including the corepressor and scaffolding protein Sin3, the histone deacetylases Rpd3/HDAC1/HDAC2, and the chromatin binding proteins RbAp46/RbAp48, are shared with the much larger 1.2–2 megadalton Rpd3L/Sin3L complex (Carrozza et al., 2005a; Carrozza et al., 2005b; Fleischer et al., 2003; Jelinic et al., 2011; Laherty et al., 1997; Nicolas et al., 2007; Yochum and Ayer, 2002; Zhang et al., 1998). In the emerging view, the larger complex is involved in promoter-based repression whereas the smaller complex is involved in repressing aberrant gene transcription from cryptic transcription initiation sites and mitigating RNA polymerase II progression in the intragenic regions of actively transcribed genes. The MRG15 and Pf1 subunits are unique to the Rpd3S/Sin3S complex and play important roles in the proper assembly and chromatin targeting of this complex (Jelinic et al., 2011; Li et al., 2007). Here we describe a structural and functional characterization of the interaction between MRG15 and Pf1 and discuss how MRG15 might be co-opted by disparate multi-protein complexes.
Results
MRG15 Binds with High-Affinity to a Conserved Region of Pf1
Early studies of the MRG15-Pf1 interaction had demonstrated a direct interaction between the MRG domain of MRG15 and the region linking the two Pf1 PHD domains (Yochum and Ayer, 2002). Pf1 constructs spanning the entire linker segment as well as two shorter segments encompassing an evolutionarily-conserved region that we described recently (Kumar et al., 2011) were evaluated for binding to the MRG15 MRG domain via isothermal titration calorimetry (ITC) and NMR spectroscopy. The shortest fragment comprising Pf1 residues 200–241 (designated the MRG-binding domain or MBD) was deemed suitable for structural analyses, as it interacted with MRG15 MRG domain with comparable affinity (in the 10–20 nM range) and induced comparable changes in the MRG spectrum as the longer Pf1 constructs (Figure S1A–C). The chemical shift perturbations in the Pf1 MBD spectrum upon MRG15 MRG binding were restricted to an internal ~30 residue segment (Figure S2), confirming that this segment was the minimal MBD.
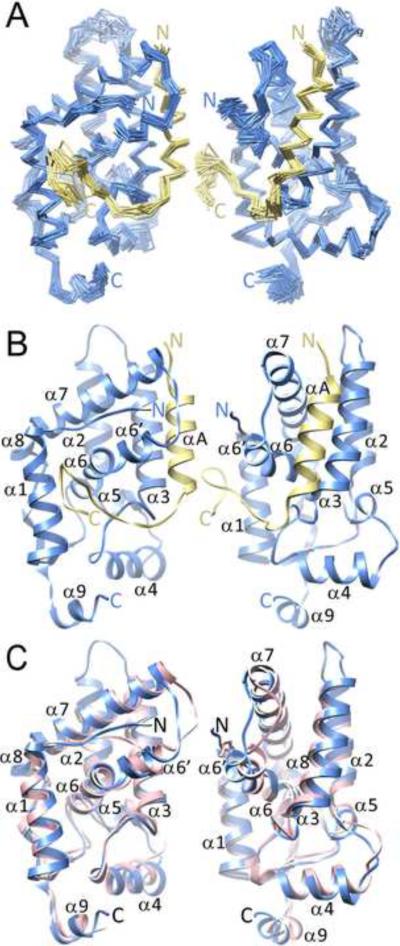
To clarify the structural basis for the high-affinity interaction between MRG15 MRG and Pf1 MBD, we determined the solution structure using NMR. Almost complete sequence-specific resonance assignments for both proteins were made using standard double- and triple-resonance approaches. The structure was determined using a combination of 1H-1H NOE-based distance restraints and backbone chemical shift-based dihedral angle restraints. Conformers of reasonable precision and covalent geometry and in good agreement with experimental data including no distance violations >0.5 Å and dihedral angle violations >5° were obtained (Figure 1A; Table 1). The structure is reasonably well-defined for the internal segments spanning residues P204 to T235 of Pf1 MBD and residues E155 to A322 of MRG15 MRG, barring loops connecting secondary structural elements in the MRG domain in general, but a loop connecting two helices spanning residues R204 to N209 in particular.
Figure 1.
Solution structure of the MRG15 MRG-Pf1 MBD complex and comparison with the crystal structure of the apo-MRG domain. (A) Cα traces of an ensemble of 20 NMR conformers following a best-fit superposition of the backbone atoms within regular secondary structural elements of MRG15 (blue) and Pf1 (yellow). (B) A ribbon representation of a representative conformer from the ensemble. Only the relatively ordered segment of Pf1 spanning residues 204–234 is shown in panels A and B. (C) A best-fit superposition of the backbone atoms of MRG15 MRG in the Pf1-bound form in solution (blue) and in the apo form in the crystal (pink; PDB ID: 2AQL). In all three panels, the views on the right are rotated by 70° along the vertical direction relative the views on the left. See also Figure S3.
Table 1.
NMR Structure Determination Statistics for the MRG15 MRG-Pf1 MBD Complex
| Restraint Statistics | |
| NOE-based distance restraints | 4012 |
| Unambiguous NOE-based restraints | 3620 |
| Intra-residue | 1783 |
| Sequential (|i − j| = 1) | 720 |
| Medium-range (1 < |i − j| ≤ 4) | 467 |
| Intra-molecular long-range (|i − j| > 4) | 181 |
| Inter-molecular restraints | 469 |
| Ambiguous NOE-based restraints | 392 |
| Hydrogen bonding distance restraints | 112 |
| Torsion angle restraints | 268 (134 ϕ, 134 ψ) |
| Structure Quality of NMR Ensemble | |
| Restraint satisfaction | |
| RMS differences for distances (Å) | 0.015 ± 0.002 |
| RMS differences for torsion angles (°) | 0.294 ± 0.072 |
| Deviations from ideal covalent geometry | |
| Bond lengths (Å) | 0.003 ± 0.000 |
| Bond angles (°) | 0.471 ± 0.008 |
| Impropers (°) | 1.259 ± 0.066 |
| Ramachandran plot statistics (%) | |
| Residues in most favored regions | 89.9 |
| Residues in additional allowed regions | 8.3 |
| Residues in generously allowed regions | 1.0 |
| Residues in disallowed regions | 0.9 |
| Average Atomic RMSDs from Average Structure (Å) | |
| All atoms | 1.95 |
| All atoms except in disordered regions | 1.45 |
| Backbone atoms (N, Ca, C′) | |
| All residues | 1.62 |
| All residues except disordered regions | 0.80 |
| All residues in secondary structure elements | 0.75 |
disordered regions include residues 200–203 and 235–241 of Pf1 and the three non-native residues at the N-terminus, residues 204–209 and residue 323 of MRG15
MRG15 and Pf1 Undergo Striking Conformational Changes upon Complex Formation
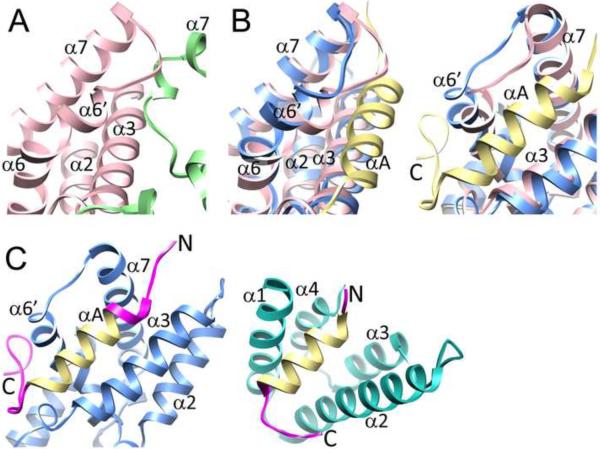
The overall structure of the MRG15 MRG domain in complex with the Pf1 MBD in solution strongly resembles those described previously for the apo-protein in crystalline environments with backbone RMSDs in the 1.2–1.3 Å range for the representative solution structure (Figure 1B & 1C; (Bowman et al., 2006; Zhang et al., 2006)). The domain contains nine helices including three short helices of less than two turns and two short, anti-parallel beta strands at virtually identical locations in the crystal and the solution structures (Figure 1B & Figure S3). The MRG domain reportedly crystallized as a dimer (Bowman et al., 2006; Zhang et al., 2006) and in our hands appeared to form higher-order oligomers at millimolar concentrations in solution judging by the severely broadened resonance linewidths in the NMR spectra, but which narrowed considerably at micromolar concentrations, consistent with a monomer (Figure S4). However, the MRG oligomerization activity was inhibited upon complex formation with the Pf1 MBD as evidenced by the narrow resonance linewidths in the NMR spectra even at millimolar concentrations (Figure S1D). Key differences between the apo- and Pf1-bound states of the MRG domain are the conformations of the α6' helix and the succeeding loop which are `open' in the bound state but are `closed' in the apo-state; residues in both the helix and the loop dominate the relatively small dimer interface in the apo-state (Figure 2A; (Bowman et al., 2006; Zhang et al., 2006)).
Figure 2.
Conformational diversity of the MRG15 MRG and Pf1 MBD domains. (A) A close-up view of the apo-MRG15 MRG dimer interface (PDB ID: 2AQL). (B) Close-up views of MRG15 MRG showing differences in backbone conformation in the absence (pink) and presence (blue) of Pf1 MBD in the vicinity of the Pf1-binding site. The two views differ by a 70° rotation in the vertical direction with the view on the left identical to the one shown in panel A. (C) Views of the Pf1 MBD when bound to MRG15 MRG (blue) and mSin3A PAH2 (green); only the ordered segments in the respective complexes are shown. Pf1 residues adopting different backbone conformations in the two complexes are colored in magenta while those with similar conformations are shown in yellow (disordered Pf1 segments in both complexes are not shown). See also Figure S4.
Pf1 MBD interacts with the MRG domain via a bipartite structural motif comprising an N-terminal α-helix spanning residues R207 to M218 and a tail segment spanning residues E219 to T234 in an extended conformation capped by a series of turns at the C-terminus (Figure 1B). Pf1 MBD is essentially unstructured in the apo-state but retains the helix at more or less the same location in the complex with mSin3A PAH2 (Kumar et al., 2011). However, the segment following the helix adopts widely different conformations in the MRG15 MRG- and mSin3A PAH2-bound states (Figure 2B). Further, whereas the segment is well-ordered up to T234 in the former complex, it is essentially disordered following F225 in the latter.
Pf1 MBD Targets two Discrete Surfaces on the MRG15 MRG Domain
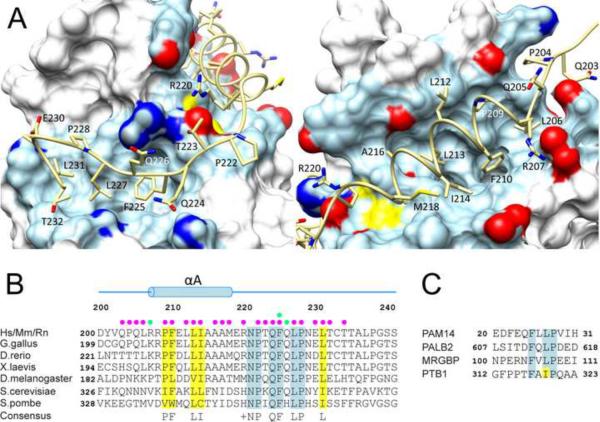
The protein-protein interface in the MRG15 MRG-Pf1 MBD complex extends ~1250 Å2, consistent with a nanomolar affinity interaction (Table 2). The bipartite structural motif of Pf1 MBD targets two discrete surfaces of the MRG domain with the Pf1 helix engaging a groove-like surface (designated site 1) defined by residues in the α3, α6', and α7 helices and the loop connecting the α6' and α7 helices of MRG while the Pf1 tail segment engages a shallower surface (designated site 2) formed by residues in the α1 and α6 helices and in the extended segment preceding and the loop segment following the α1 and α3 helices, respectively, of the MRG domain (Figure 1B & Figure S3). Several residues in the Pf1 helix and in the preceding extended segment including P204, L206, R207, P109, F110, L113, I114, A116, and A117 make significant contributions to the interface and/or are occluded from solvent (Figure 3A & Figure S5). Similarly, several residues in the Pf1 tail segment including R220, Q224, F225, L227, L231 and T232 make key contributions to the interface with the side chain of F225, in particular, engaging a deep pocket in the MRG domain. Major contributors to the interface include the MRG side chains of Y212, E216, V217, E223, Y224, V227, M228, M273, L274, Y276, T277, L279, L286, K282, and L287 (site 1) and those of I160, K165, L168, V169, W172, Y235, K236, R265, V268, and R269 (site 2). Most of the intermolecular interactions are hydrophobic in character with the aliphatic portions of polar residues such as arginines, glutamines, glutamates, and lysines making significant contributions. The complex appears to be stabilized by two well-defined intermolecular hydrogen bonding interactions between the amide nitrogen of Pf1 F225 and the carbonyl oxygen of MRG15 L234 and the carbonyl oxygen of Pf1 Q226 and the guanidinium moiety of MRG15 R269, respectively; the only well-defined electrostatic interaction involves the side chains of Pf1 R207 with MRG15 E223 (Figure S5). Additional intermolecular non-hydrophobic interactions are plausible but are not consistently detected in a majority of the NMR conformers.
Table 2.
Equilibrium Dissociation Constants for Various MRG15 MRG-Pf1 Complexes
| Reactants | KD(μM) |
|---|---|
| MRG15 MRG + Pf1 (aa 114–258) | 0.016 ± 0.004a |
| MRG15 MRG + Pf1 MBD | 0.012 ±0.002 |
| MRG15 MRG + Pf1 MBD F210A | 2.6 ± 0.7 |
| MRG15 MRG + Pf1 MBD L212A | 0.035 ± 0.017a |
| MRG15 MRG + Pf1 MBD L213A | 0.280 ± 0.063 |
| MRG15 MRG + Pf1 MBD N221E | 0.015 ± 0.001a |
| MRG15 MRG + Pf1 MBD F225A | 19 ± 14 |
| MRG15 MRG + Pf1 MBD L227A | 0.370 ± 0.087 |
| MRG15 MRG + Pf1 MBD P228A | 0.085 ± 0.017 |
| MRG15 MRG + Pf1 (aa 219–232) | 119 ± 26b |
| Pf1 MBD + MRG15 MRG W172A | 0.067 ± 0.013a |
| Pf1 MBD + MRG15 MRG R201Kc | 0.023 ± 0.006 |
| Pf1 MBD + MRG15 MRG V227A | 0.194 ± 0.073 |
| Pf1 MBD + MRG15 MRG M228A | 0.075 ± 0.013 |
| Pf1 MBD + MRG15 MRG Y235A | – d |
| Pf1 MBD + MRG15 MRG V268A | 0.184 ± 0.062 |
| Pf1 MBD + MRG15 MRG M273A | 0.096 ± 0.019 |
| Pf1 MBD + MRG15 MRG L287A | 0.407 ± 0.102 |
average values from two independent measurements; all others from at least three independent measurements
measured using NMR; all others measured using ITC
note that all studies with MRG15 MRG proteins were inadvertently conducted in a K201R mutant background
no detectable binding; hence could not be quantified
Figure 3.
The MRG15-Pf1 intermolecular interface and conservation of a sequence motif in MRG binders. (A) Non-covalent interactions at the MRG15 MRG-Pf1 MBD interface. The MRG domain is rendered as a molecular surface with residues making contacts with Pf1 shown in light blue with the side chain oxygen, nitrogen and sulfur atoms colored in red, blue and yellow, respectively. (B) A MEME-guided multiple sequence alignment of the MRG-binding domain of Pf1 orthologs. Species abbreviations: Hs: Homo sapiens; Mm: Mus musculus; Rn: Rattus norvegicus. Conserved and invariant residues are highlighted in yellow and blue, respectively. Filled circles denote intermolecular hydrophobic (magenta) and hydrogen bonding (green) interactions in the MRG15-Pf1 complex. (C) A multiple sequence alignment of various MRG interactors that harbor FxLP motifs in their MRG-binding domains. See also Figure S5.
Functional Analysis of Pf1-MRG15 Interactions
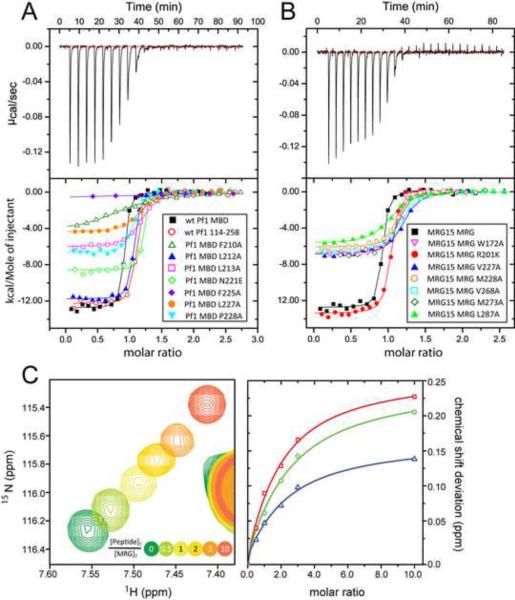
To evaluate the contributions of various residues towards the stability of the MRG15-Pf1 complex, we engineered single-site alanine mutations (with two exceptions) of selected residues at the protein-protein interface of each protein and measured the affinities of the resulting proteins in vitro using ITC; the effects of the mutations were also evaluated in cells in the context of the full-length proteins via co-immunoprecipitation (co-IP) assays. The wild-type Pf1 MBD and MRG15 MRG polypeptides interacted with 12 nM affinity (Table 2; Figure 4A & 4B). Alanine mutations of Pf1 F210 and F225 had the most debilitating effect on the interaction, as these mutations caused over 100- and 1000-fold reductions, respectively, in affinity relative to the wild-type protein. Mutations of L213 and L227 also caused between 20- and 30-fold reductions in affinity, although the mutation involving P228 produced a relatively modest effect (~7-fold reduction). Pf1 N221E and L212A bound to the MRG15 MRG polypeptide almost as well as the wild-type Pf1 MBD, consistent with their non-involvement or peripheral involvement, respectively, in MRG interactions in the NMR structure. Residues exhibiting large effects on binding are well-conserved (Figure 3B). However, not all conserved residues were important for the interaction as shown by the mutational analyses (e.g. N221E) and as implied by the lack of interactions requiring specific groups on these side chains in the NMR structure.
Figure 4.
Functional analysis of MRG15-Pf1 interactions in vitro. ITC analysis of various (A) Pf1 MBD and (B) MRG15 MRG mutants. Representative titrations for various mutant and wild-type proteins are shown. Symbols denote raw data while the continuous lines correspond to fits. The fitted equilibrium dissociation constants (KDs) for the various proteins are cataloged in Table 2. (C) NMR titrations of 15N-labeled MRG15 MRG domain with a Pf1 peptide corresponding to the tail segment (residues 219–232). The movement of a well-resolved MRG peak in the 1H-15N correlated spectrum as a function of added Pf1 peptide is shown (left). Graphs of the raw chemical shift deviations for three representative MRG peaks as a function of molar ratio and the curves resulting from the fitting procedure (right). See also Figure S6.
Among the MRG domain mutants, the largest effects (>15-fold) were witnessed for three mutants including V227A and L287A in site 1 and V268 in site 2. In each case, the residue is an integral part of the binding site in the NMR structure. Significant albeit comparably smaller effects on binding were noted for M228A and M273A in site 1 and W172 in site 2 (>5-fold but <10-fold reductions in affinity), consistent with their slightly peripheral location in the binding site. The effects of L168A and Y235A, which lines the pocket for the side chain of Pf1 F225, could not be evaluated because of poor protein expression or a lack of detectable signal in the ITC experiment, respectively.
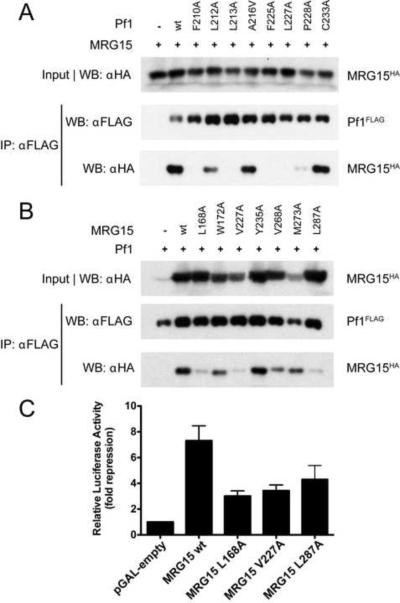
We then asked whether the MRG15-Pf1 association was similarly affected by mutations within the respective proteins inside the cells. To this end, we conducted co-IP analyses using HA-tagged and FLAG-tagged constructs of the full-length Pf1 and MRG15 proteins harboring various single-site mutations. Strongly reduced MRG15-binding was seen for Pf1 mutants including F210A, L213A, F225A, L227A and P228A whereas L212A was only slightly affected, thus mirroring the activity trends noted in vitro (Figure 5A); two additional mutants that could not be evaluated by in vitro assays including A216V and C233A (a non-interacting residue) showed comparable binding as the wild-type protein. The activity of the A216V mutant suggests that a slightly bulkier residue could be readily accommodated at this position in the complex. MRG15 mutants including L168A, V227A, and L287A showed strongly reduced Pf1-binding activity whereas the W172A, V268A, and M273A were comparatively modestly perturbed (Figure 5B). These trends were again in qualitative agreement with those observed from in vitro analyses. We further tested the transcriptional repression profiles of a subset of the MRG mutants (as GAL4 fusions) including L168A, V227A, and L287A and compared their activity relative to the wild-type protein and the empty vector. In accord with the results from co-IP and/or in vitro binding experiments (cf. above), and consistent with the notion that MRG15 represses transcription at least partly through its ability to recruit Pf1/Sin3-associated HDAC activity, each of the mutants showed reduced repression activity when compared with the wild-type protein (Figure 5C).
Figure 5.
Functional analysis of MRG15-Pf1 interactions in cells. Co-immunoprecipitation analysis of (A) various FLAG-tagged Pf1 mutants with wild-type HA-tagged MRG15 and (B) various HA-tagged MRG15 mutants with wild-type FLAG-tagged Pf1 conducted in HEK293T cells. (C) Transcriptional repression assays conducted in HEK293T cells with GAL4 DBD-fusions of wild-type and mutant MRG15 using a luciferase reporter and a thymidine kinase promoter harboring four tandem GAL4 DNA-binding sites.
An FxLP Motif is Found in Other MRG-Interactors but is not Sufficient for High-Affinity Binding
Since MRG15 was previously shown to interact via its MRG domain with a variety of proteins, we asked whether these proteins shared any conserved sequence feature(s) with Pf1. Analysis of the PAM14, MRGBP, and PALB2 sequences – all of which have been biochemically shown to interact with MRG15 – revealed that each of the proteins contained an FxLP sequence motif analogous to Pf1 (Figure 3C; (Bowman et al., 2006; Sy et al., 2009; Zhang et al., 2006)). The polypyrimidine tract binding proteins PTB1 and PTB2 have not been shown to bind to MRG15 but specific isoforms of PTB1 contain an FxIP motif, potentially implicating this protein in direct interactions (Luco et al., 2010).
Interestingly, none of the aforementioned interactors share obvious sequence similarity in the Pf1 helical region. Given the higher degree of sequence conservation for the Pf1 tail segment relative to the helix, we asked whether the tail could bind efficiently to the MRG domain. A Pf1 peptide spanning residues E219 to T232 when titrated with 15N-labeled MRG15 MRG domain produced small but significant changes in the NMR spectrum that were characteristic of a rapidly associating and dissociating complex (Figure 4C). Analysis of the chemical shift deviations yielded a KD indicative of a low affinity interaction (~120 μM; Table 2), implying that this segment alone was not sufficient for efficient interactions with MRG15.
We then sought to test the involvement of the FxLP motif of one of the MRG-interactors. A previous study had coarsely mapped the MBD of PALB2 to a segment spanning residues 562–814 (Sy et al., 2009). The same study also showed that residues 610–765 were necessary, as deletion of this segment abrogated the interaction. Since the PALB2 FxLP motif straddles residues 612–615 (Figure 3C), we generated two constructs spanning residues 562–814 and 562–629, both encompassing the FxLP motif, and evaluated the interaction with MRG15 MRG in vitro via size exclusion chromatography. Both proteins co-eluted with the MRG domain (Figure S6), implying the shorter construct harbored the necessary affinity determinants to form a stable complex.
Discussion
Previous studies of MRG domains aimed at clarifying the structure of the domain and deducing the location of the PAM14 and MRGBP interaction surface(s) (Bowman et al., 2006; Zhang et al., 2006). These studies suggested the involvement of a shallow hydrophobic surface (i.e. site 2) in these interactions. Our studies of the interaction between MRG15 and Pf1 – two evolutionarily conserved proteins with orthologs from yeast to human – reveal the involvement of two discrete surfaces on the MRG domain in these interactions with one site overlapping the surface previously identified for efficient interactions with PAM14 and the other site overlapping a surface that harbors a weak dimerization activity for the apo-protein (Bowman et al., 2006; Zhang et al., 2006). Both surfaces are enriched in hydrophobic residues and expectedly, the MRG-Pf1 interaction is dominated by hydrophobic interactions. Two phenylalanine residues, F210 and F225 of Pf1, each target a specific surface (site 1 and site 2, respectively) and serve to anchor the respective polypeptide segments giving rise to a bipartite structural motif, both of which are key to high-affinity interactions with the MRG domain.
Our studies provide insight into the sequence and structure requirements for efficient interactions with MRG domains, highlighting in particular the importance of hydrophobic residues within an amphipathic helix and an FxLP motif within a largely extended conformation. The FxLP motif is also present in other MRG-interactors including the PAM14, MRGBP, and PALB2 proteins found in the MAF1, NuA4/Tip60 and BRCA complexes, respectively; a variation of this motif (FxIP) is present in the PTB1 subunit of a co-transcriptional alternative splicing complex (interestingly, the FxIP motif is itself located within an alternatively spliced segment). The absence of a readily apparent Pf1-like helical structure immediately N- or C-terminal to the FxLP motif in these MRG-interactors is intriguing since our studies with Pf1 revealed that an FxLP motif is necessary but not sufficient for high-affinity interactions. However, our observation that PALB2 engages in high-affinity interactions with MRG15 MRG suggests that a surface other than site 2 (targeted by the FxLP motif) must also be contacted by PALB2. Additional studies are required to clarify whether PALB2 and the other interactors also employ a helical motif to target site 1 and indeed, whether this site is utilized at all, preferring instead to interact with an alternative MRG surface. A broader question is whether MRG15, by virtue of its chromodomain, serves as a general purpose `reader' of the H3 K36(me2/3) mark on histones for all of these different complexes. Indeed, besides serving as a marker for transcriptionally-active chromatin, H3 K36(me2/3) appears to be also involved in signaling DNA repair and co-transcriptional splicing (de Almeida et al., 2011; Fnu et al., 2011).
We had previously described competition between Sin3 and MRG15, two subunits of the Rpd3S/Sin3S complex, for the Pf1 subunit (Kumar et al., 2011). The structure of the MRG15 MRG-Pf1 MBD complex provides the molecular basis for this competition with the MRG domain engaging an overlapping surface of the Pf1 MBD involving the non-polar surface of the helix that is also targeted by Sin3. However, unlike the Sin3 PAH2 domain, MRG15 MRG domain engages a much larger Pf1 MBD surface, accounting for the more than two orders of magnitude higher affinity than the corresponding Sin3 PAH2-Pf1 MBD complex. Unlike Pf1, which engages in multivalent interactions with Sin3, the Pf1 MBD appears to be the sole point of contact for MRG15 with the rest of the Rpd3S/Sin3S complex, justifying the need for a high-affinity interaction.
The dual specificity (i.e. engaging two structurally dissimilar targets) of Pf1 MBD suggests that the domain might function as a molecular switch cycling between an `off' state when bound to the Sin3 PAH2 domain in the absence of MRG15, signifying an inactive complex, and an `on' state in the presence of MRG15, signifying an active, fully matured Rpd3S/Sin3S complex available for engaging chromatin targets. In so doing, the MBD might preclude misrecruitment of an incompletely assembled complex by sequence-specific DNA-binding repressors that often target the Sin3A PAH2 domain (Swanson et al., 2004). Also, given the widespread involvement of ϕ-x-x-ϕ-ϕ (ϕ: hydrophobic and x: any non-proline residue) helical motifs in transcription coregulator interactions (Plevin et al., 2005) and the occurrence of this motif in Pf1 MBD, the MBD by interacting with Sin3A PAH2 might also be protected from misrecruitment by other coregulators.
The conformational plasticity demonstrated by the Pf1 MBD in binding to diverse targets including Sin3 PAH2 and MRG15 MRG domains brings into sharp focus the role of intrinsically unstructured regions in regulating basic cellular processes. Indeed, the ability to bind diverse targets was one of the early predictions of the Wright-Dyson intrinsically unstructured protein hypothesis (Wright and Dyson, 1999).
Experimental Procedures
Production of the MRG15 MRG Polypeptide
The human MRG15 MRG polypeptide (residues 155–323) was expressed and purified as described previously (Kumar et al., 2011). Uniformly 15N- and/or 13C-labeled proteins were produced using this procedure except cells were grown in M9 minimal medium containing 15N-ammonium sulfate and/or 13C-D-glucose. Protein identity, integrity, and the extent of 15N/13C isotope incorporation were assessed by electrospray ionization-mass spectrometry (ESI-MS). NMR and ITC studies with MRG15 MRG were inadvertently conducted in a K201R mutant background; however, the Pf1 MBD-binding affinity of the R201K `mutant' is not significantly different from K201R (Table 2) and the mutation itself, besides being conserved, is not at the intermolecular interface.
Production of the Pf1 MBD Polypeptide
The coding sequence for human Pf1 MBD (residues 200–241) was sub-cloned into the pMCSG7 expression vector, but the sequence for the N-terminal 24-residue tag was removed by PCR. Pf1 MBD was co-expressed and co-purified with the MRG15 MRG polypeptide analogous to the co-expression and co-purification protocol described previously except the Pf1 MBD was selectively eluted after binding to the Ni2+-resin in the presence of 6 M guanidine hydrochloride prior to HPLC purification (Kumar et al., 2011). Uniformly 15N- or 15N,13C-labeled Pf1 MBD were produced as described above for MRG15 MRG and ESI-MS used to assess protein identity, integrity, and extent of isotope incorporation.
Isothermal Titration Calorimetry
ITC experiments were performed as previously described except a 20 mM sodium phosphate buffer (pH 7.8) containing 0.15 M NaCl was used (Kumar et al., 2011). Wild-type and mutant MRG15 MRG were in the cell while the wild-type and mutant Pf1 MBD were in the syringe at initial concentrations of 10–20 μM and 0.15–0.55 mM, respectively.
MRG15 MRG-Pf1 MBD Complex Generation and NMR Sample Preparation
The MRG15 MRG-Pf1 MBD complex was generated by mixing equimolar amounts of uniformly 15N,13C-labeled MRG15 MRG with unlabeled Pf1 MBD or vice versa at a low concentration (~20 μM) in NMR buffer (50 mM sodium phosphate (pH 6.8) containing 5 mM dithiothreitol-d10, 10% D2O and 0.2% NaN3). The samples were concentrated via ultrafiltration to ~0.9 mM for NMR studies. Protein concentrations were determined spectrophotometrically (Gill and von Hippel, 1989). Samples of the complex were lyophilized and dissolved in 99.996% D2O for experiments in D2O.
NMR Spectroscopy and Structure Determination
NMR data were acquired on a Varian Inova 600 MHz spectrometer at 25 °C. NMR data processing and analysis were performed using Felix 98.0 and Sparky (Goddard and Kneller, 2004). Backbone and side chain 1H, 15N, and 13C resonances for each protein in the MRG15 MRG-Pf1 MBD complex were assigned by analyzing 3D HNCACB, CBCA(CO)NH, HNCA, HN(CO)CA, HNCO, HCACO, HCCH-COSY and HCCH-TOCSY spectra (Bax and Grzesiek, 1993; Ferentz and Wagner, 2000). Aromatic resonances were assigned from 2D 1H-13C aromatic HSQC, 3D 13C-edited NOESY and 15N,13C-double half-filtered NOESY spectra (Otting and Wüthrich, 1990).
Backbone ϕ and ψ torsion angle restraints were derived from analysis of 1Hα, 13Cα, 13Cβ, 13C', and amide 15N chemical shifts using TALOS+ (Shen et al., 2009); only those residues with TALOS+ reliability scores of 10 were restrained. NOE restraints for each protein in the complex were derived from 3D 15N-edited NOESY (mixing time, τm=75 ms) and 3D 15N,13C-filtered, 15N,13C-edited NOESY (τ =120 ms) spectra recorded in H2O, 3D 13C-edited aliphatic NOESY (τm = 60 ms) and 2D 15N,13C-double half-filtered NOESY (τm = 60 ms) spectra recorded in D2O.
Structures were determined using ARIA 1.2 and CNS 1.1 starting from an initial structure in an extended conformation (Brünger et al., 1998; Linge et al., 2003). All NOEs were calibrated automatically and assigned iteratively by ARIA; assignments were checked manually for errors after every run. Out of 80 conformers calculated in the final iteration, the 40 conformers with the lowest restraint energies were refined in a shell of explicit water and the 20 conformers with the lowest restraint energies, restraint violations, and RMS deviations from ideal covalent geometry were analyzed using CNS (Brünger et al., 1998), PROCHECK (Laskowski et al., 1996), MONSTER (Salerno et al., 2004), and DeepView (Guex and Peitsch, 1997). Images were generated using CHIMERA (Pettersen et al., 2004). The RCSB PDB accession code for the atomic coordinates of the MRG15 MRG-Pf1 MBD complex is 2LKM. The BMRB code for NMR chemical shifts is 18000.
NMR Titrations
A 0.05 mM sample of 15N-labeled MRG15 MRG in NMR buffer was titrated with a Pf1 peptide spanning residues 219–232 at peptide:protein molar ratios of 0:1, 0.5:1, 1:1, 2:1, 3:1 and 10:1 and 2D 1H-15N HSQC spectra were recorded at each ratio. The chemical shift changes induced by the peptide for multiple well-resolved resonances were quantified and the equilibrium dissociation constant KD was determined by non-linear least-squares fitting as described previously.
Co-Immunoprecipitation Assays
Point mutations were introduced using one-step-directed mutagenesis in expression plasmids encoding for FLAG-tagged mouse Pf1 and HA-tagged human MRG15. For co-IPs, HEK293T cells were transfected with the indicated plasmids. 48 h after transfection, cells were washed twice in ice cold PBS and lysed in lysis buffer (20 mM HEPES, pH 7.9, 150 mM KCl, 5% glycerol, 1 mM DTT, 0.1 mM zinc acetate, 2 mM MgCl2, 2 mM EDTA, 0.2% NP-40) with protease inhibitors. Whole cell extracts were incubated at 4 °C with anti-FLAG M2 affinity gel (Sigma) for 4 h. After five washes in high-salt buffer (lysis buffer with 300 mM KCl), the samples were boiled in SDS-PAGE loading buffer, the proteins were resolved by SDS-PAGE and detected with Western blotting by corresponding antibodies.
Luciferase assays
Point mutations were introduced using one-step-directed mutagenesis in expression plasmids encoding human MRG15 fused to the GAL4 DNA-binding domain (pGAL0 vector). Sub-confluent HEK293T cells were transfected with various GAL4-Pf1 or GAL4-MRG15 fusion constructs, the luciferase reporter plasmid driven by four GAL4 binding sites upstream of the thymidine kinase promoter, and the β-galactosidase reporter plasmid driven by a CMV promoter, for normalization purposes. Cells were lysed 48 h post-transfection and whole cell extracts were assayed for luciferase activity using luciferase buffer and an automated luminometer. Transfection efficiencies were normalized using β-galactosidase.
Supplementary Material
Acknowledgements
This work was supported by a grant from the NIH to I.R. (R01 GM064715). We gratefully acknowledge access to resources in the WCAS Biological NMR Center and support for structural biology research from the Robert H. Lurie Comprehensive Cancer Center at Northwestern.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bax A, Grzesiek S. Methodological advances in protein NMR. Accounts Chem Res. 1993;26:131–138. [Google Scholar]
- Bertram MJ, Berube NG, Hang-Swanson X, Ran Q, Leung JK, Bryce S, Spurgers K, Bick RJ, Baldini A, Ning Y, et al. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol Cell Biol. 1999;19:1479–1485. doi: 10.1128/mcb.19.2.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman BR, Moure CM, Kirtane BM, Welschhans RL, Tominaga K, Pereira-Smith OM, Quiocho FA. Multipurpose MRG domain involved in cell senescence and proliferation exhibits structural homology to a DNA-interacting domain. Structure. 2006;14:151–158. doi: 10.1016/j.str.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Florens L, Swanson SK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim Biophys Acta. 2005a;1731:77–87. doi: 10.1016/j.bbaexp.2005.09.005. discussion 75-76. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005b;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chen M, Pereira-Smith OM, Tominaga K. Loss of the chromatin regulator MRG15 limits neural stem/progenitor cell proliferation via increased expression of the p21 Cdk inhibitor. Stem Cell Res. 2011;7:75–88. doi: 10.1016/j.scr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Takano-Maruyama M, Pereira-Smith OM, Gaufo GO, Tominaga K. MRG15, a component of HAT and HDAC complexes, is essential for proliferation and differentiation of neural precursor cells. J Neurosci Res. 2009;87:1522–1531. doi: 10.1002/jnr.21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Tominaga K, Pereira-Smith OM. Emerging role of the MORF/MRG gene family in various biological processes, including aging. Ann N Y Acad Sci. 2010;1197:134–141. doi: 10.1111/j.1749-6632.2010.05197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol. 2011;18:977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Q Rev Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- Fleischer TC, Yun UJ, Ayer DE. Identification and characterization of three new components of the mSin3A corepressor complex. Mol Cell Biol. 2003;23:3456–3467. doi: 10.1128/MCB.23.10.3456-3467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia SN, Kirtane BM, Podlutsky AJ, Pereira-Smith OM, Tominaga K. Mrg15 null and heterozygous mouse embryonic fibroblasts exhibit DNA-repair defects post exposure to gamma ionizing radiation. FEBS Lett. 2007;581:5275–5281. doi: 10.1016/j.febslet.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner KE, Allis CD, Strahl BD. OPERating ON Chromatin, a Colorful Language where Context Matters. J Mol Biol. 2011;409:36–46. doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. Sparky 3. 2004 http://www.cgl.ucsf.edu/home/sparky/
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Zhang F, Hayakawa N, Ohtani Y, Shinmyozu K, Nakayama J, Andreassen PR. MRG15 binds directly to PALB2 and stimulates homology-directed repair of chromosomal breaks. J Cell Sci. 2010;123:1124–1130. doi: 10.1242/jcs.060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinic P, Pellegrino J, David G. A novel mammalian complex containing Sin3B mitigates histone acetylation and RNA polymerase II progression within transcribed loci. Mol Cell Biol. 2011;31:54–62. doi: 10.1128/MCB.00840-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kumar GS, Xie T, Zhang Y, Radhakrishnan I. Solution structure of the mSin3A PAH2-Pf1 SID1 complex: a Mad1/Mxd1-like interaction disrupted by MRG15 in the Rpd3S/Sin3S complex. J Mol Biol. 2011;408:987–1000. doi: 10.1016/j.jmb.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SI. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol. 2007;14:372–380. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- Otting G, Wüthrich K. Heteronuclear filters in two-dimensional [1H,1H]-NMR spectroscopy: combined use with isotope labelling for studies of macromolecular conformation and intermolecular interactions. Q Rev Biophys. 1990;23:39–96. doi: 10.1017/s0033583500005412. [DOI] [PubMed] [Google Scholar]
- Pardo PS, Leung JK, Lucchesi JC, Pereira-Smith OM. MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation. J Biol Chem. 2002;277:50860–50866. doi: 10.1074/jbc.M203839200. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Plevin MJ, Mills MM, Ikura M. The LxxLL motif: a multifunctional binding sequence in transcriptional regulation. Trends Biochem Sci. 2005;30:66–69. doi: 10.1016/j.tibs.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Salerno WJ, Seaver SM, Armstrong BR, Radhakrishnan I. MONSTER: inferring non-covalent interactions in macromolecular structures from atomic coordinate data. Nucleic Acids Res. 2004;32:W566–568. doi: 10.1093/nar/gkh434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Hong J, Zhang P, Dong X, Shen X, Lin D, Ding J. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. J Biol Chem. 2008;283:36504–36512. doi: 10.1074/jbc.M806564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KA, Knoepfler PS, Huang K, Kang RS, Cowley SM, Laherty CD, Eisenman RN, Radhakrishnan I. HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat Struct Mol Biol. 2004;11:738–746. doi: 10.1038/nsmb798. [DOI] [PubMed] [Google Scholar]
- Sy SM, Huen MS, Chen J. MRG15 is a novel PALB2-interacting factor involved in homologous recombination. J Biol Chem. 2009;284:21127–21131. doi: 10.1074/jbc.C109.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga K, Kirtane B, Jackson JG, Ikeno Y, Ikeda T, Hawks C, Smith JR, Matzuk MM, Pereira-Smith OM. MRG15 regulates embryonic development and cell proliferation. Mol Cell Biol. 2005;25:2924–2937. doi: 10.1128/MCB.25.8.2924-2937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Xu C, Cui G, Botuyan MV, Mer G. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure. 2008;16:1740–1750. doi: 10.1016/j.str.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochum GS, Ayer DE. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and Transducin-Like Enhancer of Split. Mol Cell Biol. 2002;22:7868–7876. doi: 10.1128/MCB.22.22.7868-7876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhao J, Wang B, Du J, Lu Y, Chen J, Ding J. The MRG domain of human MRG15 uses a shallow hydrophobic pocket to interact with the N-terminal region of PAM14. Protein Sci. 2006;15:2423–2434. doi: 10.1110/ps.062397806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun ZW, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.