Abstract
HPV-16E7 is a major transforming protein, which has been implicated in the development of cervical cancer. The stability of E7 is thus important to ensure its fully functional status. Using the yeast two-hybrid system, we found that USP11 (ubiquitin-specific protease 11), a member of a protein family that cleaves polyubiquitin chains and/or ubiquitin precursors, interacts and forms a specific complex with HPV-16E7. Our results indicate that the USP11 can greatly increase the steady state level of HPV-16E7 by reducing ubiquitination and attenuating E7 degradation. In contrast, a catalytically inactive mutant of USP11 abolished the deubiquitinating ability and returned E7 to a normal rate of degradation. Moreover, USP11 not only protected E7 from ubiquitination but also influenced E7 function as a modulator of cell growth status. These results suggest that USP11 plays an important role in regulating the levels of E7 protein and subsequently affects the biological function of E7 as well as its contribution to cell transformation by HPV-16E7.
Human papillomaviruses (HPVs)3 have been etiologically linked to human cervical cancer (1). Much pathological evidence has indicated that persistent infections with specific high risk type HPVs (HPV-16, HPV-18, and several others) have a significant relationship with the formation of malignant tumors (2). HPVs are small, double-stranded DNA viruses that infect mucosal and cutaneous epithelial tissues. Since HPVs do not encode a viral DNA polymerase, these viruses are completely dependent on the host cell machinery for DNA replication except an origin-binding protein and a replicative DNA helicase (3). In order to provide all of the necessary cellular proteins required for viral replication, the virus has to keep its host cell in cycle, and this serves as the molecular basis for the proliferative phenotype of HPV lesions. The functions of two viral proteins, E6 and E7, from high risk type HPVs, are reflected in many of the cellular proteins with which they interact. Among a variety of cellular targets, the effects of E6 and E7 on p53 and pRb as well as on many other cellular proteins have been extensively investigated in the past. The results all point to significant alterations in the regulation of the cell cycle and apoptosis as well as chromosomal stability (4). However, a detailed mutagenesis study of E7 has also revealed that binding to Rb alone is not sufficient to cause cell transformation (5). It is clear that mutations in the carboxyl-terminal sequences of E7, outside the Rb-binding site, also severely impair the transformation function of E7 (6, 7). These results suggest that, aside from binding to Rb, other protein-protein interaction(s) are also necessary for the E7-mediated transformation.
The HPV E7 protein is a small, rather acidic, polypeptide composed of nearly 100 amino acid residues. E7 is also short lived (8), and its degradation is regulated by the ubiquitin-proteasome pathway in cervical cancer cells (9, 10). The first 11 amino acid residues of E7 is the signal responsible for ubiquitination, and deletion of the N-terminal 11 amino acid residues stabilizes the E7 protein in cells. Recently, in a study of the E7 degradation pathway, one group identified SOCS1, an E3 ubiquitin ligase, as interacting with the E7 protein and degrading E7 in a SOCS-box-dependent manner (9). Another group demonstrated that E7 interacts with the SCF (Skp-Cullin-F box) ubiquitin ligase complex containing Cullin 1 (Cul1) and Skp2 and that the protein can be ubiquitinated by the Cul1-containing ubiquitin ligase in vitro (11). These two reports provided more insight into an understanding of the E7 degradation mechanism.
In eukaryotic cells, the homeostasis for a number of cellular proteins is regulated by not only phosphorylation and dephosphorylation but also ubiquitination and deubiquitination. A number of proteins involved in the degradation of polypeptides have been isolated in various eukaryotic organisms ranging from Saccharomyces cerevisiae to humans. Ubiquitination of proteins is now considered to be an essential step for degradation by the proteasome and for internalization into the lysosomal system as well as being involved in the modification of the functions of some target proteins (12). Although much progress has been made in characterizing enzymes that link ubiquitin to proteins, our understanding of deubiquitinating enzymes is less well developed. Deubiquitination of ubiquitin from H2A was the first case described. Since then, a number of enzymes have been identified, and the results have been reviewed in several excellent articles (13–16). Deubiquitinating enzymes (DUBs) have been categorized as a group of cysteine proteases based on their sensitivity to inhibition by thiol reagents (17, 18). Ubiquitin-specific processing proteases (UBPs) are the major family of a variety of DUBs (14). The UBPs vary in size but possesses conserved domains, such as the Cys box and the His box. A variety of sequences aside from conserved catalytic domains have been suggested to function in substrate recognition, subcellular localization, and protein-protein interactions. In addition to the processing of ubiquitin precursors and the disassembly of free polyubiquitinated chains; UBPs are responsible for removing ubiquitin from specific substrate proteins, which then prevents protein degradation (19). Deubiquitination is a reversible process; that is to say the net results of ubiquitination and deubiquitination play key roles in controlling fundamental cellular activities, such as cell growth, differentiation, and oncogenesis. However, knowledge of the specific biological roles of DUBs in viral infection and of ubiquitination on viral oncoproteins is still lacking. In this study, we report the identification of another USP11 function in stabilizing HPV-16E7 and also further modulate the E7 biological activity.
EXPERIMENTAL PROCEDURES
Plasmids—The plasmids used for the yeast two-hybrid screening were pGBKT7–16E7 and pGBKT7–18E7. These two plasmids were constructed by inserting cDNA fragments encoding the full length of either HPV-16E7 or HPV-18E7 into the EcoRI and BamHI sites of pGBKT7 (Clontech). PRK5–16E7, which expresses a FLAG HPV-16E7, was cloned by inserting intact HPV-16E7 into the EcoRI and BamHI sites of pRK5 (20). To construct the USP11 expression vectors, the DNA sequences corresponding to NCBI accession number AB073597 were amplified by PCR and subcloned either into the HindIII and XhoI sites of pCMVTaq2B and pCMVTaq3B (Stratagene) or with an HA tag into the HindIII and KpnI sites of pcDNA3 (Invitrogen). The DNA sequence corresponding to the full-length protein of p53 was cloned into the EcoRI and SalI sites of pCMVTaq2C vector to generate the FLAG-p53. The catalytically deficient mutant of USP11 (FLAG-USP11 cys mt) was generated by substituting two cysteine (C275S and C283S) residues within the Cys conserved domain of USP11 with serine by PCR; then the USP11 cys mt fragment was inserted into the HindIII and XhoI sites of pCMVTaq2B. The DNA sequences of USP11 and USP11 cys mt were cloned into pGEX-5x-1 to obtain GST-USP11 and GST-USP11 cys mt, respectively, for recombinant protein expression. The HA-ubiquitin expression construct was obtained by inserting a full-length ubiquitin sequence into the EcoRV and BamHI sites of pcDNA3 with an HA tag. For small interfering RNA targeting of USP11 (si-USP11), the oligonucleotides CTA GTG ATG AGA TAA ACT GGC GCC TTCAA GAG AGG CGC CAG TTT ATC TCA TCT TTT TTG GAA A and CTA GTT TCC AAA AAA GAT GAG ATA AAC TGG CGC CTC TCT TGA AGG CGC CAG TTT ATC TCA TCA were annealed and ligated into pLEGFP-C1 (BD Biosciences) containing a U6 promoter (21) (underlining indicates RNA interference target sequences).
Yeast Two-hybrid Screening—Yeast two-hybrid screening was conducted using a pretransformed Matchmaker library (Clontech). The library is HeLa-S3 cDNAs cloned into the yeast GAL4 activation domain vector and pretransformed into S. cerevisiae host strain Y187. Then pGBKT7-16E7 was used as bait and transformed into a yeast reporter strain AH109 that can serve as a mating partner for Y187. Screening was performed according to the manufacturer's protocol. Briefly, the HeLa-S3 cDNA library in Y187 was simply combined with the bait strain and incubated for 24 h, and then the mixture was plated on selected medium. Approximately 4.15 × 106 transconjugants were screened. Proteins interacting with HPV-16E7 were identified by growth on SD/–Leu/–Trp/–His/–Ade plates and confirmed by assaying for β-galactosidase activity. Plasmids rescued from positive yeast colonies were retransformed into Y187 and incubated together with AH109 containing either pGBKT7-16E7 or pLAM5′-1 to measure the specificity of the interaction.
Cell Cultures and Transfection—Human cervical carcinoma CaSki cells and human lung carcinoma H1299 cells were cultured in RPMI1640 supplemented with 10% fetal bovine serum at 37 °C in 5% CO2. The 293T cells were cultured in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum. All three cell lines were transfected using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). pLEGFP-C1/U6 vector alone or vector containing the si-USP11 sequence was introduced into CaSki cells to generate the CaSki (si-) and CaSki (si-USP11) cell lines. G418 selection was started 24 h after transfection. The surviving cells were maintained in medium containing 200 g/ml G418.
Western Blot and Immunoprecipitation—Cell extracts were prepared in lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, 0.5% sodium deoxycholate, and 0.1% SDS) containing 1× protease inhibitor mixture (Complete™; Roche Applied Science). For Western blot analysis, cell extracts were collected and subjected to SDS-PAGE analysis. After separation, the proteins on gel were transferred onto polyvinylidene difluoride membrane and were immunostained with the antibodies as indicated. The α-HPV-16E7 monoclonal antibody was purchased from Zymed Laboratories Inc. α-FLAG M2 was obtained from Sigma; α-ubiquitin, α-pRb, and α-Bcl-2 were bought from Upstate Biotechnology; α-tubulin and α-Cdc-2 were purchased from Oncogene Science and α-HA was bought from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). For immunoprecipitation, equal amounts of protein were incubated with various antibodies as indicated for 1 h at 4 °C, followed by precipitation with protein-agarose beads (Upstate Biotechnology). All immunoprecipitates were washed three times with washing buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 0.5% Nonidet P-40, 1 mm EDTA). Bound proteins were collected and analyzed by SDS-PAGE and followed by Western blotting analysis.
In Vivo Ubiquitination Assay—293T cells were transiently co-transfected with plasmids as indicated. Then, 24 h after transfection, the cells were treated with 20 m MG132 for 1.5h. Cells were harvested in lysis buffer (see above), immunoprecipitated with α-FLAG, and resolved by SDS-PAGE. Ubiquitin-tagged proteins were detected by α-HA (Roche Applied Science) Western blot analysis.
In Vitro Deubiquitination Assay—GST-USP11 and GST-USP11 cys mt fusion proteins were expressed in BL21 (pLysS) bacteria (Novagen) with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 30 °C for 3.5 h. Cells were pelleted and resuspended in lysis buffer (50 mm Tris, 2 mm EDTA, 1 mg/ml lysozyme, and 0.25% Triton X-100) and then were incubated at room temperature for 30 min. After sonication, the insoluble material was removed by centrifugation. Supernatants were applied to glutathione-agarose beads and incubated for 3 h at 4 °C. Beads were pelleted and washed four times with washing buffer (50 mm Tris, 2 mm EDTA, and 0.25% Triton X-100). Bound proteins were eluted in 50 mm Tris, 2 mm EDTA containing 10 mm reduced glutathione at 4 °C. The concentration of purified GST proteins was determined by a BCA assay. The His-16E7 protein was in vitro transcribed and translated by the Single Tube Protein System 3, T7 (Novagen) according to the manufacturer's protocol. For preparation of ubiquitinated 16E7, in vitro translated His-16E7 (10 μl) was incubated with HeLa cell extracts (50 μl) and reticulocyte lysates (4 μl) at 30 °C in a reaction mixture (80 μl) containing ubiquitin (40 μg), an ATP-generating system (20 mm Tris, pH 7.5, 10 mm ATP, 10 mm magnesium acetate, 300 mm creatine phosphate, 0.5 mg/ml creatine phosphokinase), and ubiquitin aldehyde (4 m). After 2 h of incubation, 16E7-ubiquitin conjugates were purified using a nickel affinity column. For the deubiquitination assay, ubiquitinated 16E7 was incubated in deubiquitination buffer (50 mm Tris, pH 7.5, 50 mm NaCl, 1 mm EDTA, 5 mm dithiothreitol), and then purified GST-USP11 proteins were added. The reaction mixtures were incubated for 2 h at 30 °C, and the reaction was terminated by the addition of SDS-PAGE loading buffer. Reaction mixtures were resolved on a 10% SDS-polyacrylamide gel and probed with anti-ubiquitin antibody using Western blotting.
Colony Formation Assay—The same number of CaSki (si-) and CaSki (si-USP11) cells were seeded and maintained in the culture medium containing G418 (200 g/ml). After 2 weeks of selection, the colonies were fixed in 10% methanol, 10% acetic acid (v/v) for 10 min, stained with 1% (w/v) crystal violet in methanol for 10 min, and then rinsed three times with water. The colonies were then counted.
RESULTS
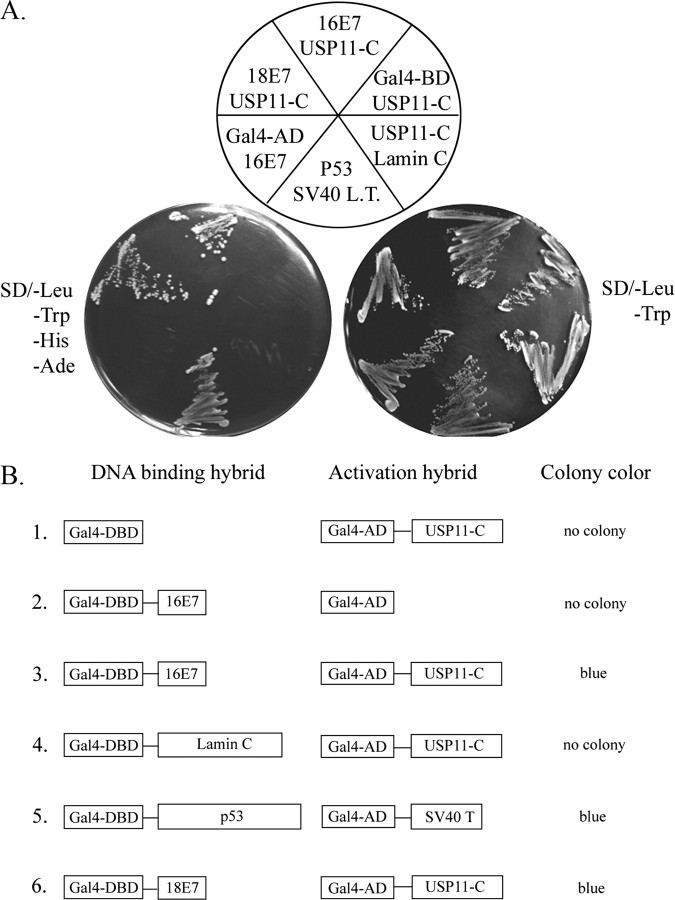
Identification of USP11 as Cellular Target for HPV-16E7—Employing a yeast two-hybrid assay allowed the identification of USP11 as a cellular target for the HPV-16E7 oncoprotein. The cDNA library from HeLa cell was screened using intact HPV-16E7 (amino acids 1–97) as bait. One of the cDNAs encoding the carboxyl-terminal domain of USP11 was repeatedly isolated (Fig. 1A). To verify the specificity of interaction between HPV-16E7 and USP11, several control experiments were conducted. As shown in Fig. 1B, co-expression of the HPV-16E7 and the C terminus of USP11 (USP11-C) rendered the yeast strain CG1945/Y187 able to grow on SD/–Leu/–Trp/–His/–Ade minimal plates (Fig. 1B, lane 3), whereas the Gal4 activation domain (Gal4-AD) failed to bind to the HPV-16E7 (Fig. 1B, lane 2), indicating that USP11 sequences are essential for binding to HPV-16E7. The inability of the Gal4 DNA binding domain (Gal4-BD) on its own (lane 1) or fused with lamin (lane 4) to bind to USP11 confirms that the C terminus of USP11 interacts specifically with HPV-16E7. Interestingly, the C-terminal of USP11 also binds to HPV-18E7, but the protein failed to bind to HPV-11E7, a strain not closely linked to cervical cancer (data not shown).
FIGURE 1.
Identification of USP11 as a novel interacting protein of HPV-16E7. A, the yeast strain AH109 containing the indicated BD plasmids (16E7, 18E7, p53, Lamin C, and Gal-BD) was co-incubated with Y187 harboring AD plasmids (USP11-C, SV40 LT, and Gal4-AD). After overnight incubation, yeast cultures were streaked on SD medium lacking leucine and tryptophan or grown on a selective minimal plate (SD medium lacking leucine, tryptophan, histidine, and adenine). The Gal4 DNA-binding domain (Gal4 BD) and the Gal4 activation domain (Gal4 AD) were incorporated as negative controls. p53 and simian virus 40 large T-antigen (SV40 LT) were used as positive controls. Laminin C was used to elucidate the specificity of the interaction between 16E7 and USP11-C. B, a schematic summary of A. Yeast colony color was observed after incubation for a standard period of time. The color blue represents USP11-C associating with HPV-16E7 and turning on the lacZ promoter.
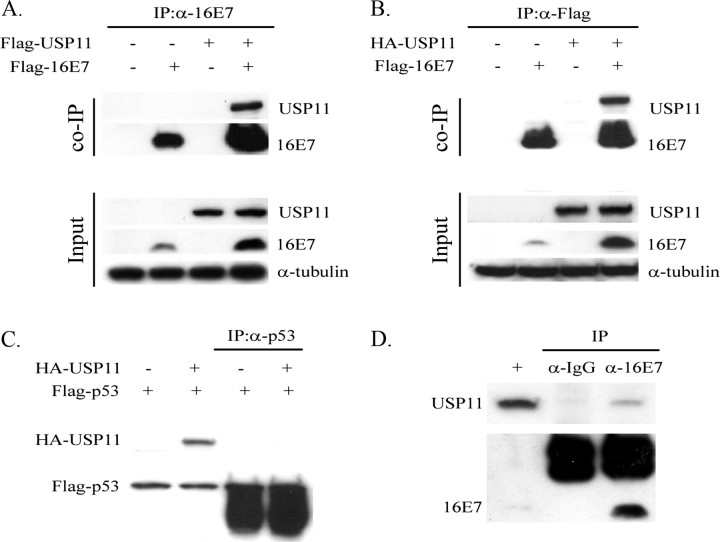
In Vivo Association between 16E7 and USP11—To address a potential interaction between HPV-16E7 and USP11 in mammalian cells, co-immunoprecipitation and Western blot experiments were performed. FLAG-HPV-16E7 and FLAG-USP11 were transiently expressed in H1299 cells, as indicated, and immunoprecipitated using anti-HPV-16E7 (Fig. 2A). The presence of FLAG-USP11 was detected by α-FLAG Western analysis. As shown in Fig. 2A, the 16E7 antibody precipitated large amounts of FLAG-16E7 from extracts of transfected cells. The result showed that FLAG-USP11 was co-precipitated with FLAG-HPV-16E7; however, no co-precipitation was detected when either one of the two plasmids was omitted. A similar experiment was repeated by replacing FLAG-USP11 with HA-USP11 (Fig. 2B). Again, the FLAG antibody coprecipitated HA-USP11 from extracts of cells transiently transfected with both FLAG-HPV-16E7 and HA-USP11. We also expressed FLAG-p53 and HA-USP11 during a control co-precipitation. FLAG-p53 was unable to bind to HA-USP11 (Fig. 2C). This result indicates a specific association of USP11 with HPV-16E7. In addition, we also have performed an in vitro binding assay to address the issue of direct interaction between the two proteins. The GST alone and GST-USP11 fusion proteins were expressed in bacteria, isolated, and then incubated with in vitro translated [35S]methionine-labeled HPV-16E7, respectively. The result showed that [35S]methionine-labeled HPV-16E7 was specifically retained on the GST-USP11-conjugated Sepharose, whereas GST alone was not (date not shown). Moreover, to confirm that the endogenous USP11 and HPV-16E7 proteins also associate, cell extracts were prepared from CaSki and then immunoprecipitated with HPV-16E7 or IgG antibodies. As shown in Fig. 2D, USP11 was co-precipitated with HPV-16E7 but not with IgG antibody. Taken together, these results strongly suggest that HPV-16E7 and USP11 interact specifically in mammalian cells.
FIGURE 2.
HPV-16E7 associates with USP11 in vivo. A, the in vivo interaction of HPV-16E7 with USP11 by immunoprecipitation assay. FLAG-USP11 and FLAG-16E7 plasmids were transfected into H1299 as indicated. After 48 h of transfection, cell extracts were prepared and subjected to immunoprecipitation (IP) by α-HPV-16E7. The proteins from the immunoprecipitation were separated using 10% SDS-PAGE and analyzed by α-FLAG to detect the associated proteins. Western blotting using α-FLAG indicates the expression of the product. Equal amounts of proteins were loaded as indicated by detection of tubulin. B, the in vivo interaction of HPV-16E7 with USP11 by immunoprecipitation assay in different combinations. An experiment similar to that in A was conducted, except that FLAG-USP11 was replaced by HA-USP11, immunoprecipitation was conducted using α-FLAG, and α-HA was used to detect the associated protein. C, to confirm the specific association, HA-USP11 and FLAG-p53 plasmids were transfected into H1299 as indicated. Immunoprecipitations were performed using α-p53, and the precipitates were detected by α-HA antibody. D, the in vivo interaction of USP11 with endogenous E7 in CaSki cells. Immunoprecipitations were performed with cell extracts of CaSki cells using eitherα-HPV-16E7 or normal mouse IgG as a negative control. Precipitates were recognized by α-HPV-16E7, and the association was detected using α-USP11.
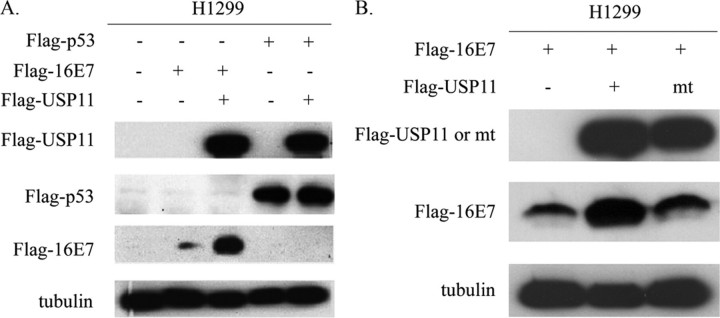
USP11 Specifically Stabilizes 16E7 through the Deubiquitinating Enzymatic Activity—USP11 is a member of the UBPs. UBPs are responsible for removing ubiquitin from polyubiquitinated proteins, and this renders target proteins able to escape proteasome degradation. Since USP11 and HPV-16E7 can form a complex in vivo, there exists the possibility that USP11 could extend the stability of HPV-16E7. To assess the functional consequence of the interaction between USP11 and HPV-16E7, we tested whether USP11 affects the stabilization of HPV-16E7. H1299 cells were transfected with FLAG-16E7 alone or together with FLAG-USP11. As shown in Fig. 3A, FLAG-USP11 expression significantly increased the steady-state levels of FLAG-16E7. To test the specificity of this USP11 function, another short lived tumor suppressor protein, p53, whose stability is also regulated by the ubiquitination pathway, was used as a control. Unlike the HPV-16E7, the FLAG-p53 level was not affected in the presence of USP11. These results lead us to conclude that USP11 specifically stabilizes HPV-16E7 in vivo.
FIGURE 3.
USP11 specifically stabilizes HPV-16E7, and the deubiquitinating activity of USP11 is required for this function. A, USP11 stabilized HPV-16E7 but not p53. FLAG-16E7 and FLAG-p53 were transfected into H1299 cells either alone or together with FLAG-USP11 as indicated. At 48 h after transfection, 100 μg of cell extracts from transfected cells were assayed by Western blot using α-FLAG. Approximately equal amounts of proteins were loaded as indicated by the tubulin signal. B, mutant USP11 (USP11 cys mt) lost the ability to stabilize HPV-16E7. The FLAG-USP11 and the mutant form, FLAG-USP11 cys mt, were transfected together with FLAG-16E7 into H1299 cells as indicated. At 48 h after transfection, 100 μg of cell extracts were subjected to Western blot using α-FLAG to detect the levels of FLAG-16E7 and FLAG-USP11 expression. The amount of proteins loaded is indicated by the tubulin signal.
Proteins in the UBP family differ in length but possess conserved domains, such as the Cys box and the His box. These two regions are responsible for the catalytic activity of UBPs (22). To investigate whether the deubiquitinating activity of USP11 is required for stabilizing HPV-16E7, we generated a mutant of USP11 (USP11 cys mt) in which two highly conserved Cys residues within the catalytic core domain were replaced by serine. As shown in Fig. 3B, co-expression of both HPV-16E7 and USP11 significantly stabilized E7. In contrast, the USP11 cys mt strain lost the ability to stabilize HPV-16E7. This result demonstrated that the deubiquitinating enzymatic activity of USP11 is critical for USP11-mediated stabilization of HPV-16E7.
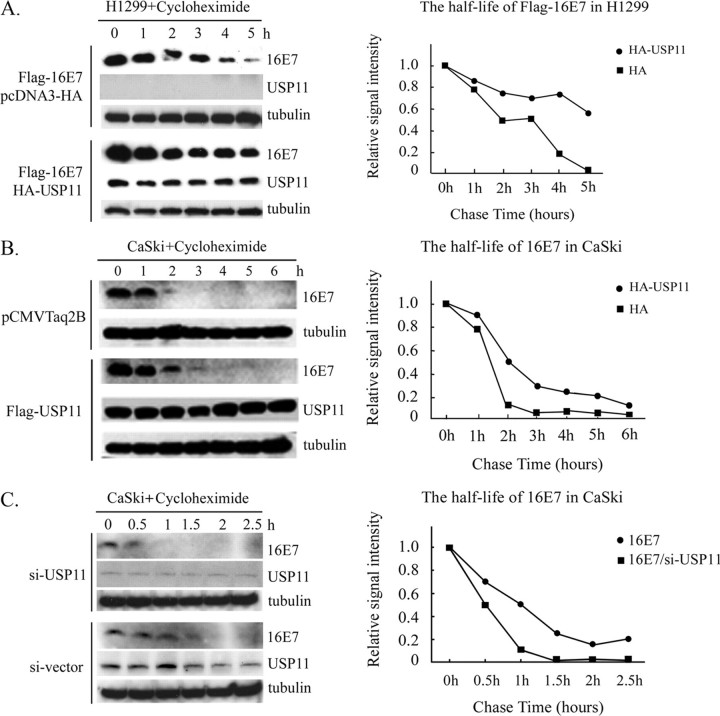
USP11 Reduces Degradation of both Exogenous and Endogenous 16E7—To explore whether the levels of HPV-16E7 are increased upon co-expression of USP11 as a result of an extended half-life, we performed a cycloheximide chase experiment. H1299 cells were transfected with FLAG-HPV-16E7 either alone or together with HA-USP11, and this was followed by continuous cycloheximide treatment. Extracts were then analyzed by Western analysis with specific antibodies. As shown in Fig. 4A, HA-USP11 expression resulted in a longer degradation time for the transfected FLAG-16E7, which was increased from around 2 h in the absence of HA-USP11 to more than 6 h in the presence of HA-USP11. It has been reported that HPV-16E7 is ubiquitinated at its amino terminus (11). In order to avoid the interference of epitope tag in the E7 ubiquitinated experiment, similar experiments using a cell line (CaSki) with endogenous 16E7 were conducted. An extended degradation time of HPV-16E7 was also observed in the presence of HA-USP11 (Fig. 4B). To further confirm the effect of endogenous USP11 on HPV-16E7, we employed an antisense strategy to reduce the level of endogenous USP11 expression. For this purpose, CaSki cells were transfected with pLEGFP-C1/U6 (si-) containing antisense USP11 sequence (si-USP11) or vector alone, and the cells then underwent G418 selection for a period. As shown in Fig. 4C, the expression of the antisense USP11 resulted in a significant reduction in USP11 protein, and this reduction in USP11 led to an apparent decrease in the half-life of HPV-16E7. These results demonstrate that USP11 attenuates the degradation of both exogenous and endogenous HPV-16E7.
FIGURE 4.
USP11 extends the half-life of both FLAG-16E7 and endogenous HPV-16E7. A, USP11 significantly extends the half-life of HPV-16E7 after cycloheximide treatment. FLAG-16E7 was introduced into H1299 together with pcDNA3-HA empty vector or pcDNA3-HA-USP11. At 48 h after transfection, the cells were treated with 50 g/ml cycloheximide to inhibit protein synthesis. Total cell extracts were collected at each indicated time point and were assayed either with α-FLAG to detect the remaining FLAG-16E7 or with α-HA to indicate HA-USP11 expression. Western blot analysis using α-tubulin showed that an equal amount of protein was loaded onto each lane. B, the same procedure also allowed us to study the ability of USP11 to stabilize endogenous E7 in CaSki cells. The amount of proteins loaded is indicated by the tubulin signals. C, USP11 significantly extends the half-life of endogenous HPV-16E7 after si-USP11 treatment. CaSki (si-) and CaSki (si-USP11) cells were cultured in medium containing 200 μg/ml G418. To assay the endogenous HPV-16E7 protein level, cell extracts were collected at the indicated time points after 50 μg/ml cycloheximide was added. The remaining HPV-16E7 was detected by α-16E7, and α-USP11 was used to detect endogenous USP11 expression.
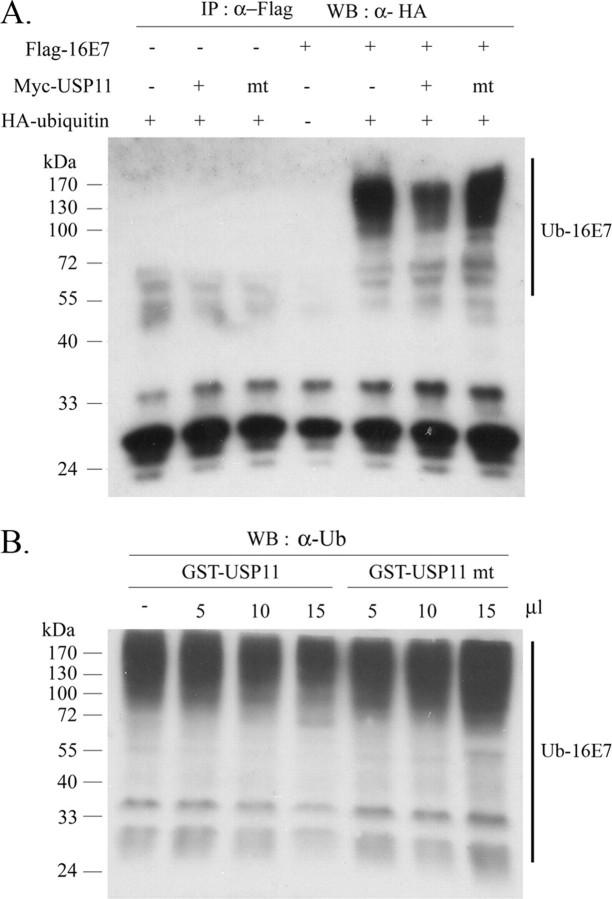
Deubiquitination of HPV-16E7 by USP11 in Vivo and in Vitro—To elucidate the molecular mechanism by which USP11 stabilizes HPV-16E7, we tested whether USP11 directly controls the levels of E7 ubiquitination in vivo. FLAG-HPV-16E7 and HA-ubiquitin were co-expressed with wild type USP11 or USP11 cys mt in 293T cells and then immunoprecipitated with anti-FLAG antibody and blotted with anti-HA for the presence of HA-ubiquitin. Translational levels of all constructions were examined and confirmed in 293T cells before immunoprecipitation (supplemental Fig. 1). As shown in Fig. 5A, wild type USP11 diminished ubiquitination of HPV-16E7, whereas USP11 cys mt removed ubiquitin from HPV-16E7 less effectively. To further confirm that USP11 deubiquitinates HPV-16E7, polyubiquitinated HPV-16E7 was incubated with purified recombinant GST-USP11 or GST-USP11 cys mt. As shown in Fig. 5B, USP11 reduced ubiquitination of HPV-16E7 in a dose-dependent manner. A significant decrease in polyubiquitinated HPV-16E7 was observed with GST-USP11, whereas GST-USP11 cys mt was unable to deubiquitinate HPV-16E7. Thus, these results indicate that the function of USP11 is to reduce the ubiquitination level of HPV-16E7 and thereby protect E7 from proteasome-mediated degradation.
FIGURE 5.
USP11 deubiquitinates HPV-16E7 both in vivo and in vitro. A, ubiquitination and deubiquitination of HPV-16E7 in vivo. 293T cells were transfected with expression plasmids for FLAG-16E7, Myc-USP11, and HA-ubiquitin, as indicated. At 20 h after transfection, cells were treated with 20 m MG132 for 2 h. Total cell extracts were first subjected to immunoprecipitation (IP) using α-FLAG, and then immune complexes were assayed by Western blot, and α-HA was used to detect the ubiquitination levels of FLAG-16E7. mt, FLAG-USP11 cys mt. B, USP11 cys mt has lost its ability to deubiquitinate HPV-16E7 in vitro. The purified polyubiquitinated His-16E7 was incubated with the indicated amounts of purified recombinant GST-USP11 or GST-USP11 cys mt in a deubiquitination buffer at 30 °C for 1 h. Ubiquitinated His-16E7 was detected by Western blot analysis using α-ubiquitin.
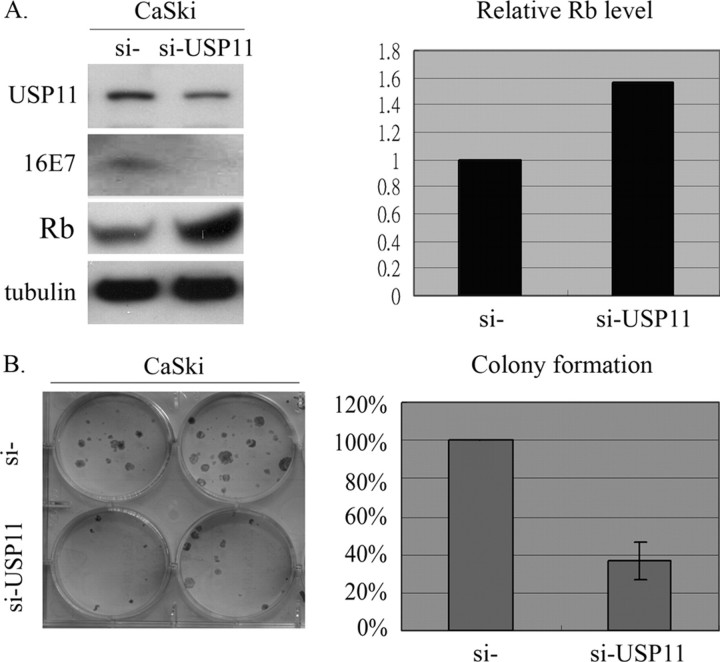
USP11 Stabilizes HPV-16E7 and Regulates the Biological Activity of HPV-16E7 Protein—One of the suggested functions of DUBs is the editing of polyubiquitinated substrates, which may rescue them from proteasome-dependent destruction and thus resume their original designated functions. So far, we have investigated and proved that USP11 binds to HPV-16E7, trims ubiquitin from HPV-16E7, and avoids targeting HPV-16E7 to proteasome degradation. We next explored, therefore, whether the degradation of the tumor suppressor retinoblastoma protein (pRb) should be increased via the interaction between E7 and USP11. As shown in Fig. 6A, antisense against USP11 reduced the protein level of USP11 and then destabilized HPV-16E7 in CaSki (si-USP11) cells. The cells containing antisense USP11 showed a remarkable elevation in Rb protein compared with cells harboring the control vector-only CaSki (si-). To further investigate the biological role of USP11 in stabilizing HPV-16E7, we examined its effect on cell growth in a colony formation assay. Both CaSki (si-) and CaSki (si-USP11) cells were cultured in the presence of G418 (200 μg/ml) for 2 weeks. The results are displayed in Fig. 6B and show that the CaSki (si-) cells demonstrate more malignant characteristics, together with increased ability to grow in plates, compared with CaSki (si-USP11). It should be noted that the cells containing the si-USP11 oligonucleotide showed a remarkable reduction in the number of drug-resistant colonies compared with cells harboring the control vector only. Hence, the association of USP11 with HPV-16E7 not only increases the stability of HPV-16E7 but also regulates E7-modulated cell growth. Similar trends of colony formation also were found in other carcinoma cells, without endogenous HPV, through a forced expression process (supplemental Fig. 2). On the contrary, in the absence of HPV-16E7, colony formation remained the same when compared between the si- and si-USP11 transfected cells (supplemental Fig. 3).
FIGURE 6.
Effects of USP11 on subsequent HPV-16E7-mediated protein activation and cell growth activation. A, the USP11 si-RNA leads to reduced HPV-16E7 and increased Rb in CaSki cells. 300-g cell extracts from CaSki (si) and CaSki (si-USP11) cell lines were subjected to Western blot analysis using α-USP11 or α-HPV-16E7 to detect the levels of endogenous HPV-16E7 and USP11 expression, respectively. Tubulin protein was assayed to monitor equal loading between individual lanes. Quantitative measurements of signals are presented on the right. B, colony formation assay. CaSki (si) and CaSki (si-USP11) cells were subcultured and kept in the medium containing G418 (200 μg/ml), and surviving colonies were counted 2 weeks later. Relative cell viability is summarized from the five separate experiments and is presented on the right.
DISCUSSION
The ubiquitin-mediated protein degradation pathway exerts a wide spectrum of effects and modulates a variety of biological processes, including cell cycle progression, transcriptional regulation, signal transduction, antigen presentation, apoptosis, oncogenesis, preimplantation, endocytosis, vesicle trafficking, and DNA repair (12, 14, 23). Ubiquitination is a dynamic and reversible process, and it has become increasingly evident that deubiquitination plays an important role in regulating the ubiquitin-dependent pathway (14). The importance of the deubiquitination mechanism has also emerged, and it has been demonstrated that the Drosophila fat facets protein, DUB-1, enhances the stabilities of NF-κB and cyclin B as an essential regulatory step to control cellular mechanisms for homeostasis (24–27). The deubiquitinating enzymes that have been identified are generally classified into ubiquitin C-terminal hydrolases (UCHs), ubiquitin-specific processing proteases (UBPs), Jab1/Pad1/MPN domain-containing metalloenzymes, Otu domain ubiquitinaldehyde-binding proteins, and Ataxin-3/Josephin domain-containing proteins (Ataxin-3/Josephin) (28). During the past few years, more and more UBPs have been identified, which implies that UBP enzymes each may carry a distinct and specialized function (29). Although a number of deubiquitinating enzymes have recently been isolated, relatively little is known about their substrates and biological functions. Thus, identifying distinct substrates and assigning specific physiological roles to each member of the UBP family is the major work going on in this area at the moment. In our present study, we have identified that USP11 specifically interacts with HPV-16E7 and, at the same time, inhibits HPV-16E7 ubiquitination and degradation. We have also tested the binding with another UBP family protein, USP7. The results, however, showed no interaction between USP7 and HPV-16E7 and thus no effect on the E7 ubiquitination (supplemental Figs. 4 and 5). One of the other well understood examples of virus-ubiquitin association is the control of gene expression by herpes simplex virus 1. Vmw110, a herpes simplex virus 1 immediate early protein, is an important regulator of viral gene expression and the onset of the viral lytic cycle. HAUSP, a UBP, when bound to Vmw110, is not only able to enhance the stability of Vmw110 but also can activate gene expression in cultured cells (30–33).
USP11 was originally isolated and characterized with the name ubiquitin C-terminal hydrolase on the X chromosome (UHX1) (34). The UBP family is so vast and complex that a systematic nomenclature for human UBPs has been proposed (19). Therefore, UHX1 was renamed as USP11. The full length of USP11 consists of 2763 bp and encodes a protein with a relative molecular mass of 105 kDa. It contains both Cys and His boxes that are responsible for its deubiquitinating activity. USP11 is a ubiquitous protein distributed across a wide range of human tissues (34) and particularly localized to the nucleus (35). In addition, it has been demonstrated recently that USP11 interacts with RanBPM, a Ran-binding protein, which is required for correct microtubule nucleation (35, 36). USP11 and USP9 can both interact with the NF-κB transcription factors RelB and p100. Although the significance of these interactions remains to be determined, these findings suggest that UBPs may play a regulatory role in their pathways.
HPV E7 has also been observed to be predominantly localized in the nucleus (37, 38). This observation is supported by the multiple nuclear functions of E7, including association with members of the pRb family, transactivation of E2F-regulated genes, and interaction with a large number of nuclear factors, such as AP-1, TBP, p21cip1, etc. (39–42). In addition, conflicting reports have described the cytoplasmic presence of E7 (43, 44). Cytoplasmic proteins, such as F-actin, α-glucosidase, and M2 pyruvate kinase, all interact with E7 in the cytoplasm (45). Recently, it was reported that in CaSki cells a major portion of the E7 protein is localized in the cytoplasm. Interestingly, exposure of CaSki cells to the proteasome inhibitor MG132 led to a significant elevation of E7 levels in the nucleus. Thus, it has been suggested that polyubiquitinated E7 accumulates in nucleus for degradation in CaSki cells (11). In the present study, we showed that USP11 reduced the ubiquitination level of E7 and attenuates degradation of E7. Since USP11 is mainly localized in nucleus and ubiquitinated E7 also accumulates in nucleus, therefore, we propose that USP11 interacts with HPV-16E7 in the nucleus. This raises a question as to what mechanism triggers ubiquitinated E7 transportation into the nucleus. Recently, proteasomal degradation modulating protein translocation has been shown to occur with other cellular proteins. For example, C-terminally phosphorylated Smad can undergo ubiquitination, and this enhances its nuclear accumulation. Subsequently, ubiquitinated Smad can interact with a Ring finger protein, Roc1, and this consequently leads to nuclear translocation (46). Therefore, further studies will be required to determine whether ubiquitination regulates the nuclear entry of E7 in vivo.
E7 is normally present at low levels within infected cells (47). Our data presented here showed that USP11 increased the steady state levels of 16E7 and hence led us to examine whether USP11 affects the 16E7-regulated genes. One of the key functions of E7 is that E7 targets pRb and induces its proteolysis through the proteasome-dependent pathway. As anticipated, our results showed that USP11 enhances the E7-induced pRb degradation. In addition, it has been reported that expression of the E7 protein of HPV-16 also up-regulates the expressions of Bcl-2 and BH3 through disruption of pRb-E2F complexes (22). Bcl-2, a gene that protects the integrity of mitochondria, prevents cytochrome c release and the subsequent activation of caspase-9. The expression of Bcl-2 appears to inhibit cells from undergoing apoptosis. It has been reported that E7 induced a significant increase in Bcl-2 expression (48). In our supplemental result, when USP11 and HPV-16E7 are co-expressed in mammalian cells, USP11 augments the ability of HPV-16E7 to induce Bcl-2 expression (supplemental Fig. 6). Moreover, the Cole-cyclin B kinase is another gene pivotal in regulating G2-M phase transition. Cdc-2 is maintained in an inactive state during G2 phase. As cells approach M phase, Cdc-2 is activated and then efficiently drives the cells into mitosis. It has been mentioned that Cdc-2 is elevated in the E7-transfected keratinocytes (48). We also observed that USP11 enhanced the elevation of Cdc-2 by E7 (supplemental Fig. 6). Taken together, USP11 not only extends E7 stability but also influences the downstream genes regulated by E7. It is also of interest to note that USP11 alone (without 16E7 co-transfection) is able to elevate Cdc-2 and Bcl-2 levels but to a much lesser degree. These observations are not in conflict with our conclusion, considering the fact that USP11 can interact and deubiquitinate many other cellular proteins and transcription factors and the significantly added effect observed when both USP11 and 16E7 are present. However, similar future investigations will be required.
Posttranslational modification of proteins by the small molecule ubiquitin is a key regulatory event, and the enzymes catalyzing these modifications have been the focal point of many studies. Deubiquitinating enzymes, which mediate the removal and processing of ubiquitin, may be functionally as important but are yet less well understood. Here, we have identified USP11 as a new cellular target of HPV-16E7. USP11 binds to HPV-16E7 and functionally extends the half-life of HPV-16E7. A mutant of USP11 lacking enzymatic activity fails to deubiquitinate and stabilize HPV-16E7. Additionally, USP11 augments the HPV-16E7 function in modulating downstream target genes, such as pRb, Bcl-2, and Cdc-2, suggesting that this interaction may contribute to cell transformation by HPV-16E7.
Supplementary Material
This work was supported by National Health Research Institutes and National Science Council, Taiwan, Grant NSC96-2320-B-400-006. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
Footnotes
The abbreviations used are: HPV, human papillomavirus; E3, ubiquitin-protein isopeptide ligase; DUB, deubiquitinating enzyme; AD, activation domain; UCH, ubiquitin C-terminal hydrolase; HA, hemagglutinin; GST, glutathione S-transferase.
References
- 1.zur Hausen, H. (1996) Biochim. Biophys. Acta 128855 –78 [DOI] [PubMed] [Google Scholar]
- 2.Mansur, C. P., and Androphy, E. J. (1993) Biochim. Biophys. Acta 1155323 –345 [DOI] [PubMed] [Google Scholar]
- 3.Hughes, F. J., and Romanos, M. A. (1993) Nucleic Acids Res. 215817 –5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheffner, M., Werness, B. A., Huibregtse, J. M., Levine, A. J., and Howley, P. M. (1990) Cell 631129 –1136 [DOI] [PubMed] [Google Scholar]
- 5.Helt, A. M., and Galloway, D. A. (2001) J. Virol. 756737 –6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jewers, R. J., Hildebrandt, P., Ludlow, J. W., Kell, B., and McCance, D. J. (1992) J. Virol. 661329 –1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McIntyre, M. C., Frattini, M. G., Grossman, S. R., and Laimins, L. A. (1993) J. Virol. 673142 –3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selvey, L. A., Dunn, L. A., Tindle, R. W., Park, D. S., and Frazer, I. H. (1994) J. Gen. Virol. 75,1647 –1653 [DOI] [PubMed] [Google Scholar]
- 9.Reinstein, E., Scheffner, M., Oren, M., Ciechanover, A., and Schwartz, A. (2000) Oncogene 195944 –5950 [DOI] [PubMed] [Google Scholar]
- 10.Wang, J., Sampath, A., Raychaudhuri, P., and Bagchi, S. (2001) Oncogene 204740 –4749 [DOI] [PubMed] [Google Scholar]
- 11.Oh, K. J., Kalinina, A., Wang, J., Nakayama, K., Nakayama, K. I., and Bagchi, S. (2004) J. Virol. 785338 –5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amerik, A. Y., and Hochstrasser, M. (2004) Biochim. Biophys. Acta 1695189 –207 [DOI] [PubMed] [Google Scholar]
- 13.Andersen, M. W., Ballal, N. R., Goldknopf, I. L., and Busch, H. (1981) Biochemistry 201100 –1104 [DOI] [PubMed] [Google Scholar]
- 14.Kim, J. H., Park, K. C., Chung, S. S., Bang, O., and Chung, C. H. (2003) J. Biochem. (Tokyo) 1349 –18 [DOI] [PubMed] [Google Scholar]
- 15.Wilkinson, K. D. (2000) Semin. Cell Dev. Biol. 11141 –148 [DOI] [PubMed] [Google Scholar]
- 16.Wing, S. S. (2003) Int. J. Biochem. Cell Biol. 35590 –605 [DOI] [PubMed] [Google Scholar]
- 17.Chung, C. H., and Baek, S. H. (1999) Biochem. Biophys. Res. Commun. 266633 –640 [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson, K. D. (1997) FASEB J. 111245 –1256 [DOI] [PubMed] [Google Scholar]
- 19.Baker, R. T., Wang, X. W., Woollatt, E., White, J. A., and Sutherland, G. R. (1999) Genomics 59264 –274 [DOI] [PubMed] [Google Scholar]
- 20.Schall, T. J., Lewis, M., Koller, K. J., Lee, A., Rice, G. C., Wong, G. H., Gatanaga, T., Granger, G. A., Lentz, R., Raab, H., Kohr, W. J., and Goeddel, D. V. (1990) Cell 61361 –370 [DOI] [PubMed] [Google Scholar]
- 21.Chang, H. S., Lin, C. H., Chen, Y. C., and Yu, W. C. (2004) Am. J. Pathol. 1651535 –1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hershko, T., and Ginsberg, D. (2004) J. Biol. Chem. 2798627 –8634 [DOI] [PubMed] [Google Scholar]
- 23.Pickart, C. M., and Eddins, M. J. (2004) Biochim. Biophys. Acta 169555 –72 [DOI] [PubMed] [Google Scholar]
- 24.Huang, Y., Baker, R. T., and Fischer-Vize, J. A. (1995) Science 2701828 –1831 [DOI] [PubMed] [Google Scholar]
- 25.Palombella, V. J., Rando, O. J., Goldberg, A. L., and Maniatis, T. (1994) Cell 78773 –785 [DOI] [PubMed] [Google Scholar]
- 26.Seufert, W., Futcher, B., and Jentsch, S. (1995) Nature 37378 –81 [DOI] [PubMed] [Google Scholar]
- 27.Zhu, Y., Carroll, M., Papa, F. R., Hochstrasser, M., and D'Andrea, A. D. (1996) Proc. Natl. Acad. Sci. U. S. A. 933275 –3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nijman, S. M., Luna-Vargas, M. P., Velds, A., Brummelkamp, T. R., Dirac, A. M., Sixma, T. K., and Bernards, R. (2005) Cell 123773 –786 [DOI] [PubMed] [Google Scholar]
- 29.Borodovsky, A., Ovaa, H., Kolli, N., Gan-Erdene, T., Wilkinson, K. D., Ploegh, H. L., and Kessler, B. M. (2002) Chem. Biol. 91149 –1159 [DOI] [PubMed] [Google Scholar]
- 30.Burch, A. D., and Weller, S. K. (2004) J. Virol. 787175 –7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chelbi-Alix, M. K., and de The, H. (1999) Oncogene 18935 –941 [DOI] [PubMed] [Google Scholar]
- 32.Everett, R. D., Orr, A., and Preston, C. M. (1998) EMBO J. 177161 –7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkinson, J., Lees-Miller, S. P., and Everett, R. D. (1999) J. Virol. 73650 –657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swanson, D. A., Freund, C. L., Ploder, L., McInnes, R. R., and Valle, D. (1996) Hum. Mol. Genet. 5533 –538 [DOI] [PubMed] [Google Scholar]
- 35.Ideguchi, H., Ueda, A., Tanaka, M., Yang, J., Tsuji, T., Ohno, S., Hagiwara, E., Aoki, A., and Ishigatsubo, Y. (2002) Biochem. J. 36787 –95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura, M., Masuda, H., Horii, J., Kuma, K., Yokoyama, N., Ohba, T., Nishitani, H., Miyata, T., Tanaka, M., and Nishimoto, T. (1998) J. Cell Biol. 1431041 –1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greenfield, I., Nickerson, J., Penman, S., and Stanley, M. (1991) Proc. Natl. Acad. Sci. U. S. A. 8811217 –11221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guccione, E., Massimi, P., Bernat, A., and Banks, L. (2002) Virology 29320 –25 [DOI] [PubMed] [Google Scholar]
- 39.Antinore, M. J., Birrer, M. J., Patel, D., Nader, L., and McCance, D. J. (1996) EMBO J. 151950 –1960 [PMC free article] [PubMed] [Google Scholar]
- 40.Funk, J. O., Waga, S., Harry, J. B., Espling, E., Stillman, B., and Galloway, D. A. (1997) Genes Dev. 112090 –2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamio, M., Yoshida, T., Ogata, H., Douchi, T., Nagata, Y., Inoue, M., Hasegawa, M., Yonemitsu, Y., and Yoshimura, A. (2004) Oncogene 233107 –3115 [DOI] [PubMed] [Google Scholar]
- 42.Phillips, A. C., and Vousden, K. H. (1997) J. Gen. Virol. 78905 –909 [DOI] [PubMed] [Google Scholar]
- 43.Rey, O., Lee, S., Baluda, M. A., Swee, J., Ackerson, B., Chiu, R., and Park, N. H. (2000) Virology 268372 –381 [DOI] [PubMed] [Google Scholar]
- 44.Zwerschke, W., Mazurek, S., Massimi, P., Banks, L., Eigenbrodt, E., and Jansen-Durr, P. (1999) Proc. Natl. Acad. Sci. U. S. A. 961291 –1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zwerschke, W., Mannhardt, B., Massimi, P., Nauenburg, S., Pim, D., Nickel, W., Banks, L., Reuser, A. J., and Jansen-Durr, P. (2000) J. Biol. Chem. 2759534 –9541 [DOI] [PubMed] [Google Scholar]
- 46.Derynck, R., and Zhang, Y. E. (2003) Nature 425577 –584 [DOI] [PubMed] [Google Scholar]
- 47.Massimi, P., Pim, D., Kuhne, C., and Banks, L. (2001) Mol. Cell Biochem. 227137 –144 [PubMed] [Google Scholar]
- 48.Pei, X. F., Sherman, L., Sun, Y. H., and Schlegel, R. (1998) Carcinogenesis 191481 –1486 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.