Abstract
Interleukin 1α (IL-1α) is capable of driving pro-inflammatory gene expression through both the initiation of transcription and by prolonging the half-life of short-lived mRNAs. Although the signaling events linking the IL-1 receptor to the activation of NFκB and the initiation of transcription have been well characterized, less is known about the signaling events linking to mRNA stabilization. As a model to study the control of mRNA stability we have used the mouse chemokine KC, expression of which requires both NFκB-driven transcription and stabilization of the constitutively unstable mRNA. We have evaluated the role of signaling adaptors known to play a role in IL-1α-driven NFκB activation in the generation of mRNA stability. Surprisingly, although TRAF6 is essential for NFκB activation, it is not required for IL-1α-induced mRNA stabilization. IRAK1, which is recognized to function upstream of TRAF6, is required for both mRNA stabilization and activation of NFκB. Consistent with the previous findings, the TRAF6 interaction sites in IRAK1 are required for NFκB activation but do not play a role in mRNA stabilization. These findings indicate that signals from the IL-1 receptor segregate into at least two separate pathways at the level of IRAK1; one couples through TRAF6 to NFκB activation while a second utilizes a TRAF6-independent pathway that is responsible for mRNA stabilization.
Regulation of the rate of mRNA decay is now recognized as a critical means for modulating expression of a variety of pro-inflammatory genes, including CXC chemokines, tumor necrosis factor α (TNFα),2 granulocyte macrophage colony-stimulating factor, and cyclooxygenase 2 (1–6). These mRNAs have very short half-lives due to the presence of AU-rich sequence elements located in their 3′-untranslated regions (7–9). In the absence of ongoing inflammation, this instability is important for silencing gene expression (10–12), although the short half-life of the message also limits expression during the initiation of an inflammatory reaction. Mechanisms that prolong the half-life of these mRNAs, however, are activated in response to pro-inflammatory stimuli, particularly ligands for members of the Toll-interleukin-1 receptor family (13–17).
The mouse chemokine CXCL1 (KC) can serve as a model to study the regulation of mRNA stability because KC expression requires both enhanced transcription and stabilization of constitutively unstable mRNA. Multiple extracellular stimuli are known to induce KC expression but differentially impact on these processes. Although stimulation with either IL-1α or lipopolysaccharide can induce both transcription and stabilization of the mRNA (17, 18), treatment with TNFα will drive transcription but does not prolong the half-life of the message (17).
Although the molecular mechanisms leading to increased transcription through the activation of NFκB have been well defined, less is known about the mechanisms responsible for stimulus-induced mRNA stabilization. The IL-1α signaling cascade leading to NFκB and MAPK activation requires the sequential involvement of the adaptors MyD88, IRAK4, IRAK1, and TRAF6. TRAF6 then activates the kinase TAK1, which in turn activates distinct kinases that lead to either NFκB or MAPK activation. IL-1α-induced stabilization of AU-rich element-containing mRNA also requires MyD88, suggesting the same pathway may be operative (18), and there are multiple reports demonstrating the participation of the p38 MAPK (14, 19).
In the present study, we have demonstrated that, surprisingly, TRAF6 is not required for the stabilization of mRNA in response to IL-1α. IRAK1, the adaptor believed to function immediately upstream of TRAF6, is required for both NFκB activation and mRNA stabilization. Although the TRAF6 interaction sites in IRAK1 are essential to link to the cascade leading to NFκB activation, they are not necessary to couple to the pathway responsible for mRNA stabilization. These data indicate that the signals leading to mRNA stabilization and NFκB activation diverge at IRAK1 and that this adaptor generates a distinct signal that links to a signaling pathway that is responsible for IL-1α-induced mRNA stabilization.
EXPERIMENTAL PROCEDURES
Reagents—G418, formamide, MOPS, salmon sperm DNA, and diethyl-pyrocarbonate were purchased from Sigma-Aldrich. Dulbecco's modified Eagle's medium, Dulbecco's phosphate-buffered saline, penicillin, and streptomycin were obtained from Central Cell Services of the Lerner Research Institute (Cleveland, OH). Fetal bovine serum was purchased from BioWhittaker (Walkersville, MA). PolyFect Transfection Reagent was obtained from Qiagen (Valencia, CA), and Tri-Reagent was purchased from Molecular Research Center (Cincinnati, OH). Recombinant human and mouse IL-1α and TNFα were purchased from R&D Systems (Minneapolis, MN). Nylon transfer membrane was purchased from Micron Separation (Westboro, MA). Luciferase assay buffer, dual luciferase assay reporter system, and passive lysis buffer were obtained from Promega (Madison, WI). PerkinElmer Life Sciences was the source of [α-32P]dCTP and western lightning chemiluminescence reagent plus. Protein assay reagents were purchased from Bio-Rad. Anti-FLAG M2 antibody was purchased from Sigma-Aldrich, anti-IκBα antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and antibody against GAPDH was purchased from Chemicon International (Temecula, CA). Antibodies recognizing p38 MAP kinase and phospho-p38 MAP kinase were purchased from Cell Signaling Technology (Boston, MA). Anti-mouse and -rabbit IgG horseradish peroxidase-linked antibody was purchased from Amersham Biosciences.
Cell Culture—HeLa tet-off cells stably transfected with the tet-repressor fused to the VP16 transactivation domain were purchased from Clontech Laboratories. The cells were cultured in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum, penicillin, and streptomycin and kept under selection with G418. Wild type and TRAF6-deficient mouse embryonic fibroblasts (MEFs) have been previously described and were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin (21).
Plasmids—Plasmids encoding KC, MIP-2, and GAPDH cDNAs, 5× κB luciferase, pTRE2 KCΔ3, MyD88, IRAK1, TRAF6, and dominant negative TRAF6-(289–522) have been previously described (18, 22–25). pTK Renilla was purchased from Promega. IRAK1-(Δ189–712) was generated by PCR and cloned into pBS-CMV. A construct encoding a version of IRAK1 incapable of interacting with TRAF6 (IRAK1 E3A) due to substitution of alanine for glutamic acid at positions 544, 587, and 706 (26) was generated using the QuikChange site-directed mutagenesis kit from Stratagene.
Transfection—HeLa tet-off cells were plated and allowed to grow for 24 h to 70% confluency in 100-mm Petri dishes. The cells were transiently transfected using PolyFect Transfection Reagent (Qiagen) according to the manufacturer's protocol for HeLa cells. Cultures were then split into smaller treatment groups and incubated for 18 h before treatment.
Western Blot, Luciferase Assay, and Northern Blot—Western blot analysis was performed as described previously (18). Luciferase activity of cell lysates was determined using luciferase assay buffer or the dual luciferase reporter assay system according to the manufacturer's protocol (Promega). Total RNA was isolated using Tri-Reagent according to the manufacturer's protocol (Molecular Research Center). mRNA levels were determined by Northern blot hybridization as previously described (18).
RESULTS
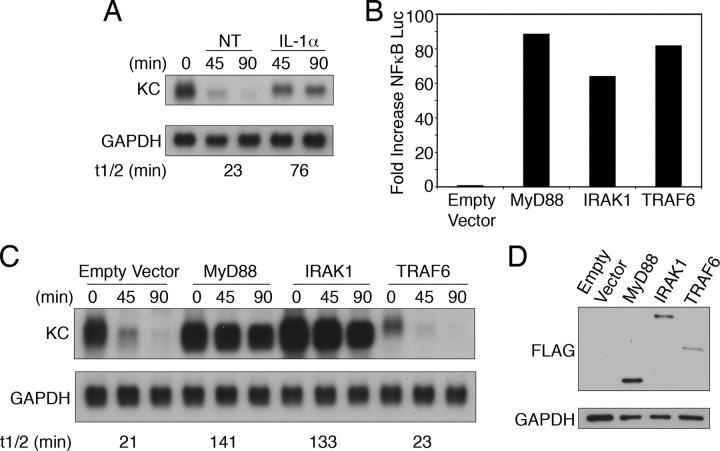
TRAF6 Overexpression Does Not Activate Stabilization of KC mRNA—KC mRNA stability was studied in a HeLa cell line stably expressing the tet-repressor-VP-16 fusion protein (HeLa tet-off). When these cells are transiently transfected with a plasmid encoding the KC 5′-untranslated region, coding region, and a fragment of the 3′-untranslated region (pTRE2 KCΔ3) the mRNA is transcribed at a high rate. Following the addition of doxycycline (dox), transcription of the plasmid is blocked and the decay of the mRNA can be assessed without use of the transcription inhibitor actinomycin D. In the absence of cytokine stimulation, KC mRNA is rapidly degraded, reflecting the constitutive instability of the message. However, addition of IL-1α along with dox is able to prolong the half-life of the message (Fig. 1A).
FIGURE 1.
Overexpression of TRAF6 cannot stimulate KC mRNA stabilization. A, HeLa tet-off cells were transfected with 2 μg of pTRE2 KCΔ3 and 4 μg of empty vector (pcDNA3) and divided into different treatment groups. Cells were treated with either dox (1 μg/ml) alone (NT) or with IL-1α (10 ng/ml). RNA was isolated at the indicated times following treatment, and KC and GAPDH mRNA levels were determined. The autoradiographs were quantified using the NIH Image software, and these values were used to calculate the mRNA half-lives. B–D, HeLa tet-off cells were transfected with 1 μg of pTRE2 KCΔ3, 1 μg of 5× κB luciferase reporter, 0.25 μg of pTK Renilla, and 4 μg of either empty vector or the indicated FLAG-tagged adaptor. B, luciferase activity was determined as described under “Experimental Procedures.” Values for firefly luciferase represent the means of duplicate samples and have been normalized to Renilla luciferase levels to control for transfection efficiency. C, cultures were treated with dox for the indicated times prior to determination of KC and GAPDH mRNA levels. D, FLAG and GAPDH expression levels were determined by Western blot analysis. All data are representative of at least three independent experiments.
We have shown previously that overexpression of the adaptor protein MyD88 is sufficient to drive stabilization of KC mRNA transcribed from co-transfected pTRE2 KCΔ3 (18, 27). To assess the contribution of downstream signaling components in the pathway(s) leading to stabilization of KC mRNA, HeLa tet-off cells were co-transfected with pTRE2 KCΔ3, an NFκB-driven luciferase reporter (5× κB luc), and either empty vector (pcDNA3) or expression plasmids encoding FLAG-tagged MyD88, IRAK1, or TRAF6. Consistent with the published literature, overexpression of any of the adaptor molecules was sufficient to drive activation of NFκB (Fig. 1B). When the half-life of KC mRNA was assessed in the same cell populations, MyD88 and IRAK1 were capable of markedly enhancing the stability and hence abundance of KC mRNA (Fig. 1C). Overexpression of TRAF6, however, had no effect on the half-life of the mRNA. Expression levels for each adaptor were determined by Western blot (Fig. 1D). This finding indicates that overexpression of TRAF6 activates signals that are adequate for NFκB activation but are not sufficient to promote mRNA stabilization.
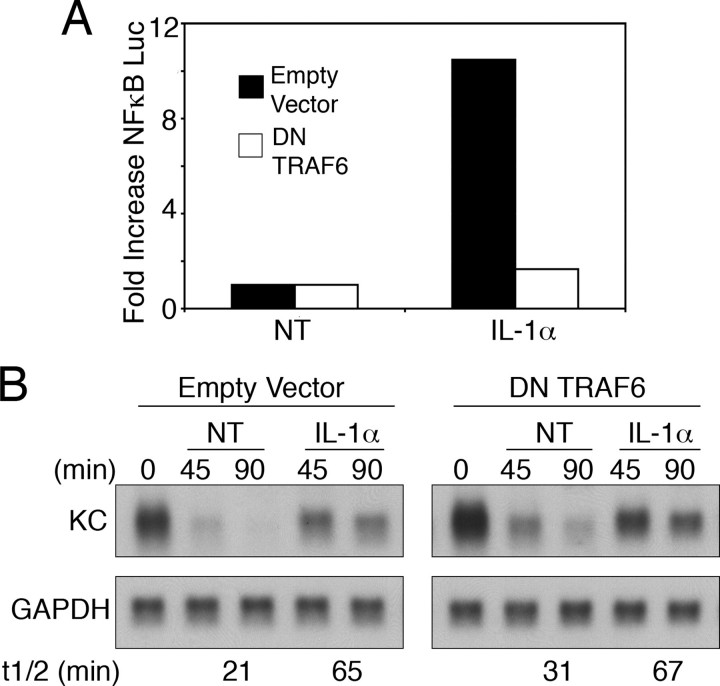
TRAF6 Is Not Required for IL-1α-induced mRNA Stabilization—To determine whether TRAF6 is required for IL-1α to induce KC mRNA stabilization, we evaluated the ability of a dominant negative version of TRAF6 (amino acids 289–522) to block the response in HeLa tet-off cells. Consistent with prior work, the dominant negative TRAF6 fully prevented the ability of IL-1α to activate NFκB (Fig. 2A) (22). In contrast to the dramatic inhibition of NFκB activation, the dominant negative TRAF6 protein did not affect the induction of mRNA stability in response to IL-1α (Fig. 2B).
FIGURE 2.
Dominant negative TRAF6 does not block IL-1α-induced mRNA stabilization. HeLa tet-off cells were transfected with 2 μgofKCΔ3, 2 μgof 5× κB luciferase reporter, and 2 μg of either empty vector or a dominant negative (DN) TRAF6. A, luciferase activity was determined following treatment with IL-1α (10 ng/ml) for 8 h. Values represent the mean of duplicate samples. -Fold increase was determined by comparing the treated samples to the untreated (NT) in the same transfection group. B, dox (1 μg/ml) was added to each sample either alone (NT) or with IL-1α (10 ng/ml). Total RNA was isolated following treatment for the indicated times, and KC and GAPDH mRNA levels were determined by northern hybridization. Data are representative of at least three independent experiments.
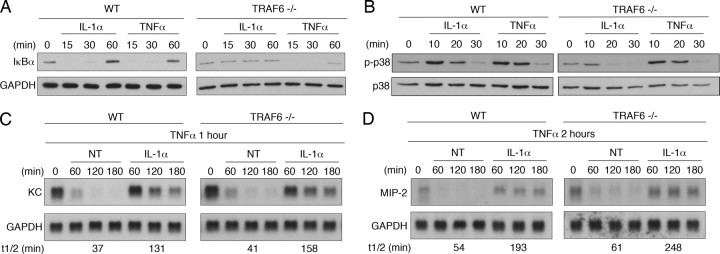
As a second strategy, we tested the effect of IL-1α treatment in TRAF6-deficient MEFs. The ability of IL-1α to activate NFκB, MAP kinases, and gene expression has been reported to be compromised in these cells (21). Consistent with these reports, IL-1α could not promote the degradation of IκBα in TRAF6–/– MEFs, indicating that IL-1α-driven NFκB activation had been lost (Fig. 3A). In addition, IL-1α was unable to activate p38 MAP kinase in TRAF6–/– MEFs (Fig. 3B). The ability of IL-1α to prolong the half-life of endogenous KC mRNA in wild type and TRAF6–/– MEFs was tested by examining the decay of KC mRNA induced by treatment with TNFα. We have reported previously that KC mRNA induced by treatment with TNFα for 1 h decays rapidly following addition of actinomycin D. However, addition of IL-1α along with actinomycin D results in a significant prolongation of the mRNA half-life (27). IL-1α was able to promote KC mRNA stabilization in wild type and TRAF6–/– MEFs (Fig. 3C). Similar results were obtained for a second CXC chemokine, MIP-2 (Fig. 3D). Hence, we conclude that TRAF6 is not a required component in the signaling pathway coupling the IL-1 receptor to mRNA stabilization.
FIGURE 3.
TRAF6 is not required for IL-1α-induced mRNA stabilization. A and B, wild type or TRAF6-deficient MEFs were treated with 10 ng/ml IL-1α or TNFα for the indicated times, and IκBα, GAPDH, phospho-p38, and total p38 levels were determined by Western blot analysis. C and D, cells were treated with TNFα (10 ng/ml) for one (C) or two (D) hours. Fresh medium was then added containing either actinomycin D (5 μg/ml) alone (NT) or with IL-1α (10 ng/ml). After further incubation for the indicated times, total RNA was isolated and KC and GAPDH mRNA levels were determined by northern hybridization. Data are representative of at least three independent experiments.
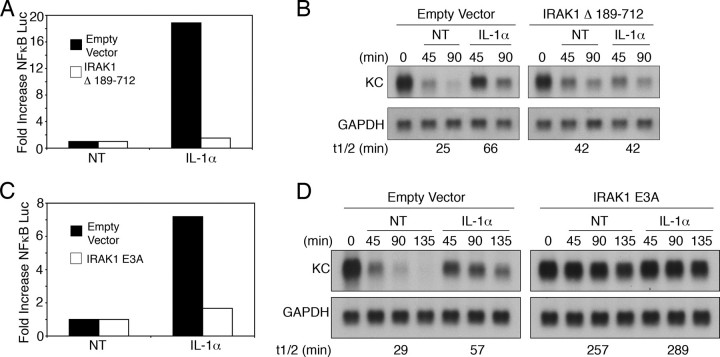
Signals Leading to IL-1α-induced NFκB Activation and mRNA Stabilization Diverge at IRAK1—Overexpression of IRAK1, the adaptor functioning directly upstream of TRAF6, was sufficient to induce both NFκB activation and stabilization of KC mRNA (Fig. 1, B and C), suggesting that IRAK1 might be necessary for IL-1α-induced KC mRNA stabilization. To test this idea, we evaluated the effect of a mutant form of IRAK1-(Δ189–712) that has been previously reported to function as a dominant negative. Overexpression of this mutant clearly blocked both IL-1α-induced NFκB activation and mRNA stabilization (Fig. 4, A and B).
FIGURE 4.
IRAK1 couples to IL-1α-induced NFκB activation and KC mRNA stabilization via distinct mechanisms. A and B, HeLa tet-off cells were transfected with 2 μg of KCΔ3, 2 μg of 5× κB luciferase reporter, and 2 μg of either empty vector or an expression plasmid encoding IRAK1-(Δ189–712). A, luciferase activity was determined after treatment with IL-1α (10 ng/ml) for 8 h. Values represent the mean of duplicate samples, and -fold increase was determined by comparing the values from treated with untreated (NT) samples in the same transfection group. B, dox (1 μg/ml) was added to each sample either alone (NT) or with IL-1α, and KC and GAPDH mRNA levels were determined at the indicated times. C and D, HeLa tet-off cells were transfected with 2 μg of KCΔ3, 1 μg of 5× κB luciferase reporter, and 3 μg of either empty vector or an expression vector encoding IRAK1-(Δ523–712). C, luciferase activities were determined after treatment with IL-1α (10 ng/ml) for 8 h. D, dox was added to each sample either alone or with IL-1α, and KC and GAPDH mRNA levels were determined at the indicated times. All data are representative of at least three independent experiments.
Although IRAK1 couples to NFκB activation through its interaction with TRAF6, the data presented earlier show that TRAF6 is not required for the modulation of mRNA decay. Thus, we reasoned that IRAK1 may be the point where the signals leading to NFκB activation and mRNA stabilization diverge and should, therefore, be capable of generating a TRAF6-independent signal leading to enhanced mRNA stability. To test this hypothesis, we generated a construct of IRAK1 in which the three TRAF6 interaction sites were mutated (IRAK1 E3A). When this mutant was overexpressed, IL-1α-induced NFκB activation was markedly inhibited (Fig. 4C). Interestingly, overexpression of the mutant was able to independently induce KC mRNA stabilization (Fig. 4D). This finding further supports the idea that TRAF6 is not required for mRNA stabilization and indicates that IRAK1 is generating a distinct signal that links to this end point.
DISCUSSION
The findings presented here show that TRAF6, although known to play an essential role in a variety of IL-1α-induced responses, including activation of NFκB, surprisingly is not required for IL-1α-induced mRNA stabilization. Rather, IRAK1, which functions directly upstream of TRAF6, is essential for both activities. Importantly, IRAK1 interacts with TRAF6 to link to NFκB activation, but not mRNA stabilization, indicating that this protein is the place where signaling pathways leading to these two end points diverge. Hence, IRAK1 links to an uncharacterized pathway that is responsible for mRNA stabilization.
The finding that IRAK1, but not TRAF6, is required for mRNA stabilization is unexpected given what is currently understood about the IL-1α signaling pathway. TRAF6 plays a critical role in many IL-1α-induced responses, functioning as an E3 ubiquitin ligase. It catalyzes formation of K63-linked polyubiquitin chains leading to the activation of the kinase TAK1 (28, 29). TAK1 not only stimulates the IKK complex leading to activation of NFκB but also phosphorylates MKK6, resulting in the activation of JNK and p38 (29). Thus, signals coupling these end points with IL-1α stimulation diverge downstream of TAK1. Consistent with this model, TRAF6 has been shown to be required for activation of extracellular signal-regulated kinase, JNK, and p38 MAP kinases in addition to its role in NFκB activation (21, 30). We are unaware of other reports that identify IL-1α-induced events leading to pro-inflammatory gene expression that do not require TRAF6. TRAF3 has been linked with MyD88 and IRAK1 in the context of signaling from several other Toll-interleukin-1 receptor family members and has been shown to be responsible for a set of end points distinct from those that require TRAF6 (31, 32). IL-1α-induced KC expression is not, however, compromised in TRAF3-deficient cells (data not shown).
The finding that TRAF6 is not essential for IL-1α-induced mRNA stabilization also suggests that TRAF6-dependent responses, such as activation of NF-κB and p38 MAP kinase, are not critical for mRNA stabilization (see Fig. 3, A and B). The lack of a requirement for p38 MAP kinase activation in mRNA stabilization is surprising because this pathway has been shown to be essential in Toll-interleukin-1 receptor-induced mRNA stabilization in a number of settings (14, 15, 19). Indeed, in mouse macrophages lipopolysaccharide-induced KC mRNA stability is heavily dependent upon p38 MAP kinase activity (34, 35). Moreover, TNFα, a potent stimulus of p38 MAP kinase activation, is unable to stabilize KC mRNA in either HeLa tet-off cells or MEFs (17, 27, 36). These findings collectively support the conclusion that p38 MAP kinase activation is not requisite for stabilization of KC mRNA under the experimental conditions employed in the present study. Although the relationship between the TRAF6-independent pathway for mRNA stabilization and those responses requiring p38 remains to be determined, the differences may result from cell type-specific mechanisms for control of mRNA decay.
IRAK1 is a multidomain protein and may be able to connect with multiple distinct downstream pathways. The three TRAF6 interaction sites in the carboxyl terminus are essential for the recruitment of TRAF6 and the subsequent activation of NFκB. Although IRAK1 contains a functional kinase domain, this activity is not required for IL-1α-induced NFκB or JNK activation (37). However, the kinase activity has been shown to be essential for IRF7 activation in the setting of stimulation via TLR7 and TLR9 (20). IRAK1 can be directly phosphorylated by IRAK4, which functions upstream of IRAK1 in the IL-1α signaling pathway. The finding that IRAK4 kinase activity is required for Toll-like receptor-induced mRNA stabilization (33) suggests that phosphorylation of IRAK1 by IRAK4 may be a required step in the signaling pathway leading to mRNA stabilization. A detailed understanding of the sequences within IRAK1 required to link to the pathway leading to mRNA stabilization will help further clarify the multiple functions this adaptor plays in signaling and should help in the characterization of this signaling pathway.
This work was supported by United States Public Health Service Grants CA39621 and T32GM007250. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TNF, tumor necrosis factor; IL-1α, interleukin 1α; tet, tetracycline; dox, doxycycline; MEF, mouse embryo fibroblast; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MAPK, mitogen-activated protein kinase; MOPS, 4-morpholinepropanesulfonic acid; JNK, c-Jun N-terminal kinase.
References
- 1.Saklatvala, J., Dean, J., and Clark, A. (2003) Biochem. Soc. Symp. 70 95–106 [DOI] [PubMed] [Google Scholar]
- 2.Fan, J., Heller, N. M., Gorospe, M., Atasoy, U., and Stellato, C. (2005) Eur. Respir. J. 26 933–947 [DOI] [PubMed] [Google Scholar]
- 3.Seko, Y., Cole, S., Kasprzak, W., Shapiro, B. A., and Ragheb, J. A. (2006) Autoimmun. Rev. 5 299–305 [DOI] [PubMed] [Google Scholar]
- 4.Newbury, S. F. (2006) Biochem. Soc. Trans. 34 Pt. 1, 30–34 [DOI] [PubMed] [Google Scholar]
- 5.Garneau, N. L., Wilusz, J., and Wilusz, C. J. (2007) Nat. Rev. Mol. Cell. Biol. 8 113–126 [DOI] [PubMed] [Google Scholar]
- 6.Shim, J., and Karin, M. (2002) Mol. Cells 14 323–331 [PubMed] [Google Scholar]
- 7.Caput, D., Beutler, B., Hartog, K., Thayer, R., Brown-Shimer, S., and Cerami, A. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 1670–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C. Y., and Shyu, A. B. (1995) Trends Biochem. Sci. 20 465–470 [DOI] [PubMed] [Google Scholar]
- 9.Shaw, G., and Kamen, R. (1986) Cell 46 659–667 [DOI] [PubMed] [Google Scholar]
- 10.Carballo, E., Lai, W. S., and Blackshear, P. J. (1998) Science 281 1001–1005 [DOI] [PubMed] [Google Scholar]
- 11.Kontoyiannis, D., Pasparakis, M., Pizarro, T. T., Cominelli, F., and Kollias, G. (1999) Immunity 10 387–398 [DOI] [PubMed] [Google Scholar]
- 12.Taylor, G. A., Carballo, E., Lee, D. M., Lai, W. S., Thompson, M. J., Patel, D. D., Schenkman, D. I., Gilkeson, G. S., Broxmeyer, H. E., Haynes, B. F., and Blackshear, P. J. (1996) Immunity 4 445–454 [DOI] [PubMed] [Google Scholar]
- 13.Biswas, R., Datta, S., Gupta, J. D., Novotny, M., Tebo, J., and Hamilton, T. A. (2003) J. Immunol. 170 6202–6208 [DOI] [PubMed] [Google Scholar]
- 14.Dean, J. L., Brook, M., Clark, A. R., and Saklatvala, J. (1999) J. Biol. Chem. 274 264–269 [DOI] [PubMed] [Google Scholar]
- 15.Holtmann, H., Winzen, R., Holland, P., Eickemeier, S., Hoffmann, E., Wallach, D., Malinin, N. L., Cooper, J. A., Resch, K., and Kracht, M. (1999) Mol. Cell. Biol. 19 6742–6753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoeckle, M. Y. (1991) Nucleic Acids Res. 19 917–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tebo, J., Datta, S., Kishore, R., Kolosov, M., Major, J. A., Ohmori, Y., and Hamilton, T. A. (2000) J. Biol. Chem. 275 12987–12993 [DOI] [PubMed] [Google Scholar]
- 18.Datta, S., Novotny, M., Li, X., Tebo, J., and Hamilton, T. A. (2004) J. Immunol. 173 2755–2761 [DOI] [PubMed] [Google Scholar]
- 19.Winzen, R., Kracht, M., and Ritter, B. (1999) EMBO J. 18 4969–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uematsu, S., Sato, S., Yamamoto, M., Hirotani, T., Kato, H., Takeshita, F., Matsuda, M., Coban, C., Ishii, K. J., Kawai, T., Takeuchi, O., and Akira, S. (2005) J. Exp. Med. 201 915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lomaga, M. A., Yeh, W. C., Sarosi, I., Duncan, G. S., Furlonger, C., Ho, A., Morony, S., Capparelli, C., Van, G., Kaufman, S., van der Heiden, A., Itie, A., Wakeham, A., Khoo, W., Sasaki, T., Cao, Z., Penninger, J. M., Paige, C. J., Lacey, D. L., Dunstan, C. R., Boyle, W. J., Goeddel, D. V., and Mak, T. W. (1999) Genes Dev. 13 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao, Z., Xiong, J., Takeuchi, M., Kurama, T., and Goeddel, D. V. (1996) Nature 383 443–446 [DOI] [PubMed] [Google Scholar]
- 23.Qian, Y., Zhao, Z., Jiang, Z., and Li, X. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 9386–9391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novotny, M., Datta, S., Biswas, R., and Hamilton, T. (2005) J. Biol. Chem. 280 30166–30174 [DOI] [PubMed] [Google Scholar]
- 25.Armstrong, D. A., Major, J. A., Chudyk, A., and Hamilton, T. A. (2004) J. Leukocyte Biol. 75 641–648 [DOI] [PubMed] [Google Scholar]
- 26.Ye, H., Arron, J. R., Lamothe, B., Cirilli, M., Kobayashi, T., Shevde, N. K., Segal, D., Dzivenu, O. K., Vologodskaia, M., Yim, M., Du, K., Singh, S., Pike, J. W., Darnay, B. G., Choi, Y., and Wu, H. (2002) Nature 418 443–447 [DOI] [PubMed] [Google Scholar]
- 27.Hartupee, J., Lu, C., Novotny, M., Li, X., and Hamilton, T.A. (2007) J. Immunol. 179 4135–4141 [DOI] [PubMed] [Google Scholar]
- 28.Deng, L., Wang, C., Spencer, E., Yang, L., Braun, A., You, J., Slaughter, C., Pickart, C., and Chen, Z. J. (2000) Cell 103 351–361 [DOI] [PubMed] [Google Scholar]
- 29.Wang, C., Deng, L., Hong, M., Akkaraju, G. R., Inoue, J., and Chen, Z. J. (2001) Nature 412 346–351 [DOI] [PubMed] [Google Scholar]
- 30.Baud, V., Liu, Z. G., Bennett, B., Suzuki, N., Xia, Y., and Karin, M. (1999) Genes Dev. 13 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hacker, H., Redecke, V., Blagoev, B., Kratchmarova, I., Hsu, L. C., Wang, G. G., Kamps, M. P., Raz, E., Wagner, H., Hacker, G., Mann, M., and Karin, M. (2006) Nature 439 204–207 [DOI] [PubMed] [Google Scholar]
- 32.Oganesyan, G., Saha, S. K., Guo, B., He, J. Q., Shahangian, A., Zarnegar, B., Perry, A., and Cheng, G. (2006) Nature 439 208–211 [DOI] [PubMed] [Google Scholar]
- 33.Kim, T. W., Staschke, K., Bulek, K., Yao, J., Peters, K., Oh, K. H., Vandenburg, Y., Xiao, H., Qian, W., Hamilton, T., Min, B., Sen, G., Gilmour, R., and Li, X. (2007) J. Exp. Med. 204 1025–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai, Y., Datta, S., Novotny, M., and Hamilton, T. A. (2003) Blood 102 1178–1185 [DOI] [PubMed] [Google Scholar]
- 35.Datta, S., Biswas, R., Novotny, M., Pavicic, P., Herjan, T., Mandal, P., and Hamilton, T. A. (2008) J. Immunol. 180 2545–2552 [DOI] [PubMed] [Google Scholar]
- 36.Tebo, J., Der, S., Frevel, M., Khabar, K. S., Williams, B. R., and Hamilton, T. A. (2003) J. Biol. Chem. 278 12085–12093 [DOI] [PubMed] [Google Scholar]
- 37.Li, X., Commane, M., Burns, C., Vithalani, K., Cao, Z., and Stark, G. R. (1999) Mol. Cell. Biol. 19 4643–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]