Abstract
Cortactin is a filamentous actin-binding protein that plays a pivotal role in translating environmental signals into coordinated rearrangement of the cytoskeleton. The dynamic reorganization of actin in the cytoskeleton drives processes including changes in cell morphology, cell migration, and phagocytosis. In general, structural proteins of the cytoskeleton bind in the N-terminal region of cortactin and regulatory proteins in the C-terminal region. Previous structural studies have reported an extended conformation for cortactin. It is therefore unclear how cortactin facilitates cross-talk between structural proteins and their regulators. In the study presented here, circular dichroism, chemical cross-linking, and small angle x-ray scattering are used to demonstrate that cortactin adopts a globular conformation, thereby bringing distant parts of the molecule into close proximity. In addition, the actin bundling activity of cortactin is characterized, showing that fully polymerized actin filaments are bundled into sheet-like structures. We present a low resolution structure that suggests how the various domains of cortactin interact to coordinate its array of binding partners at sites of actin branching.
Regulated polymerization, depolymerization, and branching of actin filaments drives fundamental cellular mechanical processes such as cell migration, morphology changes, endocytosis, and metastasis. However, the mechanisms by which environmental signals are transmitted to the cytoskeleton and direct the coordinated restructuring of actin are poorly understood. A protein that plays a key role in this process is the f-actin-binding protein cortactin (1).
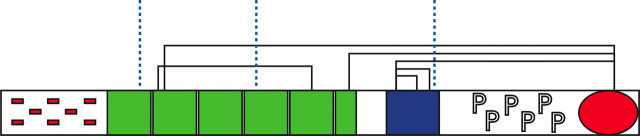
The domain structure of cortactin (Fig. 1) gives clues to its function and potential mode of action. There are five discrete regions: the N-terminal acidic region, the actin-binding repeats region, the helical region, the proline-rich region, and the SH3 domain. The N-terminal acidic region incorporates a binding site for the arp2/3 complex (2, 3), a multiprotein complex that nucleates actin filaments and can itself bind to filamentous actin (f-actin), forming branch points (4). Following the N-terminal acidic region, there are six-and-a-half repeats of a 32-amino-acid motif that together bind to f-actin. These repeats have also been reported to bundle actin into filaments (5). Three cortactin splice isoforms have been identified that lack either one or two of these repeats. These isoforms bind f-actin, although f-actin bundling activity is reduced in the 5.5-repeat isoform as compared with the 6.5-repeat isoform and absent in the 4.5-repeat isoform (6). The affinity of cortactin for both f-actin and the arp2/3 complex suggests a mode of action, whereby cortactin may localize arp2/3 to mature actin filaments, causing branching of f-actin. Indeed, cortactin has been found at actin branch points (7) and shown to stabilize branches (8).
FIGURE 1.
A schematic view of the domain structure of cortactin splice isoform 1 with 5.5 actin-binding repeats. The N-terminal acidic region is denoted by red minus symbols, the actin-binding repeats are denoted by green boxes, the helical region is denoted by a blue box, the proline-rich domain is denoted by the letters P, and the SH3 domain is denoted by a red oval. The figure also summarizes the results of the cross-linking experiments. Intramolecular cross-links are designated by black lines, and the positions of cross-links to filamentous actin are shown by blue vertical dotted lines.
The C-terminal SH3 domain of cortactin binds to protein modulators of actin polymerization, including the arp2/3-activating proteins N-Wasp (9) and WIP (10) as well as dynamin2, a GTPase associated with membrane ruffling and vesicle trafficking (11). Cortactin binds to f-actin, recruiting arp2/3 and various activators and modulators of actin dynamics. However, between the SH3 domain and the f-actin-binding repeats, there is a helical domain of unknown function and a proline-rich region that contains many sites of serine, threonine, and tyrosine phosphorylation (12). This region is targeted by kinases that transduce signals from cell surface receptors (13-15). For example, the kinase c-Src acts downstream of epidermal and platelet-derived growth factor receptor activation, resulting in rearrangement of actin stress fibers and, ultimately, cell migration (16). Cortactin phosphorylation by c-Src enhances arp2/3-mediated actin branching in vitro (17). An autoinhibition model has been proposed in which the SH3 domain makes intramolecular interactions and is therefore unavailable to potential interacting proteins. Phosphorylation events in the proline-rich region alter the affinity of the intramolecular SH3 domain interactions that regulate association with the various binding proteins. Phosphorylation in turn either up-regulates or down-regulates cortactin-mediated reorganization of the cytoskeleton (18, 19).
Structural studies are key to understanding the molecular basis of cortactin regulation of f-actin branching and rearrangement. Two such studies have been reported previously. The first study used deep-etch electron microscopy and analytical ultracentrifugation to show that cortactin was a “thread-like” molecule, 220-290 Å long and ∼20 Å wide (2), consistent with an unstructured “beads-on-a-string” arrangement of the cortactin domains. This structure contributed to a model where cortactin acts simply as a scaffolding protein. In this model, the various cortactin domains act independently, and the protein functions passively by recruiting effector proteins to the correct location. The second structural study focused on a truncated construct of cortactin including only the f-actin-binding repeats domain and used cryo-electron microscopy to reveal an approximately spherical patch of density at the common protein-binding pocket of f-actin (20), a region on the surface of actin where most actin-binding proteins interact (21).
Two important structural aspects of cortactin remain unclear. First, if the SH3 domain makes intramolecular interactions, which other parts of the cortactin molecule does it interact with? Second, how do regulatory molecules that bind at the C-terminal SH3 domain of cortactin “talk” to molecules such as actin and arp2/3 that bind at the N-terminal region? Both questions relate to the orientation of the domains of cortactin with respect to each other. In the study presented here, we analyzed the structure of cortactin, as a free molecule in solution and in complex with f-actin, using a combination of CD, chemical cross-linking, and small angle x-ray scattering (SAXS).5 Our results are not consistent with an extended conformation but rather support a globular fold in which the actin-binding repeats pack together and interact with the SH3 domain. In addition, we show that cortactin plays an active role in f-actin morphology by bundling assembled actin filaments into sheets.
EXPERIMENTAL PROCEDURES
Protein Production—Actin was extracted from 100 g of freshly dissected rabbit muscle as described by Spudich and Watt (22). Following purification, actin at 11 mg/ml in monomeric actin buffer (2 mm Tris, pH 8.0, 0.2 mm ATP, 0.2 mm CaCl2, 0.5 mm dithiothreitol) was snap-frozen in liquid nitrogen in 200-ml aliquots and stored at -80 °C. Actin was polymerized by diluting 1:1 in 2× polymerization buffer (100 mm KCl, 20 mm imidazole, pH 7.4, 0.2 mm MgCl2, 1 mm EGTA) or in a solution of 2× polymerization buffer containing cortactin at 10 mg/ml followed by incubation on ice for 5 h.
Murine cortactin, splice isoform 1, lacking the sixth actin-binding repeat (6), was chosen for this study because it retains a similar affinity for f-actin-binding as full-length cortactin but has reduced actin bundling activity (6); it was hypothesized that these properties would be advantageous for imaging cortactin on single actin fibers. Cortactin (splice isoform 1) was amplified by PCR from a mouse cDNA clone from the FANTOM2 clone set (23), clone number D230013O13, and inserted into the vector pDEST14 (Invitrogen) using a modified Gateway cloning strategy that fuses a short hexahistidine tag to the amino terminus (24). Protein was expressed in Escherichia coli strain BL21 (DE3) pLysS (Novagen) using an autoinduction protocol (25). Cells were lysed using B-PER cell lysis reagent (Pierce) and purified by Co2+ affinity and gel filtration chromatography. The Co2+ affinity resin was washed in wash buffer (25 mm HEPES, pH 7.4, 300 mm NaCl) plus 10 mm imidazole, and the bound protein was then eluted in wash buffer plus 250 mm imidazole followed by gel filtration chromatography in wash buffer. Cortactin (10 mg/ml) was snap-frozen in wash buffer using liquid nitrogen in 200-μl aliquots in thin-walled PCR tubes and stored at -80 °C. Cortactin was thawed and transferred to 2× actin polymerization buffer using a PD-10 desalting column (GE Healthcare Life Sciences) prior to use.
Circular Dichroism—Circular dichroism spectra of polymerized actin, cortactin, and the complex of the two proteins were measured on a J-810 polarimeter (Jasco). To prepare the samples, monomeric actin at 5.2 mg/ml in monomeric actin buffer was mixed 1:1 (v/v) either with 2× polymerization buffer or with cortactin (3.4 mg/ml) in 2× polymerization buffer giving a final concentration of 2.6 mg/ml (62 μm) actin and 1.7 mg/ml (29 μm) cortactin. Complex formation in this system was confirmed by actin co-sedimentation assay. Five replicates of each spectrum were made using 1-s dwell times, scanning speeds of 20 nm/min, and a data pitch of 0.2 nm. Samples were measured in a quartz suprasil demountable cuvette with a path length of 0.001 cm (Hellma). CD spectra were deconvoluted using the Selcon3, ContinLL, and CDSSTR algorithms in the CDPro package (26). Secondary structures from the CD spectra are compared with those calculated from secondary structure prediction algorithms PHD (27) or PROF (28).
Cross-linking—Solutions of cortactin alone or the actin-cortactin complex at ∼0.5 mg/ml each were incubated with the cross-linking reagent dithiobissulfosuccinimidyl propionate (DTSSP; 2.4 mm), at 25 °C for 12 min. Bands corresponding to monomeric cortactin or a cortactin/actin heterodimer were excised from non-reducing SDS-PAGE gels, digested with trypsin, and eluted for sequencing. Peptide sequences and confirmation of cross-linked molecules were determined by nano-high pressure liquid chromatography on an Agilent 1100 system followed by matrix-assisted laser desorption-time of flight/time of flight-MS using a 4700 Proteomics Analyzer mass spectrometer (Applied Biosystems).
Small Angle X-ray Scattering—Small angle x-ray scattering (SAXS) data for cortactin in solution were measured at the SIBYLS beamline 12.3.1 of the Advanced Light Source, Berkeley, CA. SAXS data for cortactin were measured in polymerization buffer using 10-μl-volume sample cells with flat mica windows at a beam energy of 12 KeV. Protein concentration and exposure time were varied to test the effects of interparticle correlation and beam damage, respectively. Images from a Mar 165 detector were integrated, blanked, converted to Q (Å-1) units, and corrected for exposure time and beam flux using software specific to this beamline. Integrated data were processed by the program GNOM (29) using the indirect transform method with Dmax of 110 Å. Ten dummy atom models of cortactin were generated by the program DAMMIN (30) using different starting parameters with good convergence. Models were averaged and contoured at the 95% confidence level with the program DAMAVER (31).
Time-resolved SAXS data of polymerizing actin and polymerizing actin in the presence of cortactin were collected at ChemMatCARS beamline (32), sector 15, Advanced Photon Source, Chicago, IL. Monomeric actin and cortactin in 2× polymerization buffer were prepared as described above. A stopped-flow apparatus was constructed consisting of two 600-μl reservoir tubes connected to a syringe pump. Monomeric actin at 2.5 mg/ml was injected into one, and either 2× polymerization buffer or 4 mg/ml cortactin in 2× polymerization buffer was injected into the other. At the start of an experiment, 300 μl of each were flowed at 100 μl/s through a hypodermic needle to ensure complete mixing into a 600-μl piece of tubing. The mixed protein was then flowed past the incident x-ray beam in quartz capillary at 0.5 μl/s to avoid radiation damage of the protein. Ten-second exposures were measured using a MAR165 CCD with a 1-s read-out time. Data were measured for 400 s after initial mixing of the sample. A wavelength of 1 Å was used with a sample to detector distance of 1.5 m giving a Q range of 0.005-0.25 Å-1.
RESULTS
Cortactin Adopts a Globular Conformation in Solution—SAXS data were collected to determine the morphology of cortactin in solution. The raw scattering data, plotted on a log scale (Fig. 2A), show a horizontal plateau at low angles of scatter (Q), a well defined “Guinier knee,” and an oscillation at high Q. These features are strong indicators of conformational mono-dispersity, implying a rigid structure. On a Guinier plot, the data are linear at angles of Q between 0.014 and 0.024 Å-1, giving a radius of gyration of 33.0 Å. The maximum dimension of the molecule calculated by GNOM is 110 Å (Fig. 2C).
FIGURE 2.
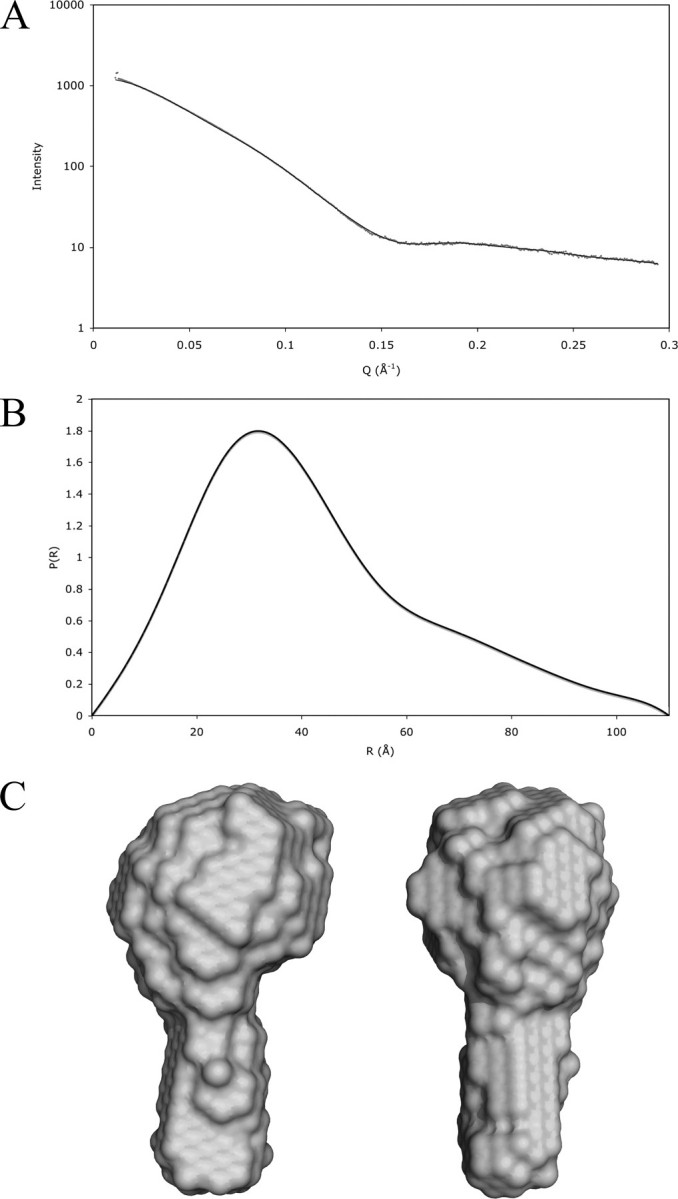
Small-angle x-ray scattering data and calculated low resolution structure of free cortactin. A shows a plot of the raw scattering data; Data points are shown in gray with the fit from the DAMMIN model overlaid as a black line. B shows the P(R) function from GNOM. C shows two views of the envelope structure of cortactin calculated from the scattering data by the program DAMMIN. Ten independent calculations were made with good convergence, and the χ-square of the fit of the most representative model is 0.0692. These were averaged together and plotted at 95% confidence interval by the program Damfilt.
The low resolution, solution structure of cortactin calculated from the SAXS data resembles a drumstick (Fig. 2C) with a globular domain and a narrow stem structure protruding from it. The globular structure suggested by SAXS data is in agreement with the cross-linking data presented below.
The Globular Region Contains the Actin-binding Repeats and the Helical Region—A chemical cross-linking strategy was used to study the spatial relationship of domains within cortactin. Free cortactin was incubated in the presence of the cross-linking reagent DTSSP, which cross-links surface-exposed lysine residues. Bands corresponding to monomeric cortactin were isolated from SDS-PAGE gels to ensure intramolecular cross-links. Tryptic digestion and mass spectrometry were used to identify pairs of cross-linked lysine residues, and the identity of these cross-links was verified by tris(2-carboxyethyl)phosphine reduction and tandem MS sequencing of the resultant modified peptides. DTSSP has a linker arm length of 12 Å. Cross-linking of lysine residues that are distant in the primary structure provides evidence that these residues are spatially close in the folded protein (33).
Five cross-links were identified between lysine residues that are separated by more than 20 amino acids (summarized in Fig. 1 and Table 1). Two of these were between lysines in the helical region. In three cases, cross-links were formed to a free cysteine residue in the SH3 domain and lysines: (i) in the second actin-binding repeat; (ii) in the final half-actin-binding repeat; and (iii) in the helical region. Cross-links between lysine and cysteine residues can arise when a disulfide exchange reaction occurs between the disulfide moiety of a DTSSP cross-linker and a nearby reactive cysteine residue. In addition, mass spectrometry identified a disulfide bond between two cysteine residues in the second and fifth actin-binding repeats. Although this disulfide bond may not be biologically relevant, it does indicate that these two repeat domains are in close proximity.
TABLE 1.
Cross-links
The top part of the table shows DTSSP cross-links and the disulfide found within cortactin; the lower part cross-links between cortactin and actin. The @ symbol represents a lysine residues conjugated to the cross-linker.
| Cross-link | Precursor (MH +) observed | Precursor (MH +) theory | MS/MS fragments (MH +) observed | MS/MS fragments (MH +) theory |
|---|---|---|---|---|
| Intramolecular cross-links | ||||
| ANFENLA@ER 333 to MA@ER 353 | 1998.8 | 1998.9 | 1245.3/1311.2 | 1245.6/1311.6 |
| 688.1/754.0 | 688.3/754.3 | |||
| ANFENLA@ER 333 to @LEEQAR 364 | 2237.9 | 2238.0 | 1245.2/1311.1 | 1245.6/1311.6 |
| 927.1/993.1 | 927.4/993.4 | |||
| ANFENLA@ER 333 to GVCK 504 | 1682.6 | 1682.7 | 1245.3/1311.8 | 1245.6/1311.6 |
| YGVQ@DR 288 to GVCK 504 | 1356.5 | 1356.5 | 919.5/985.4 | 919.4/985.4 |
|
GFGG@FGVQMDR 135 to GVCK 504
|
1789.6
|
1789.7
|
1352.8/1418.8
|
1352.6/1418.6
|
| Intramolecular disulphide | ||||
|
HCSQVDSVR 123 to CALGWDHQEK 257
|
2213.9
|
2213.9
|
997.4/1063.4
|
997.4/1063.4
|
| Intermolecular cross-links, actin cortactin | ||||
| I@IIAPPER 330 to A@K 372 | 1555.7 | 1555.9 | 1090.6/1156.6 | 1090.6/1156.6 |
| I@IIAPPER 330 to D@VDK 208 | 1855.8 | 1855.9 | 1090.5/1156.5 | 1090.6/1156.6 |
| (guanidinylated) | 189.1 Y1 ion | 189.1 | ||
| I@IIAPPER 330 to SAVGHEYQS@LSK 118 | 2643.0 | 2643.3 | 1089.9/1155.8 | 1090.6/1156.6 |
The five intramolecular cross-links and the single disulfide bond identified for cortactin indicate that: (i) the second actin-binding repeat is spatially close to the fifth repeat and (ii) the SH3 domain is spatially close to the helical region, the second actin-binding repeat, and the final half-actin-binding repeat. These observations suggest that the actin-binding repeats form a loop such that the first and last repeats are in close proximity. The interaction between the SH3 domain and helical region implies that most of the domains in the C-terminal half of cortactin may pack together to form the globular structure seen by SAXS.
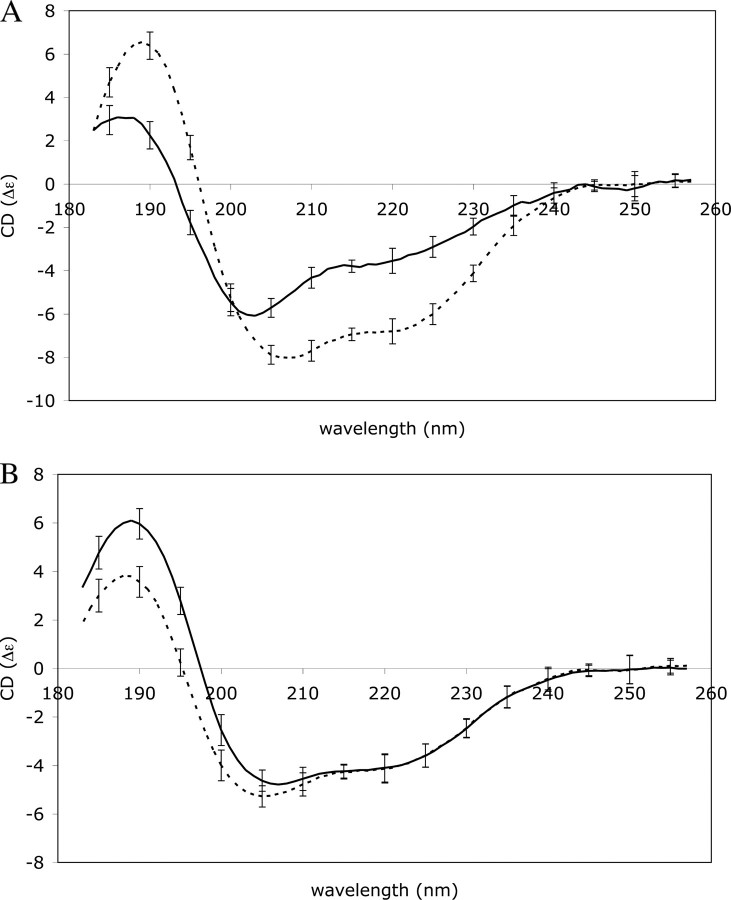
The Secondary Structure of Cortactin Does Not Change upon F-actin Binding—Secondary structure analysis based on a CD spectrum of cortactin (Fig. 3A) gave an average secondary structure composition of 32% α-helix, 6% β-sheet, and 62% random coil. This percentage of coil is high as compared with the average percentage of coil for proteins in the Protein Data Bank of around 45% (34). Secondary structure prediction algorithms predict a similar high level of random coil for cortactin (16% α-helix, 8% β-sheet, and 76% random coil), suggesting that the high percentage of coil observed by CD is not due to misfolding of the recombinant protein.
FIGURE 3.
CD spectra of cortactin, actin, and the complex measured between 260 and 180 nm on a Jasco J-810 instrument and smoothed using a 3-nm box car average. A shows the individual spectra of cortactin (solid line) and f-actin (broken line). B shows the measured spectrum of a complex between f-actin and cortactin (solid line) and the theoretical spectrum of the complex calculated by combining the spectra of free f-actin and cortactin from panel A (broken line).
To investigate whether there are changes in cortactin secondary structure upon binding to f-actin, a CD spectrum of the cortactin-f-actin complex was measured and compared with a CD spectrum calculated by combining the spectra of free f-actin and cortactin. This calculated reference spectrum represents a theoretical complex in which no structural changes occur upon binding (Fig. 3B) (35). Changes in structure during complex formation were then quantified by comparing the composition of secondary structure elements from the observed and calculated spectra. The calculated spectrum (that assumes no change in structure from free proteins to complex) gives 38% α-helix and 14% β-sheet, whereas the observed spectrum gives 37% α-helix and 14% β-sheet. The similarity in these values indicates that there is no gross change in the composition of secondary structure in either actin or cortactin upon complex formation.
The significant differences seen between the calculated and observed spectra at low wavelength can be attributed to burying of hydrophobic residues, interactions between aromatic residues, and possible tertiary/quaternary effects arising from complex formation (35, 36). However, such differences do not represent gross conformational changes upon complex formation. Cortactin, with its high percentage of random coil, does not undergo a dramatic increase in α-helix or β-sheet conformation upon binding to f-actin. Thus, cortactin does not appear to be an intrinsically unstructured protein that only adopts its native structure upon interaction with actin.
The Helical Region and the First and Fourth Repeats Are Implicated in F-actin Binding—The complex between f-actin and cortactin was also analyzed by cross-linking. Following formation of the cortactin-actin complex and incubation with the cross-linking reagent, bands corresponding to covalent cortactin-actin heterodimers were excised from SDS-PAGE gels. Three cross-links between cortactin and f-actin were identified by mass spectrometry (Fig. 1). Lysine residues from the first and fourth actin-binding repeat and from the end of the helical region were cross-linked to a single lysine situated near the common protein-binding site of actin. This result is consistent with previous reports showing that the fourth actin-binding repeat of cortactin is involved in actin binding (37), as well as with the electron microscopy data that localize the actin-binding repeats close to the common protein-binding site of actin (20). This result demonstrates that the first and fourth repeats and the end of the helical domain are likely to be spatially close to each other, supporting the intramolecular cross-linking results. In addition, the juxtaposition of the helical region with f-actin during complex formation suggests that this domain may also play an active role in f-actin-binding.
Cortactin Bundles Actin Filaments into a Sheet—Actin polymerization in the presence and absence of cortactin was studied by time-resolved SAXS. Monomeric actin in low salt buffer was rapidly mixed with a high salt polymerization buffer and flowed slowly past an incident x-ray beam collecting 10-s exposures over a period of several minutes. The earliest time points of the polymerization measurements in high salt buffer show the scattering curve of a globular molecule with a near horizontal plateau at low Q and a well defined Guinier knee. During the first 90 s of polymerization, there is an increase in the power-law slope (i.e. aq-p dependence) of the scattering curves of actin from -0.7 to -0.9. These power-law gradients with magnitude below unity are likely to have arisen from superposition of scatter from an increasing population of large aggregate particles (i.e. polymerized actin filaments) upon scatter from monomeric actin and thus represent the course of polymerization into filaments (Fig. 4A). The shape of the actin scattering curve evolves into a curve that fits well with a cylindrical actin filament (Fig. 4, A and B and supplemental data). After overnight incubation at 4 °C, the gradient of the f-actin curve in a Q range 0.01-0.02 Å-1 on a log:log plot is -0.95, and the gradient in a Q range 0.08-0.1 Å-1 is -4.1. These gradients are typical of cylindrical molecules. The inflection point between these two linear regions is at 26 Å, corresponding to the radius of the actin filament.
FIGURE 4.
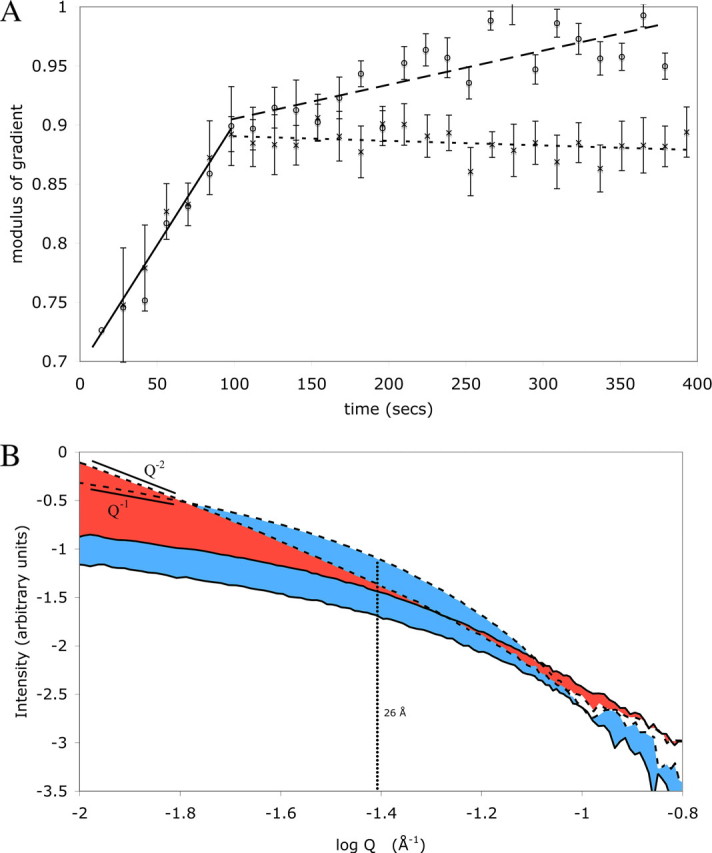
Small-angle x-ray scattering data of f-actin and f-actin in the presence of cortactin. A shows the magnitude change in power-law slope gradient versus time of the scattering curves of polymerizing actin and polymerizing actin in the presence of cortactin in the Q range from 0.01 to 0.02 Å-1 after mixing of monomeric actin with high salt polymerization buffer. Data for actin alone are represented by crosses, and data for for actin-cortactin are represented by open circles. For clarity, best-fit lines have been added to the curves, the portion of the curves below 90 s where the two data sets agree has been overlaid with a solid line, the portion above 90 s of the actin-alone data has been overlaid with a wide dashed line, and the actin-cortactin data has been overlaid with a finer dashed line. B shows the change in scattering curves of actin and actin plus cortactin during the 400 s of the polymerization experiments. The data from actin alone are shown in green, and the actin plus cortactin data are shown in light blue. The earliest time point of each data set is marked with a solid line, and the final data set of each is marked by a broken line. A linear regression fit to the region between Q 0.01 and 0.02 Å (equating to log Q of -2 to -1.7 Å-1) is shown by black broken lines with the gradient of these regions marked. The inflection point of the actin curve is marked by a vertical broken line with the corresponding real space radius indicated.
When actin is polymerized in the presence of equimolar amounts of cortactin, the scattering curves at early time points closely parallel those of actin alone (Fig. 4B). During the first 90 s of the time course, the change in power-law gradient at low Q ranges is similar to that observed for actin alone, indicating that actin polymerizes at the same initial rate in the presence or absence of cortactin (Fig. 4A). However, after ∼90 s, there is a slow increase in the power-law gradient of the cortactin-actin curves at low Q, over and above the actin alone curves (Fig. 4A). This increase continues during the course of the experiment until it reaches -1 at around 350 s. Following overnight incubation, the gradient of the actin-plus-cortactin scattering curves in this range is -2.0. The inflection point of the curve reflects the thickness of the disk; interestingly, it remains at 26 Å, the radius of a single actin filament. This is strong evidence that the actin filaments are bundled into a two-dimensional, sheet-like particle a single actin filament in thickness. These data suggest that cortactin does not affect the initial polymerization rate of f-actin but that following initial polymerization, cortactin induces slow bundling of the filaments into a sheet that is a single filament wide.
DISCUSSION
We used CD spectroscopy to determine the secondary structure composition of cortactin and showed that this protein contains a high percentage of random coil. CD measurements of free actin, cortactin, and their complex show that upon complex formation, there is no large change in secondary structure content, such as a transition from disordered to ordered structure. We conclude that the secondary structure of free cortactin approximates that of the actin-complexed protein and that cortactin is not an intrinsically unstructured protein.
The effect of cortactin on the cytoskeleton is altered by phosphorylation events on the cortactin molecule. This regulation is a key feature of cortactin and is thought to be a major mechanism by which remodeling of the cytoskeleton is tied to cell signaling events. However, the molecular mechanism by which changes in phosphorylation alter the activity of cortactin has not been determined. It has been proposed that intramolecular binding of the SH3 domain to the proline-rich region of cortactin may control the availability of this domain to its various binding partners and that phosphorylation in this region may modulate this interaction (19). In this study, we present evidence by chemical cross-linking for intramolecular interactions between the SH3 domain and the helical and actin-binding domains within the cortactin molecule (Fig. 1). This results in a molecule of globular shape and is supported by SAXS data (Fig. 2C). These observations are consistent with the autoinhibition model of SH3 domain regulation and suggest that the intramolecular interaction may be more extensive than simply with the proline-rich region as originally thought.
Previous experiments by analytical ultracentrifugation and electron microscopy of full-length (6.5 actin-binding repeats) cortactin have revealed an elongated molecule between 220 and 290 Å long and 20 Å wide (2). The SAXS data of free cortactin presented here support a molecule that is 110 Å long, approximately half the previously reported length. We speculate that the structure presented here represents the case in which the SH3 domain is bound into the body of cortactin in the autoinhibited state and that perhaps the previous study represents the SH3 domain in its free state. The major difference between the two studies is that the 220-290 Å long molecule has 6.5 actin-binding repeats, and the molecule 110 Å long used here is a splice isoform with 5.5 repeats. As there are intramolecular interactions between the SH3 domain and the actin-binding repeats, it is conceivable that the equilibrium between bound and free SH3 domain may be different between the two splice isoforms. The relationship between the availability of the SH3 domain and the structure/composition of the actin-binding repeats may be an important feature of cortactin regulation.
The effect of cortactin on actin polymerization was characterized in vitro. We demonstrated that cortactin does not significantly affect the rate of actin polymerization but that, following actin polymerization, cortactin induces bundling of the actin filaments into a two-dimensional array. This is a surprising result because electron microscopy has shown a radial pattern of cortactin binding around the actin filament (20). Cortactin-mediated filament-to-filament interactions would therefore be expected to be equally likely on all sides of the actin fiber leading to filament bundles of circular cross-section. This kind of actin bundling is observed in the case of scruin, a protein that “glues” filaments together in a hexagonal array in the actin bundles of the acrosomal process (38). The three-dimensional glue model does not rule out a nonspecific bundling effect caused by decoration of the filament with an aggregation-prone recombinant protein.
The simplest model to explain the two-dimensional bundling of actin filaments is that neighboring filaments come together side-by-side and are cross-linked top and bottom by cortactin leading to a flat sheet. Although two-dimensional actin sheets have not been reported in biology, the mechanism by which cortactin bridges two juxtaposed filaments in this system may have relevance to the way in which cortactin stabilizes the interaction between mother and daughter filaments at actin branch points in cells (7, 8).
Our results demonstrate a globular shape of cortactin, formed by interactions between the SH3 domain and the actin-binding repeats. This is in contrast to the previously described elongated form. This, together with the ordered manner with which cortactin bundles actin filaments, points to cortactin being more than an inert scaffolding protein that simply tethers several proteins to a specific site. It seems likely that understanding the mechanical and structural properties of cortactin will be crucial to determining how this protein is regulated by post-translational modification and how it coordinates its array of binding partners at sites of actin branching.
Supplementary Material
Acknowledgments
We thank Tom Alber, Ben Hankamer, Dmitri Mouradov, and Tim Geppert for helpful discussions and Cath Latham for critical review of the text.
This work was supported by an Australian Synchrotron Radiation Program Fellowship and Access to Major Research Facilities Program travel funding (to N. C.), an Australian Research Council (ARC) grant (to J. L. M. and B. K.), and a grant from the University of Queensland (to T. H. and B. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental spreadsheet.
Footnotes
The abbreviations used are: SAXS, small-angle X-ray scattering; DTSSP, dithiobissulfosuccinimidyl propionate; MS, mass spectrometry; MS/MS, tandem mass spectrometry.
References
- 1.Wu, H., and Parsons, J. T. (1993) J. Cell Biol. 120 1417-1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weaver, A. M., Heuser, J. E., Karginov, A. V., Lee, W. L., Parsons, J. T., and Cooper, J. A. (2002) Curr. Biol. 12 1270-1278 [DOI] [PubMed] [Google Scholar]
- 3.Uruno, T., Liu, J., Zhang, P., Fan, Y., Egile, C., Li, R., Mueller, S. C., and Zhan, X. (2001) Nat. Cell Biol. 3 259-266 [DOI] [PubMed] [Google Scholar]
- 4.Higgs, H. N., and Pollard, T. D. (2001) Annu. Rev. Biochem. 70 649-676 [DOI] [PubMed] [Google Scholar]
- 5.Weed, S. A., and Parsons, J. T. (2001) Oncogene 20 6418-6434 [DOI] [PubMed] [Google Scholar]
- 6.van Rossum, A. G., de Graaf, J. H., Schuuring-Scholtes, E., Kluin, P. M., Fan, Y. X., Zhan, X., Moolenaar, W. H., and Schuuring, E. (2003) J. Biol. Chem. 278 45672-45679 [DOI] [PubMed] [Google Scholar]
- 7.Egile, C., Rouiller, I., Xu, X. P., Volkmann, N., Li, R., and Hanein, D. (2005) PLoS Biol. 3 e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weaver, A. M., Karginov, A. V., Kinley, A. W., Weed, S. A., Li, Y., Parsons, J. T., and Cooper, J. A. (2001) Curr. Biol. 11 370-374 [DOI] [PubMed] [Google Scholar]
- 9.Mizutani, K., Miki, H., He, H., Maruta, H., and Takenawa, T. (2002) Cancer Res. 62 669-674 [PubMed] [Google Scholar]
- 10.Kinley, A. W., Weed, S. A., Weaver, A. M., Karginov, A. V., Bissonette, E., Cooper, J. A., and Parsons, J. T. (2003) Curr. Biol. 13 384-393 [DOI] [PubMed] [Google Scholar]
- 11.McNiven, M. A., Kim, L., Krueger, E. W., Orth, J. D., Cao, H., and Wong, T. W. (2000) J. Cell Biol. 151 187-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin, K. H., Jeffery, E. D., Grigera, P. R., Shabanowitz, J., Hunt, D. F., and Parsons, J. T. (2006) J. Cell Sci. 119 2851-2853 [DOI] [PubMed] [Google Scholar]
- 13.Sangrar, W., Gao, Y., Scott, M., Truesdell, P., and Greer, P. A. (2007) Mol. Cell. Biol. 27 6140-6152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu, J., Yu, D., Zeng, X. C., Zhou, K., and Zhan, X. (2007) J. Biol. Chem. 282 16086-16094 [DOI] [PubMed] [Google Scholar]
- 15.Boyle, S. N., Michaud, G. A., Schweitzer, B., Predki, P. F., and Koleske, A. J. (2007) Curr. Biol. 17 445-451 [DOI] [PubMed] [Google Scholar]
- 16.Liu, J., Huang, C., and Zhan, X. (1999) Oncogene 18 6700-6706 [DOI] [PubMed] [Google Scholar]
- 17.Tehrani, S., Tomasevic, N., Weed, S., Sakowicz, R., and Cooper, J. A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 11933-11938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cosen-Binker, L. I., and Kapus, A. (2006) Physiol. (Bethesda) 21 352-361 [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Quiles, N., Ho, H. Y., Kirschner, M. W., Ramesh, N., and Geha, R. S. (2004) Mol. Cell. Biol. 24 5269-5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pant, K., Chereau, D., Hatch, V., Dominguez, R., and Lehman, W. (2006) J. Mol. Biol. 359 840-847 [DOI] [PubMed] [Google Scholar]
- 21.Dominguez, R. (2004) Trends Biochem. Sci. 29 572-578 [DOI] [PubMed] [Google Scholar]
- 22.Spudich, J. A., and Watt, S. (1971) J. Biol. Chem. 246 4866-4871 [PubMed] [Google Scholar]
- 23.Bono, H., Kasukawa, T., Furuno, M., Hayashizaki, Y., and Okazaki, Y. (2002) Nucleic Acids Res. 30 116-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Listwan, P., Cowieson, N., Kurz, M., Hume, D. A., Martin, J. L., and Kobe, B. (2005) Anal. Biochem. 346 327-329 [DOI] [PubMed] [Google Scholar]
- 25.Studier, F. W. (2005) Protein Expression Purif. 41 207-234 [DOI] [PubMed] [Google Scholar]
- 26.Sreerama, N., and Woody, R. W. (2000) Anal. Biochem. 287 252-260 [DOI] [PubMed] [Google Scholar]
- 27.Rost, B., Sander, C., and Schneider, R. (1994) Comput. Appl. Biosci. 10 53-60 [DOI] [PubMed] [Google Scholar]
- 28.Ouali, M., and King, R. D. (2000) Protein Sci. 9 1162-1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svergun, D. I. (1992) J. Appl. Crystallogr. 25 495-503 [Google Scholar]
- 30.Svergun, D. I. (1999) Biophys. J. 76 2879-2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volkov, V. V., and Svergun, D. I. (2003) J. Appl. Crystallogr. 36 860-864 [Google Scholar]
- 32.Cookson, D., Kirby, N., Knott, R., Lee, M., and Schultz, D. (2006) J. Synchrotron Radiat. 13 440-444 [DOI] [PubMed] [Google Scholar]
- 33.Mouradov, D., Craven, A., Forwood, J. K., Flanagan, J. U., Garcia-Castellanos, R., Gomis-Ruth, F. X., Hume, D. A., Martin, J. L., Kobe, B., and Huber, T. (2006) Protein Eng. Des. Sel. 19 9-16 [DOI] [PubMed] [Google Scholar]
- 34.Zhang, C. T., and Zhang, R. (1999) Protein Eng. 12 807-810 [DOI] [PubMed] [Google Scholar]
- 35.Cowieson, N. P., Miles, A. J., Robin, G., Forwood, J. K., Kobe, B., Martin, J. L., and Wallace, B. A. (2007) Proteins 70 1142-1146 [DOI] [PubMed] [Google Scholar]
- 36.Woody, A. Y., and Woody, R. W. (2003) Biopolymers 72 500-513 [DOI] [PubMed] [Google Scholar]
- 37.Weed, S. A., Karginov, A. V., Schafer, D. A., Weaver, A. M., Kinley, A. W., Cooper, J. A., and Parsons, J. T. (2000) J. Cell Biol. 151 29-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherman, M. B., Jakana, J., Sun, S., Matsudaira, P., Chiu, W., and Schmid, M. F. (1999) J. Mol. Biol. 294 139-149 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.