Abstract
The small Tims chaperone hydrophobic precursors across the mitochondrial intermembrane space. Tim9 and Tim10 form the soluble TIM10 complex that binds precursors exiting from the outer membrane. Tim12 functions downstream, as the only small Tim peripherally attached on the inner membrane. We show that Tim12 has an intrinsic affinity for inner mitochondrial membrane lipids, in contrast to the other small Tims. We find that the C-terminal end of Tim12 is essential in vivo. Its deletion crucially abolishes assembly of Tim12 in complexes with the other Tims. The N-terminal end contains targeting information and also mediates direct binding of Tim12 to the transmembrane segments of the carrier substrates. These results provide a molecular basis for the concept that the essential role of Tim12 relies on its unique assembly properties that allow this subunit to bridge the soluble and membrane-embedded translocases in the carrier import pathway.
The small Tims escort hydrophobic proteins of the mitochondrial inner membrane across the intermembrane space (IMS).4 Key to their function is their organization in heterooligomeric complexes. Upon entering the outer membrane channel, the small Tims are recognized in the IMS by Mia40 (1). Site-specific oxidation of their cysteines into intramolecular disulfides releases them in an assembly-competent folded state that allows formation of specific Tim complexes (2–7).
Tim12 stands out as the only small Tim exclusively located on the inner membrane (8, 9). It functions after recognition of the substrate by the soluble small Tim complexes (8, 9), and it associates with both the Tim9-Tim10 complex and Tim22 in the membrane (10). However, the molecular basis of its interactions is still unknown. Here, we show that Tim12 has an intrinsic affinity for inner mitochondrial membrane lipids, which are enriched in cardiolipin. Deletion analysis, in vivo complementation assays, and import assays followed by analysis of native Tim complexes revealed distinct functional domains in Tim12: the N-terminal segment contains necessary import information but is dispensable for function, whereas the C-terminal end is essential in vivo, has a determining role in lipid binding and association with Tim9, and affects the assembly of Tim12 in complexes with other Tims. We show that Tim12 directly binds to transmembrane segments of its substrates, primarily via its N-terminal domain. As this “substrate sensor” domain of Tim12 overlaps in substrate binding with that of Tim10 and is dispensable, whereas on the other hand the Tim12 C-terminal “assembly domain” is essential, we propose that the key feature of Tim12 function relies on its role as a linker subunit between the soluble and membrane-embedded translocase complexes.
EXPERIMENTAL PROCEDURES
Cloning of Tim12 Variants
In Vitro Transcription/Translation pSP64 Vector—Wild-type TIM12 was PCR-amplified with Tim12 (forward BamHI) and Tim12 (reverse EcoRI) primers from a yeast genomic DNA and cloned into pSP64 with BamHI-EcoRI. The deletion mutants were then PCR-amplified as BamHI-EcoRI fragments and cloned into pSP64.
Bacterial Expression pET28 and pRSET Vectors—Wild-type TIM12 was PCR-amplified from a yeast genomic DNA and cloned as an NdeI-EcoRI fragment. The deletion mutants were extracted from pSP64 and cloned into pET28 (ΔN17 and ΔC18) or pRSETa (ΔN28 and ΔC39) by restriction digestion with BamHI-EcoRI. The extra sequence between the His tag and the ATG site in pET28 was extracted by digestion with NdeI-BamHI, and the 5′-overhangs were filled with Klenow and religated.
Yeast Vector pRS316 for in Vivo Complementation of Tim12 Variants—The Tim12 flanking regions were PCR-amplified from genomic DNA using Tim12/Prom-5′ (forward) and Tim12/Prom-3′B (reverse) for the promoter (593 bp) and Tim12/Term-5′ (forward) and Tim12/Term-3′ (reverse) for the terminator (300 bp). The fragments obtained were subcloned into pGEMTeasy (Promega) or pCR2.1 (Invitrogen). The promoter was isolated by digestion with HindIII-BamHI and cloned in front of the TIM12 open reading frame in pSP64/TIM12. The HindIII-EcoRI cassette was then subcloned into pRS316 to give the pRS316/5′B/TIM12 vector. The terminator was then cut with EcoRI-XbaI and cloned at the 3′-end of the open reading frame. This vector, pRS316/5′B/TIM12/3′, was then used to subclone the deletion mutants by replacing the TIM12 open reading frame with BamHI-EcoRI. The primers used for the cloning procedures are shown in supplemental Table 2.
Protein Purification
Recombinant proteins were expressed in the Escherichia coli BL21(DE3) strain from pET28a, pRSETa, or pGEX4T-1 constructs (see supplemental Table 1). Purification of His6-tagged or glutathione S-transferase-tagged fusion proteins was performed according to published methods (12, 13).
Yeast Methods
GAL-TIM12 Strain Generation—The cassette containing the KANMX selection marker followed by the GAL1-10 promoter was extracted from M4801 by ScaI-BamHI digestion and inserted into the pSP64TIM12prom/Tim12 plasmid, giving rise to the pSP64Tim12prom/KANMX4/GAL1-10/TIM12 plasmid. The whole cassette was amplified using the TIM12 promoter forward and Tim12 reverse primers. The PCR product was then used to transform the FT5 yeast strain (14) and selected on YPGal medium for 2 days. The cells were replicated on YPGal supplemented with 0.2 mg/ml Geneticin (Sigma). To confirm the genotype of the colonies, genomic DNA was extracted, and the correct orientation was checked by PCR using Tim12 reverse EcoRI and KANMX4 reverse primers. Depletion of Tim12 from the GAL-TIM12 strain was confirmed by growth arrest in glucose-containing medium and by Western blotting of mitochondria purified from the GAL-TIM12 strain.
Complementation Experiments—For the complementation experiments, the GAL-TIM12 strain was transformed with the pRS316 plasmid carrying the corresponding genes (different mutants of Tim12 or the fusion cytochrome b2-(1–85)/ΔN28Tim12) under the control of the endogenous TIM12 promoter (5′: 593 bp upstream of TIM12) and terminator (3′: 300 bp downstream of TIM12). The transformed clones were grown overnight at 30 °C in minimal medium SC galactose-Ura and then either shifted to SC glucose-Ura for 36 h at 30 °C or reinoculated in SC galactose-Ura. Serial 1/10 dilutions of each culture were made, and 5 μl were dropped on either SC glucose-Ura or SC galactose-Ura. Plates were then incubated at 30 °C for 2 days.
Liposome Preparation
Phosphatidylcholine (PC) and cardiolipin (CL) were purchased from Avanti Polar Lipids. 12 mg of lipids (PC or PC/CL at a 5/1 ratio) in chloroform were dried with nitrogen for 30 min; resuspended in 1 ml of 150 mm NaCl and 50 mm Tris, pH 7.4; and incubated at room temperature for 1 h to obtain multilayer vesicles. Subsequent cycles of freezing/thawing (10 s in liquid nitrogen and 50 s in a 50 °C water bath) created single layer vesicles that were then homogenized by 10 cycles of extrusion through 100-nm filters. Liposome concentration was measured according to Ref. 15.
Flotations of Liposomes
0.5 μg of recombinant protein was incubated with liposomes (lipid concentration of 5 mg/ml) in 50 μl of 100 mm NaCl and 20 mm Hepes, pH 7.4 (30 min, 22 °C). The reaction mixture was diluted with 50 μl of 2.4 m sucrose, 100 mm NaCl, and 20 mm Hepes, pH 7.4 (final sucrose concentration of 1.2 m). The reaction was overlaid with 100 μl of 0.25 m sucrose. The samples were centrifuged (100,000 × g, 3 h, 22 °C), and 50-μl fractions from the top were analyzed by SDS-PAGE and Western blotting (the “pellet” is material left in the tube and solubilized with SDS sample buffer).
Flotations of Mitoplasts
Mitoplasts derived from swelling of 50 μg of WT yeast mitochondria (either pretreated with 0.1 mg/ml protease K or left untreated) were incubated with 0.5 μg of recombinant protein in 50 μl of 0.25 m SEM buffer (0.25 m sucrose, 1 mm EDTA, and 10 mm Mops, pH 7.4), for 30 min at 22 °C. 130 μl of 2.4 m SEM buffer were added (final sucrose concentration of 1.8 m), and the samples were loaded at the bottom of a sucrose gradient (500 μl of 1.8 m SEM buffer, 1 ml of 1.6 m SEM buffer, and 500 μl of 0.25 m SEM buffer) and centrifuged (100,000 × g, 4 h, 4 °C). Fractions of 250 μl were collected from the top, precipitated with 10% trichloroacetic acid, and analyzed by SDS-PAGE and Western blotting.
Peptide Spot Arrays
Peptide spot membranes of ScAAC2 (103 13-mers overlapping by 10 residues) and ScTim9 (38 13-mers overlapping by 11 residues) were purchased from JPT Peptide Technologies. Screening of peptide spots was performed according to Ref. 16.
Miscellaneous
Purification of mitochondria, in vitro import reactions, and subsequent analyses by SDS-PAGE and BN-PAGE were performed as described previously (13, 17). Quantification of immunodetected or radioactive bands was made using Molecular Dynamics software, ImageQuant 5.2. Surface plasmon resonance analysis using a BIAcore 3000 machine was performed according to Ref. 16.
RESULTS
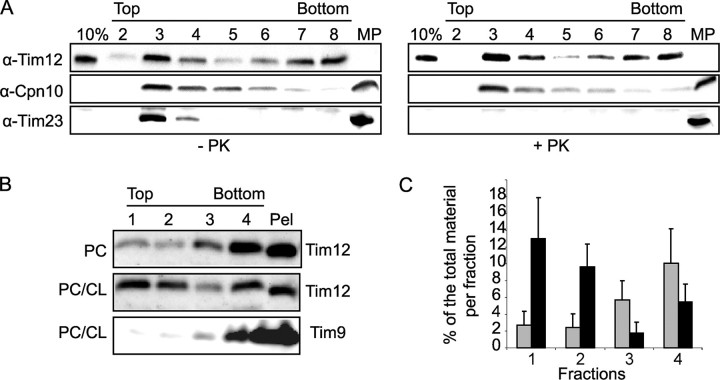
Tim12 Binds to Mitochondrial Inner Membrane Lipids—Tim12 is rather unique in terms of sequence (11), being the only small Tim that is exclusively associated with the inner membrane. As it was reported to be poorly soluble (12), in contrast to its homologues Tim9 and Tim10, we asked whether it might have an affinity for lipids. To address this, we mixed pure Tim12 with either mitoplasts or synthetic liposomes and tested their flotation in a sucrose gradient. Fractions were analyzed by Western blotting using conditions in which only the exogenous protein (His-tagged Tim12) could be detected. In these conditions, pure Tim12 floats with mitoplasts to the top fractions of the gradient (Fig. 1A). Pretreatment of mitoplasts with pyruvate kinase (which degrades protein domains facing the IMS such as Tim23 used here as a control) did not affect binding at all, suggesting Tim12 could intrinsically associate with inner membrane lipids or with a pyruvate kinase-resistant proteinaceous component. We further tested this by using synthetic liposomes instead of mitoplasts. In the presence of PC liposomes, Tim12 floated weakly to the top fractions, but this interaction increased significantly when a mixture of PC and CL (a characteristic lipid of the inner membrane) was used. As a control, Tim9 did not float with liposomes (Fig. 1, B and C). This strongly suggests that Tim12 has an intrinsic affinity for mitochondrial inner membrane lipids, a property presumably not shared by any other small Tim.
FIGURE 1.
Interaction of Tim12 with mitoplasts and liposomes. A, flotation of recombinant Tim12 with pyruvate kinase (PK)-treated or untreated WT yeast mitoplasts. 10% of the protein used was loaded in the first lane of each panel (10%). An equal amount of untreated mitoplasts (MP lane) was loaded as the control. The collected fractions of the gradient, from top to bottom, are shown. Recombinant Tim12 and endogenous mitochondrial proteins were detected by antibodies as shown. B, flotation of recombinant Tim12 with liposomes (PC alone or a PC/CL mixture; see “Experimental Procedures”). As a control, recombinant Tim9 was used with PC/CL liposomes. Numbers indicate the collected fractions that were analyzed with antibodies as shown. C, quantification of Tim12 binding from B. Error bars were derived from three independent experiments (black, PC/CL mixture; gray, PC alone).
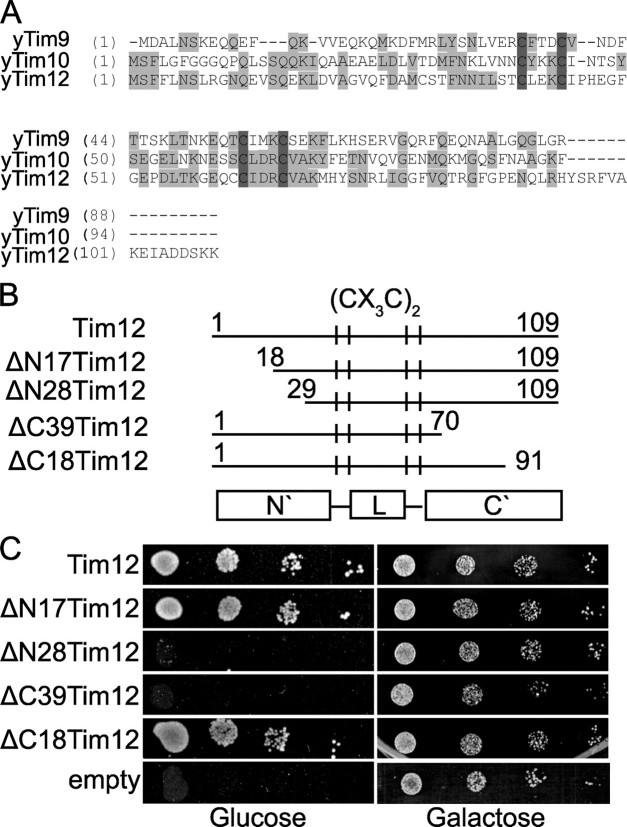
Functional Domain Mapping of Tim12: The C-terminal Segment Is Essential in Vivo—A sequence alignment of Tim12 with Tim9 and Tim10 from yeast shows that Tim12 has a longer C-terminal region (Fig. 2A). Could that be the basis for its unique properties? To address this, we created appropriate deletions of Tim12 that were used as a tool to uncover the precise function of Tim12 in translocase assembly and substrate binding. The deletions we generated for Tim12 (ΔC39, ΔC18, ΔN17, and ΔN28) are shown in Fig. 2B.
FIGURE 2.
Deletion analysis and functional domain mapping of Tim12 in vivo. A, sequence alignment of Saccharomyces cerevisiae Tim9, Tim10, and Tim12. y, yeast. B, graphic representation of the Tim12 deletion mutants. L, central Tim12 loop. C, in vivo complementation of the different constructs of Tim12 in the GALTIM12 strain.
We first performed in vivo complementation assays to test the viability of the different Tim12 deletions by generating a GAL-TIM12 strain with endogenous TIM12 (an essential gene) under the control of the GAL1-10 promoter (Fig. 2C). The GAL-TIM12 strain grows well in galactose, but cells died in glucose after 2 days of repression (Fig. 2C, empty). Growth was fully restored when the WT TIM12 gene with the endogenous TIM12 promoter and terminator was introduced (Fig. 2C, Tim12). The four deletion mutants revealed striking differences. The shorter deletions, ΔN17 and ΔC18, efficiently restored growth of the GAL-TIM12 strain on glucose, indicating that they are fully active. However, deletion of the C-terminal 39 residues (ΔC39) was lethal. Deletion of the N-terminal 28 residues (ΔN28) also caused a serious growth defect after growth for 2 days on glucose, but when the cells were left for more than 4 days, some surviving colonies could be seen (data not shown).
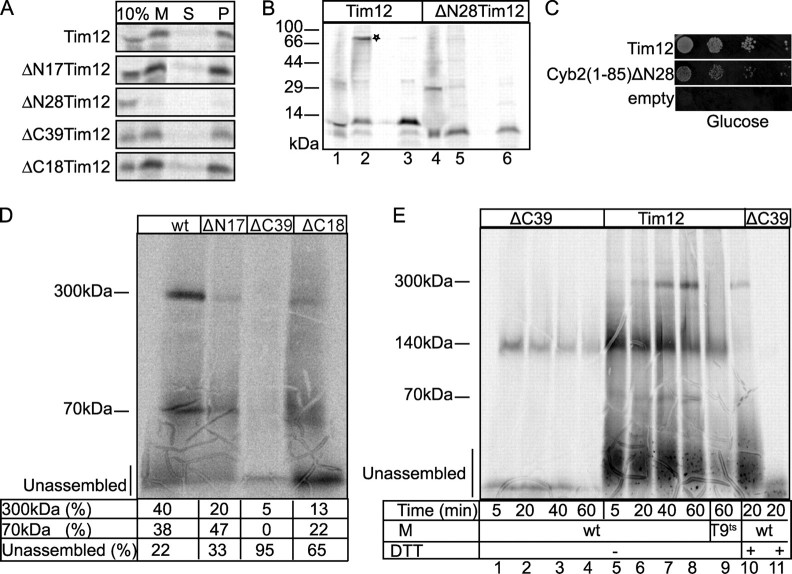
The N-terminal End of Tim12 Is Necessary for Import—To further investigate the molecular basis of the in vivo phenotypes, all Tim12 versions were tested for import into WT mitochondria (Fig. 3A). Interestingly, ΔN28 showed a clear import defect, in sharp contrast to all other versions of Tim12 that could properly cross the outer membrane (M), and localized, upon swelling, to the mitoplast pellet (P). As ΔN28 could not even cross the outer membrane, it must be abrogated at an early stage of import. Because Mia40 is the crucial import receptor that facilitates translocation of small Tims across the outer membrane, we tested this interaction. Import into mitoplasts was performed for ΔN28 and WT Tim12 under conditions that allow monitoring the intermediate between Mia40 and Tim (Fig. 3B) (5, 6). In lane 2, a proper intermediate of WT Tim12 with Mia40 is seen (shown by an asterisk) that is β-mercaptoethanol-sensitive (lane 3). By contrast, ΔN28 failed to give any such intermediate (lane 5). Although we cannot rule out that ΔN28 has additional defects in its interaction with TOM subunits, the specific defect in Mia40 interaction solidifies the point that ΔN28 is affected in import. We then fused ΔN28 to the presequence of cytochrome b2, an efficient presequence that targets proteins to the IMS (18). This should allow us to overcome the import defect and test the functionality of ΔN28 in vivo. We found that such a fusion can be properly imported in organello (albeit somewhat more slowly than the WT; data not shown), and it restored growth of the GAL-TIM12 strain on glucose (Fig. 3C). Collectively, these data suggest that segment 17–28 of the N-terminal end of Tim12 contains necessary targeting information, but the ΔN28 deletion is not abrogated in an essential function in vivo.
FIGURE 3.
Import and assembly of Tim12 deletions in mitochondria. A, import into WT mitochondria, followed by SDS-PAGE and autoradiography. 10% of the radioactive translation mixture was loaded as standard (10%). After import, the material was divided, with one half solubilized in SDS sample buffer (M) and the other half subjected to swelling followed by centrifugation to separate the mitoplast pellet (P) from the intermembrane space (S). B, import of radioactive Tim12 and ΔN28Tim12 in WT mitoplasts (10 min, 30 °C) with 10% standard as in A. To monitor the interaction with Mia40, the import reaction was stopped by adding 20 mm N-ethylmaleimide and analyzed without (lanes 2 and 5) or with (lanes 3 and 6) β-mercaptoethanol. C, complementation test of the fusion cytochrome b2-(1–85)/ΔN28Tim12 (Cyb2(1–85)ΔN28) in the GAL-TIM12 strain. D, BN-PAGE of the complex assembly for imported WT Tim12 and mutants. E, BN-PAGE kinetics of imported radioactive Tim12 and ΔC39Tim12 in WT or Tim9-ts mitochondria. The last two lanes were in the presence of 20 mm dithiothreitol (DTT). Radioactive bands were detected by autoradiography. M, mitochondria.
The C-terminal End of Tim12 Has a Crucial Role in Assembly—To further address the assembly of the other three mutants that have no apparent import defects, BN-PAGE was used (Fig. 3D). Imported WT Tim12 associates mainly with a 300- and 70-kDa complex, in agreement with previous work (10). Strikingly, ΔC39 was found essentially unassembled and incapable of forming either a 70- or 300-kDa complex. On the other hand, ΔN17 and ΔC18 had some clearly weaker assembly defects, which are presumably tolerated in vivo, as these mutants are viable (Fig. 2C). Because the amount of ΔC39 properly crossing the outer membrane is similar to WT, ΔN17, and ΔC18 (Fig. 3A), ΔC39 is presumably unaffected at an early stage of import but rather has a specific assembly defect. We extended the BN-PAGE analysis by monitoring the kinetics of formation of the different assembly intermediates (Fig. 3E). WT Tim12 associates with three major complexes: (i) a 140-kDa complex, which is dithiothreitol-sensitive (Fig. 3E, compare lanes 6 and 10) and represents the Mia40-bound intermediate (1); (ii) the 70-kDa complex, which is dependent on Tim9 and Tim10, as it is completely absent in Tim9-ts mitochondria (compare lanes 8 and 9); and (iii) the 300-kDa TIM22 complex, which appears significantly later than the 140- and 70-kDa complexes. As shown in Fig. 3E (lanes 1–4), ΔC39 is unaffected in the early steps of Mia40 interaction in the 140-kDa complex, which is dithiothreitol-sensitive (compare lanes 2 and 11). However, formation of the 70- or 300-kDa complex is completely abolished. This analysis supports a mechanism whereby Tim12 first assembles in the 140-kDa complex with Mia40. Oxidation by Mia40 releases Tim12 in an assembly-competent state, associating probably with Tim9 and/or Tim10 to a 70-kDa complex first and then to a 300-kDa complex formed with the other subunits of the TIM22 complex. Further work is needed to substantiate this model, but collectively, these data clearly support that the C-terminal end of Tim12 is a key determinant for Tim12 assembly in vivo.
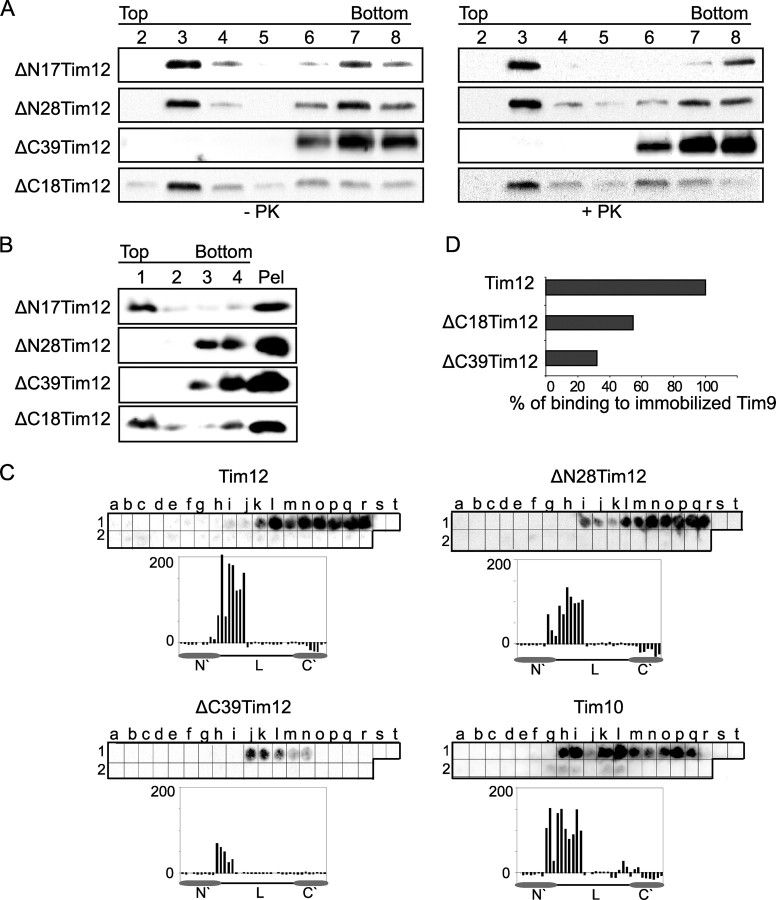
The C-terminal End Has a Dual Role Mediating Both Protein-Lipid and Protein-Protein Interactions—Which bimolecular interaction or interactions are affected by the C-terminal end that is crucial for assembly? As Tim12 was shown to interact with lipids (Fig. 1) and with Tim9 (12), we tested both of these possibilities. In Fig. 4, we first tested in flotation assays the interaction of all four Tim12 deletions (purified from bacteria) with either mitoplasts (Fig. 4A) or synthetic PC/CL liposomes (Fig. 4B). Neither of the smallest deletions (ΔN17 and ΔC18) had any effect on lipid binding. However, when mitoplasts were used, ΔN28 could associate efficiently with the membranes of the top fractions, whereas ΔC39 could not. The association of ΔN28 with the mitoplasts persisted even when these were treated with pyruvate kinase, mirroring the behavior of WT Tim12 (Fig. 1A). By contrast, both ΔN28 and ΔC39 are deficient in their association with liposomes (Fig. 4B). This indicates that both the N-terminal segment 18–29 and the C-terminal segment 70–91 are important for the intrinsic binding of Tim12 to synthetic PC/CL liposomes; however, for the interaction with the more physiological milieu of the inner membrane, the N-terminal end is dispensable, but the C-terminal end is crucial, probably reflecting an interaction of the C-terminal end with a yet unidentified pyruvate kinase-resistant proteinaceous component of the membrane.
FIGURE 4.
Interactions of Tim12 deletions with lipids and with Tim9. A, flotation of Tim12 deletion mutants with pyruvate kinase (PK)-treated or untreated WT yeast mitoplasts. The fractions of the gradient, from top to bottom, are shown in the numbered lanes. B, flotation of Tim12 deletion mutants with PC/CL liposomes. The fractions of the gradient, from top to bottom, are shown in the numbered lanes. The material left in the tube was collected with SDS sample buffer and loaded as the pellet (Pel). Recombinant proteins were detected by Western blotting with anti-Tim12 antibody. C, binding of Tim12, ΔN28Tim12, ΔC39Tim12, and Tim10 on a ScTim9 peptide array. D, a chart representing the binding capacity of Tim12 and C-terminal deletion mutants on immobilized Tim9 (BIAcore assay). The binding capacity of Tim12 was set to 100. L, central Tim12 loop.
The specific interaction recently observed in vitro between Tim12 and Tim9 (12) may be important for the assembly of Tim12 within the protein complexes of 70 and 300 kDa (Fig. 3, D and E). To investigate a putative effect of the Tim12 deletions on Tim9 binding, we performed PepSpot arrays with immobilized Tim9 peptides (Fig. 4C). Tim12 binds to Tim9 with a similar, albeit somewhat more restricted pattern than Tim10. ΔC39 displays a clear defect in binding to Tim9, in contrast to ΔN28, which appears unaffected. We confirmed this defective binding of ΔC39 with BIAcore analysis using Tim9 immobilized on the optical chip. Tim12 binding was reduced by >70% when the C-terminal end 39 amino acids were missing (ΔC39), and even ΔC18 showed a 50% decrease in binding. These results support a very crucial role of the C-terminal end of Tim12 in binding to both lipids and Tim9.
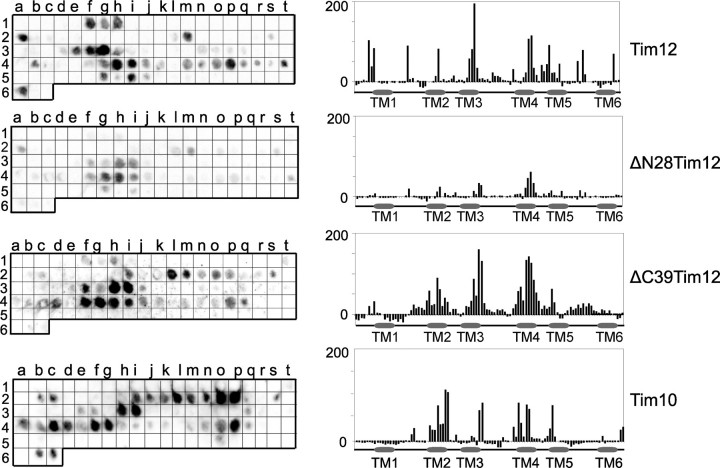
Tim12 Directly Binds the Transmembrane Segments of Substrates via Its N-terminal Segment—Once assembled properly in its native complexes, Tim12 mediates carrier insertion into the membrane. Can Tim12 directly bind the substrate like Tim10, or is it a mainly structural component like Tim9 (16)? To address this, PepSpot arrays of the carrier AAC were used (Fig. 5) (19). WT Tim12 binds to AAC with a preference for the transmembrane regions (top), similar to Tim10 binding (bottom). Binding is unaffected in the ΔN17 and ΔC18 mutants (data not shown), only slightly affected in the ΔC39 deletion, but strongly decreased in the ΔN28 deletion (middle). It therefore appears that (i) Tim12 directly binds the substrate preferably on the transmembrane regions and that (ii) its N-terminal end is important for this interaction.
FIGURE 5.
Interaction of Tim12 with the substrate ScAAC2 using peptide spot arrays. Binding of recombinant Tim12, ΔN28Tim12, ΔC39Tim12, and Tim10 on a ScAAC2 peptide array is shown. The bar charts demonstrate the quantified intensity of each spot of the peptide array.
DISCUSSION
The first novel finding of the present study is the property of Tim12 to bind lipids. This distinguishes Tim12 from the other small Tims and is specifically dependent on cardiolipin, which is enriched in the inner mitochondrial membrane. Association of peripherally attached translocase subunits with inner membrane lipids is not unprecedented: Tim44, a peripherally attached subunit of the TIM23 translocon, also binds to inner membrane lipids (20). Tim12 and Tim44 are functionally and structurally completely different and belong to different translocases. However, their lipid binding capacity may reflect a common strategy to recruit peripherally attached components (either from the IMS for Tim12 or from the matrix for Tim44) to inner membrane translocons so as to stabilize the necessary protein-protein interactions with the other translocon subunits and the substrates. As the organization of all small Tims in specific complexes is crucial for their function, we investigated the Tim12 assembly mechanism. Deletion analysis of its structural domains, coupled with in vivo complementation assays and analysis of native complexes, revealed strikingly different properties of its N- and C-terminal ends. The N-terminal end contains targeting information but is dispensable for function, as the cytochrome b2/ΔN28 fusion, which bypasses the import defect of ΔN28, restores growth of a GAL-TIM12 strain. By contrast, the ΔC39 deletion is lethal, arguing for essential functions of the C-terminal end in vivo. Indeed, we found that this deletion abrogates important molecular interactions in organello and in vitro. First, assembly of the 70- and 300-kDa complexes (reflecting interactions with small Tims and TIM22 components, respectively) is essentially abolished. Interestingly, these defects are manifested after the interaction with Mia40, which proceeds normally. Second, in vitro binding of pure ΔC39 with lipids and mitoplasts is affected, indicating that the intrinsic interaction of the folded Tim12 with the membrane is impaired. Third, the interaction with pure Tim9 is strongly decreased. These data point to a crucial role of the C-terminal end of Tim12 in assembly.
How is this relevant in terms of binding to the substrate? Here we have shown that Tim12 binds to transmembrane segments of AAC and that this interaction is mediated by the N-terminal end in the same manner as Tim10. These results identify Tim12 as an additional substrate sensor in the small Tim family. However, an important difference between Tim10 and Tim12 is that the substrate sensor N-terminal end of Tim10 is essential (21), whereas in Tim12 it is dispensable, probably because of this overlap in binding the same substrate segments. On the other hand, the C-terminal assembly domain is indispensable in vivo and ensures the unique role of Tim12 as a linker subunit in the carrier import pathway.
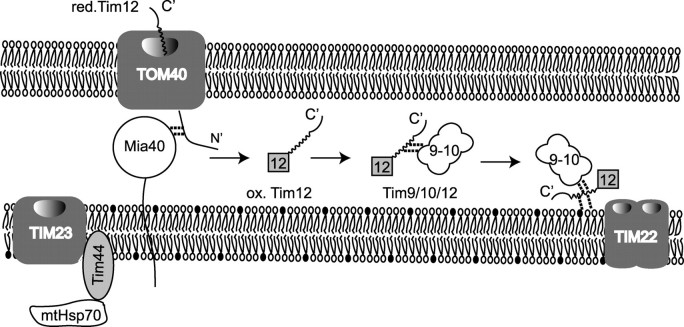
The model that we propose for the biogenesis and function of Tim12, based on our findings, is shown in Fig. 6. Tim12 is imported via the TOM channel in mitochondria in a reduced state. Mia40 acts as the receptor of Tim12, helping it to fold to its native state by oxidizing it. Oxidized Tim12 is first integrated in a 70-kDa complex that contains Tim9 (presumably the TIM10 complex). This interaction is mediated by the C terminus of Tim12. Subsequently, Tim12 is integrated in a 300-kDa complex (presumably, the TIM22 complex). Tim12 is a peripheral subunit with an intrinsic affinity for inner membrane lipids from the IMS side, just as shown for Tim44 at the matrix side of the inner membrane. We propose that Tim12 tethers the TIM10 chaperone of the intermembrane space, close to the TIM22 channel, to facilitate the transport of the substrate from the chaperone into the transporting channel, just as Tim44 brings the mutant Hsp70 chaperone close to the TIM23 channel. This linker function of Tim12 is what makes it essential for cell viability, rather than its actual interaction with the substrates, which can be overcome by the substrate binding capacity of Tim10.
FIGURE 6.
Model scheme for Tim12 biogenesis and function (see “Discussion” for details). red., reduced; mt, mutant; ox., oxidized.
Supplementary Material
Acknowledgments
We are grateful to N. Ismail and M. Vergnolle for preliminary work; S. Hadjicharalambous, K. Melzak, and E. Gizeli for help with preparation of liposomes; D. Tzamarias and T. Gligoris for the FT5 strain and help with the GAL-TIM12 strain; Antonis Makris for the M4801 strain; C. Koehler for the Tim9-ts strain; members of our laboratory for helpful discussions and comments; and Nitsa Katrakili for excellent technical assistance.
This work was supported in part by the Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology Hellas, the University of Crete, and the European Social Fund and national resources (to K. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
Footnotes
The abbreviations used are: IMS, intermembrane space; WT, wild-type; PC, phosphatidylcholine; CL, cardiolipin; Mops, 4-morpholinepropanesulfonic acid; SC, synthetic complete; BN, blue native.
References
- 1.Chacinska, A., Pfannschmidt, S., Wiedemann, N., Kozjak, V., Sanjuan Szklarz, L. K., Schulze-Specking, A., Truscott, K. N., Guiard, B., Meisinger, C., and Pfanner, N. (2004) EMBO J. 23 3735–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mesecke, N., Terziyska, N., Kozany, C., Baumann, F., Neupert, W., Hell, K., and Herrmann, J. M. (2005) Cell 121 1059–1069 [DOI] [PubMed] [Google Scholar]
- 3.Webb, C. T., Gorman, M. A., Lazarou, M., Ryan, M. T., and Gulbis, J. M. (2006) Mol. Cell 21 123–133 [DOI] [PubMed] [Google Scholar]
- 4.Allen, S., Lu, H., Thornton, D., and Tokatlidis, K. (2003) J. Biol. Chem. 278 38505–38513 [DOI] [PubMed] [Google Scholar]
- 5.Milenkovic, D., Gabriel, K., Guiard, B., Schulze-Specking, A., Pfanner, N., and Chacinska, A. (2007) J. Biol. Chem. 282 22472–22480 [DOI] [PubMed] [Google Scholar]
- 6.Sideris, D. P., and Tokatlidis, K. (2007) Mol. Microbiol. 65 1360–1373 [DOI] [PubMed] [Google Scholar]
- 7.Muller, J. M., Milenkovic, D., Guiard, B., Pfanner, N., and Chacinska, A. (2008) Mol. Biol. Cell 19 226–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koehler, C. M., Jarosch, E., Tokatlidis, K., Schmid, K., Schweyen, R. J., and Schatz G. (1998) Science 279 369–373 [DOI] [PubMed] [Google Scholar]
- 9.Sirrenberg, C., Endres, M., Folsch, H., Stuart, R. A., Neupert, W., and Brunner, M. (1998) Nature 391 912–915 [DOI] [PubMed] [Google Scholar]
- 10.Adam, A., Endres, M., Sirrenberg, C., Lottspeich, F., Neupert, W., and Brunner, M. (1999) EMBO J. 18 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentle, I. E., Perry, A. J., Alcock, F. H., Likic, V. A., Dolezal, P., Ng, E. T., Pursell, A. W., McConville, M., Naderer, T., Chanez, A. L., Charriere, F., Adchinger, C., Schneider, A., Tokatlidis, K., and Lithgow, T. (2007) Mol. Biol. Evol. 24 1149–1160 [DOI] [PubMed] [Google Scholar]
- 12.Baud, C., de Marcos Lousa, C., and Tokatlidis, K. (2007) Protein Pept. Lett. 14 597–600 [DOI] [PubMed] [Google Scholar]
- 13.Vial, S., Lu, H., Allen, S., Savory, P., Thornton, D., Sheehan, J., and Tokatlidis, K. (2002) J. Biol. Chem. 277 36100–36108 [DOI] [PubMed] [Google Scholar]
- 14.Tzamarias, D., and Struhl, K. (1994) Nature 369 758–761 [DOI] [PubMed] [Google Scholar]
- 15.Bartlett, G. R. (1959) J. Biol. Chem. 234 469–471 [PubMed] [Google Scholar]
- 16.Vergnolle, M. A., Baud, C., Golovanov, A. P., Alcock, F., Luciano, P., Lian, L. Y., and Tokatlidis, K. (2005) J. Mol. Biol. 351 839–849 [DOI] [PubMed] [Google Scholar]
- 17.Luciano, P., Vial, S., Vergnolle, M. A., Dyall, S. D., Robinson, D. R., and Tokatlidis, K. (2001) EMBO J. 20 4099–4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glick, B. S., Brandt, A., Cunningham, K., Muller, S., Hallberg, R. L., and Schatz, G. (1992) Cell 69 809–822 [DOI] [PubMed] [Google Scholar]
- 19.Curran, S. P., Leuenberger, D., Oppliger W., and Koehler, C. M. (2002) EMBO J. 21 942–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss, C., Oppliger, W., Vergeres, G., Demel, R., Jeno, P., Horst, M., de Kruijff, B., Schatz, G., and Azem, A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 8890–8894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vergnolle, M. A., Alcock, F. H., Petrakis, N., and Tokatlidis, K. (2007) J. Mol. Biol. 371 1315–1324 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.