Abstract
Purpose
Age-related macular degeneration (AMD) has a complex etiology arising from genetic and environmental influences. This past decade have seen several genes associated with the disease. Variants in five genes have been confirmed to play a major role. The objective of this study was to evaluate whether genes influence treatment response to ranibizumab for neovascular AMD. The hypothesis was that an individual’s genetic variation will determine treatment response.
Methods
The study was a two-site prospective open-label observational study of patients newly diagnosed with exudative (neovascular) AMD receiving intravitreal ranibizumab therapy. Treatment-naïve patients were enrolled at presentation and received monthly “as needed” therapy. Clinical data was collected monthly and DNA extracted. Genotyping was performed using the Illumina (San Diego, California) 660-Quad single-nucleotide polymorphism (SNP) chip. Regression analyses were performed to identify SNPs associated with treatment-response end points.
Results
Sixty-five patients were enrolled. No serious adverse events were recorded. The primary outcome measure was change in ETDRS visual acuity at 12 months. A SNP in the CFH gene was found to be associated with less improvement in visual acuity while receiving ranibizumab therapy. The C3 gene, among others, was associated with reduced thickening and improved retinal architecture. VEGFA, FLT1, and CFH were associated with requiring fewer ranibizumab injections over the 12-month study.
Conclusions
This study is one of the first prospective pharmacogenetic study of intravitreal ranibizumab. Although preliminary, the results identify a number of putative genetic variants, which will be further examined by replication and functional studies to elucidate the complete pharmacogenetic architecture of therapy for AMD.
INTRODUCTION
STUDY OVERVIEW
Objective
The objective of this study was to evaluate associations between genetic factors and treatment response to the humanized monoclonal anti–vascular endothelial growth factor (VEGF) antibody, ranibizumab (Lucentis), for neovascular age-related macular degeneration (AMD). The principal hypothesis was that an individual’s genetic variation would influence both functional visual and biological end points to this intravitreal therapy. It is hoped that such research will define the genetic biomarker spectrum to allow treatment individualization and optimize visual outcomes.
Significance of the Problem
Previous therapies for “wet,” or exudative, AMD that utilized laser therapy to destroy or occlude the choroidal neovascularization (CNV)1–3 have been largely superseded by the introduction of anti-VEGF antibodies given by injection into the vitreous cavity of the eye.4–7 Two agents are currently used: ranibizumab (Lucentis, which is approved by the US Food and Drug Administration [FDA]) and bevacizumab (Avastin, “off label”). Both have revolutionized outcomes for those with the condition; evidence is clear that the vast majority of patients benefit from therapy. However, little is known about which eyes will respond best or what might be the best treatment regimen. Since the injections are costly and need to be repeated frequently, it would be of significant benefit to design an individualized regimen to optimize the visual outcome while minimizing the number and frequency of injections.
Rationale and Key Study Design Considerations
Interactions between drugs and genes—pharmacogenetics—can be studied using a variety of in vitro and in vivo methods. In vitro studies may be most useful for drug screening and investigations of basic biology but cannot easily be extrapolated to predict treatment effects in humans. These are best evaluated in pharmacogenetic clinical trials. This thesis describes one such clinical study, the Lucentis Genotype Study, which was undertaken prospectively to avoid the limitations inherent in retrospective review (clinical heterogeneity, missing data, and variations in therapeutic administration).
The length of the study (time from enrollment to primary end point) was chosen to be 12 months. This would allow enough time for a substantial treatment effect while keeping the study to a manageable time frame. Adherence to a clinical evaluation and treatment protocol that mirrored “standard of care” was imperative to maximize the relevance of findings to clinical practice. It was considered an advantage to have more than one site so as to minimize ascertainment and treatment bias.
PHARMACOGENETICS
The theory of complex traits is based upon the idea that multiple variations in the genetic code (most frequently single-nucleotide polymorphisms [SNPs], insertions or deletions [“indels”], and copy number variants) act in concert to determine a particular phenotype. Evidence suggests that these variants result in functionally important alterations in, among other things, the activity, expression levels, stability, and splicing of the RNA and proteins they encode. The action of these variants is, however, not independent of external and environmental influences. A simple example would be obesity, which is determined by a number of genetic variants.8,9 Given the same diet, an individual with one genotype will maintain a different body mass index than someone with a different genotype. However, faced with starvation, the individual will be thinner than someone with the same genetic profile who is well-nourished. In the same way, the response to other exogenous factors, such as drugs, will be influenced by genetic variation. This forms the basis of pharmacogenetics, which attempts to define the genetic variants that influence variable response to medication. The ultimate goal of pharmacogenetic studies is to identify those who respond best and avoid adverse reactions.
History
The British physician Archibald Garrod first recognized a familial or genetic tendency to variability in drug response.10 He hypothesized that drugs were metabolized by specific pathways, and defects in their component enzymes would result in differences in drug concentrations and therefore drug effect. This was validated in the 1940s with the observation of a high incidence of hemolysis to exposure to antimalarial drugs among individuals with glucose-6-phosphate dehydrogenase deficiency.11 A decade later, Price-Evans showed in a classic twin study that metabolism of the antituberculous drug, isoniazid, was much less variable in monozygotic twins as compared with dizygotic twins, suggesting a strong heritable component.12 Subsequently, a large number of studies have defined pharmacogenetic interactions in many biomedical fields. These include therapies for neurologic and psychiatric disorders,13–15 asthma,16 cardiovascular disease,17 and cancer.18,19
PHARMACOGENETIC MECHANISMS
Pharmacokinetic Variability
The term pharmacokinetic variability refers to variability in the delivery of a drug or metabolite(s) to target molecules and is known as drug disposition. Typically, this has included drug absorption, distribution, metabolism, and elimination, but more recently recognized are intracellular molecular trafficking, chaperoning, and the regulation of gene expression. Probably the best known examples of pharmacogenetic variability are drug elimination by N-acetylation in which genetic variation forms the basis of fast and slow acetylators20 and variants in drug metabolism by the cytochrome P-450 system.21 Examples are given in Table 1. Typically, SNPs play a more significant role where the therapeutic range is narrow.
TABLE 1.
EXAMPLES OF PHARMACOGENETIC INTERACTIONS BETWEEN GENES AND DRUGS CURRENTLY IN CLINICAL PRACTICE
| GENE PRODUCT | DRUG | EFFECT OF THE MINOR ALLELE* |
|---|---|---|
| Drug disposition | ||
| CYP2C9 | Warfarin | Reduced anticoagulant effect |
| Drug targets | ||
| HERG/MiRP1 | QT-prolonging drugs | Increased risk of arrhythmia |
| Modulators of drug action | ||
| G6PD | Antimalarials | Increased risk of hemolysis |
In each case, the effect on drug activity of carrying the genetic change is shown.
Pharmacodynamic Variability
Individuals with the same drug tissue concentration of a drug vary in their response to treatment. Two mechanisms are at play: (1) genetic variability in the molecular target of the drug and (2) interactions with the molecules downstream of the target. A good example of pharmacodynamic variability is that of β-blockers, which are highly beneficial in those individuals at risk for heart failure who are homozygous for an intronic deletion in the ACE (angiotensin-converting enzyme) gene (the DD genotype).22 This gene encodes a key enzyme in the renin-angiotensin system involved in the maintenance of plasma volume even though β-blockers do not act directly on the gene itself.23
Identification of Candidate Genes: Pharmacogenomics
As with all analyses, the initial step is to determine, identify, and thoroughly evaluate the phenotype. In pharmacogenetic research, the phenotype should be a clinically relevant treatment end point and ideally one without known cause but with significant inter-individual variability (suggesting that genetic variation may be important). The next step is to accumulate a list of candidate genes based on the biology of the drug with focus on pharmacokinetic and pharmacodynamic candidates. In an era where genome-wide association is routine and Whole Exome sequencing is a reality, it is tempting to suggest that these technologies may be gainfully employed. However, problems with multiple testing and false discovery rates may limit their use unless large numbers of samples can be studied. Other classic gene identification methods, such as positional cloning, are of limited utility since these are reliant on large pedigrees, which are rare in this field. The final step is to choose which polymorphisms within each gene should be studied. In some instances, common haplotypes are known or can be constructed from such resources as the HapMap (http://www.hapmap.org), offering the opportunity to limit genotyping to only tagging SNPs and so most parsimoniously capture most of the variation in the genome. In other genes, these may not so readily be defined, and typically the choice in SNPs is focused on those which are predicted to result in amino acid changes (nonsynonymous coding SNPs).
Challenges Specific to Pharmacogenetic Analysis
The detection of genetic interactions is challenging because the large number of possible genotype combinations reduces the power to detect associations due to multiple testing and false discovery. Since this study was likely to enroll a relatively small cohort, it was designed as a carefully conducted pilot study with the specific intent to generate hypothesis about genetic signals for evaluation in future studies. Since both environmental and genetic risk factors are associated with development of neovascular AMD, it was important that analyses were appropriately performed to include all variables to avoid confounding.
Ethical, Economic, and Social Considerations
The field of pharmacogenetics has ethical considerations as well as social and economic implications beyond simply the development of the science. Patients are usually required to consent to not only the clinical trial but also research into specific genetic analyses and, more significantly, unspecified genetic tests to be used in future pharmacogenetics research. It is likely that patients entering into pharmacogenomics-related trials will not have given consideration to all potential risks and benefits of this additional research. Since there is no likely direct benefit to the patient from the immediate clinical study, this additional risk cannot be offset against potential therapeutic benefit. One potential solution might be to collate all DNA samples from pharmacogenetic studies in genetic databases with pharmaceutical companies paying for access.
Much of the pharmaceutical sector is involved in pharmacogenetics research, from “big pharma” who are interested in developing new drugs with better targets, to the smallest biotech start-ups who provide test kits and supporting technological innovations. Companies feel that such endeavors will provide them with competitive advantage for providing individualized medicines and reduced research and development costs. Not all, however, are persuaded that pharmacogenetics will bring increased profits. Concerns center on the potential for reduced market size or increased segmentation if drugs become licensed for use in specific subgroups of individuals.
There are several significant social consequences of successful pharmacogenetics research. There will be a major change in the way individuals view their health and the taking of medicines. The hope would be that the large numbers of adverse drug reactions would be reduced. There is a possibility that when whole genome sequencing becomes a routine and inexpensive undertaking, a pharmacogenetic profile might be constructed in early life in readiness for future therapeutic need.
AGE-RELATED MACULAR DEGENERATION
Clinical Features
AMD affects the macula, and individuals lose central vision in one or both eyes, affecting activities such as reading and in some cases causing legal blindness. Initially, the disease is characterized by drusen (lipid/protein deposits under the retina), which may progressively accumulate and predispose to the advanced forms of the disease24–26: geographic atrophy (“dry” AMD) of the macula and/or neovascular/exudative (“wet”) AMD, characterized by CNV that tends to bleed and result in retinal scarring.
In the United States, 1.75 million people have advanced AMD and several more million have earlier stages of the disease. The prevalence of advanced disease is estimated to be 8% in those older than 75 years.27 As the population is now aging, the prevalence of AMD will increase perhaps by as much as 50% by the year 2020.28
Although geographic atrophy accounts for the majority of cases of advanced disease, neovascular AMD causes most legal blindness. Neovascular AMD is usually rapidly progressive, resulting in loss of acuity and distortion of shapes. Examination is characterized by hemorrhage in the retina and retinal fluid.29,30 The sequelae of the neovascular process are retinal scarring and permanently reduced macular visual function. In recent years, fluorescein angiography has been replaced by optical coherence tomography (OCT) as the preferred method for monitoring the progress of treatment with monthly intravitreal anti-VEGF agents.6
ETIOLOGY OF AMD: ENVIRONMENTAL RISK FACTORS
Extensive epidemiologic and genetic analyses have led us to the conclusion that like other chronic age-related diseases, AMD results from multiple environmental and genetic factors. AMD is age-related, and tobacco smoking is the most consistently identified environmental risk factor.31–33 Studies have also implicated cardiovascular disease,34–36 hypertension,37,38 high body mass index,39 and low education level.40
GENETICS OF AMD
That genetics has a significant etiological role in AMD is now beyond question.41,42 Studies to identify causal variants initially concentrated on genome-wide linkage and association analyses.43–45 A meta-analysis46 of these and other results showed reassuring replication of similar chromosomal loci, several of which remain under investigation.47–49 A listing of replicated susceptibility variants is shown in Table 2.
TABLE 2.
GENES IMPLICATED IN ANGIOGENESIS OF AGE-RELATED MACULAR DEGENERATION
| AMD SUSCEPTIBILITY GENE | ANGIOGENESIS GENE |
|---|---|
| CFH, complement factor H | VEGFA, vascular endothelial growth factor A |
| ARMS2, age-related maculopathy susceptibility 2 | VEGFR, (FLT1), vascular endothelial growth factor receptor 1 |
| C2, complement factor 2 | FGFR1, fibroblast growth factor receptor 1 |
| C3, complement factor 3 | FGFR2, fibroblast growth factor receptor 2 |
| CFB, complement factor B | THBS1, thrombospondin-1 |
| CFI, complement factor inhibitor | PF4, platelet factor 4 |
| KDR, kinase domain receptor | |
| CTGF, connective tissue growth factor | |
| ANG1, angiopoietin 1 | |
| ANG2, angiopoietin 2 | |
| TGF, transforming growth factor | |
| HIF1α, hypoxia inducible factor 1, alpha subunit | |
| VHL, von Hippel Lindau factor | |
| CX3CR1, chemokine (C-X3-C motif) receptor 1 | |
| PDGF, platelet derived growth factor | |
| PI 3-KC A, B, C, phosphoinositide-3-kinase, catalytic | |
| ICAM-1, intracellular adhesion molecule | |
| MAPK 1, 2, 3, mitogen activated protein kinase |
The first specific replicated genetic variant to be associated with advanced AMD was the SNP rs1061170 (T1277C; Y402H) in the complement factor H (CFH) gene.50–53 Additional SNPs and haplotypes in CFH54,55 and neighboring genes56 have also been associated with drusen formation and advanced AMD.53,57 CFH is a regulator of complement, dysfunction of which has been linked to retinal pathology.58 Recently, SNPs in other complement components have been associated with advanced AMD: complement factors 2 (C2), B (CFB),59,60 3 (C3),61,62 and I (CFI).63
A major AMD-susceptibility locus has also been identified on chromosome 10q26,46,47 a region where linkage disequilibrium has made it difficult to distinguish the causal genetic variant64: the SNP, rs10490924 (A69S), is within the gene, ARM-susceptibility 2 (ARMS2).65,66 This putative gene has unknown function, and its protein product has been identified in several subcellular compartments67,68 or the cytoplasm. The SNP, rs11200638, is located in the promoter of the gene HTRA1,69,70 a serine protease found in the retina (among other tissues), and the SNP may alter gene expression.69 In complete linkage disequilibrium with this SNP is an indel in ARMS2 that may affect translation of the ARMS2 protein.68 Associations in the genes APOE,71–73 ABCA4,74,75 CX3CR1,76,77 PON1,78 TLR4,79 ERCC6,80 ELOVL4,81,82 VLDLR,83 fibulin-5,84 hemicentin-1,85 TLR,86 C1q,87 and LRP683 have also been suggested.
AMD Associations With VEGF, PEDF, Proangiogenic and Antiangiogenic Genes, and Other Pathway Genes
Associations between VEGFA and advanced AMD have been examined by a number of investigators. Several studies have suggested that there is an association between selected SNPs in the gene and AMD.83,88–92 Other studies have failed to find an association, including recent genome-wide association studies.93 Other genes in the VEGF pathway have not been studied. A couple of small Asian cohorts have examined the association with the PEDF gene, but these findings have not been confirmed.94–96
CLINICAL EXPERIENCE WITH RANIBIZUMAB
Dose Regimen
Ranibizumab has been studied in more than 5,000 subjects with neovascular AMD in a number of Phase I, I/II, II, III, and IIIb clinical trials and has been approved for use in the treatment of this condition by the FDA.6 Cumulatively, the studies show that approximately 25% of subjects show a significant improvement in vision (defined as gain of ≥15 ETDRS letters), 70% maintain or show slight improvement from their acuity at presentation (defined as a gain of ≥0 letters), and the remainder lose vision.4
The reasons for the variation are not known but are unrelated to conventional clinical descriptors of CNV, including lesion size and angiographic characteristics. In one study (PIER), patients that underwent fixed dosing every 3 months lost vision as compared with monthly treatment. It has been further concluded that OCT is important in monitoring patients with CNV. When re-treatment is guided by the presence of retinal thickening, intraretinal fluid, or subretinal fluid, visual acuity at 3 months is maintained at 12 months.
This data guided the dosing and treatment regimen employed in this study, which adhered as closely as possible to the commonly employed practice of clinical care at the time of the study.
Safety Considerations
Patient inclusion adhered to current safety criteria. Following successful completion of Phase I studies, which showed no significant adverse events, detailed safety data were collected from three randomized, 1- or 2-year follow-up, double-masked, sham- or active-controlled trials. Ranibizumab is contraindicated in patients with active ocular infections and in those with known hypersensitivity to the drug. Serious adverse events related to the injection procedure occur in <0.1% and include endophthalmitis, rhegmatogenous retinal detachment, iatrogenic traumatic cataract, intraocular inflammation, and transient increases in intraocular pressure. Ranibizumab is contraindicated in those with a history of stroke, since it appears to increase the risk for a subsequent stroke.
PHARMACOGENETICS IN AMD
Genetic variants contribute substantially to the etiology of AMD. As a result, there has been interest in examining whether these common SNPs and other candidate genes may play a pharmacogenetic role. Three treatments are currently utilized for the treatment of AMD: Age-Related Eye Disease Study (AREDS) supplementation, anti-VEGF therapy, and photodynamic therapy (PDT). Studies thus far have largely been limited to retrospective analyses.
AREDS Supplements
The AREDS study (a large prospective multicenter randomized trial) found a beneficial effect of zinc and antioxidants (beta carotene, vitamin C, and vitamin E)97 in slowing progression of disease as compared with placebo alone. Using this progression data and combining it with genetic analyses of samples from the cohort, it has been possible to suggest that those individuals taking the supplements who had the low-risk genotype in CFH experienced less disease progression. Although more studies are needed, this result may have a biological basis, since dysregulation of CFH is thought to lead to inflammation, which may be reduced by the action of antioxidant therapy.21–25 One potential conclusion is that any genetic predisposition to AMD reduces the effectiveness of the supplements that remain commonly used for dry AMD in the United States.98
Photodynamic Therapy
PDT has largely been replaced by anti-VEGF therapy, though it still has a role for certain patients with AMD, potentially in combination with other agents99–101 and in those where other treatments may be contraindicated.3 In the treatment, verteporfin is given systemically and localized to abnormal neovascular vessels in the macula, where it can be photo-activated by a laser.102,103 The beneficial effects of PDT were established in several carefully conducted clinical trials, including the Treatment of Age-Related Macular Degeneration With Photodynamic Therapy (TAP), Verteporfin (Visudyne) in Photodynamic Therapy (VIP), and Visudyne in Minimally Classic Choroidal Neovascularization studies.104
Experience has shown variability in treatment success,105 leading some observers to begin to hypothesize that effectiveness might be altered by an individual’s ability to activate coagulation factors in response to PDT. A number of single gene disorders result in coagulapathies and their prevalence was evaluated in two studies. Patients determined to be PDT responders and nonresponders were genotyped for a number of coagulation factor mutations. The results suggested that those carrying the G185T mutation of factor XIII-A, which results in a hyperfibrinolytic state, were more likely not to respond to PDT, whereas those with factor V 1691A and prothrombin 20210A, both prothrombotic state, did better.106–108
Anti-VEGF Agents
This treatment has been shown to have significant efficacy and has been the subject of interest as to whether genetics may play a role in outcomes. In a study of 86 patients treated with bevacizumab (Avastin), those with the risk CC genotype in CFH had worse visual outcomes than those with other genotypes.109 This appears to be replicated by a larger study of patients receiving ranibizumab.110 Although these are associations rather than causal findings, these studies introduce the idea that common AMD-susceptibility genes may play a role in determining treatment outcome.
PATHOPHYSIOLOGY OF AMD AND THE IDENTIFICATION OF CANDIDATES GENES
It would be optimal to employ an approach in which all possible genes and genetic variants were included in the pharmacogenetic analysis of ranibizumab in AMD. Unfortunately, there are statistical drawbacks to such an approach (false discovery rates and multiple testing issues). While a genome-wide strategy was employed in this study, a secondary, more focused evaluation of genes considered “good biological candidates” likely to harbor variants that confer susceptibility was undertaken. The purpose of this section is to provide a pertinent overview of such a list. Those genes already implicated in AMD pathogenesis (see above) were also included.
Vision is damaged in AMD because photoreceptors in the macula become dysfunctional and are ultimately lost. The hallmark of the condition are drusen, extracellular lipid/protein deposits that develops over time between the retinal pigment epithelium (RPE) and Bruch’s membrane. Drusen are seen in non-AMD aged retinas, but when they accumulate to a certain degree are considered the earliest feature of AMD. The area occupied by the drusen, their size and number, together with adjacent retinal pigmentary abnormalities, directly correlate with progression to vision loss from the two advanced forms of AMD: geographic atrophy and CNV.29 Geographic atrophy is characterized by gradually increasing areas of photoreceptor, RPE, and choroidal atrophy. CNV is characterized by the growth of new blood vessels frequently derived from the choroidal vasculature, into the retina, which then leak (producing edema), bleed, and gliose to form a macular scar.
A considerable amount is known of the pathophysiological processes that underlie AMD. Although the primary initiating defect is still to be identified, current theories support a number of interacting processes, namely, inflammation in the retina,58,111,112 mitochondrial dysfunction,113,114 oxidative stress,115–117 deficient choroidal blood flow in the macula,118,119 an abnormal Bruch’s membrane,120,121 metabolic dysfunction of the RPE,122 retinal defects,75,123–127 and chronic infection(s).128,129
PATHOGENESIS OF CHOROIDAL NEOVASCULARIZATION
Angiogenesis
Angiogenesis describes the formation of new capillaries from preexisting vessels. It is one of the most important biological processes involved in, for example, embryogenesis and wound repair.130 Abnormal angiogenesis, neovascularization, is usually harmful, and a sophisticated system of interrelated and interacting proteins has evolved to manage this. Angiogenesis is controlled by interlinked pathways of genes, transcription factors, secreted factors, receptors, and second messengers that exert either a proangiogenic or antiangiogenic influence in a tissue-specific fashion. It seems the “angiogenic switch” that initiates the process depends upon the balance between these factors.
Key Cellular and Molecular Events in Angiogenesis
The inner wall of blood vessels comprises a single layer of endothelial cells. Angiogenesis begins with vessel dilation and vessel wall permeability. The surrounding extracellular matrix is then degraded accompanied by endothelial cell proliferation and migration into adjacent tissues. The neovascular cells then organize into tubes.
Choroidal Neovascularization in AMD
It is known that choroidal capillaries proliferate and penetrate Bruch’s membrane to form a fibrovascular “membrane” external to the RPE, which may then extend through the RPE into the subretinal space. In some instances, connections are made with the retinal circulation or indeed develop from the retina.131 The new blood vessels show increased permeability, which can lead to accumulation of serous fluid or blood under the RPE or between the RPE and the sensory retina. The process is accompanied by inflammation, hemorrhage, and progressive fibrosis with resultant significant loss of vision. Involution of the new vessels is accompanied by fibrous metaplasia and organization that can result in an elevated subretinal mass called a disciform scar (disciform macular degeneration).132
The precise sequence of molecular signals that precede CNV is unknown. However, key to the immediate development and maintenance of CNV, and possibly reflecting a final common pathway for disease development, is the proangiogenic factor VEGF. Numerous online databases and resources that provide excellent visual summaries of angiogenesis are available (for example, http://www.ncbi.nlm.nih.gov/images) and are therefore not replicated here. A list of genes/proteins implicated in angiogenesis is shown in Table 2.
Vascular Endothelial Growth Factor
VEGF133–135 is now considered the major factor mediating CNV in exudative AMD. VEGF is a constitutively produced secreted protein.136,137 VEGF gene expression is controlled in large part by hypoxia-inducible factor (HIF) complex. If tissue oxygen saturation is normal, the regulatory α-subunit of the protein is efficiently degraded by HIF prolyl hydroxylase (PHD) enzymes.138,139 However, if oxygen concentrations drop, PHDs are inhibited, HIF-α is stabilized, and its levels rise,140 increasing its binding to a genomic DNA sequence, the hypoxia response element (HRE).141 The HRE up-regulates a number of other genes, including plasminogen activator inhibitor-1 and transforming growth factor-β3. Recently, a new regulator of VEGF activity has been identified.142
The proangiogenic activity of VEGF revolves around its ability to promote endothelial cell proliferation and survival. VEGF is also proinflammatory, increases vascular permeability (principally venules and capillaries),143 and orchestrates the activity of a number of downstream factors. In the retina, VEGFA appears most important.144 VEGFA is alternately spliced into four isoforms,145 which are coupled to two receptors, VEGFR-1 (Flt-1) and VEGFR-2 (kinase insert domain-containing receptor, KDR).146 Most of the angiogenic activity appears to be mediated through VEGFR-2.147 It is important to note that increased VEGF levels alone do not appear to be sufficient to encourage CNV. Instead, an accompanying defect in the Bruch’s membrane or defective RPE function is required.148
Angiopoietins
Angiopoietin-1 (ang-1) acts through the Tie2 receptor system with ang-2 to stabilize developing neovascularization in the presence of VEGF.149–152 Interestingly, ang-1 appears to antagonize the proinflammatory activity of VEGF.153
Pigment Epithelium–Derived Factor (PEDF) and Thrombospondin-1 (TSP-1)
Studies indicate a largely inverse relationship between PEDF and VEGF levels. PEDF appears to be the major inhibitor of VEGF-mediated endothelial proliferation,154 and its down-regulation appears to encourage retinal neovascularization.155 Its major physiological role appears to be in maintaining the avascular nature of such structures as the cornea and vitreous.156,157
In exudative AMD, evidence suggests that vitreous PEDF levels are lower than in normal patients.158 PEDF levels appear to be regulated by VEGF through feedback mechanisms at the protein and possibly the genomic level. Interestingly, the antiangiogenic activity of PEDF seems to be exerted only at the early stage of neovascularization.156,159,160 Paradoxically, increasing levels later in the process appear to augment neovascularization.161 TSP-1, another antiangiogenic factor,159,162 acts similarly to PEDF to encourage endothelial cell apoptosis.
Fibroblast Growth Factor, Connective Tissue Growth Factor, and Transforming Growth Factor
Basic fibroblast growth factor (bFGF), a member of the heparin-binding growth factor family, encourages endothelial cell survival and therefore persistence of neovascularization.163 During CNV, bFGF is produced by the RPE and choroid.164 bFGF works via receptors FGFR1 and 2, which in their turn are coupled to a number of downstream intermediates.165 Connective tissue growth factor (CTGF) and transforming growth factor are all secreted by RPE cells in CNV, and they act on fibroblasts to increase VEGF expression.166,167
Extracellular Matrix and Cell-Adhesion Molecules
A requirement for the development of new capillary vessels is the remodeling of the extracellular matrix.168 For example, without appropriate adhesion to the extracellular matrix, endothelial cells undergo apoptosis. A large number of investigations have identified players such as matrix metalloproteinases (MMPs) and their inhibitors, complement C3b and C5-9 complexes,169 I-CAM, integrins, and leukocyte adhesion molecules,170 vibronectin and laminin-1.171,172
Chemokines, Inflammation, Clotting
The chemokines are secreted inflammatory cytokines that recruit macrophages and other leukocytes, which in turn secrete such factors as VEGF.173 One such chemotactic protein is MCP-1, secreted by the RPE. Mice deficient in MCP-1 have a predilection for developing CNV.174,175 Microglia also become activated as part of the pathology in CNV, releasing, among other factors, CX3CR1. In animal models this protein accelerates choroidal neovascular processes.76 Platelet factor-4 (PF-4) is a platelet component secreted during activation which may interfere receptor binding of bFGF and VEGF to their receptors.176 The angiostatic proteins angiostatin and endostatin are cleavage products of plasminogen, collagen XVIII, and MMP-2. They act through different mechanisms to PEDF and TSP-1 and exhibit chemotactic activity as well.177
Nitric Oxide
In AMD, acute rises in VEGF produce increases in inducible nitric oxide species (NOS) through activation of tyrosine and PI-3K kinases. Chronic VEGF exposure increases expression of nitric oxide synthetase. Increased NOS results in vascular permeability and angiogenesis.178
METHODS
INSTITUTIONAL REVIEW BOARD/ETHICS COMMITTEE APPROVAL AND TRIAL REGISTRATION
This study was approved by the institutional review boards of the participating clinical sites. Prior to commencement, the study was submitted and approved as an Investigational New Drug application to the FDA (FDA IND 100 451) and registered with ClinicalTrials.gov (Identifier: NCT00469352).
Data accumulation conformed to all Federal and State laws and was compliant with HIPAA guidelines (http://www.hhs.gov/ocr/hipaa/privacy.html). The conduct of this study was overseen by a Data Safety Monitoring Committee.
CLINICAL TRIAL DESIGN SUMMARY AND DATA MONITORING
The Lucentis Genotype Study (LGS) was a two-site prospective open-label observational study of patients newly diagnosed with exudative or neovascular AMD undergoing intravitreal ranibizumab therapy. Participants were enrolled consecutively. There were two participating sites. Participants were enrolled exclusively in this study and were not part of any other interventional study.
Blood was drawn and DNA extracted for genotyping. The primary (change in visual acuity) and secondary outcome measures were analyzed with genotype to evaluate for potential pharmacogenetic interactions while controlling for demographic, phenotypic, and environmental factors.
Monthly visit information was documented in the patient’s chart and on custom-designed case report forms. The case report forms were stored in secure, locked offices in compliance with institutional regulations. The study was conducted as shown in Table 3. Additional testing was allowed at the discretion of the treating physician. Subjects who withdrew from the study prior to completion were asked to return for an early termination evaluation 30 days (±7 days) following the last injection/study visit to monitor all adverse events (serious and nonserious). The schedule of assessments for early termination was the same as that for the final visit.
TABLE 3.
CLINICAL ASSESSMENT PROTOCOL AND TREATMENT REGIMEN IN THE LUCENTIS GENOTYPE STUDY
| VISIT (MONTH) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ASSESSMENTS | SCREEN | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| Informed consent | X | ||||||||||||
| Demographic data | X | ||||||||||||
| Physical examination and medical history | X | ||||||||||||
| Vital signs* | X | X | X | X | X | X | X | X | X | X | X | X | X |
| Height and weight | X | ||||||||||||
| Environmental risk factor questionnaire | X | ||||||||||||
| Blood taken for genotyping | X | ||||||||||||
| EDTRS protocol visual acuity and refraction | X | X | X | X | X | X | X | X | X | X | X | X | X |
| Full dilated slit-lamp examination | X | X | X | X | X | X | X | X | X | X | X | X | X |
| Optical coherence tomography | X | X | X | X | X | X | X | X | X | X | X | X | X |
| Fluorescein angiography | X | X | X | X | X | ||||||||
| Ranibizumab treatment | X | X | X† | X† | X† | X† | X† | X† | X† | X† | X† | X† | X† |
| SAE monitoring | X | X | X | X | X | X | X | X | X | X | X | X | X |
SAE, serious adverse event.
Pulse, blood pressure, temporal and respiration rate.
As determined by re-treatment criteria.
All information collected in this study was fully monitored by an independent data monitor who was masked to the information. Data were doubled-entered onto custom Excel spreadsheets. Reanalysis of 10% of all data entries showed a 0% error rate.
INCLUSION AND EXCLUSION CRITERIA
Patients (>55 years of age) enrolled had never received treatment of any type (laser therapy, PDT, intravitreal therapy) for neovascular AMD in the study eye. Visual acuity in the study eye had to be between 20/30 and 20/320 (ETDRS). All AMD-related CNV lesion types were included. Pigment epithelial detachments without evidence of CNV were not included in this study.
Patients were excluded if the study eye had CNV from causes other than AMD, concomitant non-AMD-related maculopathy, or other visual pathology resulting in vision loss. Individuals were also excluded if ranibizumab was contraindicated or ranibizumab/bevacizumab was being used in the fellow eye at the time of enrollment. If treatment for neovascular AMD became necessary in the fellow eye during the study, this was not considered a reason for termination from the study and Lucentis was given.
RANIBIZUMAB TREATMENT REGIMEN
Table 3 shows the treatment and follow-up schedule. Participants received multiple open-label intravitreal injections of 0.5 mg ranibizumab administered according to the following re-treatment criteria, which reflected on-label recommendations and “standard of care” at the time of the study.
Lucentis Re-treatment Criteria
Criteria were as follows: presence of intraretinal fluid, subretinal fluid, or retinal thickening on OCT; presence of new subretinal hemorrhage; active, new subretinal choroidal neovascular membrane documented on fluorescein angiography; visual acuity ≥5 ETDRS letter reduction in best-corrected refracted visual acuity as compared with prior visit.
Concomitant and Excluded Therapies
Patients continued oral AREDS supplements as well as all other medications and standard treatments prescribed by their physician(s). Patients requiring therapy for AMD other than ranibizumab during the study period continued in the study but were not analyzed in the final data set. Patients that developed disease in the fellow eye received ranibizumab.
DATA COLLECTION AND MASKING
This was an open-label study. However, treating physicians and study coordinators were masked to genotype throughout the study. At each visit, visual acuity testers and photographers were masked as to whether intravitreal therapy would be given.
CLINICAL EVALUATION
Phenotyping
A full dilated ophthalmic examination was performed and documented at each visit by the treating retinal specialist investigator. Clinical appearances of both maculae were detailed.
Color fundus photography and fluorescein angiography was performed in standard fashion. Appearances of the posterior pole of both eyes were documented. Choroidal neovascularization was categorized as follows: 100% classic, predominantly classic (>50% classic), minimally classic (<50% classic), or occult.
Vertical and horizontal line scans and macular volume scans were obtained using Stratus 3 OCT. Central macular thickness, macular volume, and the presence of retinal fluid or thickening was documented. The spectral domain OCT was not available at the commencement of this study.
AMD status of the fellow eye was collected from fundus photographs and clinical examination using a modified AREDS grading system as follows: category 1 (no AMD): no drusen or drusen less than 63 μm diameter, and no pigment changes in either eye; category 2: mild to moderate drusen consisting of drusen of any size, but <393,744 μm2 in total area within 1500 μm from the fovea, with or without pigment changes; category 3: extensive large drusen (>125 μm in minimum diameter) >393,744 μm2 in area (a minimum of approximately 20 large drusen) within 1500 μm of the fovea, with or without pigment changes and no evidence of advanced AMD; category 4 (advanced AMD): presence in one or both eyes of advanced macular degeneration (CNV or geographic atrophy).
Systemic Evaluation
At each visit, pulse and blood pressure were recorded.
Environmental Factors and Personal Characteristics
Patients were administered a standardized validated questionnaire at the initial visit, which included documentation of (a) medical status, including cardiovascular assessments; (b) smoking history; (c) body mass index; (d) education level; (e) diet; (f) use of nutritional supplements, including the AREDS formulation; (g) medicinal use, including statins and nonsteroidal anti-inflammatory drugs; (h) cardiovascular history; (i) family history of AMD; (j) past ocular history, including cataract and glaucoma status. Each was reviewed at every scheduled and nonscheduled visit and updated as necessary.
GENOTYPING
At the enrollment visit, consented subjects provided a 20-mL venous blood sample for genetic analysis obtained from a peripheral vein by a trained venesector. DNA was extracted using standard methods. Genotyping was undertaken using Illumina’s Human 660W-Quad high-density SNP chip (Illumina, San Diego, California). This chip interrogates ~550,000 SNP variants per sample.
OUTCOME MEASURES
The primary outcome measure was change in best-corrected ETDRS letter score in the treated eye at the primary end point of 12 months.
Secondary end points were as follows:
Change in ETDRS letter score at 6 months;
Number of injections received at 12 months;
Change in central macular thickness (microns) at 6 and 12 months;
Angiographic evidence of persistent leakage at 6 and 12 months.
Additional parameters evaluated were:
Presenting visual acuity (best-corrected ETDRS letter score);
Fluorescein angiographic lesion characteristics at baseline;
Baseline OCT central macular thickness (microns);
High-sensitivity C-reactive protein (CRP, mg/L) level at baseline.
STATISTICAL ANALYSIS
Quality Control of Genetic Data
Missing data
Analyses were based on available cases, without imputation for missing values.
Interim analyses
No interim analyses were performed. Reports of adverse events were reviewed and summarized periodically while the study was ongoing for the purposes of Data Safety Monitoring review to ensure the safety of subjects.
Estimation of allele frequencies and elimination of problematic SNPs
Estimates of allele frequencies and their standard errors were obtained, and the Hardy-Weinberg equilibrium was calculated. Genome-wide association SNPs that failed quality control were dropped from further analysis for the following reasons: too many failed calls or mendelian errors, deviation from Hardy-Weinberg equilibrium (P<0.001), and too few copy numbers (n<5).
Imputation of missing genotypes
If it was necessary to impute missing genotypes to avoid recalculating the null distribution for each SNP, the likelihood-based imputation procedures MERLIN179 and BEAGLECALL180 were employed.
Correction for hidden stratification
We included a correction for hidden stratification that uses principal components of the observed SNP variation.181
SNPs Chosen for Analysis
All SNPs meeting the quality standards above with a minor allele frequency of 0.05 or greater were included in the analyses, which were conducted in three phases. The following databases and bioinformatics resources were utilized to select and evaluate SNPs:
| Genome Variation Server build 131beta | http://gvs.gs.washington.edu/GVS131 |
| GenGen Genetic Genome Analysis of Complex Data | http://www.openbioinformatics.org/gengen/index.html |
| Genecards | http://www.genecards.org |
| HapMap | http://hapmap.ncbi.nlm.nih.gov/ |
| NCBI Entrez Gene | http://www.ncbi.nlm.nih.gov/gene |
| NCBI Entrez SNP | http://www.ncbi.nlm.nih.gov/SNP |
Genotype analyses were performed using HelixTree by Golden Helix (http://www.goldenhelix.com), including the Whole Genome and Regression modules. SNP imputation was aided by BEAGLECALL.180
Phase I: Validation Analysis
The chi-square test was used to identify differences between the personal characteristics, environmental risk factors, and AMD-susceptibility variants (CFH rs106117050–52 and ARMS2 rs1049092446,47,64–70,182) in participants in the study and individuals in a comparable population: those with CNV in the preexisting Casey Eye Institute sporadic case-control (CEIMDC) population. Briefly, the CEIMDC cohort was ascertained over an approximately 10-year period of patients presenting to Casey Eye Institute Retina Service with advanced AMD, either geographic atrophy or CNV.
Phase II: Treatment Response Analysis Among Candidate Genes
The algorithm described in the “Introduction” section was used to construct a candidate gene list comprising known AMD-susceptibility genes and genes involved in the control of angiogenesis. SNPs within each coding region were identified (including untranslated regions and ±1000 bases from transcription initiation and termination).
Regression analysis assuming an additive model for each SNP was performed for all end points. For those variables that had only two levels, we performed logistic regression; for those phenotypes that had more than two levels, we assumed they were continuous and performed linear regression analysis.
Analyses were conditioned upon environmental risk factor (smoking, body mass index, education level) and personal characteristics (age and gender). We considered the possibility that the secondary outcome factors are likely very interrelated, and therefore pairwise correlation between these factors was determined before inclusion in our model. If two factors are highly correlated, the one that has the most clinical relevance was included in the analysis. Forward stepwise logistic regression was then used to determine which of the remaining secondary outcome factors significantly predicted the primary outcome. Multiple testing was approached conservatively with a Bonferroni correction (assuming an α=0.05), P=0.05/number of SNPs included in the analysis. This correction is considerably more conservative than probably necessary because of the linkage disequilibrium that exists between many SNPs.
Phase III: Genome-Wide Treatment Response Analyses
The genetic contribution to each treatment response end point was then determined on a genome-wide basis. To reduce the number of SNPs included, pairwise linkage disequilibrium was calculated between SNP pairs. Where a SNP pair was correlated (r2>0.80), only one SNP was included in the regression analysis. Regression analysis was then performed for each end point measure as described in Phase II. The empirical significance level for genome-wide association studies of P<10−7 was applied.183
Phase IV: Associations With Baseline Characteristics
Regression analysis was used to determine the genetic contribution to baseline characteristics on both a candidate gene and genome-wide basis. The factors examined were presenting best-corrected ETDRS visual acuity, baseline OCT central macular thickness, CNV lesion characteristics, and baseline high-sensitivity CRP level.
RESULTS
STUDY PARTICIPANTS
A total of 65 individuals met the inclusion/exclusion criteria and were enrolled in the study. Sixty-four patients completed the study (primary end point: 12 months). One participant died of a cause unrelated to the study. Enrollment took almost 1 year due to the limited number of treatment-naïve, new-onset neovascular AMD patients presenting to the participant sites, together with other studies actively enrolling neovascular AMD patients.
All participants had new-onset neovascular AMD in the study eye. None of the study eyes had previously received treatment (thermal laser, PDT, intravitreal therapy) for neovascular AMD. In each case, vision loss was determined by the investigators, at presentation, to have occurred as a result of neovascular AMD, and no other etiology was evident. Only one eye per patient was enrolled in the study.
The fluorescein angiographic lesion characteristics of study eyes were as follows: occult without classic (21, 32%), minimally classic (21, 32%), predominantly classic (19, 29%), and classic (4, 7%) CNV. At the end of the study, 38% of individuals had no angiographic or OCT evidence of persistent leakage or active CNV.
Independent of genotype, individuals in the study presented with a mean best-corrected visual acuity of 60.23 ETDRS letters (SD ±15.4). During the course of the study, visual acuity improved by 3.84 (±9.02) letters at 6 months and by 5.8 letters (±9.6) at 12 months. Mean baseline central macular thickness on OCT was 329.74 (±89.74) μm at baseline and decreased by a mean of 107.62 (±115.54) μm at 6 months and 92.26 (±140.96) μm at 12 months. The mean number of injections required was 6.19 (±3.26). CRP level at baseline was 0.31 mg/L (±0.27).
STUDY INTERVENTION AND ADVERSE EVENTS
LGS participants received a median number of six injections during the study (not including the initial injection at baseline). No injections were declined. No other therapies were required, and there were no reports from treating physicians of deviations from the re-treatment criteria.
Since patients were receiving on-label therapy with ranibizumab, strictly, the “intervention” in this study was venepuncture. There were no adverse events relating to this procedure. One patient died during the study period of a cause unrelated to his ocular status or treatment. No other adverse events (Grade 2, NCI grading system or greater) were recorded. Other mild adverse events were considered unrelated to study medication or the protocol.
GENOTYPING AND QUALITY CONTROL
The Illumina (San Diego, California) 660-Quad SNP chip genotyped >550,000 SNPs in 44 individuals, and of these, 66,405 failed quality control and were excluded from further analysis (477 were out of Hardy-Weinberg equilibrium while the rest had either a minor allele frequency <0.05 or had too many failed calls).
Analysis was conducted in four phases. In Phase I, differences between the enrolled population and a similar population were investigated to determine how representative the LGS participants were of newly diagnosed neovascular AMD in general. In Phase II, a candidate gene approach was utilized to reduce the correction needed for multiple testing. This included the primary end point analysis of change in visual acuity at the end of the study (12 months). In Phase III, all SNPs in the genome scan were included. Phase IV examined all SNPs, both candidates and genome-wide association, as to whether they influenced baseline characteristics of participants.
PHASE I: VALIDATION ANALYSIS
Table 4 shows summary demographic, environmental exposure, and genetic risk factor data for the individuals in the LGS. When compared with a similar hospital, academic practice population (the CEI sporadic cases with CNV), the LGS participants were similar in age (marginally older, P=0.04) and had achieved a higher education status. In all other respects, the LGS patients were very comparable. Genotypic comparisons between LGS and CEI populations were limited to CFH and ARMS2 because the minor allele frequencies in these two genes are high enough for meaningful evaluation in the small LGS cohort. There were no significant differences between allele frequencies for these two SNPs.
TABLE 4.
SUMMARY DATA FOR DEMOGRAPHIC AND PERSONAL FACTORS FOR THE INDIVIDUALS IN THE LUCENTIS GENOTYPE STUDY (LGS) AS COMPARED WITH THE CASEY EYE INSTITUTE SPORADIC CASES WITH CHOROIDAL NEOVASCULARIZATION (CEIMDC CNV)
| DATA | LGS | CEIMDC CNV | P VALUE* | |
|---|---|---|---|---|
| Age | 81.27 | 79.20 | 0.04 | |
| Gender | ||||
| Female | 66% | 63% | ||
| Male | 34% | 37% | 0.79 | |
| Ethnicity | 100% | 100% | 1.00 | |
| Caucasian | 100% | 100% | 1.00 | |
| Non-Caucasian | 0% | 0% | ||
| Body mass index | 26.62 | 26.45 | 0.81 | |
| Smoking | ||||
| Current | 55% | |||
| Former | 55% | |||
| Never | 40% | 0.76 | ||
| AREDS supplements | ||||
| No | 44% | |||
| Yes | 56% | 0.82 | ||
| High school graduate | ||||
| No | 32% | |||
| Yes | 68% | 7.81 × 10−5 | ||
| Family history | ||||
| No | 57% | |||
| Yes | 43% | 0.64 | ||
| Genetics | ||||
| CFH rs1061170 | C | 0.58 | 0.57 | 0.95 |
| T | 0.42 | 0.43 | ||
| ARMS2 rs10490924 | G | 0.40 | 0.42 | 0.92 |
| T | 0.60 | 0.58 | ||
AREDS, Age-Related Eye Disease Study.
Significant (chi-square test) differences are shown in bold.
PHASE II: TREATMENT RESPONSE ANALYSIS AMONG CANDIDATE GENES
The following end points were investigated: best-corrected ETDRS letter score in the treated eye at 6 and 12 months, change in central macular thickness at 6 and 12 months, angiographic evidence of persistent leakage at 6 and 12 months, and number of injections received by end of study. Logistic or linear regression, as appropriate, was used to evaluate associations between these measures and a list of candidate genes considered likely to play a role in angiogenesis. The candidate gene list (Table 2) comprised known AMD-susceptibility genes and genes involved in the control of angiogenesis. The full list of SNPs investigated is shown in the Appendix.
APPENDIX.
SINGLE-NUCLEOTIDE POLYMORPHISMS SHOWING GREATEST ASSOCIATIONS IN GENOME-WIDE ANALYSES
| MARKER | CHROMOSOME | REGRESSION P | REGRESSION BONFERRONI P | PHENOTYPE | GENOTYPE | MEAN | SE |
|---|---|---|---|---|---|---|---|
| rs13421506 | 2 | 8.37036E-09 | 0.004076733 | CRP | AC | 1.03 | 0.05 |
| CC | 0.25 | 0.01 | |||||
| rs2231153 | 4 | 8.37036E-09 | 0.004076733 | CRP | AA | 0.25 | 0.01 |
| AG | 1.03 | 0.05 | |||||
| rs2725267 | 4 | 1.33361E-08 | 0.006495255 | CRP | AA | 0.26 | 0.01 |
| AG | 1.03 | 0.05 | |||||
| rs17384909 | 10 | 1.59626E-08 | 0.007774476 | CRP | AG | 1.03 | 0.05 |
| GG | 0.25 | 0.01 | |||||
| rs9675979 | 18 | 3.76321E-08 | 0.018328929 | Baseline.VA | AG | 40.00 | 1.22 |
| GG | 67.62 | 0.38 | |||||
| rs2298515 | 21 | 3.84873E-08 | 0.01874545 | X12.Month.Dif.CMT | AA | 481.00 | 0.00 |
| AC | 76.50 | 14.17 | |||||
| CC | −126.65 | 3.14 | |||||
| rs13029479 | 2 | 3.13855E-07 | 0.152861065 | CRP | AG | 0.82 | 0.06 |
| GG | 0.22 | 0.01 | |||||
| rs1205091 | 14 | 4.81169E-07 | 0.234356416 | Baseline.VA | AA | 39.00 | 2.33 |
| AG | 57.41 | 0.74 | |||||
| GG | 72.92 | 0.63 | |||||
| rs2107332 | 20 | 5.61572E-07 | 0.273517184 | X12.Month.Dif.CMT | AA | −125.52 | 3.44 |
| AG | 70.00 | 32.53 | |||||
| GG | 481.00 | 0.00 | |||||
| rs8060546 | 16 | 5.67506E-07 | 0.2764074 | X12.Month.Dif.CMT | AA | 481.00 | 0.00 |
| AC | 27.83 | 20.84 | |||||
| CC | −130.59 | 3.17 | |||||
| rs12969428 | 18 | 9.67362E-07 | 0.47115959 | X12.Month.Dif.CMT | AA | 481.00 | 0.00 |
| AG | 59.67 | 34.55 | |||||
| GG | −118.72 | 3.23 | |||||
| rs1349916 | X | 9.77933E-07 | 0.476308099 | X12.Month.Dif.CMT | AA | −114.82 | 3.23 |
| AG | 84.50 | 22.27 | |||||
| GG | 481.00 | 0.00 | |||||
| rs3113392 | 4 | 1.33312E-06 | 0.64930648 | Baseline.VA | AA | 43.11 | 1.44 |
| AG | 60.29 | 0.77 | |||||
| GG | 74.50 | 0.73 | |||||
| rs12960119 | 18 | 1.61921E-06 | 0.788626635 | CRP | AA | 0.22 | 0.01 |
| AG | 0.75 | 0.05 | |||||
| rs4800723 | 18 | 1.61921E-06 | 0.788626635 | CRP | AA | 0.22 | 0.01 |
| AG | 0.75 | 0.05 | |||||
| rs765529 | 18 | 1.61921E-06 | 0.788626635 | CRP | AA | 0.22 | 0.01 |
| AG | 0.75 | 0.05 | |||||
| rs8095871 | 18 | 1.61921E-06 | 0.788626635 | CRP | AG | 0.75 | 0.05 |
| GG | 0.22 | 0.01 | |||||
| rs9930491 | 16 | 1.61921E-06 | 0.788626635 | CRP | AG | 0.75 | 0.05 |
| GG | 0.22 | 0.01 | |||||
| rs1358889 | 9 | 1.64206E-06 | 0.799775421 | X12.Month.Dif.CMT | AA | −156.57 | 4.62 |
| AC | −34.46 | 6.38 | |||||
| CC | 298.50 | 129.05 | |||||
| rs9302755 | 16 | 1.87949E-06 | 0.915415745 | Baseline.CMT | AA | 438.43 | 6.94 |
| AC | 343.69 | 5.19 | |||||
| rs9302755 | 16 | 1.87949E-06 | 0.915415745 | Baseline.CMT | CC | 257.85 | 4.44 |
| rs9549123 | 13 | 2.16654E-06 | 1 | X12.Month.Dif.CMT | AA | 481.00 | 0.00 |
| AC | 16.67 | 18.19 | |||||
| CC | −128.28 | 3.42 | |||||
| rs11717033 | 3 | 2.38988E-06 | 1 | X12.Month.Dif.CMT | AA | −117.94 | 3.02 |
| AG | 251.00 | 66.73 | |||||
| rs3212240 | 14 | 2.73303E-06 | 1 | Baseline.CMT | AA | 415.00 | 6.47 |
| AG | 309.94 | 4.06 | |||||
| GG | 238.71 | 7.39 | |||||
| rs3757105 | 6 | 3.23771E-06 | 1 | X12.Month.Dif.CMT | AA | 481.00 | 0.00 |
| AG | 41.33 | 36.21 | |||||
| GG | −117.00 | 3.31 | |||||
| rs3823118 | 6 | 3.23771E-06 | 1 | X12.Month.Dif.CMT | AA | 481.00 | 0.00 |
| AG | 41.33 | 36.21 | |||||
| GG | −117.00 | 3.31 | |||||
| rs1150345 | 11 | 3.82051E-06 | 1 | Baseline.VA | AA | 34.50 | 3.89 |
| AC | 50.87 | 1.03 | |||||
| CC | 69.79 | 0.47 | |||||
| rs618513 | 11 | 3.82161E-06 | 1 | X12.Month.Dif.CMT | AA | 481.00 | 0.00 |
| AG | −0.38 | 8.10 | |||||
| GG | −133.96 | 3.98 | |||||
| rs1205106 | 14 | 4.30942E-06 | 1 | Baseline.VA | AA | 72.08 | 0.67 |
| AG | 58.83 | 0.76 | |||||
| GG | 39.00 | 2.33 | |||||
| rs10126713 | X | 4.31516E-06 | 1 | X6.Month.Dif.VA | AA | −4.17 | 0.56 |
| AG | 5.50 | 0.37 | |||||
| GG | 13.83 | 1.39 | |||||
| rs1050115 | 2 | 4.35301E-06 | 1 | X12.M.Fluid..Y.N. | AA | NA | NA |
| GG | NA | NA | |||||
| AG | NA | NA | |||||
| rs2842276 | 10 | 5.92277E-06 | 1 | X12.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs7622700 | 3 | 6.06553E-06 | 1 | Leakage.Month.12 | AA | NA | NA |
| AC | NA | NA | |||||
| CC | NA | NA | |||||
| rs11997272 | 8 | 6.37223E-06 | 1 | X6.Month.Dif.CMT | AA | 222.00 | 0.00 |
| AC | −6.88 | 10.08 | |||||
| CC | −147.37 | 3.42 | |||||
| rs10188066 | 2 | 6.49761E-06 | 1 | X12.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs1469996 | 2 | 6.49761E-06 | 1 | X12.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs6430585 | 2 | 6.49761E-06 | 1 | X12.M.Fluid..Y.N. | AA | NA | NA |
| AC | NA | NA | |||||
| CC | NA | NA | |||||
| rs9287442 | 2 | 6.49761E-06 | 1 | X12.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs12662871 | 6 | 6.596E-06 | 1 | Baseline.VA | AA | 67.42 | 0.47 |
| AG | 46.27 | 1.31 | |||||
| GG | 31.00 | 0.00 | |||||
| rs7607942 | 2 | 6.69616E-06 | 1 | X6.Month.Dif.VA | AA | −2.88 | 0.38 |
| AG | 8.75 | 0.48 | |||||
| GG | 13.67 | 1.39 | |||||
| rs4429936 | 6 | 7.0178E-06 | 1 | X6.Month.Dif.VA | AA | −5.67 | 0.59 |
| AG | 1.12 | 0.47 | |||||
| GG | 11.31 | 0.52 | |||||
| rs4543241 | 5 | 7.1611E-06 | 1 | Baseline.CMT | AA | 398.81 | 4.89 |
| AG | 291.53 | 4.52 | |||||
| GG | 233.20 | 9.75 | |||||
| rs1952442 | 14 | 7.53852E-06 | 1 | Baseline.CMT | AA | 458.67 | 23.14 |
| AG | 375.80 | 5.45 | |||||
| GG | 272.61 | 3.49 | |||||
| rs883159 | 14 | 7.53852E-06 | 1 | Baseline.CMT | AA | 458.67 | 23.14 |
| AG | 375.80 | 5.45 | |||||
| GG | 272.61 | 3.49 | |||||
| rs7316876 | 12 | 7.80046E-06 | 1 | X6.Month.Dif.CMT | AA | −276.75 | 16.14 |
| AG | −145.86 | 5.82 | |||||
| GG | −36.83 | 5.64 | |||||
| rs4805784 | 19 | 7.98712E-06 | 1 | Baseline.VA | AA | 51.19 | 0.70 |
| AC | 68.70 | 0.83 | |||||
| CC | 79.20 | 0.55 | |||||
| rs4877042 | 9 | 8.20526E-06 | 1 | X6.Month.Dif.VA | AA | −0.40 | 0.28 |
| AG | 12.91 | 0.61 | |||||
| GG | 0.00 | 0.00 | |||||
| rs5920818 | X | 8.52893E-06 | 1 | X6.Month.Dif.VA | AA | 13.83 | 1.39 |
| AG | 5.05 | 0.36 | |||||
| GG | −4.27 | 0.64 | |||||
| rs5967094 | X | 8.52893E-06 | 1 | X6.Month.Dif.VA | AA | −4.27 | 0.64 |
| AG | 5.05 | 0.36 | |||||
| GG | 13.83 | 1.39 | |||||
| rs209957 | 20 | 8.54623E-06 | 1 | Baseline.CMT | AA | 287.25 | 3.16 |
| AG | 418.83 | 4.98 | |||||
| rs11022983 | 11 | 8.65993E-06 | 1 | Baseline.VA | AG | 38.14 | 1.79 |
| GG | 65.21 | 0.42 | |||||
| rs12146602 | 11 | 8.65993E-06 | 1 | Baseline.VA | AG | 38.14 | 1.79 |
| GG | 65.21 | 0.42 | |||||
| rs10828564 | 10 | 8.979E-06 | 1 | X6.Month.Dif.CMT | AA | −151.22 | 3.18 |
| AG | 30.11 | 11.49 | |||||
| rs6892289 | 5 | 9.04772E-06 | 1 | Baseline.VA | AA | 66.58 | 0.48 |
| AG | 42.70 | 1.15 | |||||
| rs1542409 | 13 | 9.62144E-06 | 1 | X12.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs2576060 | 18 | 1.00522E-05 | 1 | Leakage.Month.12 | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs2068967 | 21 | 1.0656E-05 | 1 | X12.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs4428995 | 10 | 1.13512E-05 | 1 | Leakage.Month.12 | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs7268671 | 20 | 1.1412E-05 | 1 | X12.Month.Dif.VA | AA | −0.33 | 0.26 |
| AG | 14.50 | 0.66 | |||||
| GG | 28.00 | 0.00 | |||||
| rs8115510 | 20 | 1.1412E-05 | 1 | X12.Month.Dif.VA | AA | −0.33 | 0.26 |
| AG | 14.50 | 0.66 | |||||
| GG | 28.00 | 0.00 | |||||
| rs7703021 | 5 | 1.16387E-05 | 1 | X6.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs9368215 | 6 | 1.23089E-05 | 1 | X6.Month.Dif.VA | AA | −7.25 | 0.83 |
| AG | 0.06 | 0.48 | |||||
| GG | 10.00 | 0.45 | |||||
| rs7429875 | 3 | 1.25265E-05 | 1 | Baseline.CMT | AA | 395.71 | 4.73 |
| AG | 276.39 | 3.55 | |||||
| GG | 218.00 | 0.00 | |||||
| rs10501500 | 11 | 1.26442E-05 | 1 | X6.Month.Dif.VA | AA | 6.89 | 0.27 |
| AG | −7.63 | 0.55 | |||||
| rs10211519 | 2 | 1.27628E-05 | 1 | X6.Month.Dif.CMT | AA | −171.59 | 5.24 |
| AG | −77.19 | 5.65 | |||||
| GG | 113.33 | 32.54 | |||||
| rs1157907 | 2 | 1.27628E-05 | 1 | X6.Month.Dif.CMT | AA | −171.59 | 5.24 |
| AG | −77.19 | 5.65 | |||||
| GG | 113.33 | 32.54 | |||||
| rs6432474 | 2 | 1.27628E-05 | 1 | X6.Month.Dif.CMT | AA | −171.59 | 5.24 |
| AC | −77.19 | 5.65 | |||||
| CC | 113.33 | 32.54 | |||||
| rs6724694 | 2 | 1.27628E-05 | 1 | X6.Month.Dif.CMT | AA | −171.59 | 5.24 |
| AG | −77.19 | 5.65 | |||||
| GG | 113.33 | 32.54 | |||||
| rs1459019 | 10 | 1.28914E-05 | 1 | Leakage.Month.12 | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs150892 | 1 | 1.37412E-05 | 1 | X12.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs223248 | 1 | 1.37412E-05 | 1 | X12.M.Fluid..Y.N. | AA | NA | NA |
| AC | NA | NA | |||||
| CC | NA | NA | |||||
| rs730005 | 2 | 1.37944E-05 | 1 | Leakage.Month.12 | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs7614432 | 3 | 1.41381E-05 | 1 | Baseline.CMT | AA | 305.03 | 2.52 |
| AG | 483.40 | 8.74 | |||||
| rs6056327 | 20 | 1.41852E-05 | 1 | Leakage.Month.12 | AG | NA | NA |
| GG | NA | NA | |||||
| rs12599487 | 16 | 1.454E-05 | 1 | Leakage.Month.12 | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs2932126 | 18 | 1.48876E-05 | 1 | Leakage.Month.12 | AA | NA | NA |
| AC | NA | NA | |||||
| CC | NA | NA | |||||
| rs10493631 | 1 | 1.49337E-05 | 1 | X6.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs1029236 | 3 | 1.51151E-05 | 1 | X6.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs9665534 | 10 | 1.54575E-05 | 1 | Leakage.Month.12 | AA | NA | NA |
| AC | NA | NA | |||||
| CC | NA | NA | |||||
| rs7611945 | 3 | 1.56208E-05 | 1 | X6.Month.Dif.VA | AA | 18.00 | 0 |
| AG | 11.55 | 0.58 | |||||
| GG | −0.54 | 0.31 | |||||
| rs1050115 | 2 | 1.61014E-05 | 1 | X12.M.Fluid..Y.N. | AG | NA | NA |
| AA | NA | NA | |||||
| GG | NA | NA | |||||
| rs3762096 | 10 | 1.61644E-05 | 1 | Baseline.CMT | AA | 454.00 | 41.76 |
| AG | 387.00 | 5.92 | |||||
| GG | 281.62 | 3.18 | |||||
| rs10911048 | 1 | 1.621E-05 | 1 | X12.Month.Dif.VA | AA | −8.67 | 1.39 |
| AG | 1.67 | 0.34 | |||||
| GG | 11.63 | 1.05 | |||||
| rs1697143 | 5 | 1.80828E-05 | 1 | X6.Month.Dif.CMT | AA | −207.00 | 1.41 |
| AC | −173.78 | 4.55 | |||||
| CC | −16.88 | 6.35 | |||||
| rs6007260 | 22 | 1.81646E-05 | 1 | X12.Month.Dif.VA | AA | −7.00 | 1.05 |
| AG | 3.00 | 0.31 | |||||
| GG | 15.75 | 2.16 | |||||
| rs7030915 | 9 | 1.89786E-05 | 1 | X6.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs7763304 | 6 | 2.03898E-05 | 1 | X6.Month.Dif.VA | AA | −8.00 | 1.20 |
| AG | −4.88 | 0.66 | |||||
| GG | 8.00 | 0.34 | |||||
| rs2036826 | 8 | 2.10215E-05 | 1 | X12.Month.Dif.VA | AA | 7.88 | 0.51 |
| AG | −0.07 | 0.47 | |||||
| GG | −12.25 | 1.82 | |||||
| rs10212894 | 4 | 2.22367E-05 | 1 | X6.Month.Dif.CMT | AA | 119.00 | 72.83 |
| AG | −67.78 | 5.46 | |||||
| GG | −183.67 | 5.49 | |||||
| rs1865178 | 4 | 2.24963E-05 | 1 | X12.Month.Dif.VA | AA | −2.89 | 0.45 |
| AG | 5.47 | 0.44 | |||||
| GG | 23.00 | 3.54 | |||||
| rs7671764 | 4 | 2.24963E-05 | 1 | X12.Month.Dif.VA | AA | −2.89 | 0.45 |
| AG | 5.47 | 0.44 | |||||
| GG | 23.00 | 3.54 | |||||
| rs11637483 | 15 | 2.31894E-05 | 1 | X6.Month.Dif.CMT | AA | 222.00 | 0.00 |
| AG | 14.20 | 14.77 | |||||
| GG | −136.83 | 3.21 | |||||
| rs4744628 | 9 | 2.36202E-05 | 1 | Baseline.CMT | AA | 273.70 | 3.31 |
| AG | 401.15 | 5.92 | |||||
| GG | 410.33 | 20.13 | |||||
| rs925669 | 4 | 2.53075E-05 | 1 | X6.M.Fluid..Y.N. | AA | NA | NA |
| rs925669 | 4 | 2.53075E-05 | 1 | X6.M.Fluid..Y.N. | AG | NA | NA |
| GG | NA | NA | |||||
| rs12515335 | 5 | 2.58637E-05 | 1 | X6.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs238228 | 17 | 2.80628E-05 | 1 | X6.Month.Dif.VA | AA | 0.04 | 0.27 |
| AG | 13.10 | 0.76 | |||||
| rs3791935 | 2 | 2.80735E-05 | 1 | X6.Month.Dif.VA | AA | 24.00 | 0.00 |
| AG | 10.70 | 0.50 | |||||
| GG | 0.04 | 0.32 | |||||
| rs1569660 | 6 | 2.84253E-05 | 1 | X6.Month.Dif.VA | AA | 12.55 | 0.60 |
| AG | 0.84 | 0.39 | |||||
| GG | −3.67 | 1.17 | |||||
| rs4953063 | 2 | 2.91746E-05 | 1 | X12.Month.Dif.VA | AA | −8.33 | 1.91 |
| AG | 1.86 | 0.27 | |||||
| GG | 12.14 | 1.33 | |||||
| rs2272668 | 8 | 3.12468E-05 | 1 | X12.Month.Dif.VA | AA | 8.67 | 0.54 |
| AG | −1.84 | 0.38 | |||||
| GG | −19.00 | 0.00 | |||||
| rs10911044 | 1 | 3.12894E-05 | 1 | X12.Month.Dif.VA | AA | 10.00 | 0.66 |
| AG | 0.71 | 0.53 | |||||
| GG | −6.00 | 0.88 | |||||
| rs6756245 | 2 | 3.22119E-05 | 1 | X6.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs1565684 | 8 | 3.2949E-05 | 1 | X6.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| rs1190270 | 6 | 3.91556E-05 | 1 | X6.M.Fluid..Y.N. | AC | NA | NA |
| CC | NA | NA | |||||
| rs6508497 | 18 | 4.86095E-05 | 1 | X6.M.Fluid..Y.N. | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA |
The primary end point for the study was best-corrected ETDRS letter score in the treated eye at 12 months. Two genes showed statistically significant association (after correction for multiple testing) with change in visual acuity, CFH and CTGF (Table 5). Having the minor allele (A) in rs1065489 (CFH) and rs9399005 (CTGF) conferred a worse visual outcome compared with the ancestral allele. At 6 months, CFH is also significantly associated (Table 6), suggesting a consistent influence on visual improvement of variants in this gene.
TABLE 5.
CANDIDATE GENE ANALYSIS: DIFFERENCE IN BEST-CORRECTED VISUAL ACUITY AT 12 MONTHS
| GENE | SNP | CHR | POSITION | REGRESSION P VALUE | ALLELE* | MEAN† | SE |
|---|---|---|---|---|---|---|---|
| CFH | rs1065489 | 1 | 194976397 | 0.0417 | AA | −19.00 | 1.00 |
| AC | −0.40 | 1.22 | |||||
| CC | 3.39 | 0.35 | |||||
| CTGF | rs9399005 | 6 | 132310657 | 0.0294 | AA | −8.00 | 0.10 |
| AG | −2.88 | 0.58 | |||||
| GG | 6.33 | 0.47 |
Chr, chromosome on which SNP is located; SE, standard error; SNP, single-nucleotide polymorphism.
Minor alleles are shown in bold.
Mean change in ETDRS letters.
TABLE 6.
CANDIDATE GENE ANALYSIS: DIFFERENCE IN BEST-CORRECTED VISUAL ACUITY AT 6 MONTHS
| GENE | SNP | CHR | POSITION | REGRESSION P VALUE | ALLELE* | MEAN† | SE |
|---|---|---|---|---|---|---|---|
| CFH | rs3753394 | 1 | 194887540 | 0.0209 | AA | −7.00 | 1.76 |
| AG | 1.78 | 0.77 | |||||
| GG | 5.71 | 0.40 | |||||
| FLT1 | rs9319428 | 13 | 27871621 | 0.0274 | AA | −0.33 | 2.69 |
| AG | −0.06 | 0.58 | |||||
| GG | 7.19 | 0.49 | |||||
| C3 | rs1389623 | 19 | 6635197 | 0.0493 | AA | Not observed | |
| AG | 11.20 | 1.68 | |||||
| GG | 2.45 | 0.29 | |||||
Chr, chromosome on which SNP is located; SE, standard error; SNP, single-nucleotide polymorphism.
Minor alleles are shown in bold.
Mean change in ETDRS letters.
When OCT central macular thickness is examined in the same way, different candidate genes appear significant (Table 7). The results show that the minor allele of the same SNP in the gene for complement factor 3 (rs2230205) is associated with greater reduction in retinal thickness at both 6 (Table 8) and 12 months. SNPs in two other genes, thombospondin-1 (THBS1) and fibroblast growth factor receptor-2 (FGFR2), are also associated with improved treatment response to ranibizumab therapy. A SNP in FGFR2 is also associated with persistent leakage on fluorescein angiography at 12 months (Table 9), underlining the potential role of this gene in treatment response. Other genes are also noted to be associated, including SNPs in FLT1 (Tables 9 and 10).
TABLE 8.
CANDIDATE GENE ANALYSIS: CHANGE IN CENTRAL MACULAR THICKNESS AT 6 MONTHS
| GENE | SNP | CHR | POSITION | REGRESSION P VALUE | ALLELE* | MEAN† | SE |
|---|---|---|---|---|---|---|---|
| C3196 | rs2230205 | 19 | 6660704 | 0.0056 | AA | −292.00 | 12.23 |
| AG | −196.50 | 16.61 | |||||
| GG | −80.72 | 3.84 | |||||
| FGFR2209 | rs1047100 | 10 | 123288148 | AA | Not observed | ||
| 0.0439 | AG | −147.94 | 7.34 | ||||
| GG | −68.26 | 5.45 |
Chr, chromosome on which SNP is located; SE, standard error; SNP, single-nucleotide polymorphism.
Minor alleles are shown in bold.
Mean change in central macular thickness (microns) as compared with baseline.
TABLE 9.
CANDIDATE GENE ANALYSIS: FLUORESCEIN ANGIOGRAPHIC EVIDENCE OF PERSISTENT LEAKAGE FROM CHOROIDAL NEOVASCULARIZATION AT 12 MONTHS
| GENE | SNP | CHR | POSITION | REGRESSION P VALUE | ALLELE* | MEAN | SE |
|---|---|---|---|---|---|---|---|
| FLT1 | rs9319425 | 13 | 27790985 | 0.0216 | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| FLT1 | rs622227 | 13 | 27937214 | 0.0222 | GG | NA | NA |
| AG | NA | NA | |||||
| AA | NA | NA | |||||
| FLT1 | rs2387632 | 13 | 27814343 | 0.0473 | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| ICAM1210 | rs1799969 | 19 | 10255792 | 0.0249 | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA | |||||
| FGFR2 | rs2912762 | 10 | 123266280 | 0.0380 | AA | NA | NA |
| AG | NA | NA | |||||
| GG | NA | NA |
Chr, chromosome on which SNP is located; NA, not applicable to dichotomous data (yes/no); SE, standard error; SNP, single-nucleotide polymorphism.
Minor alleles are shown in bold.
TABLE 10.
CANDIDATE GENE ANALYSIS: FLUORESCEIN ANGIOGRAPHIC EVIDENCE OF PERSISTENT LEAKAGE FROM CHOROIDAL NEOVASCULARIZATION AT 6 MONTHS
| GENE | SNP | CHR | POSITION | REGRESSION P VALUE | ALLELE* | MEAN | SE |
|---|---|---|---|---|---|---|---|
| VEGFA | rs3025033 | 6 | 43859053 | 0.0363 | AA | NA | |
| AG | |||||||
| GG | |||||||
| FLT1 | rs7995976 | 13 | 27839060 | 0.0444 | AA | ||
| AC | |||||||
| CC | |||||||
Chr, chromosome on which SNP is located; NA, not applicable to dichotomous data (yes/no); SE, standard error; SNP, single-nucleotide polymorphism.
Minor alleles are shown in bold.
Several SNPs in three genes (CFH, VEGFA, and FLT1) are strongly associated with number of injections received during the study (Table 11). This parameter was measured only at the primary end point of 1 year. In the case of VEGFA and FLT1, possessing the minor allele of each SNP resulted in the need for fewer injections. By contrast, those with the minor allele in the CFH gene needed more injections.
TABLE 11.
CANDIDATE GENE ANALYSIS: NUMBER OF MONTHLY RANIBIZUMAB INTRAVITREAL INJECTIONS OVER 12 MONTHS
| GENE | SNP | CHR | POSITION | REGRESSION P VALUE | ALLELE* | MEAN† | SE |
|---|---|---|---|---|---|---|---|
| FLT1 | rs622227 | 13 | 27937214 | GG | Not observed | ||
| AG | 3.64 | 0.20 | |||||
| AA | 6.83 | 0.13 | |||||
| FLT1 | rs10507386 | 13 | 27926554 | AA | Not observed | ||
| 0.0486 | AG | 3.88 | 0.30 | ||||
| GG | 6.41 | 0.12 | |||||
| FLT1 | rs615529 | 13 | 27944327 | GG | Not observed | ||
| 0.0486 | AG | 3.88 | 0.30 | ||||
| AA | 6.41 | 0.12 | |||||
| CFH53,197 | rs3766404 | 1 | 194918455 | GG | Not observed | ||
| 0.0091 | AG | 9.20 | 0.62 | ||||
| AA | 5.27 | 0.10 | |||||
| VEGFA | rs833068 | 6 | 43850505 | 0.0456 | AA | 2.67 | 0.29 |
| AG | 6.57 | 0.24 | |||||
| GG | 6.40 | 0.19 | |||||
| VEGFA91 | rs833069 | 6 | 43850557 | 0.0456 | GG | 2.67 | 0.29 |
| AG | 6.57 | 0.24 | |||||
| AA | 6.40 | 0.19 | |||||
Chr, chromosome on which SNP is located; SE, standard error; SNP, single-nucleotide polymorphism.
Minor alleles are shown in bold.
Mean number of injections at end of study.
PHASE III: GENOME-WIDE TREATMENT RESPONSE ANALYSES
The following end points were investigated: best-corrected ETDRS letter score in the treated eye at 6 and 12 months, change in central macular thickness at 6 and 12 months, angiographic evidence of persistent leakage at 6 and 12 months, and number of injections received by end of study. Logistic or linear regression, as appropriate, was used to evaluate associations between these measures and all SNPs successfully genotyped on the Illumina 660-Quad SNP chip.
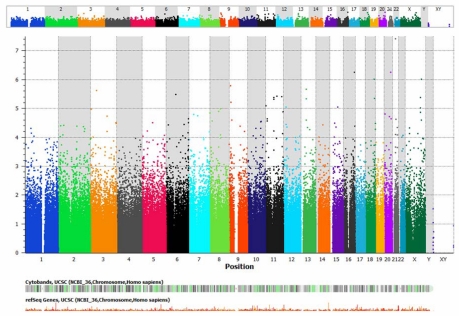
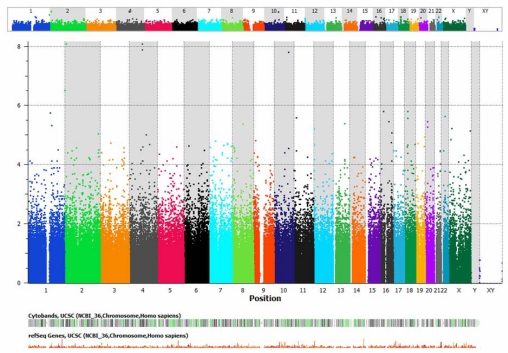
A conservative P value for significance of P<10−7 was employed, after which only one SNP near the gene noncoding RNA 158 (NCRNA00158), a member of the iRNA family, achieved genome-wide statistical significance (Table 12, Figure 1), the minor allele of which was associated with a much thicker central macula at 12 months than those with the ancestral allele. Figures 1 and 2 are a Manhattan plot whereby the y-axis shows the P value for each SNP in order along each chromosome. When a slightly less stringent correction is applied, then several SNPs achieve significance (Table 13), including several known to be expressed in the retina. TSHZ2, CCDC102B, GRIA3, PTPRD, FLJ42392, SETD2, KCNQ5, and ME3 were similarly associated with macular thickness at 12 months as NCRNA00158. The minor allele of Protocadherin-19 (PCDH19) was associated with a substantially worse visual outcome at the 6-month interim visit.
TABLE 12.
GENOME-WIDE ASSOCIATION ANALYSIS: SINGLE-NUCLEOTIDE POLYMORPHISMS ACHIEVING GENOME-WIDE SIGNIFICANCE LEVELS AFTER BONFERRONI CORRECTION FOR MULTIPLE TESTING
| MARKER | CHR | GENE* | UNCORRECTED REGRESSION P VALUE | P VALUE AFTER BONFERRONI CORRECTION | END POINT MEASURE | GENOTYPE | MEAN | SE |
|---|---|---|---|---|---|---|---|---|
| rs13421506 | 2 | Near LPIN1 | AA | Not observed | ||||
| 8.37×10−9 | 0.0041 | CRP | AC | 1.03 | 0.05 | |||
| CC | 0.25 | 0.01 | ||||||
| rs2231153 | 4 | ABCG2 | CRP | GG | Not observed | |||
| 8.37×10−9 | 0.0041 | AG | 1.03 | 0.05 | ||||
| AA | 0.25 | 0.01 | ||||||
| rs2725267 | 4 | ABCG2 | CRP | GG | Not observed | |||
| 1.33×10−8 | 0.0065 | AG | 1.03 | 0.05 | ||||
| AA | 0.26 | 0.01 | ||||||
| rs17384909 | 10 | Near ZNF518A | CRP | AA | Not observed | |||
| 1.60×10−8 | 0.0078 | AG | 1.03 | 0.05 | ||||
| GG | 0.25 | 0.01 | ||||||
| rs9675979 | 18 | Near CCDC102B | Baseline VA | AA | Not observed | |||
| 3.76×10−8 | 0.0183 | AG | 40.00 | 1.22 | ||||
| GG | 67.62 | 0.38 | ||||||
| rs2298515 | 21 | Near NCRNA00158 | 3.85×10−8 | 0.0187 | Diff CMT @12 | AA | 481.00 | 10.14 |
| AC | 76.50 | 14.17 | ||||||
| CC | −126.65 | 3.14 | ||||||
Chr, chromosome on which SNP is located; CRP, C-reactive protein; Diff CMT @ 12, change in central macular thickness between baseline and 12 months; SE, standard error; VA, best-corrected visual acuity.
Genes expressed in retina are shown in bold. “Near” refers to within 100 kbp.
FIGURE 1.
Manhattan plot of genome-wide association analysis. Change in central macular thickness at 12 months.
FIGURE 2.
Manhattan plot of genome-wide association analysis. C-reactive protein at baseline.
TABLE 13.
GENOME-WIDE ASSOCIATION ANALYSIS: SINGLE-NUCLEOTIDE POLYMORPHISMS ACHIEVING MARGINAL GENOME-WIDE SIGNIFICANCE LEVELS AFTER BONFERRONI CORRECTION FOR MULTIPLE TESTING*
| MARKER | CHR | GENE† | UNCORRECTED REGRESSION P VALUE | END POINT MEASURE | GENOTYPE | MEAN | SE |
|---|---|---|---|---|---|---|---|
| rs13029479 | 2 | TMEM18 | CRP | AA | Not observed | ||
| 3.14×10−7 | AG | 0.82 | 0.06 | ||||
| GG | 0.22 | 0.01 | |||||
| rs1205091 | 14 | RGS6 | 4.81×10−7 | Baseline VA | AA | 39.00 | 2.33 |
| AG | 57.41 | 0.74 | |||||
| GG | 72.92 | 0.63 | |||||
| rs2107332 | 20 | TSHZ2 | 5.62×10−7 | Diff CMT @ 12 | AA | −125.52 | 3.44 |
| AG | 70.00 | 32.53 | |||||
| GG | 481.00 | 6.65 | |||||
| rs8060546 | 16 | Near IRF8 | 5.68×10−7 | Diff CMT @ 12 | AA | 481.00 | 8.88 |
| AC | 27.83 | 20.84 | |||||
| CC | −130.59 | 3.17 | |||||
| rs12969428 | 18 | Near CCDC102B | 9.67×10−7 | Diff CMT @ 12 | AA | 481.00 | 7.89 |
| AG | 59.67 | 34.55 | |||||
| GG | −118.72 | 3.23 | |||||
| rs1349916 | X | Near GRIA3 | 9.78×10−7 | Diff CMT @ 12 | AA | −114.82 | 3.23 |
| AG | 84.50 | 22.27 | |||||
| GG | 481.00 | 4.87 | |||||
| rs1358889 | 9 | PTPRD | 1.64×10−6 | Diff CMT @ 12 | AA | −156.57 | 4.62 |
| AC | −34.46 | 6.38 | |||||
| CC | 298.50 | 129.05 | |||||
| rs9302755 | 16 | Near SALL1 | 1.88×10−6 | Baseline CMT | AA | 438.43 | 6.94 |
| AC | 343.69 | 5.19 | |||||
| CC | 257.85 | 4.44 | |||||
| rs9549123 | 13 | Near FLJ42392 | 2.17×10−6 | Diff CMT @ 12 | AA | 481.00 | 17.88 |
| AC | 16.67 | 18.19 | |||||
| CC | −128.28 | 3.42 | |||||
| rs11717033 | 3 | SETD2 | 2.39×10−6 | Diff CMT @ 12 | GG | Not observed | |
| AG | 251.00 | 66.73 | |||||
| AA | −117.94 | 3.02 | |||||
| rs3212240 | 14 | RIPK3 | 2.73×10−6 | Baseline CMT | AA | 415.00 | 6.47 |
| AG | 309.94 | 4.06 | |||||
| GG | 238.71 | 7.39 | |||||
| rs3757105 | 6 | KCNQ5 | 3.24×10−6 | Diff CMT @ 12 | AA | 481.00 | 10.56 |
| AG | 41.33 | 36.21 | |||||
| GG | −117.00 | 3.31 | |||||
| rs3823118 | 6 | KCNQ5 | 3.24×10−6 | Diff CMT @ 12 | AA | 481.00 | 9.78 |
| AG | 41.33 | 36.21 | |||||
| GG | −117.00 | 3.31 | |||||
| rs1150345 | 11 | Near FAM76B | 3.82×10−6 | Baseline VA | AA | 34.50 | 3.89 |
| AC | 50.87 | 1.03 | |||||
| CC | 69.79 | 0.47 | |||||
| rs618513 | 11 | ME3/Near CCDC81 | 3.82×10−6 | Diff CMT @ 12 | AA | 481.00 | 5.58 |
| AG | −0.38 | 8.10 | |||||
| GG | −133.96 | 3.98 | |||||
| rs1205106 | 14 | RGS6 | 4.32×10−6 | Baseline VA | AA | 72.08 | 0.67 |
| AG | 58.83 | 0.76 | |||||
| GG | 39.00 | 2.33 | |||||
| rs10126713 | X | PCDH19 | 4.32×10−6 | Diff VA @ 6 | AA | −4.17 | 0.56 |
| AG | 5.50 | 0.37 | |||||
| GG | 13.83 | 1.39 | |||||
CRP, C-reactive protein; Chr, chromosome on which SNP is located; Diff CMT @ 12, change in central macular thickness between baseline and 12 months; Diff VA @ 6, change in best-corrected visual acuity between baseline and 6 months; SE, standard error; VA, best-corrected visual acuity.
Bonferroni corrected P value P=0.05 – 0.10.
Genes expressed in retina are shown in bold. “Near” refers to within 100 kbp
PHASE IV: ASSOCIATIONS WITH BASELINE CHARACTERISTICS
The following baseline characteristics were evaluated: best-corrected ETDRS letter score in the treated eye, fluorescein angiographic lesion characteristics, baseline OCT central macular thickness, and CRP at baseline. When examined on a candidate gene basis, as in Phase II of the analysis, the minor alleles of SNPs in VEGFA, C3, and PF4 (platelet factor 4) were associated with a worse visual acuity at presentation (Table 14). The same SNP in C3 is also associated with having a more thickened retina at presentation (Table 15). This is the same SNP associated with less response to ranibizumab therapy throughout the study (6 and 12 months). Three SNPs in the fibroblast growth factor receptor family (FGFR-1 and -2) are also associated with worse macular thickness at presentation. FGFR2 and C3 were also associated with having a higher CRP at baseline (Table 16).
TABLE 14.
CANDIDATE GENE ANALYSIS: BASELINE BEST-CORRECTED VISUAL ACUITY (ETDRS LETTERS)
| GENE | SNP | CHR | POSITION | REGRESSION P VALUE | ALLELE* | MEAN | SE |
|---|---|---|---|---|---|---|---|
| VEGFA | rs833068 | 6 | 43850505 | 0.0045 | GG | 46.57 | 2.94 |
| AG | 59.00 | 1.02 | |||||
| AA | 67.07 | 0.81 | |||||
| VEGFA | rs833069 | 6 | 43850557 | 0.0045 | AA | 46.57 | 2.94 |
| AG | 59.00 | 1.02 | |||||
| GG | 67.07 | 0.81 | |||||
| C3 | rs2230205 | 19 | 6660704 | 0.0456 | AA | 31.00 | 1.02 |
| AG | 54.17 | 2.39 | |||||
| GG | 62.14 | 0.55 | |||||
| PF4 | rs11574452 | 4 | 75065525 | 0.0498 | AA | Not observed | |
| AC | 45.00 | 3.84 | |||||
| CC | 61.81 | 0.49 | |||||
Chr, chromosome on which SNP is located; SNP, single-nucleotide polymorphism; SE, standard error.
Minor alleles are shown in bold.
TABLE 15.
CANDIDATE GENE ANALYSIS: BASELINE CENTRAL MACULAR THICKNESS
| GENE | SNP | CHR | POSITION | REGRESSION P VALUE | ALLELE* | MEAN | SE |
|---|---|---|---|---|---|---|---|
| C3196 | rs2230205 | 19 | 6660704 | 0.0443 | AA | 456.00 | 1.02 |
| AG | 412.83 | 19.18 | |||||
| GG | 309.90 | 2.73 | |||||
| FGFR2 | rs2912762 | 10 | 123266280 | 0.0072 | AA | 379.50 | 52.68 |
| AG | 363.90 | 4.39 | |||||
| GG | 277.36 | 5.81 | |||||
| FGFR2 | rs2981451 | 10 | 123268904 | 0.0101 | AA | 236.13 | 6.01 |
| AC | 360.17 | 4.89 | |||||
| CC | 354.80 | 8.81 | |||||
| FGFR1 | rs13317 | 8 | 38388671 | 0.0458 | GG | 538.00 | 1.01 |
| AG | 347.17 | 7.21 | |||||
| AA | 313.74 | 3.88 |
Chr, chromosome on which SNP is located; SNP, single-nucleotide polymorphism; SE, standard error.
Minor alleles are shown in bold.
TABLE 16.
CANDIDATE GENE ANALYSIS: SERUM C-REACTIVE PROTEIN (MG/L) AT BASELINE
| GENE | SNP | CHR | POSITION | REGRESSION P VALUE | ALLELE* | MEAN | SE |
|---|---|---|---|---|---|---|---|
| FGFR2 | rs3135761 | 10 | 123266081 | 0.0013 | AA | 0.90 | NA |
| AG | 0.60 | 0.11 | |||||
| GG | 0.26 | 0.01 | |||||
| FGFR2 | rs1047057 | 10 | 123229102 | 0.0106 | GG | 0.57 | 0.07 |
| AG | 0.29 | 0.02 | |||||
| AA | 0.22 | 0.01 | |||||
| C3 | rs2277984 | 19 | 6630511 | 0.0120 | GG | 0.23 | 0.08 |
| AG | 0.22 | 0.01 | |||||
| AA | 0.52 | 0.03 | |||||
| CFI | rs10029485 | 4 | 110933413 | 0.0131 | GG | Not observed | |
| AG | 1.00 | 0.00 | |||||
| AA | 0.30 | 0.01 | |||||
Chr, chromosome on which SNP is located; SNP, single-nucleotide polymorphism; SE, standard error.
Minor alleles are shown in bold.
Baseline CRP was also highlighted in the genome-wide analyses where SNPs in LPIN1, ABCG2, and ZNF18A were associated (Figure 2). Worse baseline visual acuity was associated with SNPs in CCDC102B, RGS6, FAM76B, all three being robustly expressed in the retina. RIPK3 and SALL1 were associated with more thickened maculae on OCT. These two are also known to be expressed in the retina.
DISCUSSION
The hypothesis tested in this thesis is that an individual’s genetic makeup will influence response to drug therapy with ranibizumab for neovascular AMD. The design and conduct of this study had the principal aims of identifying genetic signals for future study and providing a template for other pharmacogenetic research in this area. Although a relatively small number of individuals were enrolled, the results of the study were positive and identify a number of polymorphisms in genes involved in the angiogenesis pathway as well as other novel candidates. Replication studies and functional analyses will be required before these can be confirmed as pharmacogenetic susceptibility variants.
METHODOLOGICAL CONSIDERATIONS
Clinical Trial Design
It is hoped that other pharmacogenetic studies will utilize some of the design features in this study. The strengths of this study were its prospective nature, the rigor of the entry criteria and patient follow-up, and its adherence to a real-life therapeutic regimen. It was also an advantage (not only to improve enrollment) that this study was conducted on more than one clinical site with several enrolling retinal specialists.
To date, two pharmacogenetic studies of anti-VEGF therapy for neovascular AMD have been published. Both were retrospective. The first reported the results of 86 patients treated with bevacizumab (Avastin), the humanized monoclonal antibody from which ranibizumab (Lucentis) was developed.109 The second included 156 patients who received bevacizumab.110 It was the intent of this study to overcome many of these problems with prospective enrollment and clearly stated inclusion/exclusion criteria that limit phenotypic heterogeneity while achieving a reasonable enrollment rate.
At the time of designing this study, which was early 2007, a commonly used regimen for ranibizumab was “monthly as required” as determined by the treating physician. The scenario that best fits clinical practice would be one where the treating physician would be allowed to treat according to his or her best judgment. However, for the purposes of clinical trial design and to meet regulatory requirements, re-treatment criteria had to be stipulated for this study. It is the author’s belief that these re-treatment criteria most closely matched clinical decision making and indeed were among the first to abandon the notion that a central macular thickness greater than a certain thickness, eg, 250 μm, dictated re-treatment. Combining this with the rigor of monthly patient follow-up afforded the best visual outcomes for patients while also giving the opportunity to conduct the data collection in the most robust fashion and retain all patients in the study.
Other strengths were the independence of data collection, monitoring, and analysis. Treating physicians were masked to patient genotype, and the data were collected by third parties, namely, certified experienced ophthalmic research study coordinators. Furthermore, the data were monitored (as in a standard randomized control trial) by an independent monitor. Although the data analysis plan was entirely designed by the author and principal investigator, the statistical analyses were conducted by an independent off-site statistician.
The principal weakness of this study was the small number of individuals enrolled. It is clear that the greater the number of individuals, the greater the power of the study. At best, therefore, one might hope that by a targeted analysis, relevant statistical signals and trends might be discovered. The second weakness of the study is the length of follow-up, which was 1 year. It is conceivable that a longer period of treatment may identify additional genes implicated in treatment effect. The third methodological weakness is one that is familiar to randomized control trials. The study results can likely be applied to those patients with new-onset neovascular AMD; however, the findings may be limited in what they inform us about genes that play a role in poor treatment response in those who already have the condition, recurrent disease, or those who are yet to develop CNV.
Analysis Plan
It was decided to conduct the analysis in four phases. The first was an attempt to confirm that the population studied was reflective of the population with neovascular AMD as a whole. Given the chance of gene-environment interactions, this was important. Furthermore, it was critical to know that this population showed the same genetic susceptibility to the condition and when treated with ranibizumab showed the same overall treatment response as in other studies. The second phase of the analysis sought to minimize the correction needed for multiple testing. All too often, genetic analyses are hampered because of these considerations. By prospectively defining the primary end point measure, it was possible to minimize this issue. Furthermore, by selecting key candidate genes, the number of SNPs tested was reduced, avoiding the problems of genome-wide analyses. The third phase captured much of the genetic variation present in the population studied by taking advantage of SNPs genotyped by the Illumina 660 chip. Although multiple testing was a major consideration in this analysis phase, it offered the opportunity to evaluate genes not on the candidate gene list. The fourth phase made use of participant presenting features (acuity, macular appearance, and CRP) to examine whether particular genes may be at play in determining, arguably, the severity of the disease. CRP has been linked to AMD status in other studies184–192 and was evaluated here as a potential biomarker of neovascular AMD. One weakness of the analysis plan was the SNP chip available at the time. Even though a large number of SNPs were genotyped, this includes only a minority of the >1.8 million in the human genome. Additionally, the chip (which at the time of the analysis was state-of-the-art) investigates none of those rarer alleles (minor allele frequency <0.05), and its coverage of coding variants is a mean of 4.0 per gene. Nonetheless, the chip provides a simple, robust, and cost-effective method for assessing genetic variants on a genome-wide basis.
RESULTS INTERPRETATION
There were no safety concerns in this study. There was one death; this was unrelated to the study. Other complications were mild and typical of those that accompany intravitreal injections. All the data was reviewed by a Data Safety Monitoring Committee, which made no recommendations.
Analysis Phase I
When compared with the CEI population of cases, the LGS participants appear to be representative of those with neovascular AMD. The CEI cases have been collected over the past decade from a typical hospital retina practice and carefully phenotyped with fundus photography and are known to be comparable with other case-control cohorts.53,64,193 That LGS participants had achieved a higher education level is surprising; this environmental risk factor was shown in the AREDS to be very mildly associated with AMD status.40 What impact it may have in this study is unknown but is likely to be minimal.
When outcomes are examined independent of genotype, the eyes enrolled performed similarly to other studies. On average, individuals gained almost 6 ETDRS letters, and the OCT central macular thickness reduced by about 110 μm. On average, six or seven ranibizumab injections were required over the year, which indicates that treating physicians/investigators adhered to clinical practice norms. Overall, these are typical results for eyes with neovascular AMD.6,194
Analysis Phases II and III
There are a number of different ways in which genetic variants might impact treatment outcomes in retinal and macular diseases. Patients harboring certain genotypes might experience better (or worse) vision with a given treatment. These may be correlated with, or independent of, changes in retinal anatomy or lesion activity. Additionally, certain genotypes might prolong the action of a given drug, meaning that it might need to be given less frequently. In this study, measures of all these parameters were made over the realistic time frame of 1 year, ie, one that was not too distant and yet long enough for treatment effects to likely become evident. It is quite possible to expect gene associations to change with time as different gene pathways may be involved at different stages of disease regression/treatment. Thus, it is quite possible these genes may change if the study were extended. A useful number of statistically significant genetic results were produced by the analyses. Because the number of individuals in the analyses was small, a note of caution in their interpretation is appropriate, since some of the effects might be driven by very small numbers of individuals in a particular genotypic category, especially by those homozygous for the minor alleles.
The change in visual acuity at 1 year was designated as the primary end point. In line with the two previous retrospective studies,109,110 the CFH gene was implicated in determining poorer visual outcomes together with a SNP in the CTGF gene. After correction for multiple testing, the genome-wide analysis did not identify any variants achieving statistical significance for the primary end point. It is not known why the CFH gene, which has been identified in numerous studies as one of the two major susceptibility variants for AMD development, might influence treatment response. It is tempting to speculate that this is because genotypes in CFH dictate the severity or persistence of the CNV. In a pharmacogenetic analysis of treatment effect of AREDS supplements, the same effect was noted and was presumed to be because those harboring the CFH changes had more serious disease that was less likely to be influenced by the mild beneficial effect of oral supplementation.195 A SNP in CFH is also identified as determining worse visual acuity response at the 6-month time point, suggesting that the effects of the gene on ranibizumab therapy become evident at an early interval.
Complement factor 3 (C3) is also a significant susceptibility gene for development of AMD. It is important to note that it has not been specifically implicated in the preferential development of either neovascular AMD or geographic atrophy. In analyses on the change in central macular thickness (at both 6 and 12 months), a good surrogate for improved anatomy of the central macular region and in turn crudely correlated with better vision, the minor allele of a SNP in C3 appears associated with reduced thickening and improved architecture. This same SNP has been previously implicated as an AMD susceptibility variant.196 One SNP, a few thousand base pairs upstream of the transcription initiation site of the gene NCRNA00158, is identified on the genome-wide analysis. NCRNA00158, or noncoding RNA gene 158, is transcribed into a microRNA molecule whose function has yet to be defined. The most notable of the SNPs that only marginally failed to reach genome-wide significance is rs3757105, an intronic SNP in the gene KCNQ5 (potassium voltage-gated channel, KQT-like subfamily, member 5, expressed in the retina). The SNP may result in altered splicing of the gene.
Ranibizumab directly reduces the ocular concentration of VEGF. It might be expected that the activity of the residual VEGF might be important in determining whether CNV remains active. VEGFA binds its receptor FLT1 to activate the pathways that lead to neovascularization and vascular leakage, among others. The candidate gene analysis strongly implicates FLT1 in this treatment response, identifying several SNPs associated with persistent leakage on fluorescein angiography at both 6 and 12 months. No other genes are identified on genome-wide analysis.
One of the most pertinent pharmacogenetic findings would be the identification of genetic variants that might used to predict which eyes might receive less frequent injections. In this regard, the Phase II analysis in this study reveals three biologically plausible candidates warranting further investigation: VEGFA, its receptor FLT1, and CFH. The VEGFA SNP rs833068 has been the subject of prior enquiry but no direct association with the development of advanced neovascular AMD has been confirmed.91 The CFH SNP has previously been directly implicated in AMD susceptibility.53,197
Analysis Phase IV
This final analysis took advantage of data collected at participant presentation and evaluated whether any neovascular AMD endophenotypes were influenced by genetics. The author is not aware, at the current time, that such an analysis has been performed on individuals newly identified with neovascular AMD. Although there are many determinants of how and when a patient might present with new-onset neovascular AMD, none more so than an individual’s desire and ability to access to health care, this analysis provides initial insight into initial disease severity (visual acuity and OCT central macular thickness). Additionally, blood was drawn at this point for DNA analysis, and additional consent was obtained to measure serum CRP levels. There is interest in the potential use of CRP as a biomarker of disease in AMD, and its levels have been tentatively linked to disease progression.184–192
Two SNPs in VEGFA were associated with a worse ETDRS letter score at presentation. The genome-wide analysis identified a SNP near to CCDC102B, the coiled-coil domain containing 102B gene. Little is known of its function, but it is expressed abundantly in the retina and brain and in low levels in the skin only (by SAGE analysis). The same C3 SNP rs2230205 was also associated with worse acuity and greater retinal thickening on OCT. The fibroblast growth factor receptor (FGFR) genes were also similarly associated. FGFR2 are known to be proangiogenic and also involved in the persistence of neovascularization. The genome analysis identifies several variants showing marginally nonsignificant associations, the most prominent of which is RIPK3, receptor-interacting serine-threonine kinase 3, which is expressed in the retina. The SNP rs3212240 lies in the 5′ promoter region of the gene.
Analysis of baseline CRP yielded several SNPs that achieved genome-wide significance. Potentially most prominently, two SNPs are located within with the ABCG2 gene. rs2231153 and rs2725267 are both intronic nonsynonymous SNPs. ABCG2 encodes the ATP-binding cassette, subfamily G, a member of the superfamily of ATP-binding cassette (ABC) transporters and expressed in the retina. ABC proteins transport various molecules across extracellular and intracellular membranes. ABCG2 is a xenobiotic transporter that may play an important role in the exclusion of xenobiotics from the brain and is probably involved in brain/retina-to-blood efflux.
What Is the Clinical Relevance of These Pharmacogenetic Interactions?
The findings from this type of study have several uses. Once validated, screening for these specific genetic variants can be performed quickly and easily in the clinical environment to identify patients’ response characteristics. In those with favorable genetic predisposition, in the case of ranibizumab, fewer injections need to be scheduled and perhaps the intervals between treatments and visits can be lengthened. In the case of those with worse genotypes, more frequent interventions can be considered, including potentially involving the use of other modalities. Additionally, this form of research can identify new avenues for drug development by implicating novel genes and the proteins they encode in disease pathogenesis and susceptibility.
FUTURE EXPERIMENTS
The data generated by this study warrant further analysis beyond the scope of this thesis. The first step will be to corroborate or refute each finding. This is best achieved by replication in another study. Such replication studies have become the mainstay of genetic epidemiology and are most successful in situations where the associated variants have a high allele frequency or exert a large effect. Some of the best examples are found in AMD gene-discovery research, CFH rs106117050–52 being the prime example of a common variant of large effect that can be replicated in numerous populations. The situation is much more challenging for rarer variants and/or those of smaller effect where the “noise” inherent in phenotypic variability drowns out the genetic signal. For example, in AMD, a late-onset, age-related disease, it is difficult to ascertain “true” controls who will never develop the disease. Such a challenge may well be amplified in pharmacogenetic studies where two or more variables are examined in combination, ie, phenotype and treatment response. Nonetheless, two other similar prospective studies have been identified that are currently in progress but will serve the purpose of replication. Of practical importance is which variants should be replicated. It is perfectly rational to include all those achieving significance after correction for multiple testing. However, the Bonferroni adjustment is very stringent, and although it reduces the chance of including false positives in further analyses, it may also result in truly important genetic variants being overlooked. There is no perfect answer to handling this issue with the exception of including all variants meeting the basic threshold of P<0.05 in subsequent functional studies, which is totally impractical. A pragmatic alternative is to include variants showing marginal significance, and that is the rationale behind presenting Table 13, which details variants achieving a corrected P value of between 0.10 and 0.05.
The second step will be to formally investigate the functional variant in each gene. SNP chips have been designed to include the most polymorphic of genetic variants and also tag as many known haplotypes as possible, ie, groups of SNPs that are co-inherited. With this in mind, therefore, it is more than likely that the associated SNPs are not the causal variants. The causal variants are likely to be in the same haplotype as the associated SNP or in linkage disequilibrium with the SNP. It is now possible to determine such a list of SNPs using the human genome databases such as the HapMap and target genotyping to these. An alternative strategy is to directly sequence the genomic region encompassed by the associated SNP, which will document the complete genetic variation in that region. There are advantages and disadvantages in both approaches. Targeted SNP genotyping is cheaper and quicker but might overlook the causal variant. Chances of this increase if the variant (a) is relatively rare, (b) is not encompassed by the genotyped region, or (c) lies within a highly polymorphic area, for example, an intron, promoter region, or untranslated region. Direct sequencing, by its nature, captures all sequence variations but is more time-consuming and expensive to employ, particularly when large numbers of samples are to be evaluated. Whereas the assays for targeted genotyping are robust, direct sequencing can be technically problematic to achieve in certain genomic regions. Examples would be in highly repetitive DNA or GC-rich regions, which impact the fidelity of the sequencing required.
The overarching limitation of this strategy is that the “causal variant” can only be defined statistically. Not only do nonsynonymous sequence variants imply functional, structural consequence, but there is a growing body of evidence supporting the functional consequence of synonymous sequence variants198–200 (and reviewed in201,202). Thus the second step is to evaluate any genetic variant as to its putative functional consequence. Initially, this can be achieved bioinformatically, in silico, employing existing sequencing and protein databases. Powerful software exists from which it is possible to infer what type of in vitro evaluations might yield insights into their functional consequences. For instance, annotations of cSNPs and indels can be based on the genome Web browsers of the National Center for Biotechnology Information and the University of California Santa Cruz.203,204 Variants can be aggregated into functional units based on genomic context (eg, coding regions, promoter regions, microRNAs, transcription factor binding sites, conserved regions) or their putative individual functional effects.205,206 Weighting can be introduced to the analyses based on characteristics of the variants (eg, synonymous/nonsynonymous, type of amino acid substitution). Each variant identified from the sequencing can be annotated using both existing knowledge bases and computational predictions (based on PolyPhen-2 scores) to assess the putative functional role. Further weighting schemes can be employed to assist in prioritization based on the contextual scoring and putative functional effects. Finally, pathway analysis (in which variants are summarized at the level of genes and then aggregated across a shared pathway) can be conducted, as this can aid in identification of pathway-level associations and potential gene-gene interactions. From these analyses, in vitro experiments/assays can be designed to evaluate the functional effects of identified genetic variants.
The technology of genetic research is rapidly improving, offering new opportunities for pharmacogenetic research. While the Illumina 660-Quad SNP chip was state-of-the-art when utilized in this study, it has now been superseded by much more dense SNP arrays. Currently, Illumina (San Diego, California) produces the HumanOmniExpress (OmniExpress) and the supplementary HumanOmni1S-8 (Omni1S) high-density BeadChips. The OmniExpress and the Omni1S BeadChips simultaneously interrogate >700,000 and ~1.2 million variants, respectively, using data from all three HapMap phases and the latest pilot data from the 1000 Genomes Project.207 These arrays allow interrogation of a much denser map of variants with an enhanced coverage of rarer variants (minor allele frequency ≥2.5%) than ever before.
The recent introduction and cost reductions in Next Generation sequencing offer the realistic possibility of genotyping an individual’s entire genetic variation. A very convenient technology is that of Whole Exome sequencing. For example, Illumina’s TruSeq Sample Preparation and TruSeq Exome Enrichment Kits on their GAIIX next-generation sequencing platform sequence and interrogate the complete exome that is 20,794 genes, covering an exomic target region size of ~62 Mb. Therefore, 91% of the latest RefSeq database build (Sept 2010 hg19) and 95% of the September 2010 release of the Collaborative Consensus Coding Sequence (CCDS) database can be sequenced in a single run. The GAIIX is capable of generating 95 GB of sequence data per flow cell in a pair-end read experiment, which actually sequences the same region almost 100 times for high-fidelity results.
While these technological advances are very exciting and will translate into substantial improvements in analysis, the study of pharmacogenetics is always hampered over time by changes in medical care, either changes in regimen or the introduction of new therapies. It is acknowledged, therefore, that the results of the Comparison of Age-Related Macular Degeneration Treatments Trials (CATT) (http://www.med.upenn.edu/cpob/studies/CATT.shtml) were published this year (2011), which may alter the use and frequency of ranibizumab injections. Additionally, it seems likely that Regeneron’s VEGF-trap208 will be marketed later this year.
CONCLUSION
This study is a prospective pharmacogenetic study of genetically determined treatment response to intravitreal ranibizumab for neovascular AMD. Although small in nature, the aim was principally to demonstrate the methodology needed to conduct such studies. Encouragingly, the results identify a number of putative genetic variants, which will be further examined by replication and functional studies to elucidate the complete pharmacogenetic architecture of therapy for AMD.
TABLE 7.
CANDIDATE GENE ANALYSIS: CHANGE IN CENTRAL MACULAR THICKNESS AT 12 MONTHS
| GENE | SNP | CHR | POSITION | REGRESSION P VALUE | ALLELE* | MEAN† | SE |
|---|---|---|---|---|---|---|---|
| C3196 | rs2230205 | 19 | 6660704 | 0.0171 | AA | −232.00 | NA |
| AG | −204.83 | 16.88 | |||||
| GG | −57.86 | 5.01 | |||||
| THBS1 | rs1478604 | 15 | 37660613 | 0.0460 | GG | −205.50 | 46.32 |
| AG | −125.29 | 7.13 | |||||
| AA | −41.42 | 9.08 |
Chr, chromosome on which SNP is located; SE, standard error; SNP, single-nucleotide polymorphism.
Minor alleles are shown in bold.
Mean change in central macular thickness (microns) as compared with baseline.
ACKNOWLEDGMENTS
Funding/Support: This work is supported by Research to Prevent Blindness USA and the Foundation Fighting Blindness, Columbia, Maryland. The Casey Eye Institute holds an unrestricted grant from Research to Prevent Blindness USA. Genentech Inc, as part of their Investigator Sponsored Trial (IST) program, supported data collection and genotyping.
Financial Disclosures: None.
Conformity With Author Information: The Oregon Health and Science University Institutional Review Board approved the research. Prior to commencement, the study was submitted and approved as an Investigational New Drug application to the Food and Drug Administration (FDA IND 100 451) and registered with ClinicalTrials.gov (Identifier: NCT00469352). Data accumulation conformed to all Federal and State laws and was compliant with HIPAA guidelines (http://www.hhs.gov/ocr/hipaa/privacy.html ). The conduct of this study was overseen by a Data Safety Monitoring Committee.
Other Acknowledgments: I would like to extend thanks to the patients who participated in this study; Ann Lundquist, Mitchell Schain, and Shelley Hanel (research study coordinators), who assisted with the conduct and administration of the study; and Sara Hamon, PhD, for statistical analysis. I extend thanks to my colleagues, Christina Flaxel, MD, Steven Bailey, MD, Andreas Lauer, MD, Thomas Hwang, MD, and Jill Hopkins, MD, who also enrolled patients in the study.
REFERENCES
- 1.Zimmer-Galler IE, Bressler NM, Bressler SB. Treatment of choroidal neovascularization: updated information from recent macular photocoagulation study group reports. Int Ophthalmol Clin. 1995;35(4):37–57. doi: 10.1097/00004397-199503540-00005. [DOI] [PubMed] [Google Scholar]
- 2.Custis PH, Bressler SB, Bressler NM. Laser management of subfoveal choroidal neovascularization in age-related macular degeneration. Curr Opinion Ophthalmol. 1993;4(3):7–18. doi: 10.1097/00055735-199306000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Barbazetto I, Burdan A, Bressler NM, et al. Photodynamic therapy of subfoveal choroidal neovascularization with verteporfin: fluorescein angiographic guidelines for evaluation and treatment—TAP and VIP report No. 2. Arch Ophthalmol. 2003;121(9):1253–1268. doi: 10.1001/archopht.121.9.1253. [DOI] [PubMed] [Google Scholar]
- 4.Ferrara N, Damico L, Shams N, Lowman H, Kim R. Development of ranibizumab, an anti-vascular endothelial growth factor antigen binding fragment, as therapy for neovascular age-related macular degeneration. Retina. 2006;26(8):859–870. doi: 10.1097/01.iae.0000242842.14624.e7. [DOI] [PubMed] [Google Scholar]
- 5.Heier JS, Antoszyk AN, Pavan PR, et al. Ranibizumab for treatment of neovascular age-related macular degeneration: a phase I/II multicenter, controlled, multidose study. Ophthalmology. 2006;113(4):642.e641–644. doi: 10.1016/j.ophtha.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 6.Rosenfeld PJ, Rich RM, Lalwani GA. Ranibizumab: Phase III clinical trial results. Ophthalmol Clin North Am. 2006;19(3):361–372. doi: 10.1016/j.ohc.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser PK. Verteporfin photodynamic therapy and anti-angiogenic drugs: potential for combination therapy in exudative age-related macular degeneration. Curr Med Res Opin. 2007;23(3):477–487. doi: 10.1185/030079907X167624. [DOI] [PubMed] [Google Scholar]
- 8.Cheung CY, Tso AW, Cheung BM, et al. Genetic variants associated with persistent central obesity and the metabolic syndrome in a 12-year longitudinal study. Eur J Endocrinol. 2010;164(3):381–8. doi: 10.1530/EJE-10-0902. [DOI] [PubMed] [Google Scholar]
- 9.Franks PW, Poveda A. Gene-lifestyle and gene-pharmacotherpy interactions in obesity and its cardiovascular consequences. Curr Vasc Pharmacol. 2010 Dec 14; doi: 10.2174/157016111796197233. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 10.Motulsky AG, Harper PS, Bobrow M, Scriver C. Pharmacogenetics. New York: Oxford University Press; 1997. [Google Scholar]
- 11.Beutler E, Dern RJ, Flanagan CL, Alving AS. The hemolytic effect of primaquine. VII. Biochemical studies of drug-sensitive erythrocytes. J Lab Clin Med. 1955;45(2):286–295. [PubMed] [Google Scholar]
- 12.Evans DA, Manley KA, McKusick VA. Genetic control of isoniazid metabolism in man. Br Med J. 1960;2(5197):485–491. doi: 10.1136/bmj.2.5197.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arranz MJ, Collier D, Kerwin RW. Pharmacogenetics for the individualization of psychiatric treatment. Am J Pharmacogenomics. 2001;1(1):3–10. doi: 10.2165/00129785-200101010-00001. [DOI] [PubMed] [Google Scholar]
- 14.Lerer B, Segman RH. Pharmacogenetics of antipsychotic therapy: pivotal research issues and the prospects for clinical implementation. Dialogues Clin Neurosci. 2006;8(1):85–94. doi: 10.31887/DCNS.2006.8.1/blerer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Depondt C, Shorvon SD. Genetic association studies in epilepsy pharmacogenomics: lessons learnt and potential applications. Pharmacogenomics. 2006;7(5):731–745. doi: 10.2217/14622416.7.5.731. [DOI] [PubMed] [Google Scholar]
- 16.Weiss ST, Litonjua AA, Lange C, et al. Overview of the pharmacogenetics of asthma treatment. Pharmacogenomics J. 2006;6(5):311–326. doi: 10.1038/sj.tpj.6500387. [DOI] [PubMed] [Google Scholar]
- 17.Manunta P, Bianchi G. Pharmacogenomics and pharmacogenetics of hypertension: update and perspectives—the adducin paradigm. J Am Soc Nephrol. 2006;17(4 Suppl 2):S30–35. doi: 10.1681/ASN.2005121346. [DOI] [PubMed] [Google Scholar]
- 18.Hess KR, Anderson K, Symmans WF, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24(26):4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 19.Barry EL, Baron JA, Bhat S, et al. Ornithine decarboxylase polymorphism modification of response to aspirin treatment for colorectal adenoma prevention. J Natl Cancer Inst. 2006;98(20):1494–1500. doi: 10.1093/jnci/djj398. [DOI] [PubMed] [Google Scholar]
- 20.Djordjevic N, Carrillo JA, Ueda N, et al. N-acetyltransferase-2 (NAT2) gene polymorphisms and enzyme activity in Serbs: unprecedented high prevalence of rapid acetylators in a white population. J Clin Pharmacol. 2010 doi: 10.1177/0091270010377630. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 21.Meyer UA, Zanger UM. Molecular mechanisms of genetic polymorphisms of drug metabolism. Annu Rev Pharmacol Toxicol. 1997;37:269–296. doi: 10.1146/annurev.pharmtox.37.1.269. [DOI] [PubMed] [Google Scholar]
- 22.Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86(4):1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNamara DM, Holubkov R, Janosko K, et al. Pharmacogenetic interactions between beta-blocker therapy and the angiotensin-converting enzyme deletion polymorphism in patients with congestive heart failure. Circulation. 2001;103(12):1644–1648. doi: 10.1161/01.cir.103.12.1644. [DOI] [PubMed] [Google Scholar]
- 24.Bird AC, Bressler NM, Bressler SB, et al. An international classification and grading system for age-related maculopathy and age-related macular degeneration. The International ARM Epidemiological Study Group. Surv Ophthalmol. 1995;39(5):367–374. doi: 10.1016/s0039-6257(05)80092-x. [DOI] [PubMed] [Google Scholar]
- 25.Chakravarthy U, Augood C, Bentham GC, et al. Cigarette smoking and age-related macular degeneration in the EUREYE Study. Ophthalmology. 2007;114(6):1157–1163. doi: 10.1016/j.ophtha.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 26.Klein ML, Francis PJ. Genetics of age-related macular degeneration. Ophthalmol Clin North Am. 2003;16(4):567–574. doi: 10.1016/s0896-1549(03)00063-4. [DOI] [PubMed] [Google Scholar]
- 27.Klein R, Klein BE, Linton KL. Prevalence of age-related maculopathy. The Beaver Dam Eye Study. Ophthalmology. 1992;99(6):933–943. doi: 10.1016/s0161-6420(92)31871-8. [DOI] [PubMed] [Google Scholar]
- 28.Friedman DS, O’Colmain BJ, Munoz B, et al. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122(4):564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 29.A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001;119(10):1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The Complications of Age-Related Macular Degeneration Prevention Trial (CAPT): rationale, design and methodology. Clin Trials. 2004;1(1):91–107. doi: 10.1191/1740774504cn007xx. [DOI] [PubMed] [Google Scholar]
- 31.Thornton J, Edwards R, Mitchell P, Harrison RA, Buchan I, Kelly SP. Smoking and age-related macular degeneration: a review of association. Eye. 2005;19(9):935–944. doi: 10.1038/sj.eye.6701978. [DOI] [PubMed] [Google Scholar]
- 32.Evans JR, Fletcher AE, Wormald RP. 28,000 Cases of age related macular degeneration causing visual loss in people aged 75 years and above in the United Kingdom may be attributable to smoking. Br J Ophthalmol. 2005;89(5):550–553. doi: 10.1136/bjo.2004.049726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clemons TE, Milton RC, Klein R, Seddon JM, Ferris FL., 3rd Risk factors for the incidence of Advanced Age-Related Macular Degeneration in the Age-Related Eye Disease Study (AREDS) AREDS report no. 19. Ophthalmology. 2005;112(4):533–539. doi: 10.1016/j.ophtha.2004.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein R, Deng Y, Klein BE, et al. Cardiovascular disease, its risk factors and treatment, and age-related macular degeneration: Women’s Health Initiative Sight Exam ancillary study. Am J Ophthalmol. 2007;143(3):473–483. doi: 10.1016/j.ajo.2006.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein R, Klein BE, Knudtson MD, et al. Subclinical atherosclerotic cardiovascular disease and early age-related macular degeneration in a multiracial cohort: the Multiethnic Study of Atherosclerosis. Arch Ophthalmol. 2007;125(4):534–543. doi: 10.1001/archopht.125.4.534. [DOI] [PubMed] [Google Scholar]
- 36.Duan Y, Mo J, Klein R, et al. Age-related macular degeneration is associated with incident myocardial infarction among elderly Americans. Ophthalmology. 2007;114(4):732–737. doi: 10.1016/j.ophtha.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 37.Guymer RH, Chong EW. Modifiable risk factors for age-related macular degeneration. Med J (Aust) 2006;184(9):455–458. doi: 10.5694/j.1326-5377.2006.tb00318.x. [DOI] [PubMed] [Google Scholar]
- 38.Klein R, Klein BE, Knudtson MD, et al. Prevalence of age-related macular degeneration in 4 racial/ethnic groups in the multi-ethnic study of atherosclerosis. Ophthalmology. 2006;113(3):373–380. doi: 10.1016/j.ophtha.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 39.Seddon JM, George S, Rosner B, Klein ML. CFH gene variant, Y402H, and smoking, body mass index, environmental associations with advanced age-related macular degeneration. Human Hered. 2006;61(3):157–165. doi: 10.1159/000094141. [DOI] [PubMed] [Google Scholar]
- 40.Francis PJ, George S, Schultz DW, et al. The LOC387715 gene, smoking, body mass index, environmental associations with advanced age-related macular degeneration. Hum Hered. 2007;63(3–4):212–218. doi: 10.1159/000100046. [DOI] [PubMed] [Google Scholar]
- 41.Klein ML, Mauldin WM, Stoumbos VD. Heredity and age-related macular degeneration. Observations in monozygotic twins. Arch Ophthalmol. 1994;112(7):932–937. doi: 10.1001/archopht.1994.01090190080025. [DOI] [PubMed] [Google Scholar]
- 42.Seddon JM, Ajani UA, Mitchell BD. Familial aggregation of age-related maculopathy. Am J Ophthalmol. 1997;123(2):199–206. doi: 10.1016/s0002-9394(14)71036-0. [DOI] [PubMed] [Google Scholar]
- 43.Majewski J, Schultz DW, Weleber RG, et al. Age-related macular degeneration—a genome scan in extended families. Am J Hum Genet. 2003;73(3):540–550. doi: 10.1086/377701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kenealy SJ, Schmidt S, Agarwal A, et al. Linkage analysis for age-related macular degeneration supports a gene on chromosome 10q26. Mol Vis. 2004;10:57–61. [PubMed] [Google Scholar]
- 45.Santangelo SL, Yen CH, Haddad S, Fagerness J, Huang C, Seddon JM. A discordant sib-pair linkage analysis of age-related macular degeneration. Ophthmic Genet. 2005;26(2):61–67. doi: 10.1080/13816810490967944. [DOI] [PubMed] [Google Scholar]
- 46.Fisher SA, Abecasis GR, Yashar BM, et al. Meta-analysis of genome scans of age-related macular degeneration. Hum Mol Genet. 2005;14(15):2257–2264. doi: 10.1093/hmg/ddi230. [DOI] [PubMed] [Google Scholar]
- 47.Barral S, Francis PJ, Schultz DW, et al. Expanded genome scan in extended families with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2006;47(12):5453–5459. doi: 10.1167/iovs.06-0655. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt S, Scott WK, Postel EA, et al. Ordered subset linkage analysis supports a susceptibility locus for age-related macular degeneration on chromosome 16p12. BMC Genet. 2004;5:18. doi: 10.1186/1471-2156-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schultz DW, Weleber RG, Lawrence G, et al. HEMICENTIN-1 (FIBULIN-6) and the 1q31 AMD locus in the context of complex disease: review and perspective. Ophthalmic Genet. 2005;26(2):101–105. doi: 10.1080/13816810590968023. [DOI] [PubMed] [Google Scholar]
- 50.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haines JL, Hauser MA, Schmidt S, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308(5720):419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 52.Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308(5720):421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 53.Francis PJ, Schultz DW, Hamon S, Ott J, Weleber RG, Klein ML. Haplotypes in the complement factor H (CFH) gene: associations with drusen and advanced age-related macular degeneration. PLoS One. 2007;2(11):e1197. doi: 10.1371/journal.pone.0001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hageman GS, Anderson DH, Johnson LV, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci. 2005;102(20):7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hageman GS, Hancox LS, Taiber AJ, et al. Extended haplotypes in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: characterization, ethnic distribution and evolutionary implications. Ann Med. 2006;38(8):592–604. [PMC free article] [PubMed] [Google Scholar]
- 56.Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006;38(10):1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- 57.Magnusson KP, Duan S, Sigurdsson H, et al. CFH Y402H confers similar risk of soft drusen and both forms of advanced AMD. PLoS Med. 2006;3(1):e5. doi: 10.1371/journal.pmed.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hageman GS, Luthert PJ, Victor Chong NH, Johnson LV, Anderson DH, Mullins RF. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch’s membrane interface in aging and age-related macular degeneration. Prog Ret Eye Res. 2001;20(6):705–732. doi: 10.1016/s1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- 59.Gold B, Merriam JE, Zernant J, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38(4):458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spencer KL, Hauser MA, Olson LM, et al. Protective effect of complement factor B and complement component 2 variants in age-related macular degeneration. Hum Mol Genet. 2007;16(16):1986–1992. doi: 10.1093/hmg/ddm146. [DOI] [PubMed] [Google Scholar]
- 61.Yates JR, Sepp T, Matharu BK, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357(6):553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 62.Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39(10):1200–1201. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- 63.Fagerness JA, Maller JB, Neale BM, Reynolds RC, Daly MJ, Seddon JM. Variation near complement factor I is associated with risk of advanced AMD. Eur J Hum Genet. 2008;17(1):100–104. doi: 10.1038/ejhg.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Francis PJ, Zhang H, Dewan A, Hoh J, Klein ML. Joint effects of polymorphisms in the HTRA1, LOC387715/ARMS2, and CFH genes on AMD in a Caucasian population. Mol Vis. 2008;14:1395–1400. [PMC free article] [PubMed] [Google Scholar]
- 65.Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14(21):3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 66.Jakobsdottir J, Conley YP, Weeks DE, Mah TS, Ferrell RE, Gorin MB. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet. 2005;77(3):389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanda A, Chen W, Othman M, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci. 2007;104(41):16227–16232. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fritsche LG, Loenhardt T, Janssen A, et al. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40(7):892–896. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- 69.Dewan A, Liu M, Hartman S, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314(5801):989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 70.Yang Z, Camp NJ, Sun H, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314(5801):992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 71.Baird PN, Guida E, Chu DT, Vu HT, Guymer RH. The epsilon2 and epsilon4 alleles of the apolipoprotein gene are associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2004;45(5):1311–1315. doi: 10.1167/iovs.03-1121. [DOI] [PubMed] [Google Scholar]
- 72.Baird PN, Richardson AJ, Robman LD, et al. Apolipoprotein (APOE) gene is associated with progression of age-related macular degeneration (AMD) Human Mutat. 2006;27(4):337–342. doi: 10.1002/humu.20288. [DOI] [PubMed] [Google Scholar]
- 73.Klaver CC, Kliffen M, van Duijn CM, et al. Genetic association of apolipoprotein E with age-related macular degeneration. Am J Hum Genet. 1998;63(1):200–206. doi: 10.1086/301901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Allikmets R. Further evidence for an association of ABCR alleles with age-related macular degeneration. The International ABCR Screening Consortium. Am J Hum Genet. 2000;67(2):487–491. doi: 10.1086/303018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Allikmets R, Shroyer NF, Singh N, et al. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science. 1997;277(5333):1805–1807. doi: 10.1126/science.277.5333.1805. [DOI] [PubMed] [Google Scholar]
- 76.Combadiere C, Feumi C, Raoul W, et al. CX3CR1-dependent subretinal microglia cell accumulation is associated with cardinal features of age-related macular degeneration. J Clin Invest. 2007;117(10):2920–2928. doi: 10.1172/JCI31692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tuo J, Smith BC, Bojanowski CM, et al. The involvement of sequence variation and expression of CX3CR1 in the pathogenesis of age-related macular degeneration. FASEB J. 2004;18(11):1297–1299. doi: 10.1096/fj.04-1862fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baird PN, Chu D, Guida E, Vu HT, Guymer R. Association of the M55L and Q192R paraoxonase gene polymorphisms with age-related macular degeneration. Am J Ophthalmol. 2004;138(4):665–666. doi: 10.1016/j.ajo.2004.04.053. [DOI] [PubMed] [Google Scholar]
- 79.Zareparsi S, Buraczynska M, Branham KE, et al. Toll-like receptor 4 variant D299G is associated with susceptibility to age-related macular degeneration. Hum Mol Genet. 2005;14(11):1449–1455. doi: 10.1093/hmg/ddi154. [DOI] [PubMed] [Google Scholar]
- 80.Tuo J, Ning B, Bojanowski CM, et al. Synergic effect of polymorphisms in ERCC6 5′ flanking region and complement factor H on age-related macular degeneration predisposition. Proc Natl Acad Sci U S A. 2006;103(24):9256–9261. doi: 10.1073/pnas.0603485103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Conley YP, Jakobsdottir J, Mah T, et al. CFH, ELOVL4, PLEKHA1 and LOC387715 genes and susceptibility to age-related maculopathy: AREDS and CHS cohorts and meta-analyses. Hum Mol Genet. 2006;15(21):3206–3218. doi: 10.1093/hmg/ddl396. [DOI] [PubMed] [Google Scholar]
- 82.Conley YP, Thalamuthu A, Jakobsdottir J, et al. Candidate gene analysis suggests a role for fatty acid biosynthesis and regulation of the complement system in the etiology of age-related maculopathy. Hum Mol Genet. 2005;14(14):1991–2002. doi: 10.1093/hmg/ddi204. [DOI] [PubMed] [Google Scholar]
- 83.Haines JL, Schnetz-Boutaud N, Schmidt S, et al. Functional candidate genes in age-related macular degeneration: significant association with VEGF, VLDLR, and LRP6. Invest Ophthalmol Vis Sci. 2006;47(1):329–335. doi: 10.1167/iovs.05-0116. [DOI] [PubMed] [Google Scholar]
- 84.Stone EM, Braun TA, Russell SR, et al. Missense variations in the fibulin 5 gene and age-related macular degeneration. N Engl J Med. 2004;351(4):346–353. doi: 10.1056/NEJMoa040833. [DOI] [PubMed] [Google Scholar]
- 85.Schultz DW, Klein ML, Humpert AJ, et al. Analysis of the ARMD1 locus: evidence that a mutation in HEMICENTIN-1 is associated with age-related macular degeneration in a large family. Hum Mol Genet. 2003;12(24):3315–3323. doi: 10.1093/hmg/ddg348. [DOI] [PubMed] [Google Scholar]
- 86.Yang Z, Stratton C, Francis PJ, et al. Toll-like receptor 3 and geographic atrophy in age-related macular degeneration. N Engl J Med. 2008;359(14):1456–1463. doi: 10.1056/NEJMoa0802437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ennis S, Jomary C, Mullins R, et al. Association between the SERPING1 gene and age-related macular degeneration: a two-stage case-control study. Lancet. 2008;372(9652):1828–1834. doi: 10.1016/S0140-6736(08)61348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Churchill AJ, Carter JG, Lovell HC, et al. VEGF polymorphisms are associated with neovascular age-related macular degeneration. Hum Mol Genet. 2006;15(19):2955–2961. doi: 10.1093/hmg/ddl238. [DOI] [PubMed] [Google Scholar]
- 89.Francis PJ, Hamon SC, Ott J, Weleber RG, Klein ML. Polymorphisms in C2, CFB and C3 are associated with progression to advanced age related macular degeneration associated with visual loss. J Med Genet. 2009;46(5):300–307. doi: 10.1136/jmg.2008.062737. [DOI] [PubMed] [Google Scholar]
- 90.Qu Y, Dai H, Zhou F, et al. Vascular endothelial growth factor gene polymorphisms and risk of neovascular age-related macular degeneration in a Chinese cohort. Ophthalmic Res. 2010;45(3):142–148. doi: 10.1159/000319543. [DOI] [PubMed] [Google Scholar]
- 91.Fang AM, Lee AY, Kulkarni M, Osborn MP, Brantley MA., Jr Polymorphisms in the VEGFA and VEGFR-2 genes and neovascular age-related macular degeneration. Mol Vis. 2009;15:2710–2719. [PMC free article] [PubMed] [Google Scholar]
- 92.Szaflik JP, Blasiak J, Krzyanowska A, et al. Distribution of the C-460T polymorphism of the vascular endothelial growth factor gene in age-related macular degeneration. Klinika Oczna. 2009;111(4–6):125–127. [PubMed] [Google Scholar]
- 93.Chen W, Stambolian D, Edwards AO, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010;107(16):7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bessho H, Kondo N, Honda S, Kuno S, Negi A. Coding variant Met72Thr in the PEDF gene and risk of neovascular age-related macular degeneration and polypoidal choroidal vasculopathy. Mol Vis. 2009;15:1107–1114. [PMC free article] [PubMed] [Google Scholar]
- 95.Mattes D, Haas A, Renner W, et al. Analysis of three pigment epithelium-derived factor gene polymorphisms in patients with exudative age-related macular degeneration. Mol Vis. 2009;15:343–348. [PMC free article] [PubMed] [Google Scholar]
- 96.Lin JM, Wan L, Tsai YY, et al. Pigment epithelium-derived factor gene Met72Thr polymorphism is associated with increased risk of wet age-related macular degeneration. Am J Ophthalmol. 2008;145(4):716–721. doi: 10.1016/j.ajo.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 97.Klein ML, Francis PJ, Rosner B, et al. CFH and LOC387715/ARMS2 genotypes and treatment with antioxidants and zinc for age-related macular degeneration. Ophthalmology. 2008;115(6):1019–1025. doi: 10.1016/j.ophtha.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 98.Sunness JS, Gonzalez-Baron J, Applegate CA, et al. Enlargement of atrophy and visual acuity loss in the geographic atrophy form of age-related macular degeneration. Ophthalmology. 1999;106(9):1768–1779. doi: 10.1016/S0161-6420(99)90340-8. [DOI] [PubMed] [Google Scholar]
- 99.Wan MJ, Hooper PL, Sheidow TG. Combination therapy in exudative age-related macular degeneration: visual outcomes following combined treatment with photodynamic therapy and intravitreal bevacizumab. Can J Ophthalmol. 2010;45(4):375–380. doi: 10.3129/i10-011. [DOI] [PubMed] [Google Scholar]
- 100.Hara R, Kawaji T, Inomata Y, et al. Photodynamic therapy alone versus combined with intravitreal bevacizumab for neovascular age-related macular degeneration without polypoidal choroidal vasculopathy in Japanese patients. Graefes Arch Clin Exp Ophthalmol. 2010;248(7):931–936. doi: 10.1007/s00417-010-1343-8. [DOI] [PubMed] [Google Scholar]
- 101.Shima C, Gomi F, Sawa M, Sakaguchi H, Tsujikawa M, Tano Y. One-year results of combined photodynamic therapy and intravitreal bevacizumab injection for retinal pigment epithelial detachment secondary to age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2009;247(7):899–906. doi: 10.1007/s00417-009-1067-9. [DOI] [PubMed] [Google Scholar]
- 102.Brown SB, Mellish KJ. Verteporfin: a milestone in opthalmology and photodynamic therapy. Expert Opin Pharmacother. 2001;2(2):351–361. doi: 10.1517/14656566.2.2.351. [DOI] [PubMed] [Google Scholar]
- 103.Schmidt-Erfurth U, Miller JW, Sickenberg M, et al. Photodynamic therapy with verteporfin for choroidal neovascularization caused by age-related macular degeneration: results of retreatments in a phase 1 and 2 study. Arch Ophthalmol. 1999;117(9):1177–1187. doi: 10.1001/archopht.117.9.1177. [DOI] [PubMed] [Google Scholar]
- 104.Wormald R, Evans J, Smeeth L, Henshaw K. Photodynamic therapy for neovascular age-related macular degeneration. Cochrane Database Syst Rev. 2007 July 18;(3):CD002030. doi: 10.1002/14651858.CD002030.pub3. [DOI] [PubMed] [Google Scholar]
- 105.Parmeggiani F, Costagliola C, Incorvaia C, Sebastiani A, Gemmati D. Pharmacogenetic aspects in therapeutic management of subfoveal choroidal neovascularisation: Role of factor XIII-A 185 T-allele. Curr Drug Targets. 2011;12(2):138–148. doi: 10.2174/138945011794182773. [DOI] [PubMed] [Google Scholar]
- 106.Parmeggiani F, Costagliola C, Gemmati D, et al. Coagulation gene predictors of photodynamic therapy for occult choroidal neovascularization in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49(7):3100–3106. doi: 10.1167/iovs.07-1654. [DOI] [PubMed] [Google Scholar]
- 107.Parmeggiani F, Costagliola C, Gemmati D, et al. Predictive role of coagulation-balance gene polymorphisms in the efficacy of photodynamic therapy with verteporfin for classic choroidal neovascularization secondary to age-related macular degeneration. Pharmacogenet Genomics. 2007;17(12):1039–1046. doi: 10.1097/FPC.0b013e3282f12a4e. [DOI] [PubMed] [Google Scholar]
- 108.Parmeggiani F, Gemmati D, Costagliola C, Sebastiani A, Incorvaia C. Predictive role of C677T MTHFR polymorphism in variable efficacy of photodynamic therapy for neovascular age-related macular degeneration. Pharmacogenomics. 2009;10(1):81–95. doi: 10.2217/14622416.10.1.81. [DOI] [PubMed] [Google Scholar]
- 109.Brantley MA, Jr, Fang AM, King JM, Tewari A, Kymes SM, Shiels A. Association of complement factor H and LOC387715 genotypes with response of exudative age-related macular degeneration to intravitreal bevacizumab. Ophthalmology. 2007;114(12):2168–2173. doi: 10.1016/j.ophtha.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 110.Lee AY, Raya AK, Kymes SM, Shiels A, Brantley MA., Jr Pharmacogenetics of complement factor H (Y402H) and treatment of exudative age-related macular degeneration with ranibizumab. Br J Ophthalmol. 2009;93(5):610–613. doi: 10.1136/bjo.2008.150995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Anderson DH, Mullins RF, Hageman GS, Johnson LV. A role for local inflammation in the formation of drusen in the aging eye. Am J Ophthalmol. 2002;134(3):411–431. doi: 10.1016/s0002-9394(02)01624-0. [DOI] [PubMed] [Google Scholar]
- 112.Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J. 2000;14(7):835–846. [PubMed] [Google Scholar]
- 113.Jarrett SG, Lin H, Godley BF, Boulton ME. Mitochondrial DNA damage and its potential role in retinal degeneration. Prog Retin Eye Res. 2008;27(6):596–607. doi: 10.1016/j.preteyeres.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 114.Justilien V, Pang JJ, Renganathan K, et al. SOD2 knockdown mouse model of early AMD. Invest Ophthalmol Vis Sci. 2007;48(10):4407–4420. doi: 10.1167/iovs.07-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu Z, Lauer TW, Sick A, Hackett SF, Campochiaro PA. Oxidative stress modulates complement factor H expression in retinal pigmented epithelial cells by acetylation of FOXO3. J Biol Chem. 2007;282(31):22414–22425. doi: 10.1074/jbc.M702321200. [DOI] [PubMed] [Google Scholar]
- 116.Zhang N, Peairs JJ, Yang P, et al. The importance of Bcl-xL in the survival of human RPE cells. Invest Ophthalmol Vis Sci. 2007;48(8):3846–3853. doi: 10.1167/iovs.06-1145. [DOI] [PubMed] [Google Scholar]
- 117.Zhang B, Osborne NN. Oxidative-induced retinal degeneration is attenuated by epigallocatechin gallate. Brain Res. 2006;1124(1):176–187. doi: 10.1016/j.brainres.2006.09.067. [DOI] [PubMed] [Google Scholar]
- 118.Friedman E. Update of the vascular model of AMD. Br J Ophthalmol. 2004;88(2):161–163. doi: 10.1136/bjo.2003.036277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Friedman E, Krupsky S, Lane AM, et al. Ocular blood flow velocity in age-related macular degeneration. Ophthalmology. 1995;102(4):640–646. doi: 10.1016/s0161-6420(95)30974-8. [DOI] [PubMed] [Google Scholar]
- 120.Blumenkranz MS, Russell SR, Robey MG, Kott-Blumenkranz R, Penneys N. Risk factors in age-related maculopathy complicated by choroidal neovascularization. Ophthalmology. 1986;93(5):552–558. doi: 10.1016/s0161-6420(86)33702-3. [DOI] [PubMed] [Google Scholar]
- 121.Guymer R, Luthert P, Bird A. Changes in Bruch’s membrane and related structures with age. Prog Retin Eye Res. 1999;18(1):59–90. doi: 10.1016/s1350-9462(98)00012-3. [DOI] [PubMed] [Google Scholar]
- 122.Boulton M, Dayhaw-Barker P. The role of the retinal pigment epithelium: topographical variation and ageing changes. Eye. 2001;15(Pt 3):384–389. doi: 10.1038/eye.2001.141. [DOI] [PubMed] [Google Scholar]
- 123.Allikmets R. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997;17(1):122. doi: 10.1038/ng0997-122a. [DOI] [PubMed] [Google Scholar]
- 124.Weleber RG, Carr RE, Murphey WH, Sheffield VC, Stone EM. Phenotypic variation including retinitis pigmentosa, pattern dystrophy, and fundus flavimaculatus in a single family with a deletion of codon 153 or 154 of the peripherin/RDS gene. Arch Ophthalmol. 1993;111(11):1531–1542. doi: 10.1001/archopht.1993.01090110097033. [DOI] [PubMed] [Google Scholar]
- 125.Ayyagari R, Zhang K, Hutchinson A, et al. Evaluation of the ELOVL4 gene in patients with age-related macular degeneration. Ophthalmic Genet. 2001;22(4):233–239. doi: 10.1076/opge.22.4.233.2219. [DOI] [PubMed] [Google Scholar]
- 126.Zhang K, Kniazeva M, Han M, et al. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet. 2001;27(1):89–93. doi: 10.1038/83817. [DOI] [PubMed] [Google Scholar]
- 127.Zhang K, Kniazeva M, Hutchinson A, Han M, Dean M, Allikmets R. The ABCR gene in recessive and dominant Stargardt diseases: a genetic pathway in macular degeneration. Genomics. 1999;60(2):234–237. doi: 10.1006/geno.1999.5896. [DOI] [PubMed] [Google Scholar]
- 128.Robman L, Mahdi O, McCarty C, et al. Exposure to Chlamydia pneumoniae infection and progression of age-related macular degeneration. Am J Epidemiol. 2005;161(11):1013–1019. doi: 10.1093/aje/kwi130. [DOI] [PubMed] [Google Scholar]
- 129.Miller DM, Espinosa-Heidmann DG, Legra J, et al. The association of prior cytomegalovirus infection with neovascular age-related macular degeneration. Am J Ophthalmol. 2004;138(3):323–328. doi: 10.1016/j.ajo.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 130.Griffioen AW, Molema G. Angiogenesis: potentials for pharmacologic intervention in the treatment of cancer, cardiovascular diseases, and chronic inflammation. Pharmacol Rev. 2000;52(2):237–268. [PubMed] [Google Scholar]
- 131.Klein ML, Wilson DJ. Clinicopathologic correlation of choroidal and retinal neovascular lesions in age-related macular degeneration. Am J Ophthalmol. 2011;151(1):161–169. doi: 10.1016/j.ajo.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 132.D’Amico DJ. Diseases of the retina. N Engl J Med. 1994;331(2):95–106. doi: 10.1056/NEJM199407143310207. [DOI] [PubMed] [Google Scholar]
- 133.Campochiaro PA. Ocular neovascularisation and excessive vascular permeability. Expert Opin Biol Ther. 2004;4(9):1395–1402. doi: 10.1517/14712598.4.9.1395. [DOI] [PubMed] [Google Scholar]
- 134.Bhutto IA, McLeod DS, Hasegawa T, et al. Pigment epithelium-derived factor (PEDF) and vascular endothelial growth factor (VEGF) in aged human choroid and eyes with age-related macular degeneration. Exp Eye Res. 2006;82(1):99–110. doi: 10.1016/j.exer.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bhutto IA, McLeod DS, Hasegawa T, et al. Pigment epithelium-derived factor (PEDF) and vascular endothelial growth factor (VEGF) in aged human choroid and eyes with age-related macular degeneration. Exp Eye Res. 2006;82(1):99–110. doi: 10.1016/j.exer.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Keck PJ, Hauser SD, Krivi G, et al. Vascular permeability factor, an endothelial cell mitogen related to PDGF. Science. 1989;246(4935):1309–1312. doi: 10.1126/science.2479987. [DOI] [PubMed] [Google Scholar]
- 137.Leu ng DW, Cachianes G, Kuang WJ, Goeddel DV, Ferrara N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. 1989;246(4935):1306–1309. doi: 10.1126/science.2479986. [DOI] [PubMed] [Google Scholar]
- 138.Jiang BH, Zheng JZ, Leung SW, Roe R, Semenza GL. Transactivation and inhibitory domains of hypoxia-inducible factor 1alpha. Modulation of transcriptional activity by oxygen tension. J Biol Chem. 1997;272(31):19253–19260. doi: 10.1074/jbc.272.31.19253. [DOI] [PubMed] [Google Scholar]
- 139.Epstein AC, Gleadle JM, McNeill LA, et al. C elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107(1):43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 140.Jewell UR, Kvietikova I, Scheid A, Bauer C, Wenger RH, Gassmann M. Induction of HIF-1alpha in response to hypoxia is instantaneous. FASEB J. 2001;15(7):1312–1314. [PubMed] [Google Scholar]
- 141.Caro J. Hypoxia regulation of gene transcription. High Alt Med Biol. 2001;2(2):145–154. doi: 10.1089/152702901750265251. [DOI] [PubMed] [Google Scholar]
- 142.Appukuttan B, McFarland TJ, Davies MH, et al. Identification of novel alternatively spliced isoforms of RTEF-1 within human ocular vascular endothelial cells and murine retina. Invest Ophthalmol Vis Sci. 2007;48(8):3775–3782. doi: 10.1167/iovs.06-1172. [DOI] [PubMed] [Google Scholar]
- 143.Lu M, Adamis AP. Vascular endothelial growth factor gene regulation and action in diabetic retinopathy. Ophthalmol Clin North Am. 2002;15(1):69–79. doi: 10.1016/s0896-1549(01)00010-4. [DOI] [PubMed] [Google Scholar]
- 144.Witmer AN, Vrensen GF, Van Noorden CJ, Schlingemann RO. Vascular endothelial growth factors and angiogenesis in eye disease. Prog Retin Eye Res. 2003;22(1):1–29. doi: 10.1016/s1350-9462(02)00043-5. [DOI] [PubMed] [Google Scholar]
- 145.Plate KH, Warnke PC. Vascular endothelial growth factor. J Neurooncol. 1997;35(3):365–372. doi: 10.1023/a:1005845307160. [DOI] [PubMed] [Google Scholar]
- 146.Fuh G, Garcia KC, de Vos AM. The interaction of neuropilin-1 with vascular endothelial growth factor and its receptor flt-1. J Biol Chem. 2000;275(35):26690–26695. doi: 10.1074/jbc.M003955200. [DOI] [PubMed] [Google Scholar]
- 147.Waltenberger J, Claesson-Welsh L, Siegbahn A, Shibuya M, Heldin CH. Different signal transduction properties of KDR and Flt1, two receptors for vascular endothelial growth factor. J Biol Chem. 1994;269(43):26988–26995. [PubMed] [Google Scholar]
- 148.Schwesinger C, Yee C, Rohan RM, et al. Intrachoroidal neovascularization in transgenic mice overexpressing vascular endothelial growth factor in the retinal pigment epithelium. Am J Pathol. 2001;158(3):1161–1172. doi: 10.1016/S0002-9440(10)64063-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nambu H, Umeda N, Kachi S, et al. Angiopoietin 1 prevents retinal detachment in an aggressive model of proliferative retinopathy, but has no effect on established neovascularization. J Cell Physiol. 2005;204(1):227–235. doi: 10.1002/jcp.20292. [DOI] [PubMed] [Google Scholar]
- 150.Woolf AS, Yuan HT. Angiopoietin growth factors and Tie receptor tyrosine kinases in renal vascular development. Pediatr Nephrol. 2001;16(2):177–184. doi: 10.1007/s004670000509. [DOI] [PubMed] [Google Scholar]
- 151.Liu W, Reinmuth N, Stoeltzing O, et al. Antiangiogenic therapy targeting factors that enhance endothelial cell survival. Semin Oncol. 2002;29(3 Suppl 11):96–103. doi: 10.1053/sonc.2002.34061. [DOI] [PubMed] [Google Scholar]
- 152.Jones N, Iljin K, Dumont DJ, Alitalo K. Tie receptors: new modulators of angiogenic and lymphangiogenic responses. Nat Rev Mol Cell Biol. 2001;2(4):257–267. doi: 10.1038/35067005. [DOI] [PubMed] [Google Scholar]
- 153.Kim I, Moon SO, Park SK, Chae SW, Koh GY. Angiopoietin-1 reduces VEGF-stimulated leukocyte adhesion to endothelial cells by reducing ICAM-1, VCAM-1, and E-selectin expression. Circ Res. 2001;89(6):477–479. doi: 10.1161/hh1801.097034. [DOI] [PubMed] [Google Scholar]
- 154.Mori K, Duh E, Gehlbach P, et al. Pigment epithelium-derived factor inhibits retinal and choroidal neovascularization. J Cell Physiol. 2001;188(2):253–263. doi: 10.1002/jcp.1114. [DOI] [PubMed] [Google Scholar]
- 155.Shi XH, He SZ, Zhao SH. Expression and signification of pigment epithelium-derived factor in experimental choroidal neovascularization of rat. Zhonghua Yan Ke Za Zhi. 2004;40(6):404–408. [PubMed] [Google Scholar]
- 156.Bouck N. PEDF: anti-angiogenic guardian of ocular function. Trends Mol Med. 2002;8(7):330–334. doi: 10.1016/s1471-4914(02)02362-6. [DOI] [PubMed] [Google Scholar]
- 157.Dawson DW, Volpert OV, Gillis P, et al. Pigment epithelium-derived factor: a potent inhibitor of angiogenesis. Science. 1999;285(5425):245–248. doi: 10.1126/science.285.5425.245. [DOI] [PubMed] [Google Scholar]
- 158.Tong JP, Chan WM, Liu DT, et al. Aqueous humor levels of vascular endothelial growth factor and pigment epithelium-derived factor in polypoidal choroidal vasculopathy and choroidal neovascularization. Am J Ophthalmol. 2006;141(3):456–462. doi: 10.1016/j.ajo.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 159.Volpert OV, Zaichuk T, Zhou W, et al. Inducer-stimulated Fas targets activated endothelium for destruction by anti-angiogenic thrombospondin-1 and pigment epithelium-derived factor. Nat Med. 2002;8(4):349–357. doi: 10.1038/nm0402-349. [DOI] [PubMed] [Google Scholar]
- 160.Ohno-Matsui K, Morita I, Tombran-Tink J, et al. Novel mechanism for age-related macular degeneration: an equilibrium shift between the angiogenesis factors VEGF and PEDF. J Cell Physiol. 2001;189(3):323–333. doi: 10.1002/jcp.10026. [DOI] [PubMed] [Google Scholar]
- 161.Apte RS, Barreiro RA, Duh E, Volpert O, Ferguson TA. Stimulation of neovascularization by the anti-angiogenic factor PEDF. Invest Ophthalmol Vis Sci. 2004;45(12):4491–4497. doi: 10.1167/iovs.04-0172. [DOI] [PubMed] [Google Scholar]
- 162.Febbraio M, Hajjar DP, Silverstein RL. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J Clin Invest. 2001;108(6):785–791. doi: 10.1172/JCI14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lee CH, Smits PC. Vascular growth factors for coronary angiogenesis. J Interv Cardiol. 2002;15(6):511–518. doi: 10.1111/j.1540-8183.2002.tb01097.x. [DOI] [PubMed] [Google Scholar]
- 164.Matsushima M, Ogata N, Takada Y, et al. FGF receptor 1 expression in experimental choroidal neovascularization. Jpn J Ophthalmol. 1996;40(3):329–338. [PubMed] [Google Scholar]
- 165.Zubilewicz A, Hecquet C, Jeanny JC, Soubrane G, Courtois Y, Mascarelli F. Two distinct signalling pathways are involved in FGF2-stimulated proliferation of choriocapillary endothelial cells: a comparative study with VEGF. Oncogene. 2001;20(12):1403–1413. doi: 10.1038/sj.onc.1204231. [DOI] [PubMed] [Google Scholar]
- 166.Nagineni CN, Samuel W, Nagineni S, et al. Transforming growth factor-beta induces expression of vascular endothelial growth factor in human retinal pigment epithelial cells: involvement of mitogen-activated protein kinases. J Cell Physiol. 2003;197(3):453–462. doi: 10.1002/jcp.10378. [DOI] [PubMed] [Google Scholar]
- 167.Watanabe D, Takagi H, Suzuma K, Oh H, Ohashi H, Honda Y. Expression of connective tissue growth factor and its potential role in choroidal neovascularization. Retina. 2005;25(7):911–918. doi: 10.1097/00006982-200510000-00015. [DOI] [PubMed] [Google Scholar]
- 168.Li J, Zhang YP, Kirsner RS. Angiogenesis in wound repair: angiogenic growth factors and the extracellular matrix. Microsc Res Tech. 2003;60(1):107–114. doi: 10.1002/jemt.10249. [DOI] [PubMed] [Google Scholar]
- 169.Lommatzsch A, Hermans P, Muller KD, Bornfeld N, Pauleikhoff D. Are low inflammatory reactions involved inBird AC, exudative age-related macular degeneration? Morphological and immunhistochemical analysis of AMD associated with basal deposits. Graefes Arch Clin Exp Ophthalmol. 2008;246(6):803–810. doi: 10.1007/s00417-007-0749-4. [DOI] [PubMed] [Google Scholar]
- 170.Hoffmann J, Alt A, Lin J, et al. Tenilsetam prevents early diabetic retinopathy without correcting pericyte loss. Thromb Haemost. 2006;95(4):689–695. [PubMed] [Google Scholar]
- 171.Patarroyo M, Tryggvason K, Virtanen I. Laminin isoforms in tumor invasion, angiogenesis and metastasis. Semin Cancer Biol. 2002;12(3):197–207. doi: 10.1016/S1044-579X(02)00023-8. [DOI] [PubMed] [Google Scholar]
- 172.Hynes RO. A reevaluation of integrins as regulators of angiogenesis. Nat Med. 2002;8(9):918–921. doi: 10.1038/nm0902-918. [DOI] [PubMed] [Google Scholar]
- 173.Bernardini G, Ribatti D, Spinetti G, et al. Analysis of the role of chemokines in angiogenesis. J Immunol Methods. 2003;273(1–2):83–101. doi: 10.1016/s0022-1759(02)00420-9. [DOI] [PubMed] [Google Scholar]
- 174.Grossniklaus HE, Ling JX, Wallace TM, et al. Macrophage and retinal pigment epithelium expression of angiogenic cytokines in choroidal neovascularization. Mol Vis. 2002;8:119–126. [PubMed] [Google Scholar]
- 175.Tsutsumi C, Sonoda KH, Egashira K, et al. The critical role of ocular-infiltrating macrophages in the development of choroidal neovascularization. J Leukoc Biol. 2003;74(1):25–32. doi: 10.1189/jlb.0902436. [DOI] [PubMed] [Google Scholar]
- 176.Spinetti G, Camarda G, Bernardini G, Romano Di Peppe S, Capogrossi MC, Napolitano M. The chemokine CXCL13 (BCA-1) inhibits FGF-2 effects on endothelial cells. Biochem Biophys Res Commun. 2001;289(1):19–24. doi: 10.1006/bbrc.2001.5924. [DOI] [PubMed] [Google Scholar]
- 177.Cao Y, Cao R, Veitonmaki N. Kringle structures and antiangiogenesis. Curr Med Chem Anticancer Agents. 2002;2(6):667–681. doi: 10.2174/1568011023353705. [DOI] [PubMed] [Google Scholar]
- 178.Fukumura D, Gohongi T, Kadambi A, et al. Predominant role of endothelial nitric oxide synthase in vascular endothelial growth factor-induced angiogenesis and vascular permeability. Proc Natl Acad Sci U S A. 2001;98(5):2604–2609. doi: 10.1073/pnas.041359198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 180.Browning BL, Yu Z. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am J Hum Genet. 2009;85(6):847–861. doi: 10.1016/j.ajhg.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 182.Wang G, Spencer KL, Scott WK, et al. Analysis of the indel at the ARMS2 3′UTR in age-related macular degeneration. Hum Genet. 2010;127(5):595–602. doi: 10.1007/s00439-010-0805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fanous AH, Neale MC, Webb BT, et al. A genome-wide scan for modifier loci in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(5):589–595. doi: 10.1002/ajmg.b.30442. [DOI] [PubMed] [Google Scholar]
- 184.Boekhoorn SS, Vingerling JR, Witteman JC, Hofman A, de Jong PT. C-reactive protein level and risk of aging macula disorder: The Rotterdam Study. Arch Ophthalmol. 2007;125(10):1396–1401. doi: 10.1001/archopht.125.10.1396. [DOI] [PubMed] [Google Scholar]
- 185.Despriet DD, Klaver CC, Witteman JC, et al. Complement factor H polymorphism, complement activators, and risk of age-related macular degeneration. JAMA. 2006;296(3):301–309. doi: 10.1001/jama.296.3.301. [DOI] [PubMed] [Google Scholar]
- 186.Feng X, Xiao J, Longville B, et al. Complement factor H Y402H and C-reactive protein polymorphism and photodynamic therapy response in age-related macular degeneration. Ophthalmology. 2009;116(10):1908–1912. e1901. doi: 10.1016/j.ophtha.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 187.McGwin G, Hall TA, Xie A, Owsley C. The relation between C reactive protein and age related macular degeneration in the Cardiovascular Health Study. Br J Ophthalmol. 2005;89(9):1166–1170. doi: 10.1136/bjo.2005.067397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Schaumberg DA, Christen WG, Buring JE, Glynn RJ, Rifai N, Ridker PM. High-sensitivity C-reactive protein, other markers of inflammation, and the incidence of macular degeneration in women. Arch Ophthalmol. 2007;125(3):300–305. doi: 10.1001/archopht.125.3.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Seddon JM, Gensler G, Milton RC, Klein ML, Rifai N. Association between C-reactive protein and age-related macular degeneration. JAMA. 2004;291(6):704–710. doi: 10.1001/jama.291.6.704. [DOI] [PubMed] [Google Scholar]
- 190.Seddon JM, George S, Rosner B, Rifai N. Progression of age-related macular degeneration: prospective assessment of C-reactive protein, interleukin 6, and other cardiovascular biomarkers. Arch Ophthalmol. 2005;123(6):774–782. doi: 10.1001/archopht.123.6.774. [DOI] [PubMed] [Google Scholar]
- 191.Wang JJ, Ross RJ, Tuo J, et al. The LOC387715 polymorphism, inflammatory markers, smoking, and age-related macular degeneration. A population-based case-control study. Ophthalmology. 2008;115(4):693–699. doi: 10.1016/j.ophtha.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang Y, Bian ZM, Yu WZ, Yan Z, Chen WC, Li XX. Induction of interleukin-8 gene expression and protein secretion by C-reactive protein in ARPE-19 cells. Exp Eye Res. 2010;91(2):135–142. doi: 10.1016/j.exer.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 193.Kopplin LJ, Igo RP, Jr, Wang Y, et al. Genome-wide association identifies SKIV2L and MYRIP as protective factors for age-related macular degeneration. Genes Immun. 2010;11(8):609–621. doi: 10.1038/gene.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mitchell P, Korobelnik JF, Lanzetta P, et al. Ranibizumab (Lucentis) in neovascular age-related macular degeneration: evidence from clinical trials. Br J Ophthalmol. 2010;94(1):2–13. doi: 10.1136/bjo.2009.159160. [DOI] [PubMed] [Google Scholar]
- 195.Klein ML, Francis PJ, Rosner B, et al. CFH and LOC387715/ARMS2 genotypes and treatment with antioxidants and zinc for age-related macular degeneration. Ophthalmology. 2008;115(6):1019–1025. doi: 10.1016/j.ophtha.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 196.Pei XT, Li XX, Bao YZ, et al. Association of c3 gene polymorphisms with neovascular age-related macular degeneration in a chinese population. Curr Eye Res. 2009;34(8):615–622. doi: 10.1080/02713680903003484. [DOI] [PubMed] [Google Scholar]
- 197.Lima LH, Schubert C, Ferrara DC, et al. Three major loci involved in age-related macular degeneration are also associated with polypoidal choroidal vasculopathy. Ophthalmology. 2010;117(8):1567–1570. doi: 10.1016/j.ophtha.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kimchi-Sarfaty C, Oh JM, Kim IW, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315(5811):525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 199.Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM. Silent polymorphisms speak: how they affect pharmacogenomics and the treatment of cancer. Cancer Res. 2007;67(20):9609–9612. doi: 10.1158/0008-5472.CAN-07-2377. [DOI] [PubMed] [Google Scholar]
- 200.Zhang F, Saha S, Shabalina SA, Kashina A. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science. 2010;329(5998):1534–1537. doi: 10.1126/science.1191701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3(4):285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 202.Hunt R, Sauna ZE, Ambudkar SV, Gottesman MM, Kimchi-Sarfaty C. Silent (synonymous) SNPs: should we care about them? Methods Mol Biol. 2009;578:23–39. doi: 10.1007/978-1-60327-411-1_2. [DOI] [PubMed] [Google Scholar]
- 203.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461(7261):272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Li Y, Vinckenbosch N, Tian G, et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat Genet. 2010;42(11):969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 205.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83(3):311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol. 2010;34(2):188–193. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Durbin RM, Abecasis GR, Altshuler DL, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Cao J, Zhao L, Li Y, et al. A subretinal matrigel rat choroidal neovascularization (CNV) model and inhibition of CNV and associated inflammation and fibrosis by VEGF trap. Invest Ophthalmol Vis Sci. 2010;51(11):6009–6017. doi: 10.1167/iovs.09-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Grupe A, Li Y, Rowland C, et al. A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am J Hum Genet. 2006;78(1):78–88. doi: 10.1086/498851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wang X, Cheng S, Brophy VH, et al. A meta-analysis of candidate gene polymorphisms and ischemic stroke in 6 study populations: association of lymphotoxin-alpha in nonhypertensive patients. Stroke. 2009;40(3):683–695. doi: 10.1161/STROKEAHA.108.524587. [DOI] [PMC free article] [PubMed] [Google Scholar]