Abstract
The T-box transcription factor Eomesodermin (Eomes) has been implicated in patterning and morphogenesis in frog, fish and mouse. In zebrafish, one of the two Eomes homologs, Eomesa, has been implicated in dorsal-ventral patterning, epiboly and endoderm specification in experiments employing over-expression, dominant-negative constructs and antisense morpholino oligonucleotides. Here we report for the first time the identification and characterization of an Eomesa mutant generated by TILLING. We find that Eomesa has a strictly maternal role in the initiation of epiboly, which involves doming of the yolk cell up into the overlying blastoderm. By contrast, epiboly progression is normal, demonstrating for the first time that epiboly initiation is genetically separable from progression. The yolk cell microtubules, which are required for epiboly, are defective in maternal-zygotic eomesa mutantembryos. In addition, the deep cells of the blastoderm are more tightly packed and exhibit more bleb-like protrusions than cells in control embryos. We postulate that the doming delay may be the consequence both of overly stabilized yolk cell microtubules and defects in the adhesive properties or motility of deep cells. We also show that Eomesa is required for normal expression of the endoderm markers sox32, bon and og9x; however it is not essential for endoderm formation.
Keywords: zebrafish, epiboly, eomesodermin, endoderm, T-box, gastrulation
Introduction
The T-box transcription factor Eomesodermin (Eomes) has been implicated in patterning and morphogenesis in frog, fish and mouse. In Xenopus, where Eomes was first identified, ectopic expression in animal caps leads to a concentration-dependent induction of mesodermal gene expression, with higher doses inducing expression of more dorsal mesodermal markers (Ryan et al., 1996). In zebrafish, there are two eomes genes (a and b) (Takizawa et al., 2007), with eomesa being the more intensively studied. Over-expression of Eomesa leads to ectopic expression of dorsal organizer genes and secondary axes are induced when Eomesa is expressed ventrally (Bruce et al., 2003). In addition, Eomesa has been shown to play a role in induction of the endoderm gene sox32 (Bjornson et al., 2005). More recent work has also shown that Eomesa acts in combination with the transcription factor FoxH1 to specify mesendoderm (Slagle et al., 2011). In the mouse, Eomes mutants completely lack definitive endoderm, while mesodermal patterning is relatively unaffected (Arnold et al., 2008; Russ et al., 2000).
Eomes is also important for normal gastrulation movements. Expression of dominant-negative Eomes constructs in Xenopus embryos leads to gastrulation arrest (Ryan et al., 1996). In zebrafish embryos, a similar Eomesa construct produces abnormal epiboly, which is the first coordinated cell movement during development (Bruce et al., 2005; Lepage and Bruce, 2010; Warga and Kimmel, 1990). Knock-out of Eomes in the mouse epiblast results in gastrulation defects due to blocked migration of prospective mesoderm away from the primitive streak (Arnold et al., 2008; Russ et al., 2000). This migratory defect appears to be due to the failure to down-regulate expression of the adhesion molecule E-Cadherin (Arnold et al., 2008).
In zebrafish, unlike Xenopus, Eomesa transcript and protein are maternally expressed (Bruce et al., 2003; Ryan et al., 1996). Early development in zebrafish, as in many animals, relies upon maternal stores of mRNA and protein that orchestrate development up to the midblastula transition when zygotic transcription begins (Abrams and Mullins, 2009). Previous work on Eomesa in zebrafish relied upon over-expression, morpholino oligonucleotides, and dominant-negative constructs (Bjornson et al., 2005; Bruce et al., 2005; Bruce et al., 2003; Slagle et al., 2011). Over-expression and dominant-negative constructs can have non-specific effects and morpholinos have no impact on maternal stores of protein. The caveats of these tools are reflected by the fact that there is confusion in the literature surrounding some aspects of Eomesa expression and function (Bjornson et al., 2005; Bruce et al., 2005; Bruce et al., 2003). Thus our understanding of the role of Eomesa during zebrafish development is far from complete. Here we report the phenotype of a loss of function eomesa allele generated by TILLING (Moens et al., 2008). Our characterization of embryos lacking either or both maternal and zygotic Eomesa, as well as the generation of an effective Eomesa antibody, has allowed us to gain new insights into Eomesa function. We find that Eomesa has a strictly maternal role in the initiation of epiboly, while epiboly progression is normal: demonstrating for the first time that epiboly initiation is genetically separable from progression. We also show that Eomesa plays a role in, but is not essential for, endoderm formation.
Material and Methods
Zebrafish
We generated a nonsense allele of eomesa, designated fh105, by TILLING (Moens et al., 2008). AB and eomesafh105 zebrafish were maintained and staged as described (Kimmel et al., 1995). Wild type embryos were obtained from natural matings. Homozygous eomesfh105 mutant embryos were obtained by in vitro fertilization as described (Westerfield, 1993). Heterozygous eomesfh105 mutant embryos were obtained by in vitro fertilization using homozygous eomesafh105 sperm and wild type eggs. Maternal eomesa mutant embryos were obtained by in vitro fertilization using homozygous eomesafh105 eggs and wild type sperm. Zeomesa mutant embryos were obtained by natural matings of heterozygous individuals and were confirmed by PCR genotyping (see below). Animals were treated in accordance with the policies of the University of Toronto Animal Care Committee.
Generation of Eomesa Antibody
The cDNA sequence encoding amino acids 1-661 was cloned into pETM-14 vector (EMBL Protein Expression and Purification Facility, http://www.helmholtz-muenchen.de/en/pepf/materials/vector-database/bacterial-expression-vectors/index.html) to express a His-tag fusion protein in E. coli (BL21 strain DE3). The His-tag protein was isolated according to the protocol provided by the Bjorkman Group, Howard Hughes Medical Institute, California Institute of Technology (http://www.its.caltech.edu/~bjorker/protocols.html), for the extraction of inclusion bodies (Fig. S1) and then stored in 8M urea. Two rabbits were immunized with the His-tag fusion protein, using a 3-months standard protocol by Eurogentec S.A., Seraing – Belgium.
In Situ Hybridization
In situ hybridizations were performed as described (Jowett and Lettice, 1994). Antisense riboprobes for bmp2b (Martinez-Barbera et al., 1997); bon and sox17 (Alexander and Stainier, 1999); cdh1 (Kane et al., 2005); fgf8a (Reifers et al., 1998); flh (Talbot et al., 1995); gata5 (Rodaway et al., 1999); gsc (Stachel et al., 1993); lfty1 (Bisgrove et al., 1999), mxtx2 (Hirata et al., 2000); ndr1 (Erter et al., 1998; Feldman et al., 1998), ndr2 (Rebagliati et al., 1998; Sampath et al., 1998); ntla (Schulte-Merker et al., 1994); og9x (Poulain and Lepage, 2002) and sox32 (Alexander et al., 1999) were generated as described.
Whole-mount Immunohistochemistry
Antibody and phalloidin staining were performed as described (Bruce et al., 2001; Topczewski and Solnica-Krezel, 1999). Antibodies were used as follows: anti-Eomesa (1: 500), anti–α– Tubulin (1:500; Sigma Aldrich), anti-E-Cadherin (1:2500) (Babb and Marrs, 2004) and goat-anti-mouse Alexa 488 secondary (1:1000, Invitrogen). Embryos were mounted in ProLong Gold Antifade reagent (Invitrogen) in agarose wells on a glass bottom dish (MatTek Corporation) for analysis on a Zeiss LSM510 confocal microscope or analyzed on a Leica MZ16F stereomicroscope.
Western Blots
Embryos at sphere stage were dechorionated and blastoderm caps were manually dissected off the yolk cell. Thirty blastoderm caps were homogenized in RIPA lysis buffer (Sigma Aldrich) supplemented with EDTA (1 μM) inhibition, and protease inhibitor cocktail (Roche). Two to fifteen embryo equivalents were loaded into each lane. Protein extracts were run on a 10% SDS denaturing protein gel. Gels were transferred onto nitrocellulose filter papers using the Bio-Rad semi-dry transfer apparatus and blocked in 5% milk in PBT (0.1% Tween -20 in PBS). Blots were incubated in primary antibodies diluted in 5% milk overnight at 4°C, washed in PBT (5 × 10 minutes) and incubated in secondary antibodies for 1 hr at room temperature. The following primary antibodies were used: rabbit anti-E-Cadherin (1:2500) (Babb and Marrs, 2004), anti-Eomesa antibody (1: 750), anti-α–Tubulin (1: 2000; Sigma Aldrich) and anti-β–Catenin (1:250; Sigma Aldrich).
PCR Genotyping and RT-PCR
Genotyping for the presence of the eomesfh105 allele was performed as described (Zebrafish International Resource Center, www.zebrafish.org). For RT-PCR, RNA was prepared using TRIzol reagent (Invitrogen) following the manufacturer's instructions. cDNA synthesis was performed using the AffinityScript cDNA synthesis kit (Stratagene). PCR was performed for 28 cycles using the cycling parameters and primer sequences from the genotyping assay.
Microinjections and Constructs
Microinjections were performed and RNA for eomesa-VP16, eomesa and gfp were made as described previously (Bruce et al., 2003). 50 pg of the RNAs were injected per embryo. Alexa 488 conjugated Histone H1 (Invitrogen) was injected as described (Carvalho et al., 2009). The splice blocking morpholino used to test the specificity of the Eomesa antibody targets the exon/intron 2 boundary and has the following sequence: GTAATGCTTCATTTCTTACCTGCC (GeneTools, LLC). RT-PCR using an exon 2 forward primer (GACGCGCGTAAAAGTTCTC) and an exon 3 reverse primer (CTTGATGTTGTTGTCCGCTTTC) was used to confirm the specificity of the splice blocking morpholino. In splice blocking morpholino injected embryos a band of 760 base pairs (bp) instead of the expected 600 bp was PCR amplified from cDNA. Sequencing of the product revealed retention of intron 2, which contains several stop codons. This corresponded to the truncated protein fragment of approximately 25 kDa shown in Figure 1C.
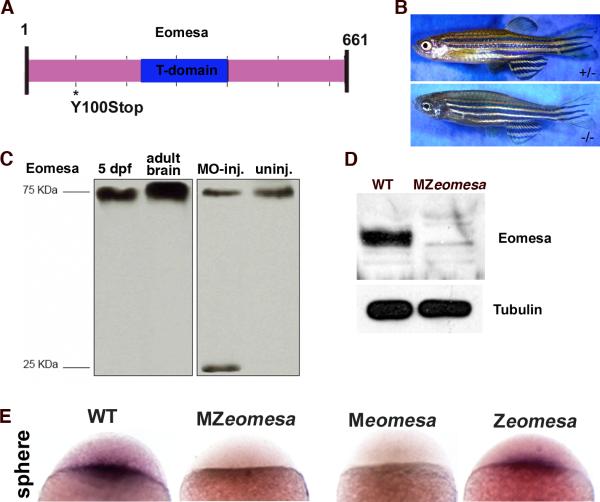
Figure 1. eomesafh105 mutant allele.
Schematic of the 661 amino acid full-length Eomesa protein with T-domain in blue. Location of the stop codon in allele fh105 marked by asterisk. (B) Images of heterozygous (top) and homozygous (bottom) eomesafh105 adult fish. (C) Control western blot for the anti-Eomesa antibody, lanes as indicated. (D)Western blot of sphere stage wild type and MZeomesa embryos, lanes as indicated. 15 embryo equivalents loaded per lane. (E) Whole-mount in situ hybridization for eomesa on sphere stage embryos of the indicated genotypes.
Imaging
Confocal data were acquired using a Zeiss LSM 510 microscope. Fixed embryos were mounted in wells made in 2% low melt agarose (Sigma Aldrich) in glass bottom culture dishes (MatTek, Ashland, MA).
Results
Characterization of the eomesafh105 mutant allele
The eomesafh105 allele was generated by TILLING and it contains a point mutation that changes a tyrosine at position 100 to a stop codon (Fig. 1A). The predicted truncated protein is unlikely to function, as it would lack most of the protein, including the critical DNA-binding T-domain. Homozygous mutant embryos are viable, although they consistently lack the dorsal fin (Fig. 1B). The lack of dorsal fin is intriguing given the known roles other T-box proteins Tbx5 and Tbx4 in forelimb and hindlimb development, respectively (Naiche et al., 2005). However, this feature of the mutant phenotype has not been investigated further.
We next wanted to examine Eomesa protein levels to assess whether the allele is a protein null. Previous reports using two antibodies made against portions of the eomesa sequence yielded contradictory results (Bjornson et al., 2005; Bruce et al., 2003), prompting us to generate an antibody against the full-length Eomesa protein. Our antiserum detected a single 75-kDa protein band on immunoblots of whole 5 days post-fertilization (dpf) larval lysates (Fig. 1C, lane 1) and a much stronger band of the same size in adult brain lysates (Fig. 1C, lane 2). The bands are slightly larger than the predicted 72-kDa and may suggest the presence of post-translational modifications in vivo. To confirm the specificity of the antibody, embryos were injected with a splice blocking morpholino targeting the second exon/intron boundary of eomesa. PCR amplification and sequencing confirmed that morpholino injection resulted in aberrant splicing of the eomesa transcript (see Materials and Methods). In protein extracts from morpholino injected embryos a much smaller band was detected, which corresponded to the truncated protein (Fig. 1C, lane 3) and was absent in uninjected embryos (Fig. 1C, lane 4).
We then performed Western blots on sphere stage wild type and mutant embryos. Eomesa was greatly reduced in embryos mutant for both maternal and zygotic Eomesa (MZeomesa) (Fig. 1D) and we did not detect the predicted truncated protein that would be generated from the mutant allele (not shown). In addition, no protein was detected by whole-mount immunohistochemistry of MZeomesa embryos (Fig. S2A). Taken together, the results strongly suggest that fh105 is a null allele. We also failed to detect eomesa transcript in sphere stage MZeomesa and Meomesa embryos by whole mount in situ hybridization, suggesting that the transcript is degraded (Fig. 1E). In previous work, we showed that ectopic eomesa leads to transcription from the endogenous eomesa gene (Bruce et al., 2003). Thus, maternal Eomesa might be required to activate the expression of zygotic eomesa. eomesa expression was normal in Zeomesa embryos at sphere stage, which could indicate activation of zygotic expression by maternal Eomesa or, alternatively, could represent the maternal transcript alone (Fig. 1E). At 60% epiboly no nuclear localized Eomesa could be detected in Zeomesa embryos, which was in contrast to control embryos in which nuclear staining was clearly visible (Fig. S2B-C). This suggests that maternal Eomesa levels decline by this stage. In addition, we failed to detect nuclear localized Eomesa protein in Zeomesa embryos at 1 dpf (Fig. S2D,E).
Eomesa is nuclear localized during blastula and gastrula stages
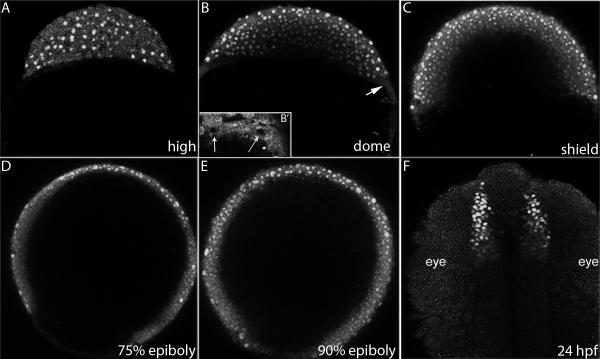
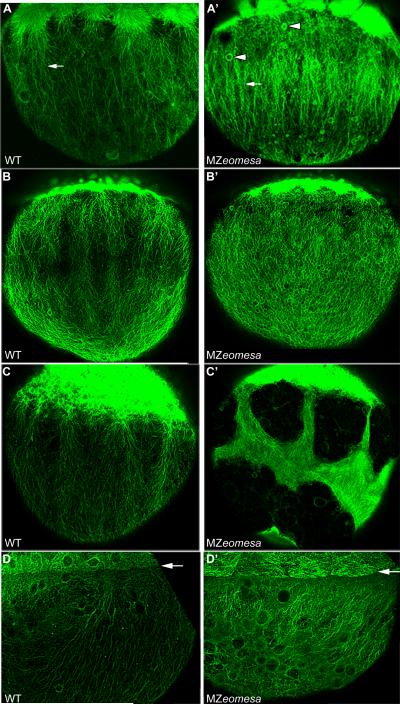
An important unanswered question from previous work is the distribution of Eomesa in the early embryo. Our previous work indicated that at late blastula and early gastrula stages Eomesa was distributed throughout the blastoderm, while work by others suggested that expression was primarily confined to the marginal mesendodermal region (Bjornson et al., 2005; Bruce et al., 2003). With our improved antibody, we were able to revisit this issue by performing whole-mount antibody staining of wild type oocytes and embryos at different stages. Eomesa protein was detected around the germinal vesicle of late stage oocytes and was distributed throughout the cytoplasm of cleavage stage embryos (data not shown), confirming the presence of maternal protein. Nuclear localized Eomesa protein was first detected throughout the blastoderm beginning at high stage, with some cytoplasmic staining still visible (Fig. 2A). During late blastula and gastrula stages, nuclear localized protein was detected throughout the blastoderm, including the enveloping layer (Fig. 2B-E, S3). Although cytoplasmic staining could be seen in the yolk syncytial layer (YSL) at sphere and dome stage (4 and 4.3 hours post-fertilization, hpf; arrow, Fig. 2B, B’), Eomesa was not detected in YSL nuclei (Fig. 2B’, S3B), suggesting that it may not function as a transcription factor in this region during epiboly initiation. Nuclear localization was also seen in the telencephalon at 24 hours post-fertilization (hpf, Fig. 2F), consistent with previous work on eomesa transcript localization (Mione et al., 2001).
Figure 2. Eomesa protein distribution.
Confocal projections of embryos stained with anti-Eomesa antibody. (A-E) lateral views (F) dorsal view. Stages indicated in lower right. Arrow in (B) indicates the YSL. (B’) Inset shows YSL of embryo at sphere stage. Arrows indicate unstained YSL-nuclei that are surrounding by Eomesa positive YSL cytoplasm.
Maternal Eomesa is required for the timely initiation of epiboly
Our previous work, using a dominant-negative construct, implicated Eomesa in the control of epiboly initiation (Bruce et al., 2005). To examine the eomesa mutant phenotype, time matched wild type (or eomesa heterozygous embryos) and MZeomesa mutant embryos were observed. Early cleavages proceeded normally in MZeomesa mutant embryos. MZeomesa embryos reached sphere stage on schedule (Fig. 3A,G) and had normal nuclear morphology (Fig. S4). When wild type embryos had domed, marking the initiation of epiboly, MZeomesa mutant embryos were still at sphere stage (Fig. 3B,H). All MZeomesa mutant embryos examined displayed a 1-hour delay in doming (n =582).
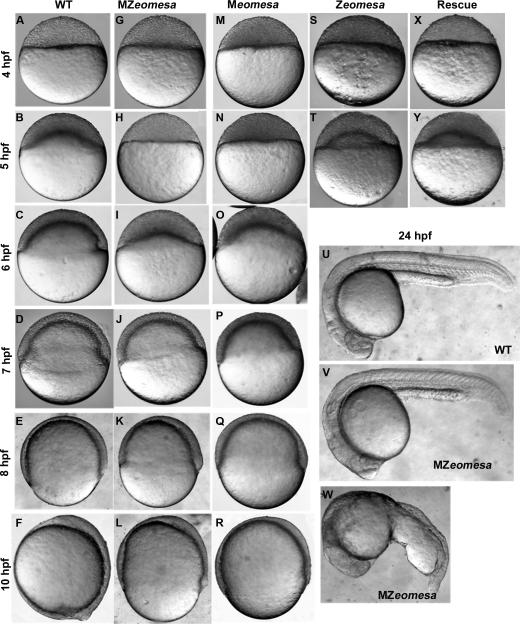
Figure 3. Phenotypes of MZeomesa, Meomesa and Zeomesa mutant embryos.
Lateral views of live embryos, stages and genotypes as indicated. Doming is delayed in MZeomesa (H) and Meomesa (N) but not Zeomesa embryos (T). (X, Y) Injection of eomesa-VP16 rescues the timing of epiboly initiation.
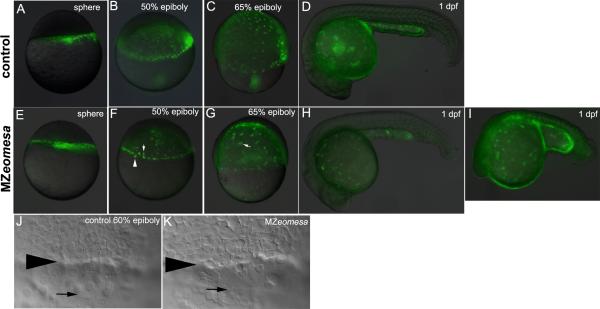
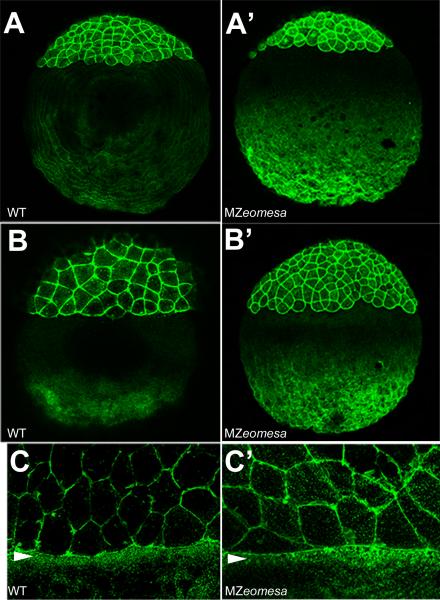
Interestingly, epiboly progression is not delayed in mutant embryos. Wild type embryos reached bud stage by 10 hpf, whereas MZeomesa embryos reached bud stage by 11 hpf. However, MZeomesa embryos often had a wider and more elongated yolk cell than wild type embryos during gastrulation (Fig. 3L). To examine the distribution of yolk syncytial layer nuclei (YSN) and the integrity of the YSL, we injected the YSL of control and MZeomesa embryos with fluorescent histone (Carvalho et al., 2009). In MZeomesa embryos epiboly of the YSL was not delayed and the YSL maintained its integrity up to 1 dpf (Fig. 4E-I). Crowding or contraction of the YSN occurred normally in mutant embryos, as seen at 50% epiboly (Fig. 4 compare F to B). YSN were present in mutant embryos although the morphology of some nuclei was abnormal (Fig. 4F) and clumping of nuclei was visible during epiboly progression (arrow, Fig. 4G). In wild type embryos, the EVL is positioned ahead of the deep cells during epiboly (Solnica-Krezel and Driever, 1994). We examined the relative positioning of EVL and deep cells during epiboly by DIC microscopy and found that it was normal in mutant embryos during epiboly progression (Fig. 4J,K).
Figure 4. Epiboly progression is normal in MZeomesa embryos.
(A-I) Overlays of fluorescent and brighfield images of control (A-D) and MZeomesa (E-I) embryos injected into the YSL with fluorescent histone to highlight the YSN. Stages as indicated. (F) Arrowhead and arrow indicate abnormally large and small nuclei, respectively. (G) Arrow indicates cluster of clumped nuclei not visible in the control embryo in (C). Wild type (H) and abnormal looking (I) MZeomesa embryos at 1 dpf have intact YSLs (J,K) DIC images of the margin of 60% epiboly control (J) and MZeomesa (K) embryo. Arrowhead indicates deep cell margin, arrow indicates EVL margin.
Taken all together, these results indicate that the epiboly delay observed in MZeomesa embryos is limited to doming, and further demonstrate that the control of epiboly initiation can be separated from that of epiboly progression. At 24 hpf, an average of 56% of mutant embryos looked wild type (with some batch to batch variation) and 44% displayed a variety of yolk defects with abnormally large yolk extensions (Fig. 3U-W). Embryos that looked normal at 24 hpf could be raised to adulthood.
To determine whether the defect in epiboly initiation was due to loss of either maternal or zygotic Eomesa alone, we examined the timing of doming in Meomesa and Zeomesa embryos and found that doming was delayed in Meomesa embryos (100%, n = 108, Fig. 3M,N), but not in Zeomesa embryos (confirmed by PCR genotyping, n = 32, Fig. 3 S,T). Thus, zygotic Eomesa is not required for epiboly initiation. However, to ensure that embryos lacked all Eomesa function during early development, we performed most of our analyses on MZeomesa embryos.
To confirm that the epiboly defect indeed reflected the loss of Eomesa function, we attempted to rescue mutant embryos by injecting eomesa or eomesa-VP16 (previously shown to mimic native Eomesa function) mRNAs into MZeomesa embryos at the 1-cell stage (Bruce et al., 2003). Both constructs rescued the doming delay and yolk morphology, though eomesa-VP16 was more effective, rescuing 70% (n = 230) of injected MZeomesa embryos (Fig. 3Y), while injection of gfp RNA had no effect (not shown). Injection of eomesa-VP16 into the YSL just after its formation was unable to rescue doming (not shown), further suggesting that Eomesa does not function in the yolk during doming.
The yolk microtubule cytoskeleton is abnormal in mutant embryos
To examine the doming delay in more detail, we performed time-lapse microscopy on mutant and wild type embryos. This analysis revealed the presence of abnormal contractions at the blastoderm/yolk interface (Supplemental Movie). When doming finally occurred, there was considerably less upward movement within the yolk as compared to control embryos. In addition, the shape of the domed yolk in mutant embryos did not resemble wild type. In wild type embryos the yolk bulges to the greatest extent at the center of the blastoderm and to a lesser extent at the periphery, whereas MZeomesa mutant embryos displayed a more uniform yolk bulge. These observations indicated that there might be defects in the yolk cytoskeleton, which has been previously implicated in driving epiboly movements (Solnica-Krezel and Driever, 1994; Strähle et al., 1993).
We first examined the microtubule cytoskeleton by anti–α–Tubulin antibody staining and confocal microscopy. Previous work showed that prior to doming there are two microtubule arrays in the yolk cell (Solnica-Krezel and Driever, 1994). Microtubules surround each of the yolk syncytial nuclei and a second array, implicated in epiboly, is organized longitudinally along the animal-vegetal axis (Solnica-Krezel and Driever, 1994). Epiboly is delayed when microtubules are disrupted using U.V. light or treatment with either the microtubule depolymerizing drug nocodazole or the stabilizing drug taxol (Solnica-Krezel and Driever, 1994; Strähle and Jesuthasan, 1993). In MZeomesa embryos microtubules were overtly normal in deep and EVL cells at all stages. At high stage, longitudinal microtubule arrays in the yolk were visible in both wild type and MZeomesa embryos (arrows, Fig. 5A,A’). While the microtubules surrounding the yolk syncytial nuclei were normal, the longitudinally organized microtubules appeared to be more dense in MZeomesa embryos and spherical structures were often visible which did not co-stain with nuclear markers (arrowheads, Fig. 5A’ and not shown). These spherical structures were also visible as early as cleavage stages.
Figure 5. Yolk cell microtubules are altered in MZeomesa embryos.
Confocal projections of lateral views of wild type (A-D) and MZeomesa (A’-D’) embryos stained for Tubulin. (A, A’) High stage, (B, B’) sphere stage (C, C’) dome stage, (D, D’) 75% epiboly. (A, A’) Arrows indicate longitudinal microtubule arrays. Arrowheads indicate spherical structures in MZeomesa embryo. (D, D’) arrows indicate blastoderm-yolk cell boundary.
At sphere stage mutant embryos lacked the longitudinal microtubule array. Instead, a dense and disorganized web of microtubules covered the yolk surface (Fig. 5B,B’). In some sphere stage mutant embryos, microtubules were abnormally bundled, leaving large regions of the yolk devoid of Tubulin (not shown). By dome stage, 100% of MZeomesa embryos examined had abnormally bundled yolk microtubules, with large regions of the yolk lacking Tubulin staining (Fig. 5C,C’). The severity of the microtubule defects at dome stage, coupled with the lack of delay in epiboly progression, led us to examine the microtubules at 75% epiboly. Although the spherical structures were still visible, most embryos no longer had large regions devoid of Tubulin staining (Fig. 5D,D’, no voids in 22/30 embryos). These results suggest that microtubule organization is restored at later stages, consistent with the lack of delay in epiboly progression.
Injection of eomesa-VP16 RNA into 1-cell stage MZeomesa embryos was able to partially rescue the microtubule defects in addition to rescuing the timing of epiboly initiation (Fig. S5). Although spherical structures were still visible in the yolk of eomesa-VP16 RNA injected MZeomesa embryos, the microtubules were more organized and void regions were no longer present. Thus, there was a correlation between the rescue of doming and yolk cell microtubule morphology.
The yolk cell actin cytoskeleton is normal in MZeomesa embryos
The actin cytoskeleton has been implicated primarily in epiboly progression (Cheng et al., 2004; Köppen et al., 2006; Zalik et al., 1999). We examined actin in wild type and mutant embryos by phalloidin staining and confocal microscopy. The actin cytoskeleton of deep cells appeared to be normal (not shown). In the yolk cell at sphere and dome stages, actin at the vegetal pole appeared to be more dense (Fig. 6A-B’). Actin at the base of the yolk has been postulated to play a role in maintaining the integrity of the yolk cell (Cheng et al., 2004). In contrast to what we observed for microtubules, we did not detect abnormal actin cables or regions of the yolk devoid of actin. Consistent with the observation that epiboly progression is unaffected in mutant embryos, the marginal actin band, which is critical for progression and closure of the blastopore (Cheng et al., 2004; Köppen et al., 2006), was normal in mutant embryos (arrowhead, Fig. 6C,C’).
Figure 6. The actin cytoskeleton is normal is MZeomesa embryos.
Confocal projections of lateral views of phalloidin stained embryos. (A-C) wild type and (A’-C’) MZeomesa embryos. (A, A’) sphere stage (B, B’) dome stage, (C, C’) close up of marginal region at 75% epiboly, arrowheads indicate actin band in the YSL.
Cellular morphology is altered in mutant embryos
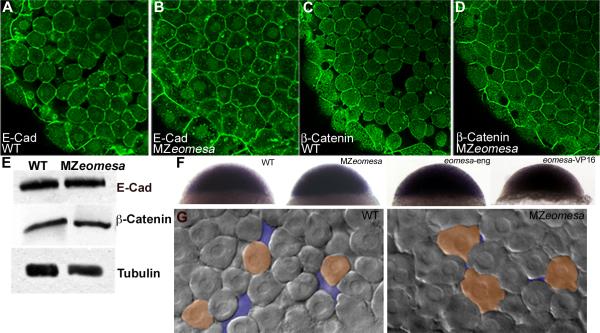
As Eomesa is expressed in the deep cells of the blastoderm, we were interested in examining these cells for possible cell autonomous defects. In the mouse, it was shown that mesodermal migration through the primitive streak is blocked in Eomes mutant embryos due to a failure to down regulate the cell adhesion molecule E-Cadherin (Cdh1) (Arnold et al., 2008). Thus, we were interested in examining whether Cdh1 was upregulated in MZeomesa mutant embryos, as this might contribute to the doming delay. If Eomesa were to negatively regulate cdh1 expression, this regulation would presumably be indirect, as Eomesa appears to be a transcriptional activator (Bjornson et al., 2005; Bruce et al., 2003). Confocal imaging of sphere stage embryos stained with Cdh1 and β-Catenin antibodies revealed that mutant cells appeared to be in greater contact with neighboring cells, with less space in between them when compared to wild type cells (Fig. 7A-D). However, Western blot analyses did not reveal obvious increases in the levels of either protein (Fig. 7E).
Figure 7. Cell morphology is altered in MZeomesa embryos.
(A-D) Confocal images of embryos at sphere stage stained for Cdh1 (A,B) and β-Catenin (C,D). (E) Western blot of wild type (lane 1) and MZeomesa mutant embryo (lane 2) sphere stage extracts. Levels of Cdh1 and β -Catenin are not obviously altered in mutant embryos. (F) Normal cdh1 expression in sphere stage wild type, MZeomesa, eomesa-eng and eomesa-VP16 injected wild type embryos, as indicated. (G) Live DIC images of wild type and MZeomesa embryos at sphere stage. Mutant cells are more tightly packed and exhibit more blebs then wild type cells. Orange highlights selected cell morphologies, purple highlights intercellular space.
In addition, in situ hybridization for cdh1 expression at sphere stage did not reveal any obvious changes in expression in MZeomesa embryos when compared to controls (Fig. 7F). As a change in global expression levels could be difficult to detect, we also injected eomesa-eng and eomesa-VP16 RNAs into 1 cell at the 8-cell stage to produce a localized region of reduced or enhanced Eomesa function, respectively. If Eomesa is required to down-regulate cdh1 expression, we would expect to see a region of increased cdh1 expression in engrailed construct injected embryos, while cdh1 expression should be reduced in a localized region in eomesa-VP16 injected embryos. However, in both cases, no changes in cdh1 expression were observed (Fig 7F), thus Eomesa does not appear to regulate cdh1 expression.
Using DIC microscopy on live embryos, we observed that deep cells in mutant embryos at sphere stage exhibited more blebs than wild type cells and that there was less space between cells (Fig. 7G), consistent with the analysis of fixed tissue. Although cellular morphology is altered in MZeomesa embryos, how this is mediated and whether it plays a role in the doming delay is currently unclear, but is suggestive of alterations in cell-cell adhesion and motility.
Expression of endoderm genes is disrupted in eomesa mutant embryos
Eomesa has been implicated in mesoderm and endoderm formation and patterning in several species (Arnold et al., 2008; Bjornson et al., 2005; Bruce et al., 2003; Ryan et al., 1996; Slagle et al., 2011). In zebrafish, Eomesa was shown to be able to directly regulate transcription of the endoderm specification gene sox32, while murine Eomes is essential for definitive endoderm formation (Arnold et al., 2008; Bjornson et al., 2005). In addition, the transcription factor mxtx2, which we previously identified as a putative downstream target of Eomesa (Bruce et al., 2005), has recently been shown to act upstream of the Nodal related factor ndr2 and to be important for expression of sox32 in the YSL (Hong et al., 2011).
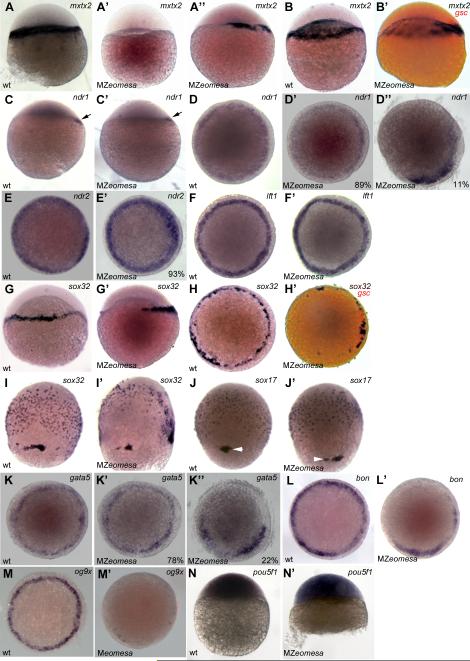
We first examined the expression of the transcription factor mxtx2. At sphere stage, mxtx2 is expressed in marginal cells and the YSL, while at dome stage expression is limited to the YSL (Hirata et al., 2000). In MZeomesa embryos, expression of mxtx2 was delayed and reduced. When MZeomesa embryos first reached sphere stage, no mxtx2 expression was detected, while 40 minutes later, when mutant embryos were still at sphere stage mxtx2 expression was detected on one side of the YSL (Fig 8A-A’’). At dome stage, mxtx2 was expressed robustly in the YSL of wild type embryos, while in mutant embryos mxtx2 was expressed dorsally (as revealed by double in situ with the dorsal marker goosecoid, gsc) and at much lower levels in the rest of the YSL (Fig. 8B, B’). This suggests that Eomesa might be involved in activating mxtx2 expression particularly in ventral-lateral regions. As Mxtx2 has been proposed to act upstream of ndr2, we next examined the expression of nodal related genes.
Figure 8. Expression of endoderm markers is reduced in MZeomesa embryos.
(A-C’, G, G’, I-J’) lateral views, (D-F’, H, H’, K-M’) animal pole views of embryos stained by whole mount in situ hybridzation. (A-B’) mxtx2 expression at sphere (A-A’’) and dome stage. (B’) gsc in red marks dorsal. (C-D’’) ndr1 expression at sphere (arrow, C,C’) and 40% epiboly (D-D’’). (E, E’) ndr2 expression at 40% epiboly. (F,F’) lft1 expression at 40% epiboly (G-H’) sox32 expression at 40% epiboly, (H’) gsc in red marks dorsal. sox32 expression (I,I’) and sox17 expression (J,J’) at 75% epiboly. Arrowheads indicate dorsal forerunner cells. (K-K’’) gata5 expression at 40% epiboly. (L,L’) bon expression at 40% epiboly (M,M’) og9x expression at 40% epiboly. (N.N’) pou5f1 at oblong/sphere. Genotypes indicated in lower left, probe in upper right and percentages of embryos that exhibited given phenotype shown in lower right where appropriate.
Two nodal-like factors ndr1 and ndr2 are expressed in the early zebrafish embryo (Feldman et al., 1998; Sampath et al., 1998). In MZeomesa embryos dorsal expression of ndr1 at sphere stage was normal (arrows, Fig. 8C, C’). At 40% epiboly, most mutant embryos had normal ndr1 expression, while a minority lacked expression in a portion of the blastoderm margin (2/18, 11%, Fig. 8D-D’’). Expression of ndr2 was mostly normal at 40% epiboly with 7% (3/41) of embryos displaying slightly patchy or reduced expression (Fig. 8E, E’) similar to ndr1 (D’’). By contrast, expression of the nodal antagonist lefty1 (lft1) was normal (Fig. 8F, F’). Thus, expression of Nodal-related factors was only slightly affected in MZeomesa embryos.
We next examined the expression of the endodermal marker sox32. In wild type embryos at 40% epiboly, sox32 is expressed in a subset of marginal cells and in the YSL (Dickmeis et al., 2001). In MZeomesa mutant embryos at 40% epiboly, sox32 expression was restricted to the dorsal YSL and marginal cells and was absent ventrally (n = 50, Fig 8G-H’). At 75% epiboly, the number of sox32 expressing endoderm precursors cells was also reduced in mutant embryos (Fig. 8I,I’). Surprisingly, at the same stage, sox17, an endoderm marker downstream of sox32, was expressed relatively normally in mutant embryos (n = 25, Fig. 8J,J’). Cell counts at 75% epiboly revealed an average of 330 sox17 positive endodermal cells in wild type embryos (n = 4) and 275 sox17 positive cells in MZeomesa mutant embryos (n = 10). The dorsal forerunner cells, which express a number of endoderm genes and give rise to Kupffer's vesicle, were often observed in separate clumps, rather than in a single cluster, as seen in wild type embryos (arrowheads, Fig. 8J,J’).
Due to the abnormal expression of sox32, we also examined the expression of zygotic transcription factors acting downstream of Nodal signaling and upstream of sox32 and sox17. In zebrafish, the major transcriptional transducers of Nodal signals that are critical for endoderm formation are Bon and Gata5 (Kikuchi et al., 2000; Reiter et al., 1999; Reiter et al., 2001). A third transcription factor, Og9x, acts partially redundantly with Bon (Poulain and Lepage, 2002). Expression of gata5 in the blastoderm margin was normal in most mutant embryos, with 22% showing reduced expression (n = 41,Fig. 8K-K’’). Expression of bon was reduced in a portion of the margin in all mutant embryos examined (Fig. 8L,L’, n = 20), while expression of og9x was nearly absent in all mutant embryos examined (Fig. 8M,M’, n = 32). We also examined expression of pou5f1 as maternal-zygotic mutant embryos for pou5f1 (spiel ohne grenzen, MZspg) have epiboly and endoderm defects (Lachnit et al., 2008; Reim and Brand, 2006). Expression at sphere stage was normal in MZeomesa embryos (Fig. 8N,N’).
Despite defects in endodermal gene expression, MZeomesa embryos contained a gut tube at 24 hpf, which stained for foxA2 (n = 24, Fig. S6). Taken together, and consistent with other findings (Bjornson et al., 2005; Slagle et al., 2011), our results suggest that Eomesa contributes to the expression of endoderm genes but that other factors can compensate for its absence such that endoderm formation occurs in most mutant embryos.
Expression of mesodermal markers is normal in MZ eomesa embryos
We previously showed that ectopic expression of Eomesa ventrally was sufficient to induce expression of the dorsal mesodermal genes goosecoid (gsc) and floating head (flh), often leading to the formation of an ectopic organizer and a secondary axis (Bruce et al., 2003). We were thus interested in examining the expression of mesodermal genes in MZeomesa embryos.
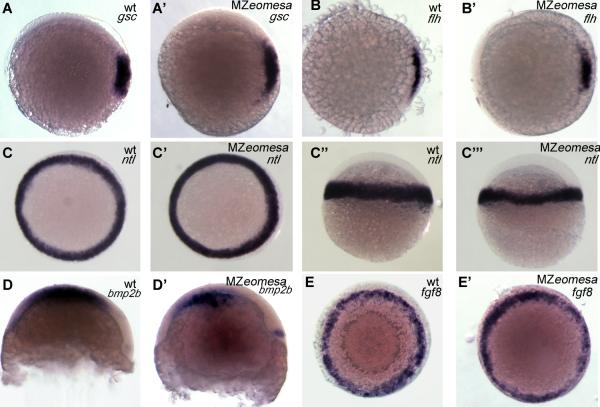
Expression of the dorsal markers gsc and flh was normal in mutant embryos (Fig 9A-B’). The pan-mesodermal marker no tail a (ntla) was expressed normally throughout the margin at 50% epiboly in mutant embryos; however, it was expressed in fewer cell tiers along the animal-vegetal axis (Fig. 9C-C’’’), which is characteristic of reduced Nodal signaling (Gritsman et al., 2000). Expression of the ventral marker bmp2b was normal in most embryos (Fig 9D-D’), with some showing a slight reduction in expression (not shown). Expression of fgf8a was normal in mutant embryos (Fig. 9E,E’). Thus, mesodermal patterning was generally unaffected in MZeomesa embryos.
Figure 9. Expression of mesodermal markers is normal in MZeomesa embryos.
(A-C’, E, E’) animal pole views (C-C’’, D, D’) lateral views. Genotypes and markers as indicated in upper right.
Discussion
Here, with the identification of the first described eomesa mutation, we are able to extend previous work using eomesa dominant negative constructs and morpholinos and to begin to clarify the role of Eomesa during zebrafish development. We show that MZeomesa mutant embryos have delayed epiboly initiation and defects in endodermal gene expression. Eomesa appears to mediate aspects of morphogenesis and endoderm specification in both zebrafish and mouse, suggesting that these activities represent the ancestral functions of this T-box gene.
Eomesa genetically separates epiboly initiation and progression
Embryos lacking the maternal supply of Eomesa have delayed epiboly initiation, while the timing of epiboly progression is normal. Previous work from many groups has shown that delays in epiboly initiation invariably lead to delays in epiboly progression, indicating that these events are coordinated. However, the extent to which initiation and progression are linked and whether the same molecular mechanisms govern these two phases of epiboly has remained unclear (Lepage and Bruce, 2010).
A small number of other maternal mutants have been described that display epiboly initiation defects. MZspg/pou5f1 mutant embryos exhibit both epiboly initiation and progression delays (Lachnit et al., 2008). screeching halt mutant embryos arrest at sphere stage, and therefore do not undergo any morphogenetic movements (Wagner et al., 2004). MZpoky embryos which carry a mutation in the conserved helix-loop-helix ubiquitous kinase gene, are delayed in epiboly due to the failure to properly differentiate the EVL (Fukazawa et al., 2010). Mutation of the maternal-effect gene mission impossible (mis) encoding the helicase Dhx16, results in embryos with defects in epiboly, involution and convergent extension (Putiri and Pelegri, 2011). Thus, the eomesa mutation is unique in that it specifically affects doming, revealing for the first time that aspects of epiboly initiation are genetically separable from epiboly progression and that distinct molecular mechanisms may govern each phase. There appears to be considerable redundancy in the mechanisms that control epiboly, which presumably explains why MZeomesa mutant embryos do eventually dome.
The cellular and molecular basis of the doming delay
Cells in the early embryo first become motile and exhibit blebs around the midblastula transition (Kane and Kimmel, 1993). At this stage, cell movement is random (Kane and Kimmel, 1993). Doming occurs shortly afterwards when the yolk cell bulges upwards into the overlying blastoderm. Deep cells move outward along radial trajectories, resulting in the thinning of the blastoderm and an increase in its surface area, thereby initiating epiboly (Warga and Kimmel, 1990). Extensive cell mixing occurs during this process but there are indications that the mixing is not entirely random, and marginal cells intercalate considerably less than central cells (Wilson et al., 1993). The current view is that radial intercalation of deep cells in response to the yolk cell doming is passive, although this has not been tested experimentally. Thus, it is not clear whether there is an active component to the movement and whether cells use blebs for directional movement at these early stages.
We observed that cells in MZeomesa embryos at sphere stage appear more tightly packed and exhibit more blebs than wild type cells. The increase in cell-cell contact, which might reflect increased adhesion, is not accompanied by obvious changes in E-Cadherin levels. As this change in cell morphology is the most obvious phenotype in the blastoderm of mutant embryos, we hypothesize that it is a factor in the doming delay. There is ample evidence in zebrafish that blebbing can play a role in directional migration and recent work has shown that excessive blebbing disrupts this process (Blaser et al., 2006; Diz-Munoz et al., 2010; Row et al., 2011; Weiser et al., 2009). In embryos carrying a mutation in another T-box gene, spadetail/tbx16, mesodermal cells exhibit increased blebbing behavior following involution and lose their ability to migrate directionally (Row et al., 2011). tbx16 mutant cells are also more adhesive, though this is not the result of changes in either E- or N-Cadherin levels (Row et al., 2011). Although the excessive blebbing that we observed in MZeomesa embryos occurs at an earlier developmental stage than in the tbx16 mutant, the work raises the possibility that the increased blebbing behavior and potentially increased cell-cell adhesion might contribute to the doming delay and is a focus of our on-going work.
Of the epiboly mutants identified, MZeomesa mutant embryos most closely resemble MZspg embryos. In MZspg mutant embryos, epiboly initiation and progression are delayed, and deep cell adhesion is altered without obvious changes in E-Cadherin expression (Lachnit et al., 2008). In the yolk, abnormal microtubule bundles are apparent starting at 50% epiboly and there are similar defects in actin organization (Lachnit et al., 2008). In MZeomesa embryos, abnormal bundling of yolk cell micrtubules is visible as early as sphere stage, while actin organization is normal. The microtubule defects in both MZspg and MZeomesa embryos resemble those observed in embryos treated with the microtubule stabilizing drug taxol, which also causes epiboly delay (Solnica-Krezel and Driever, 1994). Thus, Pouf51 and Eomesa may act redundantly to influence the regulation of microtubule dynamics in the yolk, with Eomesa having an earlier role. This redundancy could explain the remarkable recovery yolk cell microtubules in MZeomesa embryos during epiboly progression.
We previously showed that over-expression of Eomesa induces ectopic expression of the zygotic transcription factor mxtx2. Work by us and others demonstrated that mxtx2 morphants have delayed epiboly progression, often resulting in yolk cell lysis, while doming is normal (Bruce et al., 2005; Wilkins et al., 2008). Mxtx2 functions in the YSL and has been shown to be important for the formation of the marginal actin band that drives epiboly progression (Wilkins et al., 2008). Given the differences in the epiboly defects observed in MZeomesa mutant embryos and mxtx2 morphants, it is unlikely that the reduced expression of mxtx2 we observed in MZeomesa mutants accounts for the doming delay.
Eomesa was not detected in YSL nuclei at sphere and dome stages, indicating that its affects on the yolk cell doming may be non cell-autonomous. This is consistent with the fact that YSL injection of eomesa-VP16 RNA was unable to rescue doming. We note that this work is in keeping with our previous studies showing that Eomesa has other non-cell autonomous affects (Bruce et al., 2005; Bruce et al., 2003), and further suggests that Eomesa may regulate the expression or activity of an as yet unidentified signaling molecule(s) that acts on the yolk. Additional work is required to clarify the function of Eomesa in the yolk.
In MZeomesa embryos microtubules appear to be more dense than in wild type embryos, suggesting a failure in the dynamic regulation of microtubules that is required for epiboly to proceed normally. We postulate that the doming delay may be the result both of overly stabilized yolk cell microtubules and defects in the adhesive properties or motility of deep cells. Eomesa is also expressed in the EVL where it may also function in epiboly, as recent work has shown that both the differentiation of the EVL and its proper attachment to the YSL are essential for normal epiboly initiation (Fukazawa et al., 2010; Sabel et al., 2009; Siddiqui et al., 2010).
Reconciling the role of Eomesa in mesendoderm formation
Sox32 is absolutely required for expression of sox17 and, in turn, for endoderm formation (Alexander et al., 1999). Expression of sox32 is reduced but not eliminated in MZeomesa mutant embryos. Specifically, sox32 expression is relatively normal on the dorsal side of the embryo, from where the majority of the endoderm arises (Warga and Nusslein-Volhard, 1999). Previous work indicates that Bon and Gata5 interact both with Nodal activated Smads and separately with Eomesa to induce sox32 expression (Bjornson et al., 2005; Germain et al., 2000; Kunwar et al., 2003). Thus, it seems likely that sox32 expression in MZeomesa results from the remaining expression of Gata5 and Bon. MZspg mutant embryos express sox32 but fail to express sox17 and completely lack endoderm (Reim et al., 2004). Pou5f1 and Sox32 and have been shown to act together to activate sox17 expression (Reim et al., 2004). pou5f1 expression is normal in MZeomesa embryos. Thus, we postulate that in MZeomesa embryos the remaining Sox32 in combination with Pou5F1 is sufficient to activate sox17 and in turn lead to endoderm formation.
Recent work on the midway/foxh1 (mid) mutant suggested that Eomesa and FoxH1 act combinatorially downstream of Nodal signaling to pattern the mesendoderm (Slagle et al., 2011). Injection of a dominant negative eomesa-eng construct (Bruce et al., 2003) into MZmid mutant embryos phenocopies MZoep embryos (Slagle et al., 2011) which lack all Nodal signaling. This finding suggests that together FoxH1 and Eomesa can fully account for Nodal-dependent mesendodermal patterning. However, the phenotype of MZeomesa mutant embryos is milder than would be expected based upon this work. These contradictory results are likely due to the inherent differences between a loss of function mutation and a dominant-negative construct.
Similar to MZeomesa mutant embryos, Slagle and co-workers observed that injection of eomesa-eng into wild type embryos had little to no effect on expression of the dorsal markers flh, ntl (Slagle et al., 2011). They also demonstrated that FoxH1 plays a more important role in axial mesoderm patterning than Eomesa, which likely is why dorsal patterning is normal in MZeomesa mutant embryos. The authors also observed a complete loss of bon expression in eomesa-eng injected wild type embryos (Slagle et al., 2011), while we saw a consistent reduction in but not complete loss of bon in MZeomesa embryos. One possibility is that the fh105 allele is not a null, but our data convincingly show that little or no Eomesa protein is present in MZeomesa embryos. The more likely explanation is that the eomesa-eng construct, consisting of the Eomesa T-domain fused to the Drosophila engrailed repressor, has more potent affects than the loss of function allele. Genes that Eomesa and other transcription factors (for example FoxH1) regulate independently could exhibit reduced expression in eomesa mutant embryos while they might be completely silenced in eomesa-eng injected embryos. It is also possible that the eomesa-eng construct interferes with the function of other highly similar T-box genes. A strong candidate in this regard is eomesb, as the T-domain of Eomesb is 88.2% identical to the Eomesa T-box and eomesb has been reported to be expressed during early embryonic stages (Takizawa et al., 2007). It will be interesting to determine whether Eomesb has overlapping functions with Eomesa, which could explain why the eomesa mutant phenotype is not stronger.
The other marker most affected in mutant embryos is og9x, which is an immediate early gene in the Nodal pathway (Poulain and Lepage, 2002). Expression is barely detectable in mutant embryos similar to what is observed in MZoep embryos (Poulain and Lepage, 2002). Previous work demonstrated that Eomesa overexpression enhances og9x expression within its endogenous domain, providing further evidence that Eomesa regulates it (Bjornson et al., 2005). Og9x appears to act in a predominantly overlapping manner with Bon and is not essential for endoderm formation as morpholino knock-down in wild type embryos has no detectable phenotype, presumably due to the continued expression of Bon (Poulain and Lepage, 2002).
Eomesa and Nodals
The Nodal pathway plays an essential role in endoderm specification in vertebrates (Zorn and Wells, 2007). Nodal activated Smad proteins associate with transcription factor partners to regulate gene expression (Moustakas and Heldin, 2009). Work in both mouse and frog indicates that Eomes can interact with Smad2/3 to regulate target gene transcription (Picozzi et al., 2009; Teo et al., 2011). Thus, it seems likely that in zebrafish Eomesa may also interact with Smads, in addition to its interaction with Gata5 and Bon, to transduce Nodals signals. In mouse, Nodals and Eomes interact genetically, though the details of this interaction remain to be determined (Arnold et al., 2008). Several observations suggest that in zebrafish Eomesa might have an additional role upstream of Nodals and thus potentially function in a feedback loop with them. We occasionally observed reduced expression of ndr1 and ndr2 in MZeomesa mutant embryos and overexpression of Eomesa weakly induces ndr1 expression (Bjornson et al., 2005). Furthermore, Mxtx2 was recently shown to activate ndr2 expression in the YSL (Hong et al., 2011). We also showed in previous work that overexpression of Eomesa could induce ectopic expression of the dorsal organizer genes gsc and flh and that this requires Nodal signaling (Bruce et al., 2003).
Putting it together: Maternal Control of Epiboly and Endoderm Specification
Several maternal effect mutants in zebrafish play dual roles in epiboly and endoderm patterning. MZspg mutant embryos completely lack endoderm due to the failure to express sox17 (Reim et al., 2004), while Eomesa appears to act earlier to activate og9x and regulate sox32 expression. Both MZspg and MZeomesa embryos also exhibit doming delays and altered cell adhesion, but MZspg mutant embryos show an additional epiboly progression delay, which is not observed in MZeomesa embryos (Lachnit et al., 2008). Pou5f1 and Eomesa appear to act independently to regulate epiboly and endoderm formation. Interestingly, a potential link in the regulation of the two genes has been suggested by work on the mis/dhx16 mutant. mis mutant embryos displays defects in endodermal gene expression as well as defects in epiboly, involution and convergent extension (Putiri and Pelegri, 2011). It has been suggested that Dhx16 might regulate Eomesa and Pou5f1 activities post-transcriptionally (Putiri and Pelegri, 2011). This intriguing link between epiboly and endoderm patterning warrants further investigation.
Conclusion
Our results indicate that maternal Eomesa plays a conserved role in morphogenesis and endoderm patterning. Several maternal factors are involved in regulating these two processes and it remains to be seen the extent to which these factors function redundantly or in concert to control early zebrafish development. Furthermore, an open question is whether there is a functional link between endoderm specification and epiboly. The eomesa mutant phenotype also reveals for the first time that aspects of epiboly initiation and progression are separable. Additional studies of MZeomesa mutant embryos offer the exciting possibility of gaining new insights into the molecular mechanisms governing epiboly initiation.
Supplementary Material
Movie 1
Time-lapse movie of wild type embryo from sphere to 50% epiboly stage.
Movie 2
Time-lapse movie of MZeomesa embryo from oblong to 50% epiboly stage.
Supplemental Figures
S1. Eomesa inclusion bodies isolation.
Comassie staining of acrylamide gel loaded with 1, 2, and 4 μl of Eomesa recombinant protein (concentration 10 mg/ml) in lanes 2, 3 and 4.
S2. Eomesa in MZeomesa and Zeomesa embryos.
Overlay of fluorescence and bright field images of (A) wild type and (B) MZeomesa embryos stained for Eomesa at sphere stage. No nuclear or cytoplasmic fluorescence is detected in MZeomesa embryo. (B-E) Bright field, fluorescene and overlay of embryos stained for Eomesa. Stages indicated on left, genotype in upper right. (B,C) Nuclear staining visible in wild type deep cells but not Zeomesa deep cells at 60% epiboly. Background fluorescence is visible in mutant embryo. (D) Eomesa detected in the brain of wild type (arrowhead) but not Zeomesa (E) embryos at 1 dpf.
S3. Eomesa expression in the EVL.
Antibody staining for Eomesa (A) Lateral view of embryo showing Eomesa in EVL nuclei, arrow. (B) Marginal view of embryo with Eomesa expression in EVL (arrows) but not YSL nuclei (arrowheads). Black dashed lines outline selected EVL cell boundaries.
S4. Nuclear morphology is normal in MZeomesa embryos.
Lateral views of bright field (A,B) and fluorescent (A’,B’) images of fractured DAPI stained wild type (A,A’) and MZeomesa (B,B’) embryos. Nuclear morphology is normal in MZeomesa embryo.
S5. eomesa-VP16 RNA injection rescues microtubule morphology in MZeomesa embryos.
(A-C) Lateral views of confocal projections of Tubulin staining in dome stage embryos. Genotype indicated in top right and injected construct in lower right. (C) Microtubule voids are rescued in eomesa-VP16 injected MZeomesa embryo.
S6. MZeomesa embryos contain endoderm at 24 hpf.
Lateral views of deyolked wild type (A) and MZeomesa (B) embryo stained by in situ hybridization for foxA2.
Highlights.
-Zebrafish eomesa mutation was recovered by TILLING
-Maternal Eomesa is required for the timely initiation of epiboly but is not required for epiboly progression
- yolk cell microtubules are defective in maternal-zygotic eomesa mutant embryos at dome stage but recover by 75% epiboly
-Deep cell morphology is altered in maternal-zygotic mutant embryos
-expression the endoderm markers sox32, bon and og9x is abnormal in maternal-zygotic eomesa mutant embryos
Acknowledgements
A.B. thanks Scott Dougan and Stephanie Lepage for helpful discussions and comments on the manuscript, and Vicky Prince and James Marrs for reagents. M.M. thanks Gilda Nappo and Giuseppe Ossolengo. TILLING of eomesa was supported by NIH grant HG002995 to C.B.M. C.B.M. is an investigator with the Howard Hughes Medical Institute. M.M. is supported by the Fondazione Italiana Ricerca sul Cancro. A.B. is supported by NSERC and CFI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrams EW, Mullins MC. Early zebrafish development: it's in the maternal genes. Curr. Opin. Genet. Dev.. 2009;19:396–403. doi: 10.1016/j.gde.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J, Rothenberg M, Henry GL, Stainier DY. casanova plays an early and essential role in endoderm formation in zebrafish. Dev. Biol. 1999;215:343–357. doi: 10.1006/dbio.1999.9441. [DOI] [PubMed] [Google Scholar]
- Alexander J, Stainier DY. A molecular pathway leading to endoderm formation in zebrafish. Curr. Biol. 1999;9:1147–1157. doi: 10.1016/S0960-9822(00)80016-0. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Hofmann UK, Bikoff EK, Robertson EJ. Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development. 2008;135:501–511. doi: 10.1242/dev.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babb SG, Marrs JA. E-cadherin regulates cell movements and tissue formation in early zebrafish embryos. Dev. Dyn. 2004;230:263–277. doi: 10.1002/dvdy.20057. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Essner JJ, Yost HJ. Regulation of midline development by antagonism of lefty and nodal signaling. Development. 1999;126:3253–3262. doi: 10.1242/dev.126.14.3253. [DOI] [PubMed] [Google Scholar]
- Bjornson CR, Griffin KJ, Farr GH, 3rd, Terashima A, Himeda C, Kikuchi Y, Kimelman D. Eomesodermin is a localized maternal determinant required for endoderm induction in zebrafish. Dev. Cell. 2005;9:523–533. doi: 10.1016/j.devcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Blaser H, Reichman-Fried M, Castanon I, Dumstrei K, Marlow FL, Kawakami K, Solnica-Krezel L, Heisenberg CP, Raz E. Migration of zebrafish primordial germ cells: a role for myosin contraction and cytoplasmic flow. Dev. Cell. 2006;11:613–627. doi: 10.1016/j.devcel.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Bruce AE, Howley C, Dixon Fox M, Ho RK. T-box gene eomesodermin and the homeobox-containing Mix/Bix gene mtx2 regulate epiboly movements in the zebrafish. Dev. Dyn. 2005;233:105–114. doi: 10.1002/dvdy.20305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AE, Howley C, Zhou Y, Vickers SL, Silver LM, King ML, Ho RK. The maternally expressed zebrafish T-box gene eomesodermin regulates organizer formation. Development. 2003;130:5503–5517. doi: 10.1242/dev.00763. [DOI] [PubMed] [Google Scholar]
- Bruce AE, Oates AC, Prince VE, Ho RK. Additional hox clusters in the zebrafish: divergent expression patterns belie equivalent activities of duplicate hoxB5 genes. Evol. Dev. 2001;3:127–144. doi: 10.1046/j.1525-142x.2001.003003127.x. [DOI] [PubMed] [Google Scholar]
- Carvalho L, Stuhmer J, Bois JS, Kalaidzidis Y, Lecaudey V, Heisenberg CP. Control of convergent yolk syncytial layer nuclear movement in zebrafish. Development. 2009;136:1305–1315. doi: 10.1242/dev.026922. [DOI] [PubMed] [Google Scholar]
- Cheng JC, Miller AL, Webb SE. Organization and function of microfilaments during late epiboly in zebrafish embryos. Dev. Dyn. 2004;231:313–323. doi: 10.1002/dvdy.20144. [DOI] [PubMed] [Google Scholar]
- Dickmeis T, Mourrain P, Saint-Etienne L, Fischer N, Aanstad P, Clark M, Strahle U, Rosa F. A crucial component of the endoderm formation pathway, CASANOVA, is encoded by a novel sox-related gene. Genes Dev. 2001;15:1487–1492. doi: 10.1101/gad.196901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diz-Munoz A, Krieg M, Bergert M, Ibarlucea-Benitez I, Muller DJ, Paluch E, Heisenberg CP. Control of directed cell migration in vivo by membrane-to-cortex attachment. PLoS biology. 2010;8:e1000544. doi: 10.1371/journal.pbio.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erter CE, Solnica-Krezel L, Wright CV. Zebrafish nodal-related 2 encodes an early mesendodermal inducer signaling from the extraembryonic yolk syncytial layer. Dev. Biol. 1998;204:361–372. doi: 10.1006/dbio.1998.9097. [DOI] [PubMed] [Google Scholar]
- Feldman B, Gates MA, Egan ES, Dougan ST, Rennebeck G, Sirotkin HI, Schier AF, Talbot WS. Zebrafish organizer development and germ-layer formation require nodal- related signals. Nature. 1998;395:181–185. doi: 10.1038/26013. [DOI] [PubMed] [Google Scholar]
- Fukazawa C, Santiago C, Park KM, Deery WJ, Gomez de la Torre Canny S, Holterhoff CK, Wagner DS. poky/chuk/ikk1 is required for differentiation of the zebrafish embryonic epidermis. Dev. Biol. 2010;346:272–283. doi: 10.1016/j.ydbio.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain S, Howell M, Esslemont GM, Hill CS. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 2000;14:435–451. [PMC free article] [PubMed] [Google Scholar]
- Gritsman K, Talbot WS, Schier AF. Nodal signaling patterns the organizer. Development. 2000;127:921–932. doi: 10.1242/dev.127.5.921. [DOI] [PubMed] [Google Scholar]
- Hirata T, Yamanaka Y, Ryu SL, Shimizu T, Yabe T, Hibi M, Hirano T. Novel mix-family homeobox genes in zebrafish and their differential regulation. Biochem. Biophys. Res. Commun. 2000;271:603–609. doi: 10.1006/bbrc.2000.2672. [DOI] [PubMed] [Google Scholar]
- Hong SK, Jang MK, Brown JL, McBride AA, Feldman B. Embryonic mesoderm and endoderm induction requires the actions of non-embryonic Nodal-related ligands and Mxtx2. Development. 2011;138:787–795. doi: 10.1242/dev.058974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowett T, Lettice L. Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labelled probes. Trends Genet. 1994;10:73–74. doi: 10.1016/0168-9525(94)90220-8. [DOI] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Kane DA, McFarland KN, Warga RM. Mutations in half baked/E-cadherin block cell behaviors that are necessary for teleost epiboly. Development. 2005;132:1105–1116. doi: 10.1242/dev.01668. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Trinh LA, Reiter JF, Alexander J, Yelon D, Stainier DY. The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes Dev. 2000;14:1279–1289. [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Köppen M, Fernández BG, Carvalho L, Jacinto A, Heisenberg C-P. Coordinated cell-shape changes control epithelial movement in zebrafish and Drosophila. Development. 2006;133:2671–2681. doi: 10.1242/dev.02439. [DOI] [PubMed] [Google Scholar]
- Kunwar PS, Zimmerman S, Bennett JT, Chen Y, Whitman M, Schier AF. Mixer/Bon and FoxH1/Sur have overlapping and divergent roles in Nodal signaling and mesendoderm induction. Development. 2003;130:5589–5599. doi: 10.1242/dev.00803. [DOI] [PubMed] [Google Scholar]
- Lachnit M, Kur E, Driever W. Alterations of the cytoskeleton in all three embryonic lineages contribute to the epiboly defect of Pou5f1/Oct4 deficient MZspg zebrafish embryos. Dev. Biol. 2008;315:1–17. doi: 10.1016/j.ydbio.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Lepage SE, Bruce AE. Zebrafish epiboly: mechanics and mechanisms. Int. J. Dev. Biol. 2010;54:1213–1228. doi: 10.1387/ijdb.093028sl. [DOI] [PubMed] [Google Scholar]
- Martinez-Barbera JP, Toresson H, Da Rocha S, Krauss S. Cloning and expression of three members of the zebrafish Bmp family: Bmp2a, Bmp2b and Bmp4. Gene. 1997;198:53–59. doi: 10.1016/s0378-1119(97)00292-8. [DOI] [PubMed] [Google Scholar]
- Mione M, Shanmugalingam S, Kimelman D, Griffin K. Overlapping expression of zebrafish T-brain-1 and eomesodermin during forebrain development. Mech. Dev. 2001;100:93–97. doi: 10.1016/s0925-4773(00)00501-3. [DOI] [PubMed] [Google Scholar]
- Moens CB, Donn TM, Wolf-Saxon ER, Ma TP. Reverse genetics in zebrafish by TILLING. Brief. Func. Genom. Proteom. 2008;7:454–459. doi: 10.1093/bfgp/eln046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136:3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu. Rev. Genet. 2005;39:219–239. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- Picozzi P, Wang F, Cronk K, Ryan K. Eomesodermin requires transforming growth factor-beta/activin signaling and binds Smad2 to activate mesodermal genes. J. Biol. Chem. 2009;284:2397–2408. doi: 10.1074/jbc.M808704200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain M, Lepage T. Mezzo, a paired-like homeobox protein is an immediate target of Nodal signalling and regulates endoderm specification in zebrafish. Development. 2002;129:4901–4914. doi: 10.1242/dev.129.21.4901. [DOI] [PubMed] [Google Scholar]
- Putiri E, Pelegri F. The zebrafish maternal-effect gene mission impossible encodes the DEAH-box helicase Dhx16 and is essential for the expression of downstream endodermal genes. Dev. Biol. 2011;353:275–289. doi: 10.1016/j.ydbio.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebagliati MR, Toyama R, Haffter P, Dawid IB. cyclops encodes a nodal-related factor involved in midline signaling. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9932–9937. doi: 10.1073/pnas.95.17.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifers F, Bohli H, Walsh EC, Crossley PH, Stainier DY, Brand M. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125:2381–2395. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- Reim G, Brand M. Maternal control of vertebrate dorsoventral axis formation and epiboly by the POU domain protein Spg/Pou2/Oct4. Development. 2006;133:2757–2770. doi: 10.1242/dev.02391. [DOI] [PubMed] [Google Scholar]
- Reim G, Mizoguchi T, Stainier DY, Kikuchi Y, Brand M. The POU domain protein spg (pou2/Oct4) is essential for endoderm formation in cooperation with the HMG domain protein casanova. Dev. Cell. 2004;6:91–101. doi: 10.1016/s1534-5807(03)00396-4. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier DY. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Kikuchi Y, Stainier DY. Multiple roles for Gata5 in zebrafish endoderm formation. Development. 2001;128:125–135. doi: 10.1242/dev.128.1.125. [DOI] [PubMed] [Google Scholar]
- Rodaway A, Takeda H, Koshida S, Broadbent J, Price B, Smith JC, Patient R, Holder N. Induction of the mesendoderm in the zebrafish germ ring by yolk cell- derived TGF-beta family signals and discrimination of mesoderm and endoderm by FGF. Development. 1999;126:3067–3078. doi: 10.1242/dev.126.14.3067. [DOI] [PubMed] [Google Scholar]
- Row RH, Maitre JL, Martin BL, Stockinger P, Heisenberg CP, Kimelman D. Completion of the epithelial to mesenchymal transition in zebrafish mesoderm requires Spadetail. Dev. Biol. 2011;354:102–110. doi: 10.1016/j.ydbio.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, Wilson V, Evans MJ. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Ryan K, Garrett N, Mitchell A, Gurdon JB. Eomesodermin, a key early gene in Xenopus mesoderm differentiation. Cell. 1996;87:989–1000. doi: 10.1016/s0092-8674(00)81794-8. [DOI] [PubMed] [Google Scholar]
- Sabel JL, d'Alencon C, O'Brien EK, Van Otterloo E, Lutz K, Cuykendall TN, Schutte BC, Houston DW, Cornell RA. Maternal Interferon Regulatory Factor 6 is required for the differentiation of primary superficial epithelia in Danio and Xenopus embryos. Dev. Biol. 2009;325:249–262. doi: 10.1016/j.ydbio.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath K, Rubinstein AL, Cheng AM, Liang JO, Fekany K, Solnica-Krezel L, Korzh V, Halpern ME, Wright CV. Induction of the zebrafish ventral brain and floorplate requires cyclops/nodal signalling. Nature. 1998;395:185–189. doi: 10.1038/26020. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, Hammerschmidt M, Beuchle D, Cho KW, De Robertis EM, Nusslein-Volhard C. Expression of zebrafish goosecoid and no tail gene products in wild- type and mutant no tail embryos. Development. 1994;120:843–852. doi: 10.1242/dev.120.4.843. [DOI] [PubMed] [Google Scholar]
- Siddiqui M, Sheikh H, Tran C, Bruce AE. The tight junction component Claudin E is required for zebrafish epiboly. Dev. Dyn. 2010;239:715–722. doi: 10.1002/dvdy.22172. [DOI] [PubMed] [Google Scholar]
- Slagle CE, Aoki T, Burdine RD. Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin. PLoS genetics. 2011;7:e1002072. doi: 10.1371/journal.pgen.1002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnica-Krezel L, Driever W. Microtubule arrays of the zebrafish yolk cell: organization and function during epiboly. Development. 1994;120:2443–2455. doi: 10.1242/dev.120.9.2443. [DOI] [PubMed] [Google Scholar]
- Stachel SE, Grunwald DJ, Myers PZ. Lithium perturbation and goosecoid expression identify a dorsal specification pathway in the pregastrula zebrafish. Development. 1993;117:1261–1274. doi: 10.1242/dev.117.4.1261. [DOI] [PubMed] [Google Scholar]
- Strähle U, Blader P, Henrique D, Ingham PW. Axial, a zebrafish gene expressed along the developing body axis, shows altered expression in cyclops mutant embryos. Genes Dev. 1993;7:1436–1446. doi: 10.1101/gad.7.7b.1436. [DOI] [PubMed] [Google Scholar]
- Strähle U, Jesuthasan S. Ultraviolet irradiation impairs epiboly in zebrafish embryos: evidence for a microtubule-dependent mechanism of epiboly. Development. 1993;119:909–919. doi: 10.1242/dev.119.3.909. [DOI] [PubMed] [Google Scholar]
- Takizawa F, Araki K, Ito K, Moritomo T, Nakanishi T. Expression analysis of two Eomesodermin homologues in zebrafish lymphoid tissues and cells. Molec. Imm. 2007;44:2324–2331. doi: 10.1016/j.molimm.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Talbot WS, Trevarrow B, Halpern ME, Melby AE, Farr G, Postlethwait JH, Jowett T, Kimmel CB, Kimelman D. A homeobox gene essential for zebrafish notochord development. Nature. 1995;378:150–157. doi: 10.1038/378150a0. [DOI] [PubMed] [Google Scholar]
- Teo AK, Arnold SJ, Trotter MW, Brown S, Ang LT, Chng Z, Robertson EJ, Dunn NR, Vallier L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topczewski J, Solnica-Krezel L. Cytoskeletal dynamics of the zebrafish embryo. In: Detrich WH, Westerfield M, Zon LI, editors. Methods in Cell Biology The Zebrafish: Biology. Academic Press; London: 1999. pp. 206–226. [DOI] [PubMed] [Google Scholar]
- Wagner DS, Dosch R, Mintzer KA, Wiemelt AP, Mullins MC. Maternal control of development at the midblastula transition and beyond: mutants from the zebrafish II. Dev. Cell. 2004;6:781–790. doi: 10.1016/j.devcel.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Warga RM, Kimmel CB. Cell movements during epiboly and gastrulation in zebrafish. Development. 1990;108:569–580. doi: 10.1242/dev.108.4.569. [DOI] [PubMed] [Google Scholar]
- Warga RM, Nusslein-Volhard C. Origin and development of the zebrafish endoderm. Development. 1999;126:827–838. doi: 10.1242/dev.126.4.827. [DOI] [PubMed] [Google Scholar]
- Weiser DC, Row RH, Kimelman D. Rho-regulated myosin phosphatase establishes the level of protrusive activity required for cell movements during zebrafish gastrulation. Development. 2009;136:2375–2384. doi: 10.1242/dev.034892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. University of Oregon Press; Eugene: 1993. [Google Scholar]
- Wilkins SJ, Yoong S, Verkade H, Mizoguchi T, Plowman SJ, Hancock JF, Kikuchi Y, Heath JK, Perkins AC. Mtx2 directs zebrafish morphogenetic movements during epiboly by regulating microfilament formation. Dev. Biol. 2008;314:12–22. doi: 10.1016/j.ydbio.2007.10.050. [DOI] [PubMed] [Google Scholar]
- Wilson ET, Helde KA, Grunwald DJ. Something's fishy here--rethinking cell movements and cell fate in the zebrafish embryo. Trends Genet. 1993;9:348–352. doi: 10.1016/0168-9525(93)90039-k. [DOI] [PubMed] [Google Scholar]
- Zalik SE, Lewandowski E, Kam Z, Geiger B. Cell adhesion and the actin cytoskeleton of the enveloping layer in the zebrafish embryo during epiboly. Biochem. Cell Biol. 1999;77:527–542. [PubMed] [Google Scholar]
- Zorn AM, Wells JM. Molecular basis of vertebrate endoderm development. Int. Rev. Cyto. 2007;259:49–111. doi: 10.1016/S0074-7696(06)59002-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1
Time-lapse movie of wild type embryo from sphere to 50% epiboly stage.
Movie 2
Time-lapse movie of MZeomesa embryo from oblong to 50% epiboly stage.
Supplemental Figures
S1. Eomesa inclusion bodies isolation.
Comassie staining of acrylamide gel loaded with 1, 2, and 4 μl of Eomesa recombinant protein (concentration 10 mg/ml) in lanes 2, 3 and 4.
S2. Eomesa in MZeomesa and Zeomesa embryos.
Overlay of fluorescence and bright field images of (A) wild type and (B) MZeomesa embryos stained for Eomesa at sphere stage. No nuclear or cytoplasmic fluorescence is detected in MZeomesa embryo. (B-E) Bright field, fluorescene and overlay of embryos stained for Eomesa. Stages indicated on left, genotype in upper right. (B,C) Nuclear staining visible in wild type deep cells but not Zeomesa deep cells at 60% epiboly. Background fluorescence is visible in mutant embryo. (D) Eomesa detected in the brain of wild type (arrowhead) but not Zeomesa (E) embryos at 1 dpf.
S3. Eomesa expression in the EVL.
Antibody staining for Eomesa (A) Lateral view of embryo showing Eomesa in EVL nuclei, arrow. (B) Marginal view of embryo with Eomesa expression in EVL (arrows) but not YSL nuclei (arrowheads). Black dashed lines outline selected EVL cell boundaries.
S4. Nuclear morphology is normal in MZeomesa embryos.
Lateral views of bright field (A,B) and fluorescent (A’,B’) images of fractured DAPI stained wild type (A,A’) and MZeomesa (B,B’) embryos. Nuclear morphology is normal in MZeomesa embryo.
S5. eomesa-VP16 RNA injection rescues microtubule morphology in MZeomesa embryos.
(A-C) Lateral views of confocal projections of Tubulin staining in dome stage embryos. Genotype indicated in top right and injected construct in lower right. (C) Microtubule voids are rescued in eomesa-VP16 injected MZeomesa embryo.
S6. MZeomesa embryos contain endoderm at 24 hpf.
Lateral views of deyolked wild type (A) and MZeomesa (B) embryo stained by in situ hybridization for foxA2.