Abstract
Circadian rhythms occur with a periodicity of about 24 hours and regulate a wide array of metabolic and physiologic functions. Accumulating epidemiological and genetic evidence indicates that disruption of circadian rhythms can be directly linked to many pathological conditions, including sleep disorders, depression, metabolic syndrome and cancer. Intriguingly, a number of molecular gears constituting the clock machinery have been found to establish functional interplays with regulators of cellular metabolism. While the circadian clock regulates multiple metabolic pathways, metabolite availability and feeding behavior can in turn regulate the circadian clock. An in-depth understanding of this reciprocal regulation of circadian rhythms and cellular metabolism may provide insights in the development of therapeutic intervention against specific metabolic disorders.
Clock and the control of cellular metabolic processes
The cycling of day and night, caused by the earth’s rotation around its axis, has far reaching affects on our physiology. Circadian (from the Latin circa diem meaning “about a day”) clocks have evolved to enable organisms to anticipate environmental changes (such as availability of food or activity of predators) so that the organisms can adapt their behavior and physiology to the appropriate time of day. Although it has been known for several decades that many metabolic processes, such as glucose and cholesterol metabolism, or renal function are regulated by the circadian clock, it is only in the past few years that the severe metabolic consequences of circadian disruption have emerged [1]. Metabolic syndrome and obesity have been observed in mice harboring mutations in the clock genes, which encode for transcription factors that affect both the persistence and period of circadian rhythms [2,3]. Epidemiological studies have identified a correlation between metabolic syndrome and shift work [4,5]. Forced misalignment of behavioral and circadian cycles in human subjects, caused by being active and eating at biological night time, was recently shown to cause a decrease in leptin and an increase in glucose and insulin levels [6]. Moreover, food intake during rest phase [7] and exposure to light at night [8] have been shown to cause weight gain in mice. Although a clear molecular understanding of these observations is lacking, recent findings point to some highly specialized links between unique clock components and the control of cellular metabolism. There appears to be a reciprocal relationship between circadian rhythms and metabolism. While the circadian clock regulates multiple metabolic pathways, metabolites and feeding behavior can regulate the circadian clock [9]. In this review we will discuss how this reciprocal relationship is regulated and what the metabolic consequences of disruption of circadian rhythms are (Box 1).
BOX 1.
Molecular Organization of the Circadian Clock
Circadian clocks are present in almost all of the tissues in mammals. The master or “central” clock is located in the hypothalamic suprachiasmatic nucleus (SCN), which contains 10–15,000 neurons. “Peripheral clocks” are present in almost all other mammalian tissues such as liver, heart, lung and kidney, where they maintain circadian rhythms and regulate tissue-specific gene expression. These peripheral clocks are synchronized by the central clock to ensure temporally coordinated physiology. The synchronization mechanisms implicate various humoral signals, including circulating entraining factors such as glucocorticoids. The SCN clock can function autonomously, without any external input, but can be reset by environmental cues such as light. The molecular machinery that regulates these circadian rhythms comprises of a set of genes, known as “clock” genes, the products of which interact to generate and maintain the rhythms.
A conserved feature among many organisms is the regulation of the circadian clock by a negative feedback loop [72]. Positive regulators induce the transcription of clock-controlled genes (CCGs), some of which encode proteins that feedback on their own expression by repressing the activity of positive regulators. CLOCK and BMAL1 are the positive regulators of the mammalian clock machinery which regulate the expression of the negative regulators: cryptochrome (CRY1 and CRY2) and period (PER1, PER2, PER3) families (Figure 2). CLOCK and BMAL1 are transcription factors that heterodimerize through the PAS domain and induce the expression of clock-controlled genes by binding to their promoters at E-boxes [CACGTG]. Once a critical concentration of the Per and Cry proteins is accumulated, these proteins translocate into the nucleus and form a complex to inhibit CLOCK-BMAL1 mediated transcription, thereby closing the negative feedback loop (Figure 2). In order to start a new transcriptional cycle, the CLOCK-BMAL1 complex needs to be de-repressed through the proteolytic degradation of PER and CRY. Core clock genes (such as Clock, Bmal1, Period, Cryptochrome) are necessary for generation of circadian rhythms, whereas CCGs (such as Nampt, Alas1 etc.) are regulated by the core clock genes.
Some CCGs are transcription factors, such as albumin D-box binding protein (DBP), RORα and REV-ERBα, which can then regulate cyclic expression of other genes. DBP binds to D-boxes [TTA(T/C)GTAA], whereas RORα and REV-ERBα bind to the Rev-Erb/ROR-binding element, or RRE [(A/T)A(A/T)NT(A/G)GGTCA]. Approximately 10% of the transcriptome displays robust circadian rhythmicity [10,73]. Interestingly, most transcripts that oscillate in one tissue do not oscillate in another [10,73,74].
I. Circadian Regulation of Metabolism
How does the circadian clock exert its control over metabolism?
To ensure a strict temporal regulation of various metabolic pathways, many key steps and players involved in these pathways are regulated by the circadian clock machinery (Figure 1). This regulation is achieved by controlling the expression of enzymes that regulate rate-limiting steps of a metabolic pathway, or by integrating proteins such as nuclear receptors and nutrient-sensors with the clock machinery, and by regulating the abundance of several metabolites. Some of these mechanisms are discussed in detail here.
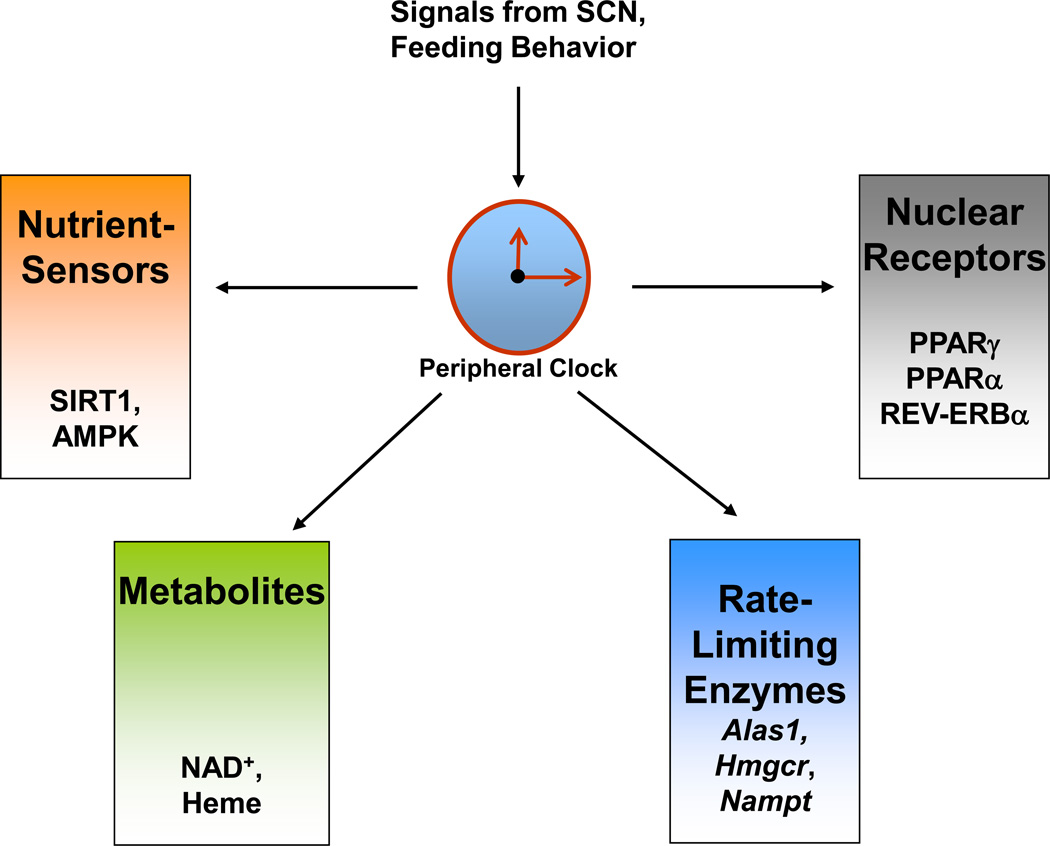
Figure 1. Regulation of metabolism by the circadian clock.
Peripheral clocks, such as the one in liver, are regulated by the master clock present in the suprachiasmatic nucleus (SCN). The liver clock can regulate multiple metabolic pathways by various mechanisms. These mechanisms include regulation of rate-limiting steps, control of metabolite levels, interaction with nutrient sensors, and modulation of nuclear receptors.
i) Regulation of Rate-Limiting Steps
An interesting way by which nature can regulate many metabolic pathways in a circadian manner is by controlling their rate limiting steps [10]. For example, the activity of the rate-limiting enzyme in cholesterol biosynthesis HMG-CoA reductase (Hmgcr), displays circadian rhythmicity [11], and is found to be highest at night [12]. Interestingly, this information is exploited in the treatment of Hypercholesterolemia. Cholesterol-lowering drugs such as Statins that act by inhibiting HMG-CoA reductase, are most effective when administered before bedtime [13]. Another example where the circadian clock can regulate an entire metabolic pathway by regulating the expression of one rate-limiting enzyme, is demonstrated in the regulation of the NAD+-salvage pathway [14,15]. Circadian clock controls the expression of nicotinamide phosphoribosyltransferase (NAMPT), a key rate-limiting enzyme in the salvage pathway of NAD+ biosynthesis [14,15]. The rhythmicity in the expression of this enzyme drives the oscillation in NAD+ levels. The oscillatory expression of Nampt is abolished in Clock/Clock mice that harbor a point mutation causing a 51 amino acid deletion in the CLOCK protein. This results in drastically reduced levels of NAD+ in mouse embryonic fibroblasts (MEFs) derived from these mice (Figure 1). Similarly, the circadian clock controls the cellular heme levels by regulating the expression of Aminolevulinate synthase 1 (ALAS1), the rate limiting enzyme in heme biosynthesis [16] (Figure 1).
ii) Circadian Control of Nuclear Receptors
Nuclear receptors constitute a superfamily of ligand-activated transcription factors which regulate a number of biological processes, including growth, development, endocrine signaling, reproduction, and energy metabolism [17]. A number of specialized nuclear receptors act as sensors of metabolites such as fat-soluble hormones, vitamins, lipids, oxysterols, bile acids and xenobiotics. The expression of several nuclear receptors is known to be directed by CLOCK and BMAL1 (transcription factors and the master regulators of the circadian clock machinery that heterodimerize to regulate circadian gene expression) (Box 1). These receptors include the Retinoic acid-related orphan receptor α (RORα) and REV-ERBα, as well as the Peroxisome proliferator-activated receptor (PPAR)α [18]. PPARα acts as sensor for polyunsaturated fatty acids, regulates fatty acid oxidation and apolipoprotein synthesis, and is a target for drugs such as fenofibrates [17]. While CLOCK and BMAL1 can directly regulate Pparα expression via binding to an E-box element on its promoter, PPARα can also directly regulate BMAL1 expression by binding to a PPARα response element (PPRE) on the Bmal1 promoter [19] (Box 1). This reciprocal regulation suggests a very close link between nuclear receptors, metabolism and the circadian clock. Further evidence of this was provided by a detailed study involving a temporal mRNA expression profiling of all 45 nuclear receptors in various metabolic tissues [20]. The expression of 25 nuclear receptors, including PPAR family members (α, γ, δ) and estrogen-related receptor (ERR) family members (α, β, γ), is rhythmic in a tissue-specific manner. This could well be a possible explanation for the observed daily oscillations in glucose and lipid metabolism.
Another mode of regulation of nuclear receptor function by the circadian clock was established by recent studies demonstrating that PERIOD proteins (PER 1–3) physically associate with several nuclear receptors [21,22,23,24]. Circadian expression of the PER proteins is directly regulated by CLOCK and BMAL1, and the PER proteins negatively feedback on their own expression by inhibiting CLOCK:BMAL1 heterodimer (Box 1). PER2 was shown to interact with a plethora of nuclear receptors including PPARγ [22], ERα [24], PPARα, REV-ERBα, HNF4α, TRα, NURR1 and RORα [21]. PER3 was also shown to interact with PPARγ [23]. These interactions had one of two consequences:
Modulation of the core clock gene expression. E.g.: PER2, in co-ordination with PPARα and REV-ERBα, regulates Bmal1 expression.
Regulation of the activity or stability of these nuclear receptors, thereby fine-tuning several metabolic pathways. E.g.: (a) PER2 inhibits the recruitment of PPARγ to its target promoters, specifically in white adipose tissue (WAT) [22]. Absence of PER2 caused de-repression of several PPARγ target genes and led to the expression of brown adipose tissue (BAT)-specific genes in WAT. This altered gene expression could be responsible for the observation that the Per2−/− mice have altered lipid metabolism [22]. (b) PER2 interaction with REV-ERBα and other nuclear receptors is critical for glycogen metabolism [21]. Rhythmic expression of Glucose-6-phosphatase, a key enzyme in glycogen degradation to produce glucose, was blunted in the Rev-Erbα−/−/Per2Brdm1 mutant mice. This leads to low and non-rhythmic levels of glycogen in the liver of these mutant animals. (c) PER3 inhibits PPARγ activity by binding to PPARγ target sites and blocks adipogenesis [23]. Per3−/− mice were hence shown to display altered body composition with increased adipose tissue and decreased muscle tissue.
Thus, interaction of PER proteins with various nuclear receptors not only influences several metabolic pathways (such as lipid and glycogen metabolism), it also modulates the expression of core clock genes (such as Bmal1) that are necessary for generation and maintenance of circadian rhythms. These observations further support the notion that the circadian clock regulates metabolism at various levels and, therefore, disruptions in circadian rhythms could manifest in metabolic disorders.
iii) Nutrient Sensors as Regulators of Clock
In order for the circadian clock to efficiently control cellular metabolism, there must be a way through which the clock machinery determines the energy status of the cell. That requirement is fulfilled by the association of nutrient-sensors, Sirtuin 1 (SIRT1) and AMP-activated protein kinase (AMPK), with the clock machinery (Figure 2).
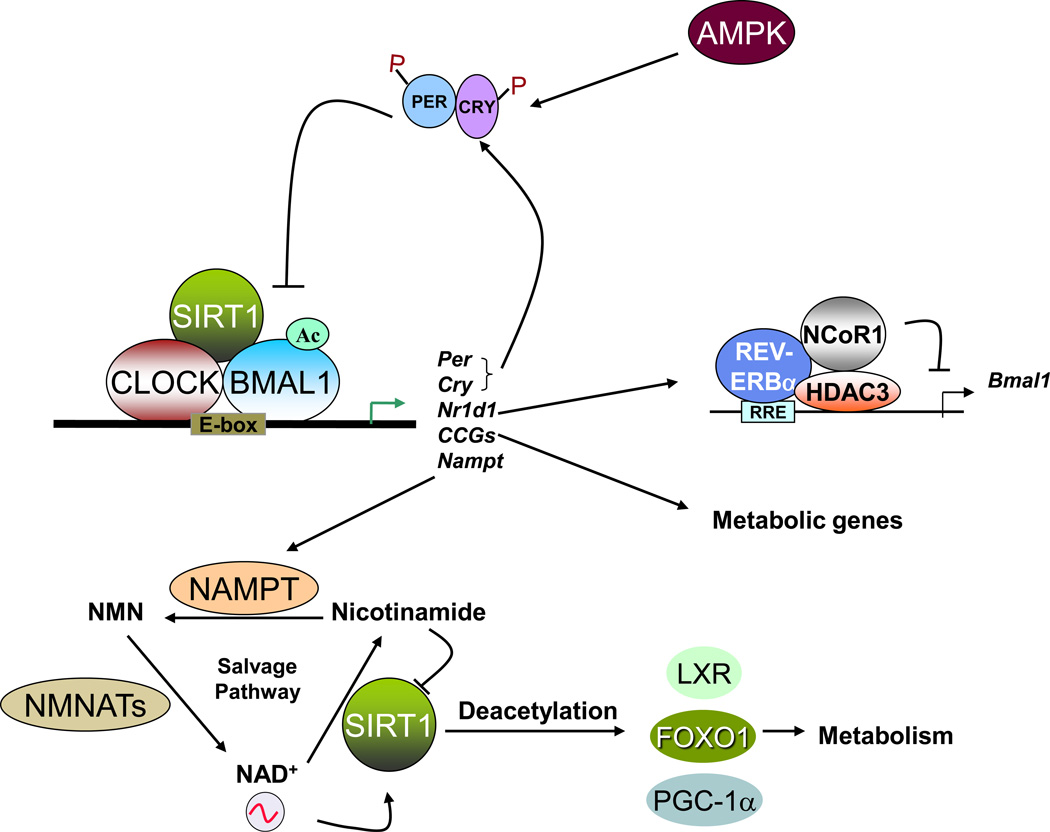
Figure 2. Interplay between regulators of circadian clock and metabolism.
CLOCK and BMAL1 regulate circadian gene expression by binding to E-box elements within the promoters of clock controlled genes (CCGs). Some of the CCGs are regulators of metabolic pathways (e.g. Pepck, Glut2, Hmgcr), while some can influence metabolite levels (e.g. Nampt, Alas1). Nutrient sensors AMPK and SIRT1 are regulated in a circadian manner, and they can modulate circadian rhythms and metabolism by post-translational modifications of key regulators.
a) SIRT1
The role of SIRT1 in metabolism is well studied [25]. SIRT1 is a deacetylase and it plays crucial roles in metabolism by (a) deacetylating several proteins that participate in metabolic pathways, and (b) regulating gene expression by histone deacetylation. A fascinating hallmark of SIRT1 is that its enzymatic activity is NAD+-dependent [26]. Since the NAD+/NADH ratio is a direct measure of the energy status of a cell, the NAD+ dependence of SIRT1 directly links cellular energy metabolism and deacetylation of target proteins. Recently, two independent studies identified SIRT1 to be a critical modulator of the circadian clock machinery [27,28]. While Asher et. al. observed oscillations in SIRT1 protein levels [28], Nakahata et.al. demonstrated that SIRT1 activity, and not its protein levels, oscillate in a circadian manner [27]. Circadian oscillations in NAD+ levels were later shown to drive SIRT1 rhythmic activity [14]. SIRT1 modulates circadian rhythms by deacetylating histones (histone H3 Lys9 and Lys14 at promoters of rhythmic genes) and non-histone proteins (BMAL1 and PER2). The CLOCK-BMAL1 complex interacts with SIRT1 and recruits it to the promoters of rhythmic genes. Importantly, circadian gene expression and BMAL1 acetylation are compromised in liver-specific SIRT1 mutant mice [27]. While BMAL1 acetylation acts as a signal for CRY recruitment [29], PER2 acetylation enhances its stability [28]. These findings led to the concept that SIRT1 operates as a rheostat of the circadian machinery, modulating the amplitude and ‘tightness’ of CLOCK-mediated acetylation and consequent transcription cycles in metabolic tissues [27] (Figure 2).
SIRT1 also deacetylates and thereby regulates several metabolic proteins. For example, SIRT1 regulates gluconeogenesis by deacetylating and activating PPARγ-coactivator α (PGC1α) and Forkhead box O1 (FOXO1) [30]. FOXO1 directly regulates expression of several gluconeogenic genes [31], whereas PGC1α coactivates glucocorticoid receptors and hepatic nuclear factor 4-alpha (HNF-4α) to induce the expression of gluconeogenic genes [32]. SIRT1 also regulates cholesterol metabolism by deacetylating, and thus activating, Liver X receptor (LXR) [33] (Figure 2). LXR regulates cholesterol metabolism by inducing the expression of the ATP-binding cassette transporter A1 (ABCA1), which mediates cholesterol efflux from peripheral tissues to the blood. Since SIRT1 activity is regulated in a circadian manner, it would be interesting to determine if the acetylation of other SIRT1 targets oscillate in a circadian manner.
b) AMPK
AMPK is activated when the cellular energy status is low, meaning the AMP/ATP ratio is high. Activation of AMPK turns on catabolic processes that produce ATP and turns off ATP consuming processes. The activity of AMPK was found to be rhythmic in the mouse liver, hypothalamus and in mouse fibroblasts [34,35]. AMPK can modulate the circadian clock by phosphorylating CRY1 [34] and Casein Kinase 1 (CK1) ε [36]. CK1ε plays an important role in the regulation of circadian rhythms by phosphorylating PER proteins and controlling their degradation [37,38]. Phosphorylation by AMPK makes CRY1 a target of SCF (Skp1/cullin/F-box protein) ubiquitin ligase F-box and leucine-rich repeat protein 3 (FBXL3), which leads to the ubiquitination and degradation of CRY1 [34]. AMPK mediated activation of CK1ε leads to degradation of PER2 [36]. Under conditions of glucose deprivation and low energy status, AMPK activation alters circadian gene expression in MEFs [34]. Deletion of AMPK isoforms in mice, leads to tissue-specific alterations in circadian gene expression [35]. Interestingly, AMPK activation also leads to an increase in NAD+ levels [39]. Thus, AMPK could also modulate circadian gene expression indirectly through SIRT1 activation.
iv) Circadian Orchestration of the Metabolome: When output can also be an input!
Metabolites, such as glucose and amino acids, have been known to display circadian rhythmicity in their abundance in plasma. Minami et.al. have recently shown that the levels of hundreds of metabolites exhibit circadian oscillations in mouse plasma [40]. These metabolites include phospholipids, amino acids, and urea cycle metabolites. In fact, the abundance of these metabolites is so precisely gated by the circadian clock that just by measuring the concentrations of these metabolites, the biological time can be determined. Moreover, the circadian oscillation in these metabolites is independent of the age, sex or genetic background of the mice [40].
An interesting concept is emerging in the regulation of circadian rhythms wherein some of these oscillating metabolites (which are the outputs of the circadian clock) might also act as inputs for subsequent cycles. For example, cyclic AMP (cAMP) and NAD+ have recently been shown to be both the outputs and regulators of the circadian clock [14,15,41]. cAMP is a second messenger that is made from ATP by the action of Adenylate Cyclase. cAMP can act as a hunger signal in several tissues such as liver and muscle where glucagon can promote glycogen breakdown by activating cAMP signaling [42]. Interestingly, cAMP levels oscillate in the suprachiasmatic nucleus (SCN) and also in fibroblasts [41]. Altering the concentrations of cAMP by pharmacological means lead to changes in the amplitude, phase and period of circadian transcription.
NAD+ levels have been reported to oscillate in fibroblasts and liver through the circadian regulation of the expression of Nampt [14,15]. Inhibition of NAMPT by FK866, a highly specific inhibitor of the enzyme, alters both the phase and the amplitude of circadian gene expression [14]. Interestingly, defects in behavioral and metabolic circadian rhythms were observed in mice deficient of NAD+ hydrolase CD38, that display elevated levels of NAD+ in many tissues [43,44]. CD38-null mice display a shortened period length of locomotor activity, alteration in the rest-activity rhythm, and alterations in the circadian levels of several amino acids [43].
These results demonstrated that cAMP and NAD+ are not only a clock output but can also regulate the clock by acting as an input signal. It would be important to identify other metabolites that can feedback into the clock machinery.
II. Regulation of the Circadian Functions by Metabolism: When Metabolism Can Twist the Hands of the Clock
Mice entrained to a 12 hr light: 12 hr dark cycle, demonstrate circadian rhythmicity in their feeding behavior. Almost 80% of food is consumed at night, when the mice are active. Recent research has shown that alterations in the timing of food availability or the composition of food can have a significant impact on the functioning of the clock. Restricting food availability to only during the day time can uncouple the circadian phases of gene expression between peripheral and central clock (Box 1) [9]. While day time feeding completely inversed the phase of the expression of circadian genes in the liver (peripheral clock), the same genes oscillated with an unaltered phase in the SCN (central clock). These results confirmed that food is a potent Zeitgeber (time giver) for peripheral clocks [9]. Interestingly, restricting food access to a certain time period could also restore rhythmicity in the expression of some genes in the livers of the arrhythmic Cry1−/−Cry2−/− mice [45]. In addition, restricting food access to day time in Drosophila, causes desynchrony between clocks located in the fat body (mammalian equivalent of liver) and the brain, and affects reproductive fitness [46].
Shift work, and accompanying light exposure at night, has been implicated in the development of metabolic syndrome and cardiovascular diseases [4,5]. A recent study showed that mice exposed to light at night gained more weight, had reduced glucose tolerance and ate more during the light phase. Interestingly, when food was restricted to the dark phase, weight gain was prevented [8]. In another study, mice fed a high-fat diet only during the light-phase gained more weight when compared to mice that ate the same high-fat diet but only during the dark-phase, an observation that highlights the importance in the timing of food intake [7]. These results raise an interesting question: Could adjusting our food intake to only during the active phase (daytime for humans) be an effective way of weight control? Since humans have evolved for thousands of years without artificial light, our internal clock still functions the best when natural light is the only source of light, so it is probable that restricting food intake to daytime, may help control weight gain.
Not just the timing, but the quality of the diet might also affect the clock. Mice fed a high-fat diet had altered circadian rhythms and displayed a lengthening of the period of locomotor activity [47]. Interestingly, these mice also consumed a higher than normal percentage of food during the light phase. Moreover, the expression of core clock genes and the clock-controlled genes (CCGs) was altered in the mice that were fed a high-fat diet [47]. These studies have clearly established that metabolism can also control the peripheral clock.
III. Role of clock components in metabolic homeostasis
If circadian machinery is critical for metabolic homeostasis, deletion or mutation of individual core clock components or of CCGs should lead to metabolic disorders. This is indeed the case as illustrated by examples discussed below and in table 1.
Table 1. Metabolic defects in mice harboring mutations in clock genes.
“Core clock genes” are represented in blue and “Clock-controlled genes” are in Red.
| Clock Gene | Function | Metabolic Defects in mutant mice |
|---|---|---|
| Clock | bHLH-PAS domain containing transcription factor, Positive Regulator, Histone Acetyltransferase | |
| Bmal1 | bHLH-PAS domain containing transcription factor, Positive Regulator | |
| Per1 | PAS-domain containing negative regulator |
|
| Per2 | PAS-domain containing negative regulator |
|
| Cry1,Cry2 | Negative Regulator | |
| Rev-erbα | Nuclear Receptor, Negative Regulator |
|
| Rorα | Nuclear Receptor, Positive Regulator | |
| Pgc-1α | Transcriptional coactivator | |
| Nocturnin | mRNA Deadenylase |
A) CLOCK and BMAL1
Loss of function of CLOCK and BMAL1, the central transcription factors that regulate circadian rhythms, leads to several metabolic anomalies. Clock/Clock mutant mice, which are arrhythmic when placed in constant darkness, become hyperphagic and obese, and develop classical signs of “metabolic syndrome” such as hyperglycemia, dyslipidemia and hepatic steatosis (fatty liver) [2]. In addition, the mRNA levels of the neuropeptides orexin and ghrelin – both involved in the neuroendocrine regulation of food intake [48,49] – are also reduced in these mice. Furthermore, renal sodium reabsorption is compromised and arterial blood pressure is reduced in the Clock−/− mice [50] (Box 2).
BOX 2.
Circadian Control of Kidney Function
The major functions of the kidney, such as urinary excretion of water and electrolytes, have long been known to display robust circadian rhythms [75]. Recent studies have demonstrated that the circadian regulation of renal function is achieved in two ways: (i) through the intrinsic renal clock [50] and (ii) through humoral factors (e.g.: aldosterone [76]). Many genes that control water and solute homeostasis in kidney display circadian oscillations in their expression [50]. For example, V2 vasopressin receptor and aquaporin-2, two critical genes that regulate water reabsorption in the kidney, display circadian rhythmicity in the abundance of their transcripts. Their expression pattern is altered in the kidney from Clock−/− mice, which might be responsible for the excretion of diluted urine in these mice [50]. Moreover, the expression of the α-subunit of the epithelial sodium channel (αENaC), a key regulator of sodium reabsorption, is reduced in the kidney of Clock−/− mice. Since renal sodium reabsorption plays a critical role in maintaining blood pressure, the Clock−/− mice display reduced arterial blood pressure [50]. αENaC expression is also reduced in the Per1-mutant mice, and correlates with increased urinary sodium excretion and total urine volume [77]. Aldosterone was shown to induce the expression of Per1, which, in turn, induces the expression of αENaC [77]. Aldosterone, a steroid hormone produced by the adrenal gland, regulates sodium reabsorption in the kidney. Doi et. al. recently demonstrated that the plasma aldosterone concentration is rhythmic in WT mice, but is highly elevated in Cry1−/−Cry2−/− mice [76]. A key enzyme in aldosterone biosynthesis, type VI 3β-hydroxyl-steroid dehydrogenase (Hsd3b6), displays circadian rhythmicity in its expression in the aldosterone producing zona glomerulosa of the adrenal gland [76]. Hsd3b6 expression is under the control of the circadian clock. Its transcription is positively regulated by DBP and its expression is elevated in the Cry1−/−Cry2−/− mice. These Cry1−/−Cry2−/− mice demonstrated high blood pressure when fed a high salt diet (salt-sensitive hypertension) [76].
Loss of BMAL1, which renders mice completely arrhythmic [51], also leads to disruption of oscillations in glucose and triglyceride levels [52]. To address the question of whether the metabolic defects are due to a loss of rhythmicity in the SCN or in the peripheral clocks, mice with tissue specific deletion of Bmal1 in the liver or pancreas have been generated. Even though these mice show normal locomotor activity, they display disturbances in the maintenance of blood glucose levels. In liver-specific Bmal1 KO mice, the circadian expression of key metabolic genes, such as glucose transporter 2 (glut2), is abolished. This results in the mice being hypoglycemic during the fasting phase of the feeding cycle [53]. Further illustrating the importance of peripheral circadian clocks, deletion of BMAL1 in the pancreas leads to diabetes [3,54]. These mice display elevated blood glucose levels, impaired glucose tolerance, and decreased insulin secretion.
Another important metabolic function of BMAL1 is its role in adipogenesis [55]. BMAL1 mRNA levels are high during adipocyte differentiation and Bmal1−/− MEFs do not differentiate into adipocytes.
B) REV-ERBα
REV-ERBα was originally identified as a nuclear receptor that regulates lipid metabolism and adipogenesis [56]. Thus, the role of Rev-Erbα in controlling Bmal1 expression – a function that provides robustness to circadian oscillations [57] – established a critical link between the molecular machinery that regulates circadian oscillations and metabolism. Although Rev-Erbα −/− mice are not arrhythmic, the rhythmicity in their locomotor activity is altered (a shorter period length under constant light or constant dark conditions) [57].
REV-ERBα appears to act downstream of PPARγ, a key regulator of fat metabolism and adipocyte differentiation [56]. The regulatory function of REV-ERBα is controlled by the nuclear receptor corepressor 1 (NCoR1), a corepressor that recruits HDAC3, a histone deacetylase, to mediate transcriptional repression of target genes, such as Bmal1. When the NCoR1-HDAC3 association is genetically disrupted in mice, circadian and metabolic defects develop [58]. These mice demonstrate a shorter period, increased energy expenditure, and are resistant to diet-induced obesity. Moreover, these animals exhibit altered cyclic expression of genes involved in lipid metabolism in the liver [58]. HDAC3 recruitment to the genome was recently shown to be rhythmic (high during the day and low at night)[59]. At these HDAC3 binding sites, REV-ERBα and NCoR1 recruitment were in phase with HDAC3 recruitment, whereas histone acetylation and RNA Polymerase II recruitment were anti-phasic. Interestingly, genes involved in lipid metabolism in the liver also appear to be major targets of HDAC3 and REV-ERBα. Depletion of either HDAC3 or REV-ERBα was shown to cause fatty liver phenotype, such as increased hepatic lipid and triglyceride content [59].
Recent reports show that heme functions as a ligand for REV-ERBα [60,61] (Figure 1). The binding of heme increases the thermal stability of REV-ERBα and enhances its interaction with the co-repressor complex, and hence is required for its repressor function. The requirement for heme places REV-ERBα in a pivotal position in the regulation of circadian rhythms and metabolism. Importantly, the circadian clock controls the cellular heme levels. Aminolevulinate synthase 1 (ALAS1), the rate limiting enzyme in heme biosynthesis, is expressed in a circadian manner and is a specific target gene for the NPAS2 (a paralogue of CLOCK)/BMAL1 heterodimer [16] (Figure 1). Heme, in turn, binds to NPAS2 and inhibits its transactivation ability [62]. ALAS1 expression is also regulated by PGC-1α [63]. The metabolic roles attributed to heme are well known. Heme is a cofactor for enzymes such as catalases, peroxidases and cytochrome p450 enzymes, playing a role in oxygen and drug metabolism [64]. Importantly, heme promotes adipocyte differentiation [65], a function also associated with REV-ERBα and BMAL1. Thus, heme appears to be a key player in the precise coordination of metabolism and circadian function, connecting a transcriptional circadian loop to an enzymatic pathway.
C) PGC-1α and PGC-1β
PGC-1α is a transcriptional coactivator, inducible by metabolic stimuli such as fasting, exercise, or low temperature, and functions as a metabolic regulator [66]. PGC-1α regulates oxidative phosphorylation, adaptive thermogenesis in brown adipose tissue, and expression of reactive oxygen species (ROS) detoxifying enzymes. By coactivating PPARα, it induces fatty acid oxidation. It also stimulates hepatic gluconeogenesis by enhancing the activities of HNF-4α and glucocorticoid recpetors [32,66,67]. Besides these critical metabolic functions, PGC-1α was recently shown to play a crucial role in the regulation of circadian rhythms [68]. PGC-1α itself displays a circadian expression pattern in metabolic tissues such as liver and skeletal muscle. It coactivates RORα and RORγ and stimulates expression of Bmal1 and Rev-erbα. Mice lacking the PGC-1α gene have defects in locomotor activity, body temperature, and metabolic rate [68]. These defects are due to aberrant expression of clock and metabolic genes. Mice lacking another PGC-1 family member, PGC-1β, display reduced activity during the dark phase [69]. PGC-1β also shows circadian rhythmicity in its expression [68], however, it is not understood whether it directly regulates the core clock machinery.
These results indicate that PGC-1α is regulated by the circadian clock and that in turn it might regulate the clock machinery. Interestingly, PGC-1α is deacetylated, and thus activated by SIRT1 [70]. It is, therefore, conceivable that PGC-1α might be a very important integrator of metabolic status and circadian function.
CONCLUSIONS
The coupling of circadian and metabolic cycles has been evident in many organisms ranging from humans to yeast [71]. Circadian control not only provides a fine-tuning mechanism to the metabolic pathways, but it also ensures that energetically incompatible reactions are temporally segregated. Mutations in clock genes lead to anomalies in metabolism of glucose and lipids, and also effect the functioning of the kidney. As more data accumulates describing specific mechanistic roles of these clock genes in regulating metabolic pathways, new therapeutic targets are emerging. It is conceivable that drugs that modulate the clock function could be an effective treatment against certain metabolic disorders. Furthermore, chronotherapy (administration of drugs at a certain time of the day when its efficacy is the highest and the side-effects are the lowest) could also prove to be an effective therapeutic option.
Acknowledgements
We wish to apologize to all colleagues whose work, because of lack of space, could not be cited. We thank all the members of the Sassone-Corsi laboratory for helpful discussions. Work in our laboratory is supported by the National Institute of Health and the Institut de la Sante et de la Recherche Medicale (France) and Sirtris Pharmaceutical Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Eckel-Mahan K, Sassone-Corsi P. Metabolism control by the circadian clock and vice versa. Nat Struct Mol Biol. 2009;16:462–467. doi: 10.1038/nsmb.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlsson B, Knutsson A, Lindahl B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occup Environ Med. 2001;58:747–752. doi: 10.1136/oem.58.11.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Bacquer D, Van Risseghem M, Clays E, Kittel F, De Backer G, et al. Rotating shift work and the metabolic syndrome: a prospective study. Int J Epidemiol. 2009;38:848–854. doi: 10.1093/ije/dyn360. [DOI] [PubMed] [Google Scholar]
- 6.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 2009;17:2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fonken LK, Workman JL, Walton JC, Weil ZM, Morris JS, et al. Light at night increases body mass by shifting the time of food intake. Proc Natl Acad Sci U S A. 2010;107:18664–18669. doi: 10.1073/pnas.1008734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 11.Demierre MF, Higgins PD, Gruber SB, Hawk E, Lippman SM. Statins and cancer prevention. Nat Rev Cancer. 2005;5:930–942. doi: 10.1038/nrc1751. [DOI] [PubMed] [Google Scholar]
- 12.Brown MS, Goldstein JL, Dietschy JM. Active and inactive forms of 3-hydroxy-3-methylglutaryl coenzyme A reductase in the liver of the rat. Comparison with the rate of cholesterol synthesis in different physiological states. J Biol Chem. 1979;254:5144–5149. [PubMed] [Google Scholar]
- 13.Saito Y, Yoshida S, Nakaya N, Hata Y, Goto Y. Comparison between morning and evening doses of simvastatin in hyperlipidemic subjects. A double-blind comparative study. Arterioscler Thromb. 1991;11:816–826. doi: 10.1161/01.atv.11.4.816. [DOI] [PubMed] [Google Scholar]
- 14.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 17.Sonoda J, Pei L, Evans RM. Nuclear receptors: decoding metabolic disease. FEBS Lett. 2008;582:2–9. doi: 10.1016/j.febslet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oishi K, Shirai H, Ishida N. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor alpha (PPARalpha) in mice. Biochem J. 2005;386:575–581. doi: 10.1042/BJ20041150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, et al. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol. 2006;20:1715–1727. doi: 10.1210/me.2006-0052. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Downes M, Yu RT, Bookout AL, He W, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 21.Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimaldi B, Bellet MM, Katada S, Astarita G, Hirayama J, et al. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab. 2010;12:509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costa MJ, So AY, Kaasik K, Krueger KC, Pillsbury ML, et al. Circadian rhythm gene period 3 is an inhibitor of the adipocyte cell fate. J Biol Chem. 2011;286:9063–9070. doi: 10.1074/jbc.M110.164558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007;26:7916–7920. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- 25.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 27.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 29.Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 30.Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 32.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Zhang S, Blander G, Tse JG, Krieger M, et al. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 34.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Um JH, Pendergast JS, Springer DA, Foretz M, Viollet B, et al. AMPK regulates circadian rhythms in a tissue- and isoform-specific manner. PLoS One. 2011;6:e18450. doi: 10.1371/journal.pone.0018450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Um JH, Yang S, Yamazaki S, Kang H, Viollet B, et al. Activation of 5'-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem. 2007;282:20794–20798. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- 37.Akashi M, Tsuchiya Y, Yoshino T, Nishida E. Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells. Mol Cell Biol. 2002;22:1693–1703. doi: 10.1128/MCB.22.6.1693-1703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirogane T, Jin J, Ang XL, Harper JW. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- 39.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minami Y, Kasukawa T, Kakazu Y, Iigo M, Sugimoto M, et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci U S A. 2009;106:9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang G, Zhang BB. Glucagon and regulation of glucose metabolism. Am J Physiol Endocrinol Metab. 2003;284:E671–E678. doi: 10.1152/ajpendo.00492.2002. [DOI] [PubMed] [Google Scholar]
- 43.Sahar S, Nin V, Barbosa MT, Chini EN, Sassone-Corsi P. Altered behavioral and metabolic circadian rhythms in mice with disrupted NAD+ oscillation. Aging (Albany NY) 2011;3:794–802. doi: 10.18632/aging.100368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aksoy P, White TA, Thompson M, Chini EN. Regulation of intracellular levels of NAD: a novel role for CD38. Biochem Biophys Res Commun. 2006;345:1386–1392. doi: 10.1016/j.bbrc.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 45.Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu K, Diangelo JR, Hughes ME, Hogenesch JB, Sehgal A. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 2011;13:639–654. doi: 10.1016/j.cmet.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 48.Adamantidis A, de Lecea L. The hypocretins as sensors for metabolism and arousal. J Physiol. 2009;587:33–40. doi: 10.1113/jphysiol.2008.164400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saper CB, Chou TC, Elmquist JK. The need to feed: homeostatic and hedonic control of eating. Neuron. 2002;36:199–211. doi: 10.1016/s0896-6273(02)00969-8. [DOI] [PubMed] [Google Scholar]
- 50.Zuber AM, Centeno G, Pradervand S, Nikolaeva S, Maquelin L, et al. Molecular clock is involved in predictive circadian adjustment of renal function. Proc Natl Acad Sci U S A. 2009;106:16523–16528. doi: 10.1073/pnas.0904890106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sadacca LA, Lamia KA, deLemos AS, Blum B, Weitz CJ. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia. 2011;54:120–124. doi: 10.1007/s00125-010-1920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, et al. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci U S A. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fontaine C, Dubois G, Duguay Y, Helledie T, Vu-Dac N, et al. The orphan nuclear receptor Rev-Erbalpha is a peroxisome proliferator-activated receptor (PPAR) gamma target gene and promotes PPARgamma-induced adipocyte differentiation. J Biol Chem. 2003;278:37672–37680. doi: 10.1074/jbc.M304664200. [DOI] [PubMed] [Google Scholar]
- 57.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 58.Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 62.Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, et al. NPAS2: a gas-responsive transcription factor. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 63.Handschin C, Lin J, Rhee J, Peyer AK, Chin S, et al. Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1alpha. Cell. 2005;122:505–515. doi: 10.1016/j.cell.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 64.Ponka P. Cell biology of heme. Am J Med Sci. 1999;318:241–256. doi: 10.1097/00000441-199910000-00004. [DOI] [PubMed] [Google Scholar]
- 65.Chen JJ, London IM. Hemin enhances the differentiation of mouse 3T3 cells to adipocytes. Cell. 1981;26:117–122. doi: 10.1016/0092-8674(81)90039-8. [DOI] [PubMed] [Google Scholar]
- 66.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 67.Feige JN, Auwerx J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007;17:292–301. doi: 10.1016/j.tcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 68.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 69.Sonoda J, Mehl IR, Chong LW, Nofsinger RR, Evans RM. PGC-1beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc Natl Acad Sci U S A. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 71.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 72.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 73.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 74.Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Firsov D, Bonny O. Circadian regulation of renal function. Kidney Int. 2010;78:640–645. doi: 10.1038/ki.2010.227. [DOI] [PubMed] [Google Scholar]
- 76.Doi M, Takahashi Y, Komatsu R, Yamazaki F, Yamada H, et al. Salt-sensitive hypertension in circadian clock-deficient Cry-null mice involves dysregulated adrenal Hsd3b6. Nat Med. 2010;16:67–74. doi: 10.1038/nm.2061. [DOI] [PubMed] [Google Scholar]
- 77.Gumz ML, Stow LR, Lynch IJ, Greenlee MM, Rudin A, et al. The circadian clock protein Period 1 regulates expression of the renal epithelial sodium channel in mice. J Clin Invest. 2009;119:2423–2434. doi: 10.1172/JCI36908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang EE, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nature medicine. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Raspe E, et al. Identification of Rev-erbalpha as a physiological repressor of apoC-III gene transcription. Journal of lipid research. 2002;43:2172–2179. doi: 10.1194/jlr.m200386-jlr200. [DOI] [PubMed] [Google Scholar]
- 80.Mamontova A, et al. Severe atherosclerosis and hypoalphalipoproteinemia in the staggerer mouse, a mutant of the nuclear receptor RORalpha. Circulation. 1998;98:2738–2743. doi: 10.1161/01.cir.98.24.2738. [DOI] [PubMed] [Google Scholar]
- 81.Lin J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 82.Leone TC, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS biology. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Green CB, et al. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Douris N, et al. Nocturnin regulates circadian trafficking of dietary lipid in intestinal enterocytes. Curr Biol. 2011;21:1347–1355. doi: 10.1016/j.cub.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]