Abstract
hSnm1B is member of the SNM family of exonucleases involved in DNA processing and is known to be localized to telomeres via binding to the telomere-binding protein TRF2. Here we demonstrate that the C terminus of hSnm1B facilitates the concentration of hSnm1B on telomeres by promoting ubiquitin-mediated degradation of hSnm1B that is not localized to telomeres, as well as by blocking protein degradation and fostering localization to telomeres via binding of TRF2. Finally, a mutant of hSnm1B stabilized independently of exogenous TRF2-induced cell death. Taken together, we speculate that sequestering hSnm1B at telomeres by a combination of stabilizing the protein when bound to telomeres and degrading it when not bound to telomeres may be a means to prevent potentially lethal effects of unregulated hSnm1B activity.
Telomeres are DNA/protein structures that protect the ends of chromosomes from illegitimate recombination and degradation (1). The DNA portion of human telomeres is composed of a G-rich repeat (TTAGGG) that extends past the complementary C-rich strand, forming a 3′ extension. This extension loops back and invades the double-stranded region, forming a lariat structure termed the t-loop that is thought to play a role in telomere function (2). The core set of telomere proteins in humans that bind directly to telomeric DNA is composed of one protein, POT1, that binds directly to single-stranded telomeric DNA, and two proteins, TRF1 and TRF2, that bind directly to double-stranded telomeric DNA (3, 4). The latter protein, TRF2, binds double-stranded telomeric DNA as a homodimer and has been shown to promote t-loop formation in vitro (2). Disruption of TRF2 function leads to telomere fusions and a DNA damage response (5, 6). As such, TRF2 plays a critical role in the structure and function of telomeres. TRF2 also recruits a number of proteins to the telomere (4), and hence understanding the relationship of these proteins to TRF2 could shed light on the central role TRF2 plays at the telomere.
One such protein that binds directly to TRF2 in human cells is hSnm1B (7–9). hSnm1B is one of three human homologues of the yeast Saccharomyces cerevisiae protein Snm1 (PSO2), so named because mutations in this gene render yeast sensitive to nitrogen mustard (10–12). Snm1 is a member of the β-CASP family of proteins that contain a metallo-β-lactamase domain and possess nuclease activity (13–16). In humans, the three proteins with homology to Snm1 are Snm1A, Artemis (Snm1C), and Snm1B (Apollo) (8, 9, 17). Snm1A is a mitotic checkpoint protein (18), and Snm1A–/– mice have a predisposition to cancer and are susceptible to infection (19). Artemis (Snm1C) is critical for processing single-strand intermediates formed during V(D)J recombination and is involved in the maintenance of telomere length and genomic stability (20–22). Finally, knockdown of hSnm1B by RNAi sensitizes cells to interstrand cross-linking agents (23), induces DNA damage foci formation at telomeres in S-phase (8), and further increases the number of these foci in cells in which TRF2 function is disrupted (8, 9). The direct binding of hSnm1B to TRF2 (7–9), the telomere localization of hSnm1B (7–9), and formation of DNA damage foci at telomeres upon knockdown of hSnm1B (8, 9) indicate that hSnm1B is a bona fide telomere-associated protein.
We previously found that ectopic expression of TRF2 increased the stability of hSnm1B, suggesting that protein stability may be a means of regulating hSnm1B function (7). A common mechanism by which protein stability is regulated is polyubiquitination and subsequent degradation by the proteasome (24). Moreover, the stability and function of at least two other telomere-associated proteins, hTERT and TRF1, are regulated by polyubiquitination and degradation via the proteasome (25–27). This prompted us to investigate the relationship between TRF2 and hSnm1B protein stability.
MATERIALS AND METHODS
Cell Culture—Human cell lines 293T and HeLa were cultured in Dulbecco's modified Eagle's medium supplemented with 15% fetal calf serum.
Plasmids—pcDNA3-myc-TRF2 was a kind gift from Dominique Broccoli, pHA-ubiquitin was a kind gift from Xiao-Fan Wang. pCMV-myc encoding Myc epitope-tagged TRF22–300 (encoding amino acids 2–300) and TRF2246–445 (encoding amino acids 246–445), and pEGFP (-C1) and pBabePuro alone or encoding N-terminally green fluorescent protein (GFP)2-tagged hSnm1B (GFP-hSnm1B) were previously described (7). pCMV5-Flag-hSnm1B was engineered by subcloning N-terminally Flag epitope-tagged hSnm1B (Flag-hSnm1B) into pCMV5 (28). pCMV5-Flag-hSnm1B2–462, pEGFP-hSnm1B2–495, and pBabepuro-GFP-STOP-hSnm1B were generated by introducing a STOP codon by site-directed mutagenesis after amino acid 462 of hSnm1B, 495 of hSnm1B, and between GFP and hSnm1B, respectively. The six pBabePuro-GFP-hSnm1B496–532-N1-N6 plasmids were generated by substituting every consecutive six amino acids beginning with mutant N1 at amino acid 496 (corresponding to the amino acid of full-length hSnm1B) with the sequence NAAIRS by site-directed mutagenesis, as previously described (29), in the terminal 37-amino acid region of hSnm1B of the previously described plasmid pBabepuro-GFP-hSnm1B496–532 (7). pCMV5-Flag-hSnm1BN2, pBabepuro-GFP-hSnm1BN2, and pBabepuro-GFP-hSnm1BN3 were generated by substituting amino acids 502LKYLLT (N2 mutation) or 508PVNFFQ (N3 mutation) in hSnm1B with the sequence NAAIRS by site-directed mutagenesis. All mutations were confirmed by direct sequencing.
Visualization of GFP-tagged hSnm1B and hSnm1B2–495— 293T cells grown on cover slips coated with 100 mg/ml poly-d-lysine, Mr >300,000 (Sigma) were transiently transfected with either pEGFP-hSnm1B or pEGFP-hSnm1B2–495 and/or pcDNA3-myc-TRF2 using FuGENE 6 (Roche Applied Sciences) according to the manufacturer's protocols. After 48 h, cells were fixed in 3.7% formaldehyde in phosphate-buffered saline for 10 min at room temperature. The cells were then washed twice with phosphate-buffered saline, mounted, and observed using the ×100 objective lens on an Olympus IX70 confocal microscope.
Immunoblot—293T cells were seeded in 10-cm tissue culture dishes and transiently transfected with 2.5 μg of each of the combinations of the plasmids encoding the indicated transgenes using the FuGENE 6 reagent as above. Whole cell lysates were collected in radioimmune precipitation assay buffer 48 h later and immunoblotted with primary antibodies α-Flag (M2) (Sigma), α-Myc (Invitrogen), and α-GFP (B-2) (Santa Cruz Biotechnology), α-β-tubulin (2.1) (Sigma) to detect Flag-hSnm1B and derived mutants, Myc-TRF2 and derived mutants, GFP-tagged hSnm1B proteins and β-tubulin, respectively. Where indicated, film was scanned and bands quantitated using Microsoft PhotoShop.
Immunoprecipitations—293T cells were seeded in 10-cm tissue culture dishes and transfected with 2.5 μg of each of the combinations of the plasmids encoding the indicated transgenes using the FuGENE 6 reagent as above. Whole cell lysates were collected in radioimmune precipitation assay buffer 48 h later and immunoprecipitated for 2 h using anti-Flag M2 agarose (Sigma), resolved by SDS-PAGE, and immunoblotted with primary antibodies α-Flag (M2) (Sigma), α-HA (Roche Applied Sciences), and α-polyubiquitin (P4D1) (Santa Cruz Biotechnology) to detect hSnm1B and polyubiquitinated hSnm1B, respectively.
Colony Formation, Cell Counting, and DAPI Staining—HeLa cells were infected with retrovirus generated from the aforementioned pBabePuro constructs as previously described (30), 48 h later, medium was supplemented with 1.0 μg/ml of puromycin (Sigma). For colony formation, cells were cultured another 2 weeks, after which plates were stained with crystal violet to visualize puromycin-resistant colonies. For cell counting, cells were cultured another 5 days after which two samplings of cells from duplicate plates were stained with trypan blue and trypan blue-negative cells counted on a hemocytometer. For DAPI staining, cells were cultured another 5 days on coverslips, fixed with 3.7% formaldehyde, and incubated with 0.5% Nonidet P-40 and stained with 0.1 μg/ml DAPI. A minimum of 10 random fields were examined for the presence of cells with apoptotic DAPI staining.
Reverse Transcriptase PCR—293T cells were transiently transfected with 1 μg of the plasmids pBabePuro, pBabePuro-GFP-STOP-hSnm1B, -GFP-hSnm1B, -GFP-hSnm1BN2, and -GFP-hSnm1BN3. After 48 h, RNA was purified from the cells using the RNA-Bee reagent (Tel-Test) according to the manufacturer's instructions and reverse-transcribed with an oligo(dT) primer as previously described. Resultant cDNA was PCR-amplified with primers 5′-AACAGCCACAACGTCTATATC-3′ and 5′-AAGAAGAGACGTGCGGTGCCAG-3′ to specifically detected ectopic GFP-hSnm1B or primers 5′-GAGGTGCAGAGCGACTAC and 5′-GCTGTTCACCTGCAAATCCA to detect glyceraldehyde-3-phosphate dehydrogenase as a control.
RESULTS
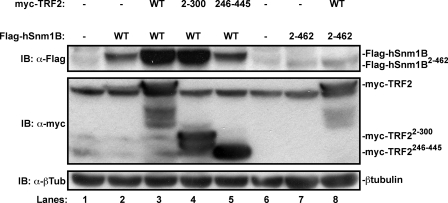
Binding to TRF2 Stabilizes hSnm1B—Given that hSnm1B binds directly to TRF2 (7–9) and that hSnm1B is more highly expressed in the presence of ectopic TRF2 (7), we investigated the mechanism by which TRF2 regulates hSnm1B levels. To begin, we tested whether hSnm1B must bind TRF2 to be stabilized. To this end, 293T cells were transiently transfected with expression plasmids encoding Flag epitope-tagged hSnm1B (Flag-hSnm1B) and Myc epitope-tagged TRF2 (Myc-TRF2) as a positive control, Flag-hSnm1B without exogenous Myc-TRF2 as a negative control, and Flag-hSnm1B2–462 lacking the last 70 amino acids that facilitate TRF2 binding, in the absence or presence of Myc-TRF2. Cells were then lysed, and immunoblotted with an α-Myc antibody to validate appropriate Myc-TRF2 expression and an α-Flag antibody to assay for Flag-hSnm1B protein levels. As already noted (7), Flag-hSnm1B levels were increased in the presence of Myc-TRF2 (Fig. 1, lanes 2 and 3). This stabilization was lost if the TRF2 binding domain was deleted in hSnm1B, evidenced by the low levels of Flag-hSnm1B2–462 detected in both the presence and absence of Myc-TRF2 (Fig. 1, lanes 7 and 8). To address the possibility that deletion of the last 70 amino acids simply led to a misfolded protein, we tested if the levels of full-length hSnm1B were reduced in the presence of the mutant Myc-TRF2246–445 that lacks the N-terminal region responsible for binding hSnm1B and the C-terminal DNA binding region (7). In contrast to the high levels of Flag-hSnm1B detected in the presence of Myc-TRF2 (Fig. 1, lane 3), the levels of Flag-hSnm1B in the presence of Myc-TRF2246–445 remained as low as Flag-hSnm1B in the absence of exogenous TRF2 (Fig. 1, lanes 2 and 5). As the TRF2 binding domain has been mapped even further to the most terminal 37 amino acids of hSnm1B (7), we validated that an hSnm1B mutant deleted for this smaller region also failed to be stabilized in the presence of Myc-TRF2 (not shown). The enhanced levels of Flag-hSnm1B did not depend upon the DNA binding activity of TRF2, as a similar experiment demonstrated that Flag-hSnm1B was still stabilized upon co-expression of the mutant Myc-TRF22–300 (31) that lacks the C-terminal Myb DNA binding domain (Fig. 1, lane 4). hSnm1B protein levels are thus elevated in the presence of TRF2, and moreover, the binding domains responsible for this interaction on the two proteins are required for this effect. Taken together, these results support the conclusion that TRF2 binding stabilizes hSnm1B.
FIGURE 1.
TRF2 binding stabilizes hSnm1B. Lysates from 293T cells transiently transfected with expression plasmids encoding the indicated combinations of WT or mutant versions lacking both DNA and hSnm1B binding regions (246–445), or the DNA binding region (2–300) of Myc-TRF2 and WT or a mutant version lacking the C-terminal 70 amino acids (2–462) of Flag-hSnm1B were immunoblotted (IB) with α-Flag and α-Myc antibodies to detect Flag-hSnm1B and Myc-TRF2, respectively. The immunoblot for β-tubulin serves as a loading control.
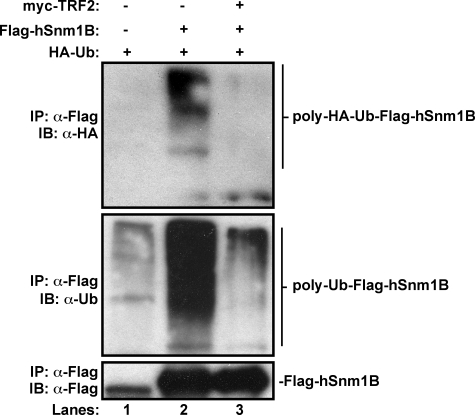
TRF2 Inhibits Polyubiquitination of hSnm1B—As TRF2 binding to hSnm1B stabilized the protein (Fig. 1) and the telomere-associated proteins TRF1 and hTERT are known to be polyubiquitinated and degraded (25–27), we hypothesized that binding of TRF2 prevents the polyubiquitination and subsequent degradation of hSnm1B. To first address whether hSnm1B is polyubiquitinated, 293T cells were transiently transfected with expression plasmids encoding HA-ubiquitin alone as a negative control or HA-ubiquitin and Flag-hSnm1B. Cells were lysed, Flag-hSnm1B was immunoprecipitated by virtue of the Flag epitope, and the immunoprecipitate was immunoblotted with α-Flag antibody to detect Flag-hSnm1B, and α-HA or α-ubiquitin antibodies to detect polyubiquitinated Flag-hSnm1B. Immunoprecipitated Flag-hSnm1B was readily detected by both the α-HA and α-ubiquitin antibodies as a smear, but was absent in negative control cells lacking Flag-hSnm1B (Fig. 2, lanes 1 and 2). Thus, hSnm1B is polyubiquitinated. To explore the influence of TRF2 on hSnm1B polyubiquitination, the status of ubiquitinated Flag-hSnm1B was assessed. 293T cells were transiently transfected with expression plasmids encoding HA-ubiquitin, Flag-hSnm1B, and Myc-TRF2, and then immunoblotted to detect ubiquitinated and total Flag-hSnm1B. hSnm1B protein levels were normalized at loading to account for stabilization of hSnm1B by TRF2. Ectopically expressed Myc-TRF2 reduced the ratio of polyubiquitinated to total Flag-hSnm1B protein (Fig. 2, lane 3). Such data are consistent with a model by which TRF2 binding blocks polyubiquitination and subsequent degradation of hSnm1B.
FIGURE 2.
hSnm1B polyubiquitination is blocked by TRF2 binding. Flag-hSnm1B was immunoprecipitated (IP) with α-Flag antibody from lysates of 293T cells transiently transfected with expression plasmids encoding the indicated combinations of Flag-hSnm1B, HA-ubiquitin, and Myc-TRF2 and immunoblotted (IB) with α-HA or α-ubiquitin antibodies to detect polyubiquitinated hSnm1B or α-Flag antibody to detect Flag-hSnm1B. Lysates were loaded to normalize protein levels of Flag-hSnm1B. Mock Flag immunoprecipitation serves as a control for nonspecific immunoprecipitation of ubiquitin proteins.
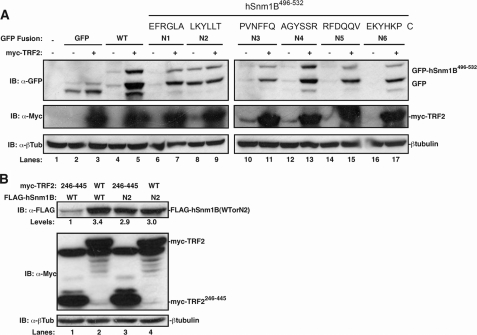
The C Terminus of hSnm1B Regulates Protein Stability—As stabilization of hSnm1B required an interaction with TRF2 (Fig. 1) and this interaction blocked polyubiquitination of hSnm1B (Fig. 2), we sought to determine whether the C-terminal TRF2-interaction domain of hSnm1B was sufficient for stabilization. As the most terminal 37 amino acids of hSnm1B was the smallest region responsible for TRF2 binding and hSnm1B stabilization, we used this region for analysis. We first assessed whether this region alone imparted protein instability when fused to GFP, and whether this instability could be rescued by binding TRF2. 293T cells were therefore transiently transfected with expression plasmids encoding GFP alone or with Myc-TRF2 as a control versus the C-terminal 37 amino acids of hSnm1B fused to GFP (GFP-hSnm1B496–532) alone or in the presence of Myc-TRF2. Lysates were then isolated and immunoblotted with an α-Myc antibody to validate appropriate Myc-TRF2 expression and an α-GFP antibody to assess the stability of GFP or GFP-hSnm1B496–532. GFP alone was readily detected by immunoblot, and ectopic expression of TRF2 had only a minor effect on the level of this protein (Fig. 3A, lanes 2 and 3). However, GFP-hSnm1B496–532 was nearly undetectable (Fig. 3A, lane 4), indicating that the last 37 amino acids of hSnm1B induced protein instability. Finally, co-expression of Myc-TRF2 stabilized the expression levels of GFP-hSnm1B496–532 protein (Fig. 3A, lane 5). Taken together, we conclude that the very C terminus of hSnm1B imparts protein instability, and moreover, that binding of TRF2 to the same region inhibits this effect.
FIGURE 3.
The C terminus of hSnm1B regulates protein stability. A, top, amino acid sequence of the terminal 37 amino acids of hSnm1B. Position of the six N1-N6 mutations in which the indicated amino acids were substituted with the sequence NAAIRS are noted below with a line. Bottom, cell lysates from 293T cells transiently transfected with expression plasmids encoding the indicated combination of GFP alone (GFP) or GFP fused to the last 37 amino acids of hSnm1B (hSnm1B496–532) in the WT or NAAIRS substitution mutation (N1-N6) configuration in the absence or presence of Myc-TRF2 were immunoblotted (IB) with an α-GFP antibody to detect GFP or GFP-hSnm1B496–532 proteins or an α-Myc antibody to detect Myc-TRF2. Mock transfection (lane 1) serves as a control for nonspecific signal. Immunoblot for β-tubulin serves as a loading control. B, cell lysates from 293T cells transiently transfected with expression plasmids encoding Flag-hSnm1B or Flag-hSnm1BN2 in the presence of either Myc-TRF2246–445 or Myc-TRF2, were immunoblotted with anα-Flag antibody to detect Flag-hSnm1B and Flag-hSnm1BN2 or an α-Myc antibody to detect Myc-TRF2. The immunoblot for β-tubulin served as a loading control. Levels: Flag-hSnm1B levels normalized to wild-type hSnm1B in the presence of Myc-TRF2246–445.
We propose that the binding of TRF2 to the C terminus of hSnm1B may physically block ubiquitination of hSnm1B as a means to stabilize the protein. Indeed, ectopic expression of TRF2 reduced polyubiquitination of hSnm1B (Fig. 2). A prediction of this hypothesis is that there exists a sequence within the terminal 37 amino acids of hSnm1B that promotes protein instability independent of exogenous TRF2, which if mutated would stabilize hSnm1B. Thus, we screened for mutants that would stabilize GFP-hSnm1B496–532 in the absence of TRF2 by substituting every consecutive six amino acids of the encoded C-terminal 37-amino acid region of hSnm1B with the sequence NAAIRS (32) and assayed the resultant mutants for stability in the absence and presence of exogenous TRF2. 293T cells were transiently transfected with expression plasmids encoding GFP-hSnm1B496–532 harboring one of the six NAAIRS mutants, which together span the C-terminal 37 amino acids of hSnm1B, in the absence or presence of exogenous Myc-TRF2. Resultant cell lysates were immunoblotted with an α-GFP antibody to monitor the stability of GFP-hSnm1B496–532 or mutants thereof, and an α-Myc antibody to confirm Myc-TRF2 expression. Five of these NAAIRS mutants behaved similar to the control GFP-hSnm1B496–532, being unstable unless exogenous Myc-TRF2 was present (Fig. 3A, lanes 6, 7, and 10–17). However, none of the mutants was as stable in the presence of Myc-TRF2 as the wild-type peptide, and correspondingly exhibited reduced Myc-TRF2 binding (not shown). There was one mutant termed GFP-hSnm1B496–532-N2, in which the sequence 502LKYLLT was mutated to NAAIRS, which was stable in both the absence and presence of Myc-TRF2 (Fig. 3A, lanes 8 and 9). The simplest interpretation of these data is that the sequences within the N2 region induce hSnm1B protein instability, and moreover, that binding of TRF2 to the C terminus of hSnm1B blocks the ability of this region to promote protein instability.
To address whether the N2 mutation would similarly promote protein stability when introduced into full-length hSnm1B, 293T cells were transiently transfected with expression plasmids encoding Flag-hSnm1B and Myc-TRF2246–445 that failed to bind hSnm1B as a negative control, Flag-hSnm1B and Myc-TRF2 as a positive control, and Flag-hSnm1B with the N2 mutation (Flag-hSnm1BN2), in the presence of either Myc-TRF2246–445 or Myc-TRF2, followed by immunoblot of resultant cell lysates with an α-Flag antibody to detect Flag-hSnm1B or Flag-hSnm1BN2 and α-Myc to validate appropriate Myc-TRF2 expression. As noted previously (Fig. 1), control Flag-hSnm1B was barely detectable, and this level was elevated in the presence of wild-type Myc-TRF2 (Fig. 3B, lanes 1 and 2). Flag-hSnm1BN2 was expressed in the presence of Myc-TRF2246–445 at levels approaching Flag-hSnm1B stabilized by wild-type Myc-TRF2 (Fig. 3B, lane 1 versus 3). Furthermore, Flag-hSnm1BN2 stability was not enhanced in the presence of wild-type Myc-TRF2 (Fig. 3B, lane 3 versus 4). Thus, the N2 mutation increased the stability of the full-length hSnm1B in the absence of TRF2.
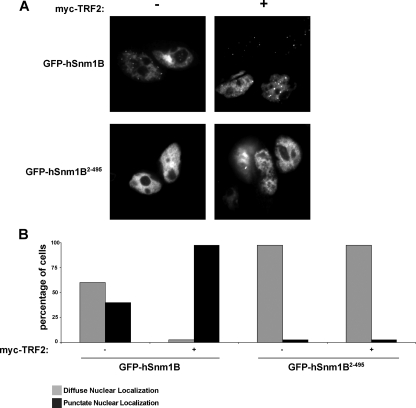
TRF2 Promotes Localization of hSnm1B to Telomeres—hSnm1B was unstable unless bound to TRF2, a telomere-binding protein (31). We thus speculated that hSnm1B not bound to TRF2 is degraded as a means of ensuring that only telomere-bound hSnm1B accumulates within cells. A prediction of this idea is that TRF2 binding should concentrate hSnm1B at telomeres. To test this, the subcellular localization of GFP-hSnm1B and GFP-hSnm1B2–495 lacking the C-terminal 37 amino acids was assessed by direct fluorescence in the absence and presence of exogenous TRF2. Specifically, 293T cells were transiently transfected with expression plasmids encoding GFP-hSnm1B or GFP-hSnm1B2–495 in the absence or presence of Myc-TRF2, and 48-h later, punctate nuclear staining indicative of hSnm1B telomere localization (7) was assayed. ∼40% of cells transfected with GFP-hSnm1B alone displayed nuclear punctate localization characteristic of telomeric localization, while the remaining cells displayed diffuse nuclear localization. However, in the presence of Myc-TRF2, this balance shifted to nearly 100% punctate with very little diffuse staining, indicating that TRF2 promotes the association of hSnm1B to telomeres. GFP-hSnm1B2–495, which cannot bind TRF2, displayed almost exclusively diffuse nuclear localization irrespective of whether Myc-TRF2 was present or not (Fig. 4, A and B). Given that TRF2 binding increases hSnm1B protein stability (Fig. 1) and telomere staining (Fig. 4), we propose that TRF2 concentrates hSnm1B at telomeres by both stabilizing hSnm1B and tethering it to telomeres.
FIGURE 4.
TRF2 promotes localization of hSnm1B to the telomere. A, representative direct fluorescence image of 293T cells transiently transfected with the indicated combinations of expression plasmids encoding GFP-hSnm1B, GFP-hSnm1B2–495, and Myc-TRF2. B, graphical quantification of the localization of GFP-hSnm1B and GFP-hSnm1B2–495 in the absence and presence of Myc-TRF2. Cells defined as having punctate nuclear localization contained at least two punctate nuclear foci per cell.
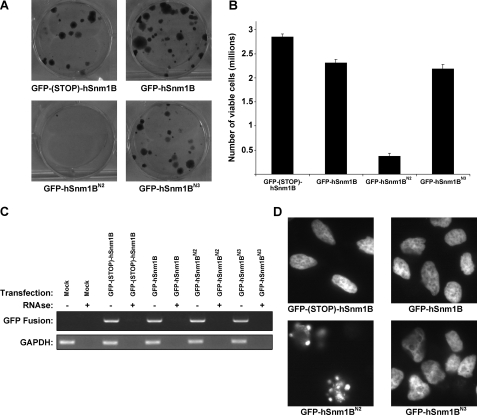
Stabilization of hSnm1B Independent of TRF2 Inhibits Colony Formation and Cell Viability—Overexpression of the related protein hSnm1A in human cells has been reported to induce cell death (33, 34). We thus speculated that hSnm1B may be targeted to telomeres or degraded, otherwise unregulated levels of hSnm1B could induce cell death. To explore this possibility we tested whether the full-length hSnm1B harboring the N2 mutation (hSnm1BN2) that is stable in the absence of exogenous TRF2 reduced colony formation, a measure of cell proliferation and viability. Specifically, HeLa cells were infected with a retrovirus encoding both the puromycin selectable marker and GFP-hSnm1BN2, which is stable in the absence of exogenous TRF2, to assess the effect of inappropriate stabilization of hSnm1B on cell viability. HeLa cells were also infected with the same retrovirus encoding both the puromycin selectable marker and GFP-hSnm1B or GFP-hSnm1BN3 (a mutation that does not effect protein stability) as controls for effects of ectopic hSnm1B on cell viability, and GFP-(STOP)-hSnm1B, in which a stop codon was inserted between GFP and hSnm1B to ensure that only GFP is expressed as a control for the effects of infection on cell viability. 48-h later, medium was supplemented with puromycin and cells cultured for 2 weeks to select for puromycin-resistant colonies or counted after 5 days after puromycin selection. Infection of cells with the GFP-(STOP)-hSnm1B-, GFP-hSnm1B-, or GFP-hSnm1BN3-expressing retrovirus all resulted in robust colony formation and cell viability, indicating a relatively minor effect of stable low expression of hSnm1B on cell viability. However, almost no colonies and few viable cells infected with retrovirus encoding GFP-hSnm1BN2 were detected (Fig. 5, A and B). This effect was not due to altered expression of the retroviral constructs as all four viral constructs expressed equivalent levels of mRNA as assayed by RT-PCR (Fig. 5C) and protein as assessed by GFP levels by fluorescence microscopy (not shown) when transiently transfected into 293T cells. The reduction in colony and cell number corresponded to an increase in cells characterized by an apoptotic phenotype. Specifically, ∼90% of HeLa cells stably expressing GFP-hSnm1BN2 displayed globular DAPI-positive staining, as compared with ∼10% of HeLa cells stably expressing GFP-(STOP)-hSnm1B, GFP-hSnm1B, or GFP-hSnm1BN3 (Fig. 5D). We thus speculate that stabilized hSnm1B produced by the GFP-hSnm1BN2 construct is deleterious to cells and that this effect is averted by either destabilization or sequestration to the telomere by binding to TRF2.
FIGURE 5.
The N2 mutation that stabilizes hSnm1B independent of exogenous TRF2 reduces colony formation and leads to apoptotic cell morphology. A, representative images of crystal violet stained plates of HeLa cells stably infected with retroviruses encoding GFP-hSnm1B, GFP-hSnm1BN2, GFP-hSnm1BN3, or GFP-(STOP)-hSnm1B 14 days after selection in puromycin. B, average number ± S.E. of viable HeLa cells stably infected with retroviruses encoding GFP-hSnm1B, GFP-hSnm1BN2, GFP-hSnm1BN3, or GFP-(STOP)-hSnm1B 5 days after selection in puromycin from two separate samples isolated from duplicate plates. C, total RNA isolated from 293T cells transiently transfected with retroviral vectors used above encoding the indicated versions of GFP-hSnm1B were RT-PCR-amplified to validate transgene expression. Addition of RNase validates that the reaction products were derived from RNA. RT-PCR of RNA isolated from mock-infected cells serves as a control for nonspecific RT-PCR amplification. Glyceraldehyde-3-phosphate dehydrogenase serves as a loading control. D, representative images of DAPI-stained HeLa cells stably infected with retroviruses encoding GFP-hSnm1B, GFP-hSnm1BN2, GFP-hSnm1BN3, or GFP-(STOP)-hSnm1B 5 days after selection in puromycin.
DISCUSSION
The C-terminal 37 amino acids help regulate hSnm1B. First, this region promotes protein instability, evidenced by the fact that fusing this region to GFP induced protein instability, and moreover, mutating the sequence responsible for this activity (N2 mutation) reversed this effect. Second, this instability is regulated by TRF2. Ectopic expression of TRF2 reversed the instability imparted by fusing the last 37 amino acids of hSnm1B to GFP, and full-length hSnm1B lacking this region was instable in the presence of TRF2. Third, hSnm1B is polyubiquitinated and this ubiquitination as well as protein instability is reduced upon binding TRF2. Fourth, binding of TRF2 and stabilization of hSnm1B is associated with elevated telomere localization of hSnm1B. We also found that ectopic expression of an hSnm1B mutant that is stable in the absence of exogenous TRF2 reduced colony formation and induced cell death, perhaps a result of unregulated exonuclease activity of the protein. It is interesting to note that the related hSnm1A protein is also toxic to cells when inappropriately expressed (33, 34), and that the levels of hSnm1A are also tightly regulated, only in this case by an IRIS sequence that regulates protein translation primarily at mitosis (18). Taken together, we speculate that sequestering hSnm1B at telomeres by a combination of stabilizing the protein when bound to telomeres and degrading the protein when not bound to telomeres, may be a means to prevent potentially lethal effects of unregulated hSnm1B activity.
Acknowledgments
We thank the Counter laboratory, Shawn Ahmed, Katherine Swenson, and Joshua Sandquist for thoughtful discussions, Ben Lampson for technical assistance, and Dominique Broccoli and Xiao-Fan Wang for reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant CA082481. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GFP, green fluorescent protein; HA, hemagglutinin; DAPI, 4′,6-diamidino-2-phenylindole; WT, wild type.
References
- 1.Blackburn, E. H. (2001) Cell 106661 –673 [DOI] [PubMed] [Google Scholar]
- 2.Griffith, J. D., Comeau, L., Rosenfield, S., Stansel, R. M., Bianchi, A., Moss, H., and de Lange, T. (1999) Cell 97503 –514 [DOI] [PubMed] [Google Scholar]
- 3.Liu, D., O'Connor, M. S., Qin, J., and Songyang, Z. (2004) J. Biol. Chem. 27951338 –51342 [DOI] [PubMed] [Google Scholar]
- 4.de Lange, T. (2005) Genes Dev. 192100 –2110 [DOI] [PubMed] [Google Scholar]
- 5.van Steensel, B., Smogorzewska, A., and de Lange, T. (1998) Cell 92401 –413 [DOI] [PubMed] [Google Scholar]
- 6.Karlseder, J., Broccoli, D., Dai, Y., Hardy, S., and de Lange, T. (1999) Science 2831321 –1325 [DOI] [PubMed] [Google Scholar]
- 7.Freibaum, B. D., and Counter, C. M. (2006) J. Biol. Chem. 28115033 –15036 [DOI] [PubMed] [Google Scholar]
- 8.van Overbeek, M., and de Lange, T. (2006) Curr. Biol. 161295 –1302 [DOI] [PubMed] [Google Scholar]
- 9.Lenain, C., Bauwens, S., Amiard, S., Brunori, M., Giraud-Panis, M. J., and Gilson, E. (2006) Curr. Biol. 161303 –1310 [DOI] [PubMed] [Google Scholar]
- 10.Henriques, J. A., and Moustacchi, E. (1980) Genetics 95273 –288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kircher, M., and Brendel, M. (1983) Chem. Biol. Interact. 4427 –39 [DOI] [PubMed] [Google Scholar]
- 12.Henriques, J. A., and Brendel, M. (1990) Curr. Genet. 18387 –393 [DOI] [PubMed] [Google Scholar]
- 13.Christofori, G., and Keller, W. (1988) Cell 54875 –889 [DOI] [PubMed] [Google Scholar]
- 14.Takagaki, Y., Ryner, L. C., and Manley, J. L. (1989) Genes Dev. 31711 –1724 [DOI] [PubMed] [Google Scholar]
- 15.Gilmartin, G. M., and Nevins, J. R. (1989) Genes Dev. 32180 –2190 [DOI] [PubMed] [Google Scholar]
- 16.Takaku, H., Minagawa, A., Takagi, M., and Nashimoto, M. (2003) Nucleic Acids Res. 312272 –2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonatto, D., Brendel, M., and Henriques, J. A. (2005) Comp. Biol. Chem. 29420 –433 [DOI] [PubMed] [Google Scholar]
- 18.Zhang, X., Richie, C., and Legerski, R. J. (2002) DNA Repair 1379 –390 [PubMed] [Google Scholar]
- 19.Ahkter, S., Richie, C. T., Zhang, N., Behringer, R. R., Zhu, C., and Legerski, R. J. (2005) Mol. Cell. Biol. 2510071 –10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, Y., Pannicke, U., Schwarz, K., and Lieber, M. R. (2002) Cell 108781 –794 [DOI] [PubMed] [Google Scholar]
- 21.Rooney, S., Alt, F. W., Lombard, D., Whitlow, S., Eckersdorff, M., Fleming, J., Fugmann, S., Ferguson, D. O., Schatz, D. G., and Sekiguchi, J. (2003) J. Exp. Med. 197553 –565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabuy, E., Newton, C., Joksic, G., Woodbine, L., Koller, B., Jeggo, P. A., and Slijepcevic, P. (2005) Radiat. Res. 16453 –62 [DOI] [PubMed] [Google Scholar]
- 23.Demuth, I., Digweed, M., and Concannon, P. (2004) Oncogene 238611 –8618 [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson, K. D. (2000) Semin. Cell Dev. Biol. 11141 –148 [DOI] [PubMed] [Google Scholar]
- 25.Chang, W., Dynek, J. N., and Smith, S. (2003) Genes Dev. 171328 –1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, T. H., Perrem, K., Harper, J. W., Lu, K. P., and Zhou, X. Z. (2006) J. Biol. Chem. 281759 –768 [DOI] [PubMed] [Google Scholar]
- 27.Kim, J. H., Park, S. M., Kang, M. R., Oh, S. Y., Lee, T. H., Muller, M. T., and Chung, I. K. (2005) Genes Dev. 19776 –781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersson, S., Davis, D. L., Dahlback, H., Jornvall, H., and Russell, D. W. (1989) J. Biol. Chem. 2648222 –8229 [PubMed] [Google Scholar]
- 29.Armbruster, B. N., Banik, S. S., Guo, C., Smith, A. C., and Counter, C. M. (2001) Mol. Cell. Biol. 217775 –7786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Hayer K, M., and Counter, C. M. (2005) Methods Enzymol. 407637 –647 [DOI] [PubMed] [Google Scholar]
- 31.Broccoli, D., Smogorzewska, A., Chong, L., and de Lange, T. (1997) Nat. Genet. 17231 –235 [DOI] [PubMed] [Google Scholar]
- 32.Wilson, I. A., Haft, D. H., Getzoff, E. D., Tainer, J. A., Lerner, R. A., and Brenner, S. (1985) Proc. Natl. Acad. Sci. U. S. A. 825255 –5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dronkert, M. L., de Wit, J., Boeve, M., Vasconcelos, M. L., van Steeg, H., Tan, T. L., Hoeijmakers, J. H., and Kanaar, R. (2000) Mol. Cell. Biol. 204553 –4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richie, C. T., Peterson, C., Lu, T., Hittelman, W. N., Carpenter, P. B., and Legerski, R. J. (2002) Mol. Cell. Biol. 228635 –8647 [DOI] [PMC free article] [PubMed] [Google Scholar]