Abstract
Venomous animals from distinct phyla such as spiders, scorpions, snakes, cone snails, or sea anemones produce small toxic proteins interacting with a variety of cell targets. Their bites often cause pain. One of the ways of pain generation is the activation of TRPV1 channels. Screening of 30 different venoms from spiders and sea anemones for modulation of TRPV1 activity revealed inhibitors in tropical sea anemone Heteractis crispa venom. Several separation steps resulted in isolation of an inhibiting compound. This is a 56-residue-long polypeptide named APHC1 that has a Bos taurus trypsin inhibitor (BPTI)/Kunitz-type fold, mostly represented by serine protease inhibitors and ion channel blockers. APHC1 acted as a partial antagonist of capsaicin-induced currents (32 ± 9% inhibition) with half-maximal effective concentration (EC50) 54 ± 4 nm. In vivo, a 0.1 mg/kg dose of APHC1 significantly prolonged tail-flick latency and reduced capsaicin-induced acute pain. Therefore, our results can make an important contribution to the research into molecular mechanisms of TRPV1 modulation and help to solve the problem of overactivity of this receptor during a number of pathological processes in the organism.
During the evolutionary process, different poisonous animals combined a set of bioactive compounds in their venoms used mainly to paralyze prey and/or as a defense against predators (1, 2). Bites of these creatures may induce inflammation, pain, tissue necrosis, allergic reactions, and neurotoxic effects such as convulsions, paralysis, respiratory failure, and cardiovascular stroke (3). Numerous toxic peptides are found within these venoms, and some of them can discriminate between closely related cellular targets that make them attractive for drug development and scientific use (4). Molecules accounting for lethal and inflammation effects of venoms have been extensively characterized, but less is known about the properties of other compounds. We concentrated on searching the compounds able to reduce TRPV12 conductivity. These receptors are expressed in mammalians in small and medium size dorsal root ganglion neurons and are localized in peripheral and central neuronal system (5–7). At present, it is accepted that TRPV1 receptors are molecular integrators of pain stimulus and initiate neuronal response during inflammation. Experiments with knock-out mice lacking the gene of vanilloid receptor clearly demonstrate its role in pain perception (8, 9). Since vanilloid receptor had been disclosed and cloned in 1997, it became an object of numerous investigations as a potential target for novel drugs against pain of different origin (10). As recently reported, vanillotoxins from a tarantula Psalmopoeus cambridgei directly activate TRPV1 in micromolar concentrations, causing pain effect in the same way as capsaicin does (11). Venoms of several jellyfish also seem to interact with TRPV1, knocking down its desensitization (12). A number of small molecules were synthesized that selectively inhibit TRPV1 in nanomolar concentration in vitro, and some of them have significant in vivo effects (13). Two acylpolyamine toxins from Agelenopsis aperta spider venom were shown to inhibit TRPV1 channels from the extracellular side (14).
The search of selective and potent polypeptide antagonists acting extracellularly has not been successful until now. We have found a sea anemone polypeptide representing the first polypeptide inhibitor of TRPV1. This compound, named analgesic polypeptide HC1 (APHC1), had analgesic effect during in vivo experiments.
Various Conus peptides have reached human clinical trials, and one is already approved as a commercial drug for intractable pain. All these peptides act through distinct mechanisms, none of which is opioid-based (15). It was also reported that peptide APETx2 from the sea anemone Anthopleura elegantissima inhibits acid-sensing ion channel 3 (ASIC3), which takes part in the transduction of acid-induced pain and hyperalgesia (16). Now, novel TRPV1 peptide inhibitor APHC1 from sea anemone Heteractis crispa enriches the toolbox for pain and inflammation study and control.
EXPERIMENTAL PROCEDURES
Polypeptide Purification—Polypeptide isolation was performed from lyophilized 70% ethanol extract of H. crispa nematocysts collected on a littoral zone of Seychelles islands. Crude polypeptide fraction was produced by hydrophobic chromatography on a Polychrom-1 (ChromLab, Moscow, Russia) 7 × 30-cm column by stepwise gradient of ethanol. Chromatography profile, gradient condition, and active fraction elution time are shown on Fig. 1, 1st stage. Further separation steps included cation exchange chromatography, first on a Bio-Rex 70 (Bio-Rad) 2.5 × 60-cm column equilibrated with 5 mm ammonium acetate buffer, pH 4.5, in a linear gradient NaCl (Fig. 1, 2nd stage) and then on an SP-Sephadex C-25 (Amersham Biosciences) 2.5 × 40-cm column in the same ammonium acetate buffer (Fig. 1, 3rd stage). Elution of compounds on stage 3 was performed in the ascent NaCl concentration accompanied with a linear gradient of pH from 4.5 to 7.3 just after half of separation process. The next stage of active compound isolation was carried out on a reverse-phase column Jupiter C5 (Phenomenex) 4.6 × 150 mm in a linear gradient of acetonitrile concentration in the presence of 0.1% trifluoroacetic acid (Fig. 1, 4th stage).
FIGURE 1.
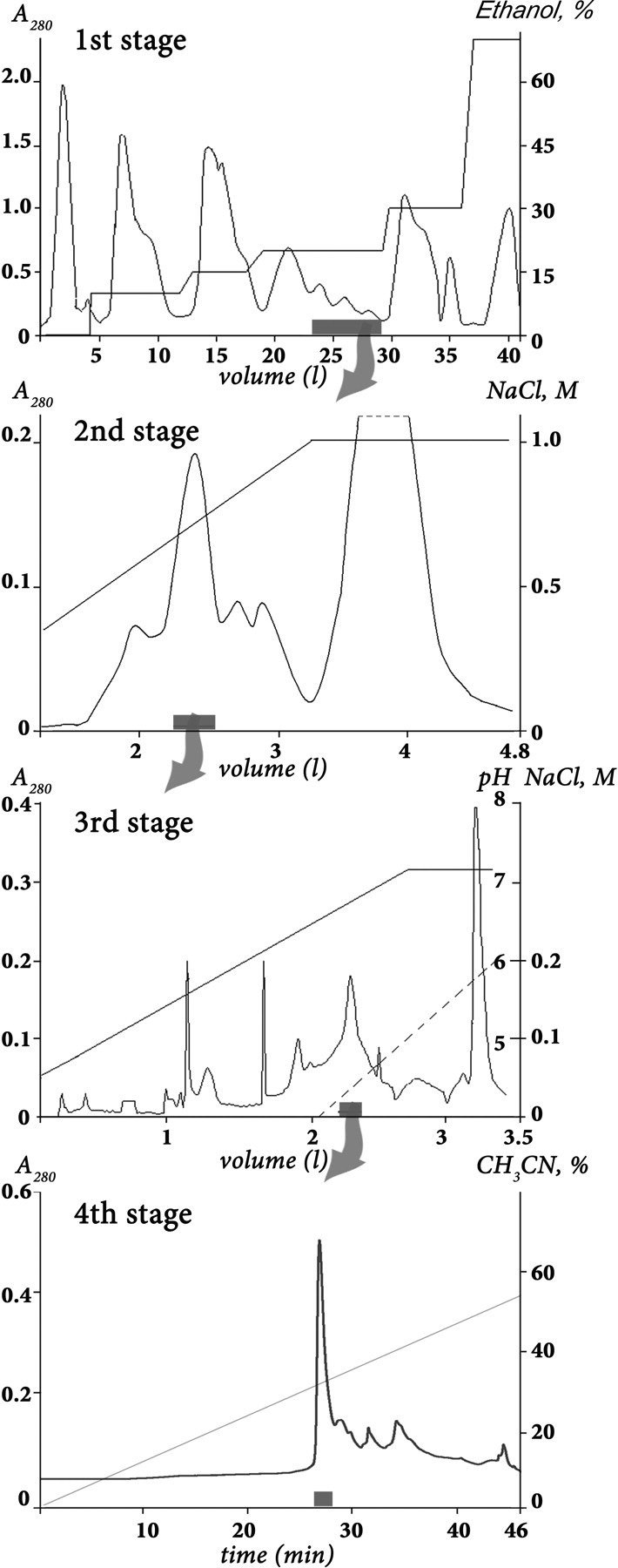
Purification of APHC1. The first separation stage of a dried ethanol extract of sea anemone H. crispa nematocysts was done on a water-equilibrated hydrophobic column Polychrom-1 (7 × 30 cm). Fractions were eluted by stepwise ethanol gradient with a flow rate of 1.2 liters/h. Active fraction (marked as a gray box on overall separation steps) has been separated on the second stage by ion exchange chromatography on Bio-Rex 70 column (2.5 × 60 cm). The separation was done in 5 mm ammonium acetate buffer (pH 4.5) by flow rate 22 ml/h in a linear gradient of NaCl concentration. The third stage of purification was performed with a flow rate 70 ml/h on the ion exchange column SP-Sephadex C-25 (2.5 × 40 cm), with the same 5 mm ammonium acetate buffer (start buffer, pH 4.5) in combined gradient of NaCl concentration and pH value. Final purification (stage 4) was achieved on a reverse-phase column Jupiter C5 (4.6 × 150 mm) in 0.1% trifluoroacetic acid with a flow rate of 1 ml/min using a linear gradient of acetonitrile concentration.
Mass Spectrometry—Separated fractions, as well as purified polypeptides, were analyzed by matrix-assisted laser desorption ionization (MALDI) time-of-flight mass spectrometry to determine fraction composition or final product purity. MALDI-LR (Waters Corp.) and Ultraflex TOF-TOF (Bruker Daltonik GmbH, Bremen, Germany) spectrometers were used. Tandem mass spectrometric (MS/MS) analysis was performed according to the manufacturer's guidelines.
Amino Acid Sequence Analysis—N-terminal sequencing was carried out by automated stepwise Edman degradation using a Procise model 492 protein sequencer (Applied Biosystems) according to the manufacturer's protocol. Cysteine residues were reduced and alkylated by 4-vinyl pyridine to determine their sequence position and improve a yield of conversion prior to analysis.
Determination of APHC1 Precursor—Total RNA was purified using RNAwiz (Ambion, Canada) as directed by the manufacturer's protocols. First strand cDNA was synthesized from 5 μg of total RNA by reverse transcriptase SuperScript II (Invitrogen) following the protocol recommended (17). Rapid amplification of cDNA ends was carried out using the universal primer T7cap (5′-GTA ATA CGA CTC ACT ATA GGG CAA GCA GTG GTA ACA ACG CAG AGT-3′) and a gene-specific degenerated primers Ing1 (5′-ATA TGT CTA GAA CCT AAR GTN GTN GG-3′), Ing2 (5′-CCT AAG GTT GTA GGA CCN TGY CAN GC-3′) for 3′-terminus determination (3′-RACE), and IngR (5′-GGC ATG CAC GCA GGG TCTC-3′) for 5′-terminus determination (5′-RACE). Taq Platinum polymerase (Invitrogen) was used for chain amplification. DNA sequencing was carried out on ABI PRISM 3100-Avant.
APHC1 Gene Synthesis—The DNA encoding APHC1 was constructed from five synthetic oligonucleotides using the PCR technique. The target PCR fragment was amplified using forward primer AL (5′-GGAATTCC ATG GGT TCT ATC TGC CTG GAA CCG AAA GTT GTTG-3′) containing EcoRI restriction enzyme site (underlined), Met-codon for BrCN cleavage, and reverse primer AR (5′-CTCTCGAG TCA AGC ACG GCA GAT AGC ACG GCA AGC ACG CAG-3′) containing XhoI restriction enzyme site (underlined) and stop codon. The PCR fragment encoding mature APHC1 was gel-purified, digested by EcoRI/XhoI, and cloned into the expression vector pET32b+ (Novagen). The resulted construct was checked by sequencing.
Production of Recombinant APHC1—Recombinant APHC1 was produced as a fusion with a thioredoxin domain. Escherichia coli BL21(DE3) cells transformed with the expression vector were cultured at 37 °C in LB medium containing 100 μg/ml ampicillin up to reaching the culture density of A600 ∼0.4–0.8. Expression was induced by adding isopropyl-1-thio-β-d-galactopyranoside up to 0.1 mm. The cells were cultured at 25 °C for 12–14 h, harvested, resuspended in the start buffer for affinity chromatography (300 mm NaCl, 50 mm sodium phosphate buffer, pH 8.0), and ultrasonicated. Then lysed cells were centrifuged for 15 min at 15,000 rpm to remove all insoluble particles. The supernatant was applied to a TALON Superflow metal affinity resin (Clontech), and the fusion protein was purified according to the protocol supplied by the manufacturer. The hybrid protein was quickly desalted on a Jupiter C5 column (Phenomenex) 4.6 × 150 mm, using a stepwise gradient of acetonitrile in 0.1% trifluoroacetic acid. The collected fusion protein was vacuum-dried and dissolved in 0.1 m HCl solution. Protein cleavage by CNBr was performed overnight at room temperature with molar ratio CNBr to protein, 600:1. Recombinant APHC1 was purified from reaction mixture on reverse-phase column Jupiter C4 (Phenomenex) 250 × 10 mm. The purity of the target polypeptide was checked by MALDI-TOF mass-spectrometry, as well as by N-terminal sequencing.
Oocyte Electrophysiology—Xenopus laevis oocytes were removed surgically, defolliculated, and injected with 2.5–10 ng of human TRPV1 cRNA (AJ272063). cRNA transcripts were synthesized from NotI-linearized TRPV1 cDNA template (EX-W1312-B02 from RZPD) using a RiboMAX™ large scale RNA production system T7 (Promega) according to a protocol for capped transcripts supplied by the manufacturer. After injection, oocytes were kept for 2–7 days at 18 °C in ND-96 medium containing (in mm) 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, 5 HEPES titrated to pH 7.4 with NaOH supplemented with gentamycin (50 μg/ml). Two-electrode voltage clamp recordings were performed using a GeneClamp 500 amplifier (Axon Instruments, Union City, CA), and data were filtered at 500 Hz and digitized at 100 Hz by an AD converter L780 (LCard, Moscow, Russia) using software created in our laboratory. Microelectrodes were filled with 3 m KCl solution. Ca2+-free ND-96 containing 0.1 mm BaCl2 was used as a bath solution. To induce ligand-activated currents, short application (20–40 s) of 2 μm capsaicin (Sigma) solution in Ca2+-free ND-96 supplemented with bovine serum albumin (0.1%) was used. Each oocyte was tested first by applying capsaicin solution 3–4 times, and only the ones with appropriate current amplitude (200–1000 nA) were used in further experiments.
Animals—Adult male wild-type mice (19–21 g) were used. The animals were housed at room temperature of 23 ± 2°C subjected to a 12-h light-dark cycle with food and water available ad libitum. Each animal was used for experimentation once only.
Tail-flick Test—Lyophilized APHC1 was dissolved in 0.9% sterile NaCl solution and administered intramuscularly into the root of the tail 100 μl/mouse. Control animals received 0.9% sterile NaCl solution 100 μl/mouse. For the tail-flick test, the mouse was restrained in a soft tissue pocket, and the distal half of the tail was immersed into water heated up to 50 °C. Latency for tail-flick was measured with a 10-s cutoff time to avoid animal injury.
Capsaicin-induced Acute Pain—Intraplantar injection of capsaicin (3 μg/10 μl in 10% ethanol/90% saline) was used to produce a capsaicin-induced acute pain. Immediately after the injection of capsaicin, mice were placed inside glass cylinders. Intraplantar injection of capsaicin evoked a licking and shaking of the injected paw in mice. The number and duration of episodes of licking and shaking the paw in response to the injection were recorded. APHC1 was administered intravenously 15 min before capsaicin. Control mice received an equal volume of 0.9% sterile NaCl solution.
Statistic—The significance of the data was determined by analysis of variance followed by Tukey's test. Data are presented as mean ± S.E.
Serine Protease Inhibitory Activity—The enzymatic activity of trypsin and chymotrypsin as well as their inhibition by APHC1 peptide, was measured spectrophotometrically in 96-well plates. Aliquots of trypsin dissolved in 90 μl in 50 mm Tris-HCl, pH 8.0, at the final concentration of about 0.5 μm were incubated for 10 min at 37 °C, in the presence of various concentrations of the peptide. After incubation, the remaining tryptic activity was determined by the addition of 10 μl of 3 mm N-a-benzoyl-dl-arginin-p-nitroanilid solution. Kinetics of paranitroaniline release was measured at 405 nm. For chymotrypsin the same incubation buffer was used. After incubation, the remaining proteolytic activity was determined by adding 1.3 mm N-benzoyl-l-tyrosine ethyl ester solution. Kinetics of tyrosine release was measured at 250 nm. Bos taurus trypsin inhibitor (BPTI) was used as control in all experiments. Inhibition constants for APHC1 were calculated for trypsin and chymotrypsin by the method described in Ref. 18.
RESULTS
Isolation of APHC1—Thirty different venoms from poisonous animals were tested for TRPV1 inhibition activity. The screening was performed by two-electrode voltage clamp standard technique on Xenopus oocytes expressing vanilloid receptors. The most attractive inhibitory action was noted for nematocyst ethanol extract from tropical sea anemone H. crispa. After the inhibitory action was evaluated, the next step of fractionation was performed up to single active component purification.
Polypeptide fraction was separated from a lot of protein compounds, lipids, pigments, and various organic molecules on Polychrom-1 chromatographic medium in accordance with the method developed earlier for purification of sea anemone neurotoxins or protease inhibitors (19, 20). Then the cation exchange separation technique in median acidic pH was used for gentle purification of active compound. As a result, the TRPV1 inhibitory fraction, which consisted of about 20 related components with measured molecular mass around 6 kDa, was isolated. The next purification step on a reverse-phase column was performed in 0.1% trifluoroacetic acid solution. The total purification procedure is shown on Fig. 1.
Novel sea anemone polypeptide, able to reduce the capsaicin-induced response of Xenopus oocytes expressed TRPV1 channels, was named as APHC1. The average molecular mass estimated by MALDI mass spectrometry was equal to 6187.0 Da.
Primary Structure Determination—Since vinylpyridine alkylated APHC1 was produced, the sequence of the first 15 amino acid residues from the N terminus was recognized by an automatic method: GSICLEPKVVGPCTA. The complete structure definition was performed by 3-RACE with degenerate primers, which were synthesized according to the structural information of N-terminal peptide, and universal primer T7Cap (as described in Ref. 17). PCR fragments (∼350 bp) were cloned into pBS-SK+. Further clone sequencing revealed a variety of closely relative sequences that had diverse point mutation at 19 positions in ∼300-bp length consensus; 11 of them are in coding part. One sequence was chosen as putative APHC1 since it encoded six Cys family polypeptides. with a calculated average molecular mass 6187.08 Da and the same N terminus. To confirm the sequence, the polypeptide chain of natural APHC1 was subjected to limited enzymatic digestion and MS/MS fragmentation. Both MS/MS spectrum and molecular weight analysis of peptide map verify the accuracy of APHC1 mature structure determination.
5-RACE performed on IngR and T7cap primers followed by isolation and sequencing of a PCR fragment about 300 bp. This fragment was aligned with sequence of 3-RACE fragment so that the complete precursor structure of APHC1 was deduced together with the 3′- and 5′-untranslated regions (Fig. 2). The precursor has a very simple organization consisting of signal peptide of 22 amino acids residues and a mature chain. No posttranslational modifications were determined.
FIGURE 2.
Structure of APHC1 gene determined from cDNA clones sequences. The signal peptide sequence deduced in the precursor structure is underlined, and partial N-terminal fragment determined by Edman degradation is shown in bold.
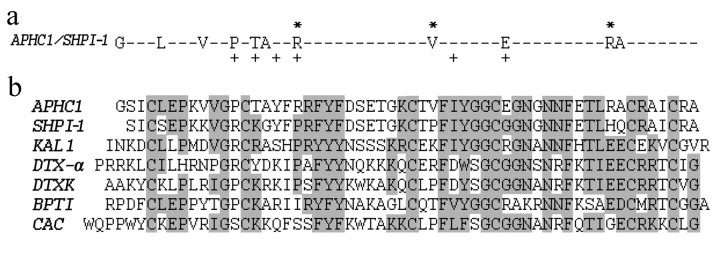
Primary Structure Homology—APHC1 has a primary structure highly homologous to BPTI/Kunitz-type trypsin inhibitors from sea anemones, such as Kunitz-type trypsin inhibitor IV from H. crispa (Uniprot ID P16344) (85% identities) and SHPI-1 from Stichodactyla helianthus (Uniprot ID P31713) (81% identities) (20, 21). Homology with sea anemones is evident, but what is more important is that APHC1 has the features of structure firstly described for protease inhibitor from bovine pancreas. This disulfide-rich α/β structure is termed BPTI/Kunitz-type fold. The special features of this fold group are the length of about 60 amino acid residues, the presence of 6 cysteine residues, glycine in the -2 position to the 2nd cysteine, a triplet of aromatic residues close to molecule center, and a typical location of several aromatic, glycine, and asparagine residues around the 4th cysteine (Pfam database 00014, cd00109). These structural features correlate with the spatial polypeptide chain fold in the ellipsoid globule but not with its functions. A number of polypeptide molecules of different functions have this fold. Thus, the alignment on Fig. 3 includes not only the structures of the protease inhibitors but also the structure of the K+ channel blockers kalicludine 1 (KAL1), DTX-α, DTXK (50–62% homology with APHC1), and Ca2+ channel inhibitor calcicludine (52% homology with APHC1).
FIGURE 3.
Alignment of APHC1 primary structure and BPTI/Kunitz type polypeptides. a, substitution plot of pairwise residues among APHC1 and sea anemone trypsin inhibitor SHPI-1 from S. helianthus (21). Amino acid residues essential for inhibitory activity of SHPI-1 as described in Ref. 22 are marked with a plus sign, and arginine/valine residues that are probably important in APHC1 are marked with asterisks. b, multiple alignment of APHC1 amino acid sequence with serine protease inhibitor SHPI-1 from sea anemone S. helianthus (21); kalicludine 1 (KAL1), K+ channel and trypsin inhibitor from sea anemone Anemonia sulcata (23); two dendrotoxins active on K+ channel DTX α from snake D. angusticeps (24) and DTXK from Dendroaspis polylepis polylepis (25); BPTI (26); and calcicludine (CAC), a blocker of high threshold Ca2+ channels from snake D. angusticeps (27). Related or similar residues are shadowed.
Recombinant Polypeptide Production—To provide sufficient material for functional investigations, recombinant APHC1 was produced in the prokaryotic expression system. Thioredoxin was chosen as the fusion partner for expression since it is known to ensure high yields of cysteine-containing polypeptides with native conformation. A synthetic gene coding for APHC1 was constructed and cloned into pET-32b(+) expression vector, and the resulting plasmid (pET-32b+APHC1) was used to transform E. coli BL21(DE3) cells. Thioredoxin-APHC1 fusion protein production and purification were followed by CNBr cleavage. The recombinant APHC1 was purified by reverse-phase high performance liquid chromatography. The final yield of purified recombinant APHC1 was estimated to be ∼0.5 mg/l of cell culture. The molecular weight of recombinant product was equal to native molecule, and the amino acid sequence of 5 N-terminal residues was determined as well. The proper peptide folding was checked by experiments on serine protease inhibition and preliminary electrophysiology tests. In both tests, the recombinant APHC1 and the natural polypeptide were equally active. Moreover, both polypeptides had the same retention time when they were co-injected on reverse-phase column. Therefore, the obtained recombinant polypeptide was widely utilized in all experiments.
Electrophysiological Study—Capsaicin (2 μm) was used as channel activator for human TRPV1 channels expressed in X. laevis oocytes. The inhibition activity was calculated as I/I0, where I is the ionic current evoked by the co-application of diluted venom, fractions, or APHC1 with agonist, and I0 is the ionic current evoked by agonist alone on the same oocyte. Fractions of the first and the second purification steps were able to reduce capsaicin-induced currents up to 25%, whereas the active fraction of the third stage was able to reduce capsaicin-induced currents up to 50%.
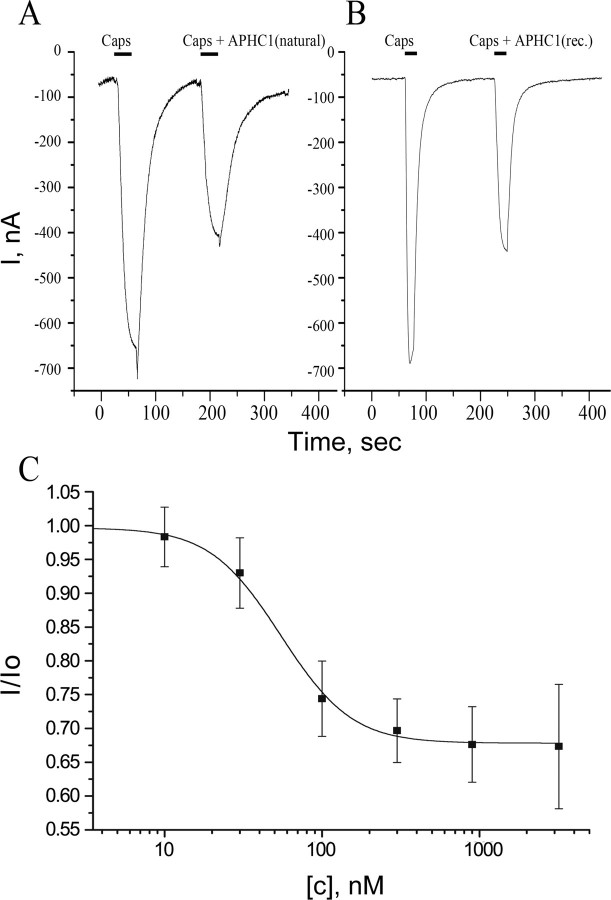
Purified APHC1 was shown to be a partial inhibitor of TRPV1 capsaicin-induced ionic currents in most of the oocytes (Fig. 4). There was no difference in APHC1 action on TRPV1 when it was co-applied with capsaicin or when a 2-min preincubation with APHC1 was performed before the capsaicin/APHC1 application. APHC1 alone (without capsaicin) did not affect TRPV1.
FIGURE 4.
Action of APHC1 on TRPV1 channels. A and B, application traces of capsaicin (Caps) alone and capsaicin mixed with purified natural APHC1 (final concentration 500 nm) (A) or with recombinant (rec.) APHC1 (final concentration 300 nm) (B). Channels were activated by 2 μm capsaicin 35 s (A) or 20 s (B) before washing, and oocytes were held at -50 mV. C, dose-response curve for APHC1 inhibitor activity on capsaicin-activated TRPV1 channels. The abscissa axis represents the ratio of ion current evoked by co-application of agonist and APHC1 (I) to ion current evoked by agonist alone (Io) on the same oocyte. Each point represents the mean ± S.E. with n = 4–8.
APHC1 binding seems to be completely reversible since a 2-min wash recovered capsaicin-induced currents to the control level. Usually the difference between the control and test applications was observed only in the response amplitude. Dose-response analysis of an inhibitory activity of recombinant APHC1 estimates the maximal inhibitory effect 32 ± 9%, half-maximal effective concentration (EC50) 54 ± 4 nm, and Hill coefficient 2.12 ± 0.19. Increasing APHC1 concentrations (up to 3.2 μm) or crude fractions of venom did not provide complete inhibition of capsaicin induced currents. The maximal observed inhibition was about 50% at concentrations ≥300 nm but did not occur in every oocyte.
Thus, APHC1 takes an effect on TRPV1 as a modulator and not as a real blocker. So far as APHC1 has significant homology with serine protease inhibitors and also has a weak protease-inhibiting activity, we carried out additional study of protease inhibitor BPTI influence on capsaicin-induced currents. Since no effect was detected, indirect action of APHC1 on TRPV1 via intermediate systems was excluded.
In Vivo Studies—Lyophilized APHC1 dissolved in NaCl saline did not provide any toxic effects on mice up to 1 mg/kg.
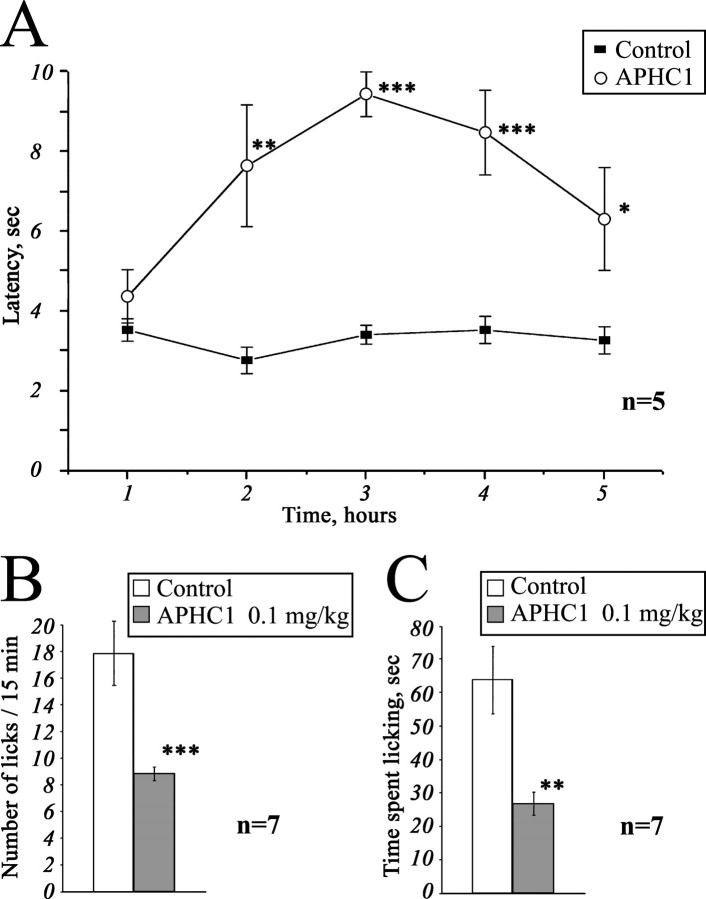
Tail-flick Test—The basal reaction time of tested animals was in 2.5–4-s range. Intramuscular administration of APHC1 (0.1 mg/kg) in root of tail considerably increased the tail-flick latency when compared with the saline-treated control (Fig. 5A). Significant antinociception effect began within 2 h following the injection, lasted all the remaining observation time (3 h), and reached maximal effect 3 h after administration. The effect began to grow weak at the time between the 4th and 5th hour of observation, and in the 5th hour, the experimental data the degree of the difference of the experimental data from the control one was not high enough (p < 0.06).
FIGURE 5.
Analgesic (antinociception) effect of a recombinant APHC1. A, tail-flick test. Latency indicates the delay of tail-flick response after one intramuscular administration of APHC1 (0.1 mg/kg) into wild-type mice, n = 5. B and C, attenuation of mice capsaicin-induced behavior by intravenous administered APHC1 (0.1 mg/kg), n = 7. Results are presented as mean ± S.E., ***, p < 0.005, **, p < 0.01, *, p < 0.06 versus control (analysis of variance and Tukey's test).
Capsaicin-induced Pain-related Behavior—Intraplantar injection of capsaicin rapidly produced distinct paw-licking behavior, mediated by direct activation of local TRPV1 receptors. Intravenous administration of APHC1 (0.1 mg/kg) 15 min before capsaicin injection significantly reduced capsaicin-induced pain-related behavior (Fig. 5, B and C).
Serine Protease Inhibition—APHC1 shows a very weak inhibition of trypsin and chymotrypsin activity. Only 8-fold molar excess of APHC1 over trypsin provides complete inhibition of paranitroaniline release from N-a-benzoyl-dl-arginin-p-nitroanilid substrate. The same APHC1 excess over chymotrypsin reduces enzyme activity only to 50%. At the same time, BPTI completely blocks trypsin/chymotrypsin in molar ratio 1:1. Constants (Ki) and inhibition types of APHC1 were determined with the Dixson method (18); trypsin inhibition was competitive with Ki = 1 × 10-6, but chymotrypsin inhibition was noncompetitive with Ki ≈ 5 × 10-6.
DISCUSSION
Stings of poisonous animals usually evoke a sensation of pain. In some cases, this sensation is connected with venom peptides. So peptide vanillotoxins isolated from tarantula venom were shown to activate the TRPV1 channels and produce pain (11). Tentacle extract of jellyfish also provokes TRPV1 activation comparable with the action of capsaicin (12). Along with activators, two polyamine inhibitors of TRPV1 channels were identified in the spider venom (14), whereas polyamines such as spermine, spermidine, and putrescine directly activated TRPV1 in a charge-dependent manner (28).
Sea anemones also produce a sensation of pain after contact with a victim. These coelenterates have large tentacles that contain venom-producing nematocysts, which are well known sources for the isolation of toxic proteins including pore-forming cytolysins (29, 30), phospholipases (31), Na+-channel toxins (32, 33, 34), K+-channel inhibitors (23, 35, 36, 37), acid-sensing ion channels inhibitor (16) and proteinase inhibitors (20, 21, 38).
It was suggested that the mixture of sea anemone toxins acting on Na+ and K+ channels has destructive neurotoxic effects due to a massive release of neurotransmitters and a resulting break of the signal transduction process. In addition, sea anemone cytolysins and phospholipases produce massive cytolytic effects on different cells (30, 39) that cause tissue damage and inflammation. In view of the pain-producing effect of sea anemone, it was interesting to find in the sea anemone extract a molecule that down-regulates such an important integrator of various pain and inflammation stimuli as TRPV1. There are several possible explanations for this phenomenon. One of them is that sea anemone nematocysts can provide an array of peptide components with distinguished activity. Another possibility is that a single peptide can activate or inhibit more than one ion channel.
APHC1 as Analgesic Agent—TRPV1 antagonists can apparently produce pain relief during a number of pathological states. Extensive research of small molecule inhibitors of TRPV1 provides intriguing evidence that TRPV1 blockade can be a useful therapeutic approach for inflammation, cancer, and possibly neuropathic pain (13, 40).
APHC1 showed significant antinociception to noxious thermal stimuli since it efficiently prolonged the latencies of tail withdrawal in a 50 °C tail-flick test (Fig. 5A). This anesthetic effect has been observed in a minimal dose of 0.05 mg/kg (data not shown). At a dose of 0.1 mg/kg, the difference of tail-flick latency for APHC1-treated and control mice was close to the reported latency for TRPV1 knock-out mice (8). However, the possible additional influence of APHC1 on other thermal nociception pathways should not be excluded. Only several small molecule TRPV1 antagonists have been reported to provide antinociception in the tail-flick test (N-(4-chlorobenzyl)-N-(4-hydroxy-3-iodo-5-methoxybenzyl) thiourea (41)) and/or in the hot plate test (DD161515, DD191515 (42)). So N-(4-chlorobenzyl)-N-(4-hydroxy-3-iodo-5-methoxybenzyl) thiourea is essential for prolongation of tail-flick latency at about ∼30–60 mg/kg, and DD161515 and DD191515 are also active in high dose (0.2 mmol/kg). The analgesic effect produced by intramuscular administration of APHC1 appears slowly (within 1–2 h after injection) but is extended for up to more then 2 h, suggesting that APHC1 may have rather a long pharmacokinetic half-life, and its diffusion in organism is quite slow, which is typical for the most part of polypeptide molecules.
Capsaicin is a selective agonist of TRPV1 receptor (8). Therefore, we tested whether APHC1 was capable of blocking capsaicin-induced behavior. Intravenous administration of APHC1 was used to provide quick distribution of the polypeptide in the organism. Doses of 0.1 mg/kg APHC1 significantly suppressed the capsaicin-induced behavior. Significant antinociception to capsaicin and noxious thermal stimuli could be an evidence of the fact that APHC1 in small doses essentially blocks TRPV1 in vivo.
Polypeptide ligands specific to TRPV1 can be of great importance as research tools or pro-drugs. To our knowledge, no selective polypeptides inhibiting TRPV1 have been reported so far. It is known that high positively charged lysine/arginine-rich peptides interacting with some Ca2+ channels also provide nonselective binding to TRPV1 (43).
Mode of Action—To date, TRPV1 is considered as a polymodal detector of noxious physical and chemical stimuli (44). Vanilloid receptor can be directly activated by various ligands (exogenous (capsaicin, EC50 = 0.7 μm, resiniferatoxin, EC50 = 40 nm and others) as well as endogenous (anandamide and others)), by heat (>43 °C), and by extracellular protons (pHout < 6.5) (13). TRPV1 is sensitive to some chemical compounds of various nature, including inflammatory agents (13), polyamines (28), and venoms of jellyfish (12) and spiders (11). APHC1 interaction with TRPV1 channels results in reduction of capsaicin-induced responses. It is most likely that APHC1 action occurs through receptor extracellular domains, which form a channel vestibule. It is also highly probable that the pore of the channel remains free, otherwise complete blockade could occur, as reported for channel blocker Ruthenium Red (45).
APHC1 was found to be high affinity (EC50 = 54 ± 4
 m) but not very potent
(maximum observed inhibition ∼50%, maximum mean inhibition 32 ±
9%). Rabbit anti-rat TRPV1 polyclonal antibodies (Ab-156H) specific to the
TRPV1 loop (considered as pH sensor) have something similar in inhibition
result to APHC1. These antibodies were not able to block completely capsaicin-
and anandamide-induced response in saturating concentrations and had maximum
inhibition of ∼55% (46).
Most likely, APHC1 shares the mechanism of action suggested to Ab-156H that
probably partially locks the channel conformation in the closed state. Other
molecules that are likely to affect TRPV1 through extracellular prepore
regions are vanillotoxins (11)
DD161515, DD191515, H-Arg-15-15C, and arginine/lysine-rich peptides
(42,
43,
47). The ability of APHC1 to
inhibit the activity of serine proteases is apparently not able to influence
channel response that was indirectly proved in experiments with BPTI-TRPV1
interaction.
m) but not very potent
(maximum observed inhibition ∼50%, maximum mean inhibition 32 ±
9%). Rabbit anti-rat TRPV1 polyclonal antibodies (Ab-156H) specific to the
TRPV1 loop (considered as pH sensor) have something similar in inhibition
result to APHC1. These antibodies were not able to block completely capsaicin-
and anandamide-induced response in saturating concentrations and had maximum
inhibition of ∼55% (46).
Most likely, APHC1 shares the mechanism of action suggested to Ab-156H that
probably partially locks the channel conformation in the closed state. Other
molecules that are likely to affect TRPV1 through extracellular prepore
regions are vanillotoxins (11)
DD161515, DD191515, H-Arg-15-15C, and arginine/lysine-rich peptides
(42,
43,
47). The ability of APHC1 to
inhibit the activity of serine proteases is apparently not able to influence
channel response that was indirectly proved in experiments with BPTI-TRPV1
interaction.
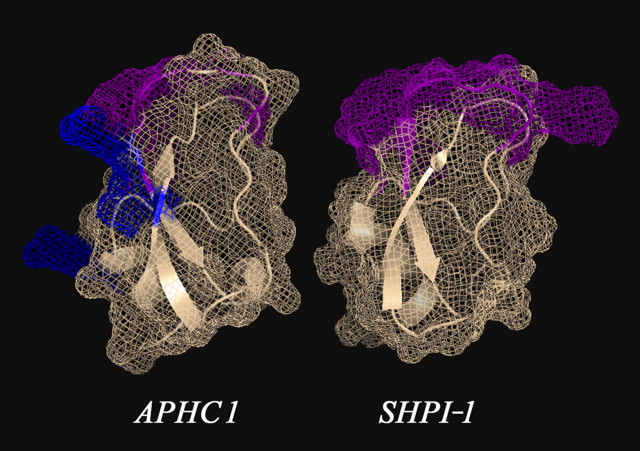
Spatial Structure and Functional Important Residues—The structure of APHC1, like proteinase inhibitors or K+-channel inhibitors, has a BPTI/Kunitz-type fold that was extensively studied on BPTI and dendrotoxin models (48, 49, 50). As mentioned above, various molecules with different functions from diverse species have this fold. BPTI is the most potent trypsin inhibitor; snake dendrotoxins (DTX I, DTXK, DTXα) are some of the most potent K+-channel blockers (49, 50). Calcicludine from Dendroaspis angusticeps snake venom is a potent blocker of high voltage-activated calcium L-type ion channels (27), as are protease inhibitors from artropoda, vertebrata, and sea anemones K+-channel blockers from sea anemone (23). The fact that distinct phyla adopt the same fold for the similar functions underlines that this fold gives significant advantages in a compact variable binding site construction. APHC1, which modulates activity of TRPV1, should also be added to the list of molecules with this fold. Despite the high primary structure homology with trypsin inhibitor SHPI-1, most of the functional important residues of APHC1 are changed. So 4 out of 6 amino acid residues, apparently important for protease inhibition (Fig. 3), differ in APHC1 and SHPI-1. On SHPI-1, NMR-determined spatial structure (Fig. 6) shows that these 6 residues form a mutual surface that interacts with the enzyme cleft and its environment. It is likely that two residues of APHC1, which remained unchanged, retain, together with other residues, an ability of the molecule to interact with proteolytic enzymes. Thus, inhibition constants for trypsin and chymotrypsin are decreased more than 1000 times: APHC1 ∼10-6; 5 × 10-6, respectively (measured in this work) in comparison with SHPI-1 ∼10-10, 2,3 × 10-9 (51).
FIGURE 6.
Three-dimensional structural models of sea anemones polypeptides. The structure of SHPI-1-trypsin inhibitor from sea anemone S. helianthus (Protein Data Bank ID 1SHP) is shown. The spatial structure of APHC1 was calculated from SHPI-1 data by molecular modeling. Amino acid residues essential for serine protease inhibition are painted in magenta on both models, and two arginine residues (Arg-18 and Arg-48) and one valine residue (Val-31), presumably important for APHC1 activity, are shown in blue.
Alignment and spatial structure analysis revealed potential residues necessary for TRPV1 binding. We are of the opinion that these are Arg-18, Arg-48, and Val-31 (shown in Fig. 3 as an asterisk and shown in Fig. 6 as blue). Their side chains are located on the same molecule interface and oriented outside. It is known that a combination of positively charged and hydrophobic residues often form active sites of toxin, for example, K+ channel blockers (49, 50). Some authors also reported that arginine/lysine-rich peptides and peptoid molecule have moderate blockade potency on TRPV1 channel in in vitro and in vivo experiments (42, 43, 47). In our opinion, one or several positively charged residues may be in an active site of APHC1, too.
We want to underscore that active sites of two highly homologous molecules SHPI-1 and APHC1 are probably located on different molecular interfaces. The same fact was reported for active sites of kalicludines that were found to have K+ channel blocker interfaces together with the trypsin inhibitor site. Finally, we would like to highlight that APHC1 is the first polypeptide inhibitor of TRPV1 channels and can play a great role in further studies of TRPV1 properties, as well as a model for designing a new generation of analgesic drugs.
Acknowledgments
We gratefully acknowledge the scientific support of S. D. Grebelny (Zoological Institute of the Russian Academy of Sciences, Sankt-Peterburg); E. I. Vozhova, E. V. Leychenko, and I. N. Lie (Pacific Institute of Bioorganic Chemistry, Far East Branch of the Russian Academy of Sciences, Vladivostok), and T. A. Egorov, and A. Kh. Musolyamov (Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences) for technical assistance.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBank™/EBI Data Bank with accession number(s) AM933240.
This work was supported by the Russian Federation Federal Agency for Science and Innovations (the State Contract 02.467.11.3003 of 20.04.2005), the Russian Foundation for Basic Research, and the Cellular and Molecular Biology program of Russian Academy of Science. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TRPV1, transient receptor potential vanilloid type 1; APHC1, analgesic polypeptide HC1; ASIC, proton-gated sodium channel; BPTI, bovine pancreatic trypsin inhibitor; DTX, dendrotoxin; MALDI, matrix-assisted laser desorption/ionization; TOF, time-of-flight; TOF-TOF, tandem time-of-flight; MS, mass spectrometry; MS/MS, tandem mass spectrometry; RACE, rapid amplification of cDNA ends.
References
- 1.Kawai, N., and Nakajima, T. (1993) in Natural and Synthetic Neurotoxins (Harvey, A. L., ed), pp. 319-345, Academic Press, London
- 2.Miljanich, G. P. (1997) Sci. Med. (Phila.) 4 6-15 [Google Scholar]
- 3.Mortaria, M. R., Cunhaa, A. O., Ferreiraa, L. B., and Ferreira dos Santos, W. (2007) Pharmacol. Ther. 114 171-183 [DOI] [PubMed] [Google Scholar]
- 4.Watters, M. R. (2005) Semin. Neurol. 25 278-289 [DOI] [PubMed] [Google Scholar]
- 5.Guo, A., Vulchanova, L., Wang, J., Li, X., and Elde, R. (1999) Eur. J. Neurosci. 11 946-958 [DOI] [PubMed] [Google Scholar]
- 6.Carlton, S. M., and Coggeshall, R. E. (2001) Neurosci. Lett. 310 53-56 [DOI] [PubMed] [Google Scholar]
- 7.Valtschanoff, J. G., Rustioni, A., Guo, A., and Hwang, S. J. (2001) J. Comp. Neurol. 436 225-235 [PubMed] [Google Scholar]
- 8.Caterina, M. J., Leffler, A., Malmberg, A. B., Martin, W. J., Trafton, J., Petersen-Zeitz, K. R., Koltzenburg, M., Basbaum, A. I., and Julius, D. (2000) Science 288 306-313 [DOI] [PubMed] [Google Scholar]
- 9.Davis, J. B., Gray, J., Gunthorpe, M. J., Hatcher, J. P., Davey, P. T., Overend, P., Harries, M. H., Latcham, J., Clapham, C., Atkinson, K., Hughes, S. A., Rance, K., Grau, E., Harper, A. J., Pugh, P. L., Rogers, D. C., Bingham, S., Randall, A., and Sheardown, S. A. (2000) Nature 405 183-187 [DOI] [PubMed] [Google Scholar]
- 10.Szallasi, A., and Fowler, C. J. (2002) Lancet Neurol. 1 167-172 [DOI] [PubMed] [Google Scholar]
- 11.Siemens, J., Zhou, S., Piskorowski, R., Nikai, T., Lumpkin, E. A., Basbaum, A. I., King, D., and Julius, D. (2006) Nature 444 208-212 [DOI] [PubMed] [Google Scholar]
- 12.Cuypers, E., Yanagihara, A., Karlsson, E., and Tytgat, J. (2006) FEBS Lett. 580 5728-5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szallasi, A., Cortright, D. N., Blum, C. A., and Eid, S. R. (2007) Nat. Rev. Drug Discov. 6 357-370 [DOI] [PubMed] [Google Scholar]
- 14.Kitaguchi, T., and Swartz, K. J. (2005) Biochemistry 44 15544-15549 [DOI] [PubMed] [Google Scholar]
- 15.Olivera, B. M. (2006) J. Biol. Chem. 281 31173-31177 [DOI] [PubMed] [Google Scholar]
- 16.Diochot, S., Baron, A., Rash, L. D., Deval, E., Escoubas, P., Scarzello, S., Salinas, M., and Lazdunski, M. (2004) EMBO J. 23 1516-1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matz, M., Shagin, D., Bogdanova, E., Britanova, O., Lukyanov, S., Diatchenko, L., and Chenchik, A. (1999) Nucleic Acids Res. 27 1558-1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon, M. (1953) Biochem. J. 55 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zykova, T. A., Vinokurov, L. M., Kozlovskaya, E. P., and Elyakov, G. B. (1985) Bioorg. Chim. 11 302-310 [Google Scholar]
- 20.Zykova, T. A., Vinokurov, L. M., Markova, L. F., Kozlovskaya, E. P., and Elyakov, G. B. (1985) Bioorg. Khim. 11 293-301 [Google Scholar]
- 21.Antuch, W., Berndt, K. D., Chávez, M. A., Delfín, J., and Wüthrich, K. (1993) Eur. J. Biochem. 212 675-684 [DOI] [PubMed] [Google Scholar]
- 22.Perona, J. J., and Craik, C. S. (1995) Protein Sci. 4 337-360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schweitz, H., Bruhn, T., Guillemare, E., Moinier, D., Lancelin, J. M., Béress, L., and Lazdunski, M. (1995) J. Biol. Chem. 270 25121-25126 [DOI] [PubMed] [Google Scholar]
- 24.Joubert, F. J., and Taljaard, N. (1980) Hoppe-Seyler's Z. Physiol. Chem. 361 661-674 [DOI] [PubMed] [Google Scholar]
- 25.Strydom, D. J. (1977) Biochim. Biophys. Acta 491 361-369 [DOI] [PubMed] [Google Scholar]
- 26.Creighton, T. E., and Charles, I. G. (1987) J. Mol. Biol. 194 11-22 [DOI] [PubMed] [Google Scholar]
- 27.Schweitz, H., Heurteaux, C., Bois, P., Moinier, D., Romey, G., and Lazdunski, M. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 878-882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahern, G. P., Wang, X., and Miyares, R. L. (2006) J. Biol. Chem. 281 8991-8995 [DOI] [PubMed] [Google Scholar]
- 29.Wang, Y., Chua, K. L., and Khoo, H. E. (2000) Biochim. Biophys. Acta 1478 9-18 [DOI] [PubMed] [Google Scholar]
- 30.Anderluh, G., and Macek, P. (2002) Toxicon 40 111-124 [DOI] [PubMed] [Google Scholar]
- 31.Grotendorst, G. R., and Hessinger, D. A. (1999) Toxicon 37 1779-1796 [DOI] [PubMed] [Google Scholar]
- 32.Norton, R. S. (1991) Toxicon 29 1051-1084 [DOI] [PubMed] [Google Scholar]
- 33.Béress, L. (2004) Toxin Rev. 23 451-466 [Google Scholar]
- 34.Ständker, L., Béress, L., Garateix, A., Christ, T., Ravens, U., Salceda, E., Soto, E., John, H., Forssmann, W. G., and Aneiros, A. (2006) Toxicon 48 211-220 [DOI] [PubMed] [Google Scholar]
- 35.Aneiros, A., Garcia, I., Martinez, J. R., Harvey, A. L., Anderson, A. J., Marshall, D. L., Engstrom, A., Hellman, U., and Karlsson, E. (1993) Biochim. Biophys. Acta 1157 86-92 [DOI] [PubMed] [Google Scholar]
- 36.Castañeda, O., Sotolongo, V., Amor, A. M., Stocklin, R., Anderson, A., Harvey, A. L., Engstrom, A., Weinstedt, C., and Karlsson, E. (1995) Toxicon 33 605-613 [DOI] [PubMed] [Google Scholar]
- 37.Diochot, S., Schweitz, H., Béress, L., and Lazdunski, M. (1998) J. Biol. Chem. 273 6744-6749 [DOI] [PubMed] [Google Scholar]
- 38.Fritz, H., Brey, B., and Béress, L. (1972) Hoppe-Seyler's Z. Physiol. Chem. 353 19-30 [DOI] [PubMed] [Google Scholar]
- 39.Šuput, D., Frangež, R., and Bunc, M. (2001) Toxicon 39 1421-1427 [DOI] [PubMed] [Google Scholar]
- 40.Krause, J. E., Chenard, B. L., and Cortright, D. N. (2005) Curr. Opin. Investig. Drugs 6 48-57 [PubMed] [Google Scholar]
- 41.Tang, L., Chen, Y., Chen, Z., Blumberg, P. M., Kozikowski, A. P., and Wang, Z. J. (2007) J. Pharmacol. Exp. Ther. 321 791-798 [DOI] [PubMed] [Google Scholar]
- 42.García-Martinez, C., Humet, M., Planells-Cases, R., Gomis, A., Caprini, M., Viana, F., De La Pena, E., Sanchez-Baeza, F., Carbonell, T., De Felipe, C., Pérez-Paya, E., Belmonte, C., Messeguer, A., and Ferrer-Montiel, A. (2002) Proc. Natl. Acad. Sc.i U. S. A. 99 2374-2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Planells-Cases, R., Aracil, A., Merino, J. M., Gallar, J., Pérez-Payá, E., Belmonte, C., González-Ros, J. M., and Ferrer-Montiel, A. V. (2000) FEBS Lett. 481 131-136 [DOI] [PubMed] [Google Scholar]
- 44.Caterina, M. J., and Julius, D. (2001) Annu. Rev. Neurosci. 24 487-517 [DOI] [PubMed] [Google Scholar]
- 45.Caterina, M. J., Schumacher, M. A., Tominaga, M., Rosen, T. A., Levine, J. D., and Julius, D. (1997) Nature 389 816-824 [DOI] [PubMed] [Google Scholar]
- 46.Klionsky, L., Tamir, R., Holzinger, B., Bi, X., Talvenheimo, J., Kim, H., Martin, F., Louis, J. C., Treanor, J. J., and Gavva, N. R. (2006) J. Pharmacol. Exp. Ther. 319 192-198 [DOI] [PubMed] [Google Scholar]
- 47.García-Martínez, C., Fernández-Carvajal, A., Valenzuela, B., Gomis, A., Van Den Nest, W., Ferroni, S., Carreño, C., Belmonte, C., and Ferrer-Montiel, A. (2006) J. Pain 7 735-746 [DOI] [PubMed] [Google Scholar]
- 48.Scheidig, A. J., Hynes, T. R., Pelletier, L. A., Wells, J. A., and Kossiakoff, A. A. (1997) Protein Sci. 6 1806-1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gasparini, S., Danse, J. M., Lecoq, A., Pinkasfeld, S., Zinn-Justin, S., Young, L. C., de Medeiros, C. C., Rowan, E. G., Harvey, A. L., and Ménez, A. (1998) J. Biol. Chem. 273 25393-25403 [DOI] [PubMed] [Google Scholar]
- 50.Smith, L. A., Reid, P. F., Wang, F. C., Parcej, D. N., Schmidt, J. J., Olson, M. A., and Dolly, J. O. (1997) Biochemistry 36 7690-7696 [DOI] [PubMed] [Google Scholar]
- 51.Delfín, J., Martínez, I., Antuch, W., Morera, V., González, Y., Rodríguez, R., Márquez, M., Saroyán, A., Larionova, N., Díaz, J., Padrón, G., and Chávez, M. (1996) Toxicon 34 1367-1376 [DOI] [PubMed] [Google Scholar]