Abstract
The mitogen-activated protein kinases (MAPKs) are key signal transduction molecules, which respond to various external stimuli. The MAPK phosphatases (MKPs) are known to be negative regulators of MAPKs in eukaryotes. We screened an Arabidopsis cDNA library using horseradish peroxidase-conjugated calmodulin (CaM), and isolated AtMKP1 as a CaM-binding protein. Recently, tobacco NtMKP1 and rice OsMKP1, two orthologs of Arabidopsis AtMKP1, were reported to bind CaM via a single putative CaM binding domain (CaMBD). However, little is known about the regulation of phosphatase activity of plant MKP1s by CaM binding. In this study, we identified two Ca2+-dependent CaMBDs within AtMKP1. Specific binding of CaM to two different CaMBDs was verified using a gel mobility shift assay, a competition assay with a Ca2+/CaM-dependent enzyme, and a split-ubiquitin assay. The peptides for two CaMBDs, CaMBDI and CaMBDII, bound CaM in a Ca2+-dependent manner, and the binding affinity of CaMBDII was found to be higher than that of CaMBDI. CaM overlay assays using mutated CaMBDs showed that four amino acids, Trp453 and Leu456 in CaMBDI and Trp678 and Ile684 in CaMBDII, play a pivotal role in CaM binding. Moreover, the phosphatase activity of AtMKP1 was increased by CaM in a Ca2+-dependent manner. Our results suggest that two important signaling pathways, Ca2+ signaling and the MAPK signaling cascade, are connected in plants via the regulation of AtMKP1 activity. To our knowledge, this is the first report to show that the biochemical activity of MKP1 in plants is regulated by CaM.
Ca2+ is a highly versatile intracellular second messenger in eukaryotes, and it mediates responses to numerous stimuli with different spatial and temporal dynamics (1–5). A variety of biotic and abiotic external stimuli promote transient changes in the cytosolic concentration of free Ca2+ in plant cells (6–8). Such changes are recognized by Ca2+ sensors or Ca2+-binding proteins (9–11). Calmodulin (CaM)4 is a well characterized primary sensor of the Ca2+ signature and contains four EF-hands in all eukaryotes. CaM regulates a variety of downstream target proteins, including metabolic enzymes, transcription factors, ion channels, protein kinases/phosphatases, and structural proteins, in a Ca2+-dependent manner (12, 13). Investigation of CaM-target interactions is crucial for our understanding of how Ca2+ signals exert downstream events.
In addition to Ca2+ signal transduction, mitogen-activated protein kinase (MAPK) signaling cascades also play a central role in the transduction of various signals in eukaryotes. Each cascade is composed of a functionally interlinked three-kinase module: an MAPK, an MAPK kinase (MEK), and an MAPK kinase kinase (MEKK). The MAPKs are activated by MEK via the phosphorylation of threonine and tyrosine residues within a conserved TXY motif (14, 15). MAPKs are inactivated by protein phosphatases such as tyrosine-specific phosphatase, serine/threonine-specific phosphatase, and dual specificity phosphatase, through specific dephosphorylation of threonine and tyrosine residues within the TXY consensus sequence (16, 17). MAPK phosphatases (MKPs) belong to the dual specificity phosphatase (DsPTP) family and dephosphorylate both phosphotyrosine and phosphoserine/phosphothreonine residues (18). Only five members of the MKPs, including the previously described AtDsPTP1, IBR5, PHS1, and AtMKP1, are predicted to exist in the Arabidopsis genome, and these five MKPs are responsible for the modulation of 20 potential MAPKs (18, 19). The MKP/MAPK ratio in Arabidopsis is disproportionately low compared with other eukaryotes, implying that plants may possess integrated and complicated mechanisms for the regulation of MAPK signaling through MKPs.
Plant MKPs play important roles in various biotic and abiotic stress responses. AtMKP1 was reported to be involved in genotoxic stress signaling (20) and AtMKP2 in the regulation of the cellular response to oxidative stress in Arabidopsis (21). NtMKP1 (22, 23) and OsMKP1 (24) are known to regulate wound responses in tobacco and rice, respectively. Mutation of the IBR5 gene decreased the sensitivity to the phytohormones auxin and abscisic acid (ABA), and PHS1 has been described as a negative regulator of abscisic acid signaling (25, 26). However, the biochemical characteristics of plant MKPs remain largely unclear.
In this study we identified a CaM-binding AtMKP1 in Arabidopsis, which showed biochemical characteristics distinct from MKP1 of other plants. Two CaMBDs of AtMKP1, CaMBDI and CaMBDII, were identified, and were found to bind CaM with different affinities. The interaction of CaMBDII with CaM was stronger than that of CaMBDI. The synthetic peptides for the two CaMBDs in AtMKP1 were able to bind CaM in a Ca2+-dependent manner. Using a split ubiquitin system, we also verified the existence of an in vivo interaction between CaM and AtMKP1. AtMKP1's dephosphorylation activity against OMFP, a common synthetic protein phosphatase substrate, was increased by CaM.
EXPERIMENTAL PROCEDURES
Screening of the Arabidopsis cDNA Expression Library—Screening of the Arabidopsis cDNA expression library with HRP-conjugated AtCaM2 (AtCaM2:HRP) was performed as described in previous reports (27, 28).
Construction of Deletion Mutants of AtMKP1 and Site-directed Mutagenesis—For mapping of the CaM binding domain, several serial fragment constructs were ligated into a pGEX-5X series vector (GE Healthcare). The full-length MKP1 cDNA clone was amplified by PCR with a forward (5′) primer containing a BamHI site (underlined) 5′-GGA TCC ATG GTG GGA AGA GAG GAT GCG ATG-3′ and a reverse (3′) primer containing an XhoI site (underlined): 5′-CTC GAG TTA TAG CGC GCT CAG CAG TGC TAG-3′. The PCR product was cloned with the pGEM-T Easy Vector (Promega) and sequenced to verify that the correct construct was present. The construct was subcloned into the pGEX-5X expression vector and digested with BamHI and XhoI. The full-length glutathione S-transferase (GST) fusion construct was designated D0 (encompassing amino acids 1–784). Serial fragment constructs were additionally generated by PCR using the following forward (F) and reverse (R) primer sequences: for D1 (amino acids 1–100), F, containing an EcoRI site (underlined): 5′-GAA TTC GTC GGT GAG TGG CCT CAT CCA CCA-3′ and R, containing a XhoI site (underlined): 5′-CTC GAG ATC ATC CGA ACC CGC CTT AGG CCA-3′; for D2 (amino acids 101–200) F, containing an EcoRI site: 5′-GAA TTC GTC GGT GAG TGG CTT CAT CCA CCA-3′ and R, containing a XhoI site: 5′-CTC GAG CAA GGA TCT GTA ACA AAA ATC AGA-3′; for D3 (amino acids 201–300) F, containing an EcoRI site: 5′-GAA TTC TGG TTA CAG GAT AGT CCG TCA GAG-3′ and R, containing a XhoI site: 5′-CTC GAG AAG TAA GGA GGT AGG GCT AAC GCC-3′; for D4 (amino acids 301–400) F, containing an EcoRI site: 5′-GAA TTC AGA ATG TAC AAA ATG TCT CCA CAC-3′ and R, containing a XhoI site: 5′-CTC GAG CCC AAT CAT AGG CAA AAT GCT TGC-3′; for D5 (amino acids 401–500) F, containing an EcoRI site: 5′-GAA TTC GGC TCG GTA ATT AAA GTT CAA CCA-3′ and R, containing a XhoI site: 5′-CTC GAG TGA TGA AGA AGT TGT TGA GGA AGG-3′; for D6 (amino acids 501–600) F, containing an EcoRI site: 5′-GAA TTC TCC ACT GCG TCG CCT CCT TTT CTC-3′ and R, containing a XhoI site: 5′-CTC GAG GGA ATG AGC CAA ACC TGG CAA CTT-3′; for D7 (amino acids 601–700) F, containing an EcoRI site: 5′-GAA TTC AAC AGA GGC ACA CCT GCT TTT ACT-3′ and R, containing a XhoI site: 5′-CTC GAG GAT TGC TAT AAC AGA TTC TGA ATC-3′; and finally for D8 (amino acids 701–784) F, containing an EcoRI site: 5′-GAA TTC CCG TTG CCA AGT GAT GCT GTA GGA-3′ and R, containing a XhoI site: 5′-CTC GAG TTA TAG CGC GCT CAG CAG TGC TAG-3′. Amplified products were cloned into pGEM-T and subcloned into a pGEX-5X expression vector using the appropriate restriction enzyme sites. To identify the critical residues in the interactions between CaM and AtMKP1, we introduced several point mutations into the GST:AtMKP1, GST:D5 (CaMBD I), and GST:D7 (CaMBD II) clones. Substitution of single amino acids was performed using a Site-Directed Mutagenesis Kit (Stratagene). The following forward (F) and reverse (R) primers were employed; for W453R F: 5′-CAT CTT CCT GCA AGG GAA AAC AGT AGG AGC TCA CTG-3′ and R: 5′-TGC AAA CTT ACA TTT CAG TGA GCT CCT ACT GTT TTC-3′. For L456R F: 5′-GCA AGG GAA AAC AGT TGG AGC TCA CGG AAA TGT AAG-3′ and R: 5′-AAA CCT TGA TGC AAA CTT ACA TTT CCG TGA GCT CCA-3′. For W678R F: 5′-TCT GGC CAG CCT TTA GCA TCT CGT AGA CCA AGT ATG-3′ and R: 5′-TTT TGT AAT CAT CTC CAT ACT TTG TCT ACG ACA TGC-3′. For I684R F: 5′-TGT CGT TGG CCA AGT ATG GAG ATG AGA ACA AAA CTG-3′ and R: 5′-TAA GTA AGC TCT GCT CAG TTT TGT TCT CAT CTC CAT-3′.
Expression of Recombinant Proteins in E. coli and CaM Binding Assay—All clones were individually introduced into Escherichia coli BL21(DE3) pLysS and expressed. Expression of the GST fusion proteins was induced by application of 1 mm isopropyl 1-thio-β-d-galactopyranoside for 5 h at 25 °C. Cells were harvested, resuspended in lysis buffer (50 mm Tris-HCl (pH 7.5), 200 mm NaCl, 1% Triton X-100 (v/v), 0.1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 0.5 mm dithiothreitol, and 5% glycerol (v/v)), and then sonicated for 2 min using a 40% pulse. Recombinant proteins were purified on a glutathione-agarose 4B column (GE Healthcare). One microgram of purified protein was separated on a 10% SDS-polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane (Millipore) after which expressed GST fusion proteins were detected using a polyclonal GST-specific antiserum. To examine the CaM binding ability of the recombinant proteins, a duplicate blot was probed with an AtCaM2:HRP conjugate in the presence of 1 mm CaCl2 or 5 mm EGTA. The CaM:HRP overlay assay was carried out as described above. The bound CaM was visualized using an ECL detection system (GE Healthcare).
CaM Mobility Shift Assay with a Synthetic Peptide—Peptides corresponding to a stretch of 25 amino acids within CaMBDI (445HLPARENSWSSLKCKFASRFDKGFR469) and 24 amino acids in CaMBDII of AtMKP1 (669SSGQPLACRWPSMEMITKLSRAYL692) were generated at a peptide synthesis facility (TaKaRa and Peptipharm). The CaM binding abilities of the synthetic peptides were determined from the relative mobility shift of CaM in the presence of the peptide (29). CaM (303 pmol) was incubated with increasing concentrations of peptide (molar ratios: 0, 0.25, 0.5, 1.0, 1.5, 2.0, and 3.0) in binding buffer (100 mm Tris-HCl (pH 7.2), plus 0.1 mm CaCl2 or 2 mm EGTA) at room temperature for 1 h. Mixtures were electrophoresed on 18% non-denaturing polyacrylamide gels containing 0.375 m Tris-HCl (pH 8.8), and either 0.1 mm CaCl2 or 2 mm EGTA. Gels were run at a constant voltage of 100 V in electrode buffer (25 mm Tris-HCl (pH 8.3), 192 mm glycine, and either 0.1 mm CaCl2 or 2 mm EGTA) and visualized by staining with Coomassie Brilliant Blue.
Phosphodiesterase Competition Assay—Cyclic nucleotide phosphodiesterase (PDE) assays were performed using commercially available bovine heart CaM-deficient phosphodiesterase (Sigma), as previously described (27). The initial reaction mixture (100 μl) contained buffer (100 mm imidazole-HCl, 2.56 mm cAMP, 5.13 mm MgSO4, 1.28 mm CaCl2) with varying amounts of AtCaM2 (1–200 nm) and a fixed concentration of synthetic peptide (100 nm). The reaction was initiated by the addition of PDE (0.5 milliunit/μl). The basal level of enzyme activity was determined in the absence of AtCaM2, and stimulated activity was determined in the presence of AtCaM2 and CaCl2. After incubation at 30 °C for 30 min, the reaction was terminated by placing the reaction tubes into a boiling water bath for 5 min and then on ice for 2 min. Following brief centrifugation, 50 μl of alkaline phosphatase (10 units) was added, and the mixture was incubated at 37 °C for 10 min. The reaction was stopped by adding 500 μl of 10% trichloroacetic acid. After vortexing, precipitates were spun down, and the supernatant (400 μl) was transferred to a new tube. After adding 1 ml of phosphate reagent (30), the supernatant was incubated at 37 °C for 30 min and assayed for Pi at A660. The dissociation constant (Kd) of AtCaM2 for the peptide was calculated from the concentration of AtCaM2 (nm) required to obtain the half-maximal (50%) PDE activity, either in the presence (100 nm) or absence of peptide. The following equation was used to calculate dissociation constants (29): Kd = ([Pt] + K – [CaM])K/([CaM] – K), where [Pt] is the total concentration of peptide added, and [CaM] and K represent the concentrations of CaM required to obtain half-maximal activation of PDE in the presence or absence of peptides, respectively. The errors are indicated by the standard deviation values of over three measurements.
Yeast Split Ubiquitin Assay—The yeast split ubiquitin assay was performed as described previously (32, 33). Saccharomyces cerevisiae strain JD53 was used for all experiments. AtCaM2 and AtMKP1 (wild type and mutant) cDNA were cloned into pMet-Ste14-Cub-RUra3, replacing yeast Ste14. AtCaM2 and AtMKP1 (WT and mutant) cDNA were cloned into modified versions of the pCup-Nub-Sec62 vector, replacing yeast Sec62 (34). Interactions between each pair of proteins were tested on selective medium containing 1 mg/ml 5-fluoroorotic acid (5-FOA) and selective medium lacking uracil. Plates were incubated at 30 °C for 3–5 days, unless specified otherwise.
Phosphatase Assay—To analyze the generic phosphatase activity of AtMKP1, 3-O-methylfluorescein phosphate (OMFP, Sigma) was used as a substrate. Phosphatase activity at various enzyme concentrations (0, 10, 20, 30, 40, 50, 60, 80, and 100 μg) of WT or mutant AtMKP1 proteins (W453R, W678R, or W453R+W678R) was examined in 200 μl of phosphatase buffer (50 mm Tris-HCl (pH 8.0), 15 mm NaCl, 1 mm EDTA) containing 500 μm OMFP, and the mixtures were incubated at 30 °C for 1 h. The absorbance of 3-O-methylfluorescein was determined at 477 nm using a spectrophotometer. To analyze phosphatase activity at the various indicated concentrations (0, 1, 2, 3, 5, 7, and 10 μg) of AtCaM2, reactions were allowed to proceed at 30 °C for 1 h using 80 μg of WT or mutant AtMKP1 (W453R, W678R, or W453R+W678R) in the presence (1 mm CaCl2) or absence of Ca2+ (5 mm EGTA). Data are presented as the mean values of over four independent assays.
RESULTS
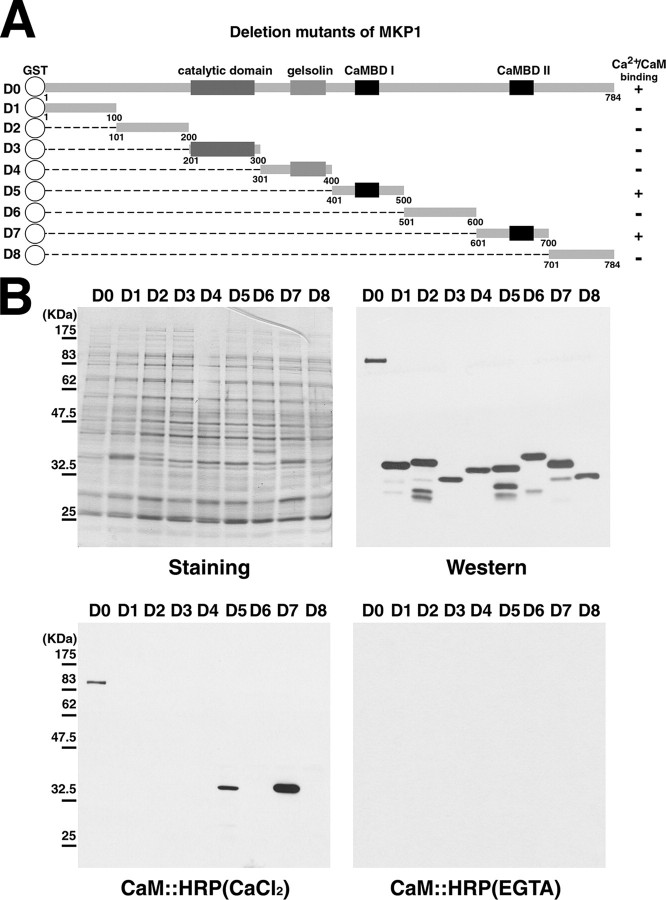
Isolation of a CaM-binding AtMKP1 in Arabidopsis—To identify the molecular components of the CaM-mediated signaling pathways, we previously screened an Arabidopsis cDNA expression library using HRP-conjugated CaM as a probe (27, 28). One of the isolated clones that bound CaM was a mitogen-activated protein kinase phosphatase 1 (MKP1). MKP1 is known to be an active protein phosphatase that hydrolyzes the phosphate group on both phosphoserine/threonine and phosphotyrosine residues of substrates in Arabidopsis (20). Moreover, the catalytic mechanism of MKP1 involves the MAPK signal pathway, as evident from data showing that the overexpressed MKP1 eliminates the kinase activity of MPK6 (20). MKP1 has a highly conserved dual specificity phosphatase catalytic domain, a gelsolin homology domain, and two putative CaMBDs (Fig. 1A).
FIGURE 1.
Identification of the CaM-binding domain of AtMKP1. A, schematic representation of AtMKP1 (D0) and serial fragment constructs (D1–D8). The putative CaM-binding domain (CaMBD) is represented by a black box, whereas the catalytic domain and gelsolin domain are represented by a gray box. Amino acid positions of each fragment are indicated. D0–D8 represent GST fusion constructs containing the specified fragments of AtMKP1. The CaM-binding ability is specified as + (CaM binding) or – (no CaM binding). B, CaM binding overlay analysis. GST and GST fusion proteins of serial fragment mutants (D0–D8) of AtMKP1 were expressed in E. coli. Recombinant proteins were analyzed by Western blotting with an anti-GST antibody (Western). The CaM:HRP overlay assay was performed using ACaM2:HRP in the presence of 1 mm CaCl2 or 5 mm EGTA.
Mapping of a CaMBD of MPK1—A large number of CaMBPs have been shown to possess one or more sequence motifs for formation of CaM complexes (35). Based on the structural characteristics of CaMBDs, two putative CaMBDs are predicted in MKP1. Both are located in the C terminus, from His445 to Arg469 in CaMBDI and from Ser669 to Lys692 in CaMBDII (Fig. 1A). These two stretches of amino acids include hydrophobic amino acids that are separated by several basic residues. The two putative CaMBDs residing in MKP1 fit well with a consensus Ca2+-dependent CaM-binding motif (36).
To confirm the presence of the putative CaMBDs of AtMKP1, we generated GST-fused constructs containing full-length cDNA (designated D0) and eight serial fragment constructs (D1, D2, D3, D4, D5, D6, D7, and D8) as shown in Fig. 1A. Recombinant fusion proteins were bacterially produced, separated by SDS-PAGE, and transferred to polyvinylidene difluoride membranes for Western blotting and CaM overlay assays. Expression of the GST fusion proteins was verified by probing the blot with an anti-GST antibody. Three recombinant proteins (D0, D5, and D7) that contained the putative CaMBD interacted with HRP-conjugated CaM (CaM:HRP), whereas GST fusion proteins lacking the predicted CaM binding region (D1, D2, D3, D4, D6, and D8) did not interact with CaM:HRP. CaM bound to AtMKP1 in the presence, but not the absence of Ca2+ (Fig. 1B). These results indicate that CaM binds to AtMKP1 at two distinct regions in a Ca2+-dependent manner.
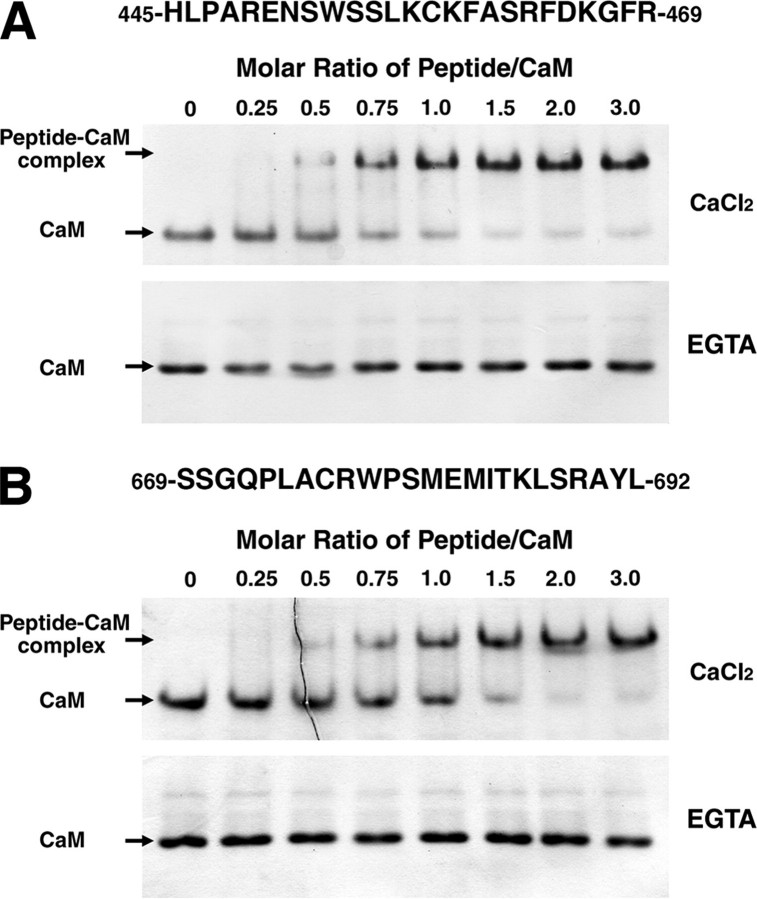
Binding of a Synthetic Peptide to CaM—To study the binding of CaMBDI and CaMBDII to CaM, peptides corresponding to these two domains were used in a CaM mobility shift assay on non-denaturing polyacrylamide gels. As shown in Fig. 2, the amount of the peptide-CaM complex increased with increasing concentrations of the synthetic peptide in the presence of 0.1 mm Ca2+, whereas the complex was undetectable in the presence of 2 mm EGTA. This finding is consistent with the data from a previous CaM overlay assay under similar Ca2+ and EGTA conditions. In the case of CaMBDI, ∼80% of the CaM was shifted at a molar ratio of 1:1 (peptide:CaM), and almost all of the CaM was shifted at molar ratios of 2:1 and 3:1 (peptide: CaM) (Fig. 2A). For CaMBDII, ∼50% of the CaM was shifted at molar ratio of 1:1 (peptide:CaM), and all of the CaM was completely shifted at a ratio of 2:1 and 3:1 (peptide:CaM) (Fig. 2B). These results suggest that the 25-mer peptide corresponding to CaMBDI (His445 to Arg469) and the 24-mer correspond to CaMBDII (Ser669 to Leu692) are sufficient for binding CaM in a Ca2+-dependent manner.
FIGURE 2.
Binding of CaM to the synthetic peptides for two CaMBDs. The CaM-binding activity of the CaMBDI peptide (A) corresponding to amino acids 445–469 and the CaMBDII peptide (B) corresponding to amino acids 669–692 in AtMKP1 was analyzed using a gel mobility shift assay. AtCaM2 (303 pmol) was incubated with increasing amounts of peptide (peptide/CaM molar ratios are indicated) in the presence of 0.1 mm CaCl2 or 2 mm EGTA. Samples were separated by nondenaturing PAGE. The positions of free CaM and the peptide-CaM complex are indicated by arrows.
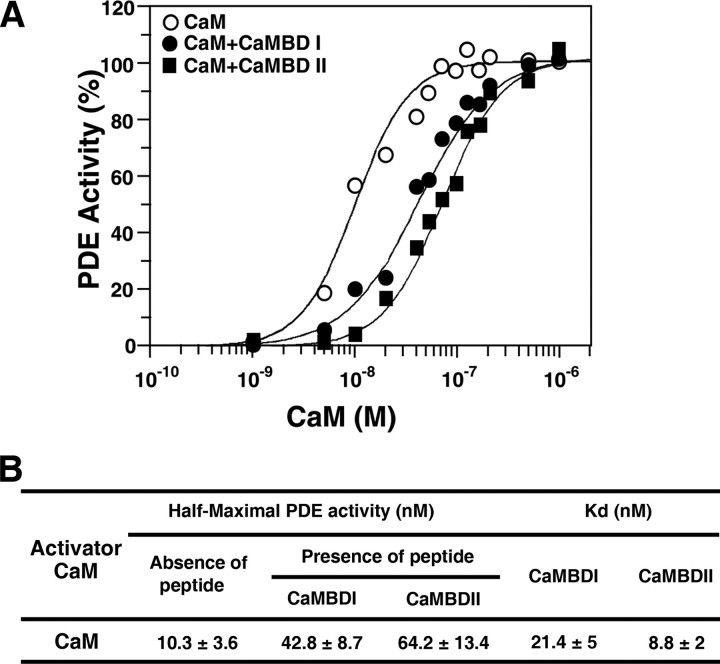
To analyze the binding of the synthetic peptides to CaM, we also performed a competition assay with PDE, a Ca2+/CaM-activated enzyme, in the presence of the two CaMBD peptides. The peptides were able to inhibit competitively the activation of PDE by CaM (Fig. 3). To determine Kd values, dose-dependent activation of PDE by CaM was monitored either in the presence (100 nm) or absence of peptides (Fig. 3A). The activation curves shifted to the right in the presence of the peptides, indicating competition between PDE and the peptides for binding to CaM. The concentration required to achieve half-maximal activation of PDE was 10.3 nm in the absence of peptides and was 42.8 nm (a 4-fold difference) and 64.2 nm (6-fold) in the presence of the two peptides, CaMBDI and CaMBDII, respectively. The Kd values of the peptide for PDE activation by AtCaM2 were determined to be 21.4 nm (CaMBDI) and 8.8 nm (CaMBDII) (Fig. 3B). These data imply that the binding affinity of CaMBDII for CaM is higher than that of CaMBDI.
FIGURE 3.
Analysis of the binding affinity between CaMBDs and CaM. A, effect of synthetic peptides derived from CaMBDs on the activation of PDE by CaM. Peptides competed with PDE for binding to CaM. PDE activity was measured in the presence of varying concentrations of AtCaM2, either in the presence or absence of a fixed concentration (100 nm) of two synthetic CaMBD peptides. B, Km and Kd values for the CaMBD peptides of AtMKP1.
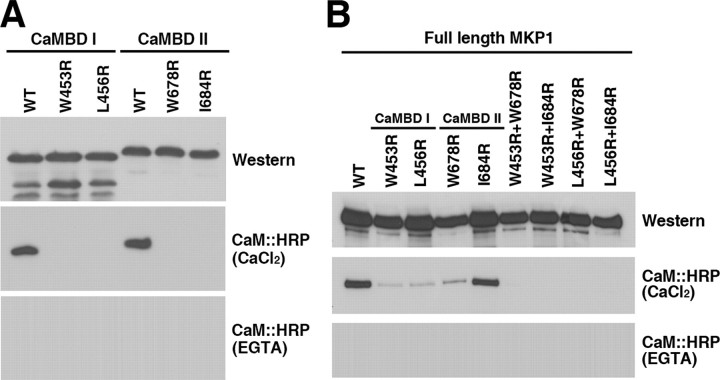
Identification of Critical Residues in the CaM-binding Motifs—Many CaM-binding proteins were found to contain a basic amphiphilic α-helical CaM-binding domain (37, 38). Although the sequence of CaM is highly conserved, and is identical among vertebrates, the Ca2+/CaM-binding regions of targets exhibit low sequence homology (36, 39). We performed site-directed mutagenesis to identify the residues in CaMBDI and CaMBDII that are critical for CaM binding. Within CaMBDI, hydrophobic residues important for CaM binding (specifically, Trp453 and Leu456) were separately converted to Arg and denoted W453R and L456R, respectively (Fig. 4A). Each mutation was introduced into the GST:D5 fusion construct containing CaMBDI. Fusion proteins were expressed in Escherichia coli and subjected to a CaM binding overlay assay. In CaMBDII, we substituted each of the hydrophobic amino acids (Trp678 and Ile684) to Arg and designated these substitutions W678R and I684R, respectively, within the GST:D7 fusion construct containing CaMBDII (Fig. 4A). CaM was found to bind to the wild-type D5 protein, but not to W453R and L456R of CaMBDI in AtMKP1. Moreover, CaM was found to bind to a GST:D7 protein, but not to W678R and I684R of CaMBDII (Fig. 4A). Mutagenesis analysis demonstrated that four key residues, specifically, Trp453 and Leu456 in CaMBDI and Trp678 and Ile684 in CaMBDII, are indispensable for binding of CaM to AtMKP1.
FIGURE 4.
Identification of critical residues for the interaction of AtMKP1 with CaM. A, CaM binding analysis of WT and mutated CaMBD constructs of the D5 and D7 domains shown in Fig. 1A. CaMBDI WT, W453R, and L456R indicate wild-type CaMBDI (D5 domain), and CaMBDI mutants containing single amino acid substitutions, respectively. CaMBDII WT, W678R, and I684R represent wild-type CaMBDII (D7 domain), and CaMBDII mutants containing single amino acid substitutions, respectively. B, interaction of WT and CaMBD mutants of full-length AtMKP1 (D0 described in Fig. 1A) with CaM. Wild-type and CaMBD mutants were fused to the C terminus of GST and expressed in E. coli. Expressed recombinant proteins were analyzed by Western blotting with an anti-GST antibody (Western). CaM binding was analyzed using a CaM: HRP overlay assay in the presence of 1 mm CaCl2 or 5 mm EGTA.
To determine the effect of CaM binding to full-length AtMKP1, we constructed a CaM-binding negative mutant, GST:AtMKP1. Based on previous mutagenesis results, we substituted Trp453 and Leu456 in CaMBDI, and Trp678 and Ile684 in CaMBDII with Arg, resulting in CaM-binding negative mutants (denoted W453R+W678R, W453R+I684R, L456R+W678R, and L456R+I684R, respectively). GST fusion proteins were produced and subjected to a CaM binding overlay assay. As shown in Fig. 4B, CaM was able to bind to AtMKP1 mutants containing a single amino acid substitution (W453R, L456R, W678R, and I684R) due to the presence of the remaining CaMBD, but could not bind to the double mutants (W453R+W678R, W453R+I684R, L456R+W678R, and L456R+I684R) of AtMKP1.
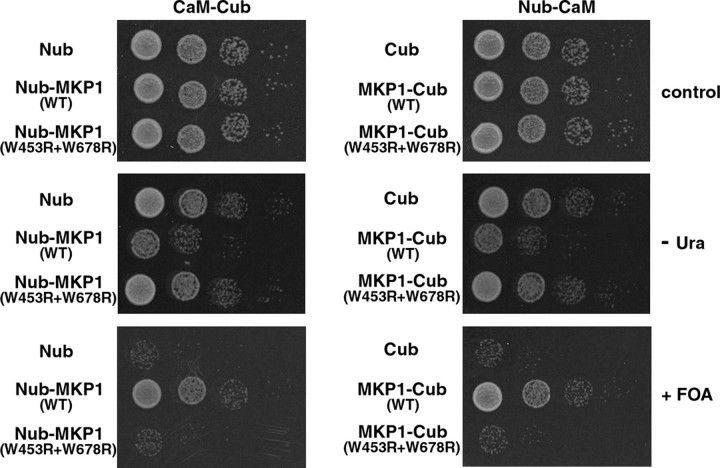
In Vivo Interactions between AtMKP1 and CaM in Yeast—To detect the direct interactions between full-length AtMKP1 and CaM, we used the yeast split-ubiquitin assay system, which is based on the reassembly of the N- and C-terminal halves (Nub and Cub) of ubiquitin (Ub) (32, 34). AtMKP1 and CaM were fused to the C terminus of Nub and the N terminus of Cub, respectively. The Cub of ubiquitin was linked to an N-terminally modified Ura3p reporter containing Arg at position 1 (RUra3p). If CaM interacts with the AtMKP1 protein, Nub and Cub should associate into native-like Ub, and cleavage of RUra3p by ubiquitin-specific proteases should follow. Released RUra3p is then rapidly degraded though the N-end rule pathway of protein degradation (40). Consequently, cells containing CaM-Cub-RUra3p and Nub-AtMKP1 or AtMKP1-Cub-RUra3p and Nub-CaM are unable to grow on plates lacking uracil, but can grow on plates containing 5-FOA, which is converted into toxic 5-fluorouracil by RUra3p. In the opposite cases, yeast cells are uracil prototrophs and are sensitive to 5-FOA.
As shown in Fig. 5, cells co-expressing CaM-Cub-RUra3p and Nub-AtMKP1 or AtMKP1-Cub-RUra3p and Nub-CaM were unable to grow on plates lacking uracil, but grew on plates containing 5-FOA, indicating that AtMKP1 effectively forms stable complexes with CaM in vivo. In negative controls, no interactions were observed between CaM-Cub-RUra3p and Nub or Cub-RUra3p and Nub-CaM. To test the specificity and CaMBD sequence dependence of the interactions, experiments were repeated with AtMKP1 CaM-binding negative mutant (W453R+W678R) instead of AtMKP1. CaM/AtMKP1 mutant (W453R+W678R)-transformed cells displayed markedly increased 5-FOA sensitivity and grew well on plates lacking uracil, indicating that CaM binding was absent in this mutant. Additionally, based on similar experiments, CaM was found to be unable to bind to other CaM-binding negative mutants (W453R+I684R, L456R+W678R, and L456R+I684R) of AtMKP1. These results are consistent with data from the previous CaM binding overlay assay (Fig. 5).
FIGURE 5.
Split ubiquitin assay to analyze the interaction between CaM and AtMKP1 in vivo. Shown are serial dilutions of cells co-expressing Nub or the Nub-AtMKP1 (WT and CaMBD mutant) fusion protein together with CaM (AtCaM2-Cub-RUra3), and Cub or the AtMKP1-Cub-RUra3 (WT and CaMBD mutant) fusion protein together with CaM (Nub-AtCaM2), on plates lacking tryptophan and histidine (control), or additionally lacking uracil (-Ura) or containing 5-FOA (+FOA). All proteins were expressed from single-copy vectors.
Effect of CaM on AtMKP1 Phosphatase Activity—To determine whether AtMKP1 possesses Ca2+/CaM-dependent phosphatase activity, we expressed recombinant GST:AtMKP1 and CaM binding negative mutant proteins in E. coli and purified the fusion proteins. We first tried to measure the phosphatase activity using pyronitrophenyl phosphate (pNPP) as an artificial substrate, but could detect no significant activity in the recombinant proteins of AtMKP1, unlike AtDsPTP1 (28). Dual-specificity phosphatases often prefer bulky polycyclic aryl phosphates and show weak enzyme activity toward simple aryl phosphates, such as pNPP (41, 42), probably because they contain shallower active site pockets (43, 44). Therefore, we next employed a bulky polycyclic substrate, OMFP, which is known to boost the second order rate constant kcat/Km of the mammalian MAPK phosphatase MKP3 by >280-fold compared with pNPP (45). The amount of OMF released was calculated using the absorbance at 477 nm. The wild-type and CaM binding negative mutant proteins of AtMKP1 indeed dephosphorylated OMFP at a lower rate, compared with already reported phosphatases such as AtMKP2 (21) or AtDsPTP1 (28). The phosphatase activities of AtMKP1 and its negative CaM binding mutants (W453R, W678R, and W453R+W678R) increased proportionally with protein concentration. Although their activities were slightly different, all CaM binding negative mutants still displayed phosphatase activity, except W456R, for which no phosphatase activity was observed (data not shown).
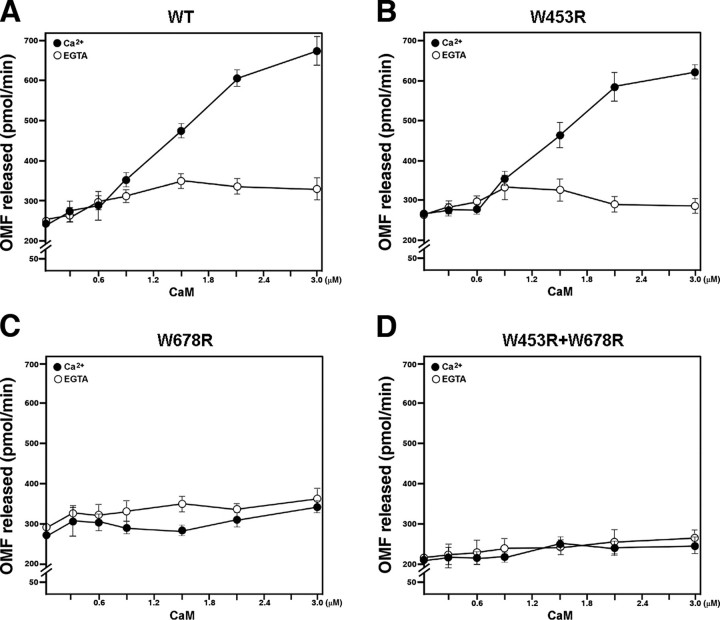
To measure the CaM-dependent activity of AtMKP1, we combined a constant amount of AtMKP1 (80 μg) with increasing concentrations of CaM (1–10 μg, corresponding to 297 nm to 2.97 μm). The phosphatase activity was found to increase in wild-type AtMKP1 in proportion to the concentration of CaM. The molar ratio of CaM to AtMKP1 (CaM:AtMKP1) ranged from 0.083 to 0.83. To determine the CaMBD critical for CaM-dependent activation of AtMKP1, we performed similar experiments using CaM binding mutants. The phosphatase activity of the CaMBDI mutant (W453R) increased with the concentration of CaM, similar to wild-type AtMKP1 (Fig. 6, A and B). However, the phosphatase activities of CaMBDII mutants (W678R) and a mutant with a double substitution (W453R+W678R) were not affected by the addition of CaM (Fig. 6, C and D). These results indicate that CaMBDII, rather than CaMBDI, is crucial for the CaM-dependent activation of AtMKP1. In the absence of Ca2+ (5 mm EGTA), the phosphatase activity of AtMKP1 was not affected by CaM.
FIGURE 6.
Regulation of AtMKP1 phosphatase activity by CaM using OMFP. Phosphatase activity was measured at varying concentrations of CaM, with a fixed amount (80 μg) of each recombinant protein and OMFP (500 μm) in the presence of 1 mm CaCl2 or 5 mm EGTA. A, wild type (WT); B, CaMBDI mutant (W453R); C, CaMBDII mutant, (W678R); D, CaMBDI and CaMBSII mutants (W453R+W678R). Activity was presented as picomoles/min of the OMF released during the reaction. Data are presented as mean values from at least four independent assays.
DISCUSSION
AtMKP1 Is a CaM Binding Phosphatase Containing Two CaMBDs—CaM serves as the primary Ca2+ sensor for a variety of external stimuli and plays an important role in transducing Ca2+ signals by modulating the activities of structurally and functionally different target proteins (46, 47). A diverse set of CaMBPs participate in Ca2+/CaM-mediated signal transduction, to trigger cellular responses. We screened an Arabidopsis cDNA expression library using HRP-conjugated CaM as a probe and isolated AtMKP1 as a plant-specific CaMBP. The AtMKP1 functions as a regulator of genotoxic stress responses. AtMKP1 dephosphorylates and inactivates MPK6, which is commonly involved the stress response (20). Two orthologs of AtMKP1, NtMKP1 and OsMKP1, were previously reported to be involved in the regulation of wound responses in tobacco (22, 23) and rice (24), respectively. It has been shown that NtMKP1 and OsMKP1 contain a single CaMBD and bind to CaM (22–24). As described in Fig. 1A, AtMKP1 from Arabidopsis has four conserved domains; a dual specificity phosphatase catalytic domain, a gelsolin homology domain, and two different CaMBDs. Interestingly, NtMKP1 and AtMKP1 show high similarity in their amino acid sequences but have different protein structures, especially at their CaMBDs. The CaMBD of NtMKP1 corresponds to the CaMBDI of AtMKP1, but the CaMBDII of AtMKP1 is absent in NtMKP1 (22, 23).
It is well known that the CaMBDs of many CaM-regulated proteins contain a short peptide of 16–35 residues, which displays separated basic and polar residues on one side and hydrophobic amino acids on the other, in a helical wheel projection (37–39). Ca2+-dependent CaM-binding motifs can be classified into two major groups, specifically, 1-8-14 and 1-5-10 motifs (35), whereby the numbers indicate the positions of the conserved hydrophobic residues. The hydrophobic and basic residues in the CaMBD are critical for binding of CaMBP to CaM (39). Based on the conserved structural features of CaMBDs, two CaM-binding motifs are predicted in the C terminus of AtMKP1, from His445 to Arg469 and from Ser669 to Lys692. The positions of the CaMBDs were identified by domain mapping of serial fragments (Fig. 1), and confirmed by CaM mobility shift and PDE enzyme competition assays using two CaMBD synthetic peptides (Figs. 2 and 3). We further confirmed the predicted CaM-binding motif by showing that the substitution of a single amino acid resulted in the loss of the Ca2+-dependent CaM-binding ability of AtMKP1 (Fig. 4). In vivo interactions between AtMKP1 and CaM were verified using a split-ubiquitin system in yeast, which is a useful detector of protein-protein interactions in a living cell (48) and is widely used for membrane proteins (47, 49) and transcriptional regulators (32). Determining the stoichiometry of two interacting proteins is important for our understanding of their enzyme activities. Recently it was reported that a soybean CaM could bind to two CaMBD peptides of NtMKP1 independently, leading to an overall stoichiometry of 1:2 (50). Although no data is presented here regarding the stoichiometry of CaM binding to AtMKP1, we suspect that two CaM molecules would bind to and activate a single AtMKP1 molecule, because AtMKP1 has two CaMBDs. Further biophysical studies for the stoichiometry of CaM binding to AtMKP1 should further advance our understanding of the binding mechanism between AtMKP1 and CaM.
AtMKP1 Is Catalytically Activated by CaM—NtMKP1 and OsMKP1 bind CaM (22–24), but CaM has no effect on NtMKP1 activity (23) and its effect on OsMKP1 has not been reported. Thus, we investigated whether CaM enzymatically regulates the activity of AtMKP1. The phosphatase activity of AtMKP1 was stimulated ∼2-fold after the addition of CaM (Fig. 6). The activity of AtMKP1 began to be stimulated by CaM at 600 nm (the molar ratio of CaM/AtMKP1 = 0.17). Interestingly, although the CaMBDI mutant of AtMKP1 was still activated by CaM, the CaMBDII mutant was not, which indicates that CaMBDII is more important than CaMBDI for the regulation of AtMKP1. A much higher level of CaM was required to stimulate the phosphatase activity of AtMKP1 than the level required for its interaction with the CaMBD peptides of AtMKP1 in the PDE competition assays (Fig. 3B). One possible explanation for the different CaM affinities of AtMKP1-GST fusion protein and CaMBD peptides could be that the bacterially expressed fusion protein employed in the phosphatase assays are not fully functional.
In contrast to activation of AtMKP1 by CaM, up to 1 μm NtCaM had no effect on the activity of NtMKP1 when OMFP was used as substrate (23). This difference in effect of their CaM's activation could be explained in two ways. First, AtMKP1 contains a CaMBDII domain, which is not present in NtMKP1. As shown in Fig. 6 (A and B), the phosphatase activity increased in proportion to the concentration of CaM in wild-type AtMKP1 and in the CaMBDI mutant (W453R) corresponding to a CaMBD of NtMKP1, implying that the CaMBDI of AtMKP1 and the CaMBD of NtMKP1 may not play important roles in CaM-dependent activation. In contrast, a CaMBDII mutant and a CaMBDI/CaMBDII double mutant did not result in the activation of OMFP phosphatase activity upon the addition of CaM (Fig. 6, C and D). These results indicate that CaMBDII plays a pivotal role in the activation of AtMKP1. Second, it is also possible that binding of CaM to AtMKP1 and NtMKP1 exerts different effects depending on the in vitro substrate or on external stimuli in vivo. AtDsPTP1, another MKP of Arabidopsis, contains two CaMBDs, and its phosphatase activity is controlled by CaM binding (28). Addition of CaM stimulated the dephosphorylation of pNPP by AtDsPTP1, but inhibited the tyrosine dephosphorylation activity of AtDsPTP1 when phosphorylated MBP was used as a substrate, suggesting that CaM regulates phosphatase activity differently, depending on its substrate (28).
It has been suggested that plant MKPs, including AtMKP1, NtMKP1, and OsMKP1, interact with or regulate the activities of MAPKs (20, 22–24). AtMKP1 has been found to play a critical role in the inactivation of MAPK6 in Arabidopsis (20), whereas NtMKP1 is critical for the inactivation of salicylic acid-induced MAPK in tobacco (22, 23), as is OsMKP1 for the inactivation of OsMPK3 and OsMPK6 in rice (24). It has previously been shown that the activation of MAPKs is dependent on the cytosolic Ca2+ concentration (31, 51). However, there has been no report yet about how Ca2+ activates MAPKs. In this report, we showed that the phosphatase activity of AtMKP1 was regulated by direct binding with CaM. As such, further analysis of the mechanisms governing CaM-regulated AtMKP1 and MAPK activity should further our understanding of the crosstalk occurring between Ca2+ signaling and MAPK signaling in Arabidopsis.
This work was supported in part by the Plant Diversity Research Center of the 21st Century Frontier Research Program (Grant #PF06303-01) and the Environmental Biotechnology National Core Research Center (Grant #R15-2003-012-02003-0) funded by the Ministry of Education and Science Technology in Korea. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CaM, calmodulin; CaMBP, calmodulin-binding protein; AtCaM, Arabidopsis calmodulin; HRP, horseradish peroxidase; GST, glutathione S-transferase; Nub and Cub, N- and C-terminal halves of ubiquitin, respectively; MAPK, mitogen-activated protein kinase; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; MEKK, MEK kinase; MKP, MAPK phosphatase; PDE, 3′,5′-cyclic nucleotide phosphodiesterase; OMFP, 3-O-methylfluorescein phosphate; 5-FOA, 5-fluoroorotic acid; pNPP, pyronitrophenyl phosphate; DsPTP, dual specificity phosphatase.
References
- 1.Knight, M. R., Campbell, A. K., Smith, S. M., and Trewavas, A. J. (1991) Nature 352 524–526 [DOI] [PubMed] [Google Scholar]
- 2.Price, A. H., Taylor, A., Ripley, S. J., Griffiths, A., Trewavas, A. J., and Knight, M. R. (1994) Plant Cell 6 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berridge, M. J., Bootman, M. D., and Roderick, H. L. (2003) Nat. Rev. Mol. Cell. Biol. 4 517–529 [DOI] [PubMed] [Google Scholar]
- 4.Reddy, V. S., and Reddy, A. S. (2004) Phytochemistry 65 1745–1776 [DOI] [PubMed] [Google Scholar]
- 5.Carafoli, E. (2005) FEBS J. 272 1073–1089 [DOI] [PubMed] [Google Scholar]
- 6.Blume, B., Nurnberger, T., Nass, N., and Scheel, D. (2000) Plant Cell 12 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grant, M., Brown, I., Adams, S., Knight, M., Ainslie, A., and Mansfield, J. (2000) Plant J. 23 441–450 [DOI] [PubMed] [Google Scholar]
- 8.White, P. J., and Broadley, M. R. (2003) Ann. Bot. (Lond.) 92 487–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders, D., Brownlee, C., and Harper, J. F. (1999) Plant Cell 11 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders, D., Pelloux, J., Brownlee, C., and Harper, J. F. (2002) Plant Cell 14 S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy, A. S. (2001) Plant Sci. 160 381–404 [DOI] [PubMed] [Google Scholar]
- 12.Snedden, W. A., and Fromm, H. (2001) New Phytol. 151 35–66 [DOI] [PubMed] [Google Scholar]
- 13.Hoeflich, K. P., and Ikura, M. (2002) Cell, 108 739–742 [DOI] [PubMed] [Google Scholar]
- 14.Widmann, C., Gibson, S., Jarpe, M. B., and Johnson, G. L. (1999) Physiol. Rev. 79 143–180 [DOI] [PubMed] [Google Scholar]
- 15.Kyriakis, J. M., and Avruch, J. (2001) Physiol. Rev. 81 807–869 [DOI] [PubMed] [Google Scholar]
- 16.Keyse, S. M. (2000) Curr. Opin. Cell Biol. 12 186–192 [DOI] [PubMed] [Google Scholar]
- 17.Theodosiou, A., and Ashworth, A. (2002) Genome Biol. 3 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerk, D., Bulgrien, J., Smith, D. W., Barsam, B., Veretnik, S., and Gribskov, M. (2002) Plant Physiol. 129 908–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fauman, E. B., and Saper, M. A. (1996) Trends Biochem. Sci. 21 413–417 [DOI] [PubMed] [Google Scholar]
- 20.Ulm, R., Ichimura, K., Mizoguchi, T., Peck, S. C., Zhu, T., Wang, X., Shinozaki, K., and Paszkowski, J. (2002) EMBO J. 21 6483–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, J. S., and Ellis, B. E. (2007) J. Biol. Chem. 282 25020–25029 [DOI] [PubMed] [Google Scholar]
- 22.Yamakawa, H., Katou, S., Seo, S., Mitsuhara, I., Kamada, H., and Ohashi, Y. (2004) J. Biol. Chem. 279 928–936 [DOI] [PubMed] [Google Scholar]
- 23.Katou, S., Katrita, E., Yamakawa, H., Seo, S., Mitsuhara, I., Kuchitsu, K., and Ohashi, Y. (2005) J. Biol. Chem. 280 39569–39581 [DOI] [PubMed] [Google Scholar]
- 24.Katou, S., Kuroda, K., Seo, S., Yanagawa, Y., Tsuge, T., Yamazaki, M., Miyao, A., Hirochika, H., and Ohashi, Y. (2007) Plant Cell Physiol. 48 332–344 [DOI] [PubMed] [Google Scholar]
- 25.Monroe-Augustus, M., Zolman, B. K., and Bartel, B. (2003) Plant Cell 15 2979–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quettier, A. L., Bertrand, C., Habricot, Y., Miginiac, E., Agnes, C., Jeannette, E., and Maldiney, R. (2006) Plant J. 47 711–719 [DOI] [PubMed] [Google Scholar]
- 27.Lee, S. H., Kim, M. C., Heo, W. D., Kim, J. C., Chung, W. S., Park, C. Y., Park, H. C., Cheong, Y. H., Kim, C. Y., Lee, K. J., Bahk, J. D., Lee, S. Y., and Cho, M. J. (1999) Biochim. Biophys. Acta 1433 56–67 [DOI] [PubMed] [Google Scholar]
- 28.Yoo, J. H., Cheong, M. S., Park, C. Y., Moon, B. C., Kim, M. C., Kang, Y. H., Park, H. C., Choi, M. S., Lee, J. H., Jung, W. Y., Yoon, H. W., Chung, W. S., Lim, C. O., Lee, S. Y., and Cho, M. J. (2004) J. Biol. Chem. 279 848–858 [DOI] [PubMed] [Google Scholar]
- 29.Erickson-Viitanen, S., and DeGrado, W. F. (1987) Methods Enzymol. 139 455–478 [DOI] [PubMed] [Google Scholar]
- 30.Lee, S. H., Kim, J. C., Lee, M. S., Heo, W. D., Seo, H. Y., Yoon, H. W., Hong, J. C., Lee, S. Y., Bahk, J. D., Hwang, I., and Cho, M. J. (1995) J. Biol. Chem. 270 21806–21812 [DOI] [PubMed] [Google Scholar]
- 31.Takahashi, Y., Berberich, T., Miyazaki, A., Seo, S., Ohashi, Y., and Kusano, T. (2003) Plant J. 36 820–829 [DOI] [PubMed] [Google Scholar]
- 32.Laser, H., Bongards, C., Schuller, J., Heck, S., Johnsson, N., and Lehming, N. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 13732–13737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, H. S., Park, B. O., Yoo, J. H., Jung, M. S., Lee, S. M., Han, H. J., Kim, K. E., Kim, S. H., Lim, C. O., Yun, D. J., Lee, S. Y., and Chung, W. S. (2007) J. Biol. Chem. 282 36292–36302 [DOI] [PubMed] [Google Scholar]
- 34.Stagljar, I., Korostensky, C., Johnsson, N., and te Heesen, S. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 5187–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhoads, A. R., and Friedberg, F. (1997) FASEB J. 11 331–340 [DOI] [PubMed] [Google Scholar]
- 36.Kudla, J., Xu, Q., Harter, K., Gruissem, W., and Luan, S. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Neil, K. T., and DeGrado, W. F. (1990) Trends Biol. Sci. 15 59–64 [DOI] [PubMed] [Google Scholar]
- 38.Meador, W. E., Means, A. R., and Quiocho, F. A. (1992) Science 257 1251–1255 [DOI] [PubMed] [Google Scholar]
- 39.Crivici, A., and Ikura, M. (1995) Annu. Rev. Biophys. Biomol. Struct. 24 85–116 [DOI] [PubMed] [Google Scholar]
- 40.Varshavsky, A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, L., Montserat, J., Lawrence, D. S., and Zhang, Z. Y. (1996) Biochemistry, 35 9349–9354 [DOI] [PubMed] [Google Scholar]
- 42.Gottlin, E. B., Xu, X., Epstein, D. M., Burke, S. P., Eckstein, J. W., Ballou, D. P., and Dixon, J. E. (1996) J. Biol. Chem. 271 27445–27449 [DOI] [PubMed] [Google Scholar]
- 43.Yuvaniyama, J., Denu, J. M., Dixon, J. E., and Saper, M. A. (1996) Science, 272 1328–1331 [DOI] [PubMed] [Google Scholar]
- 44.Stewart, A. E., Dowd, S., Keyse, S. M., and McDonald, N. Q. (1999) Nat. Struct. Biol. 6 174–181 [DOI] [PubMed] [Google Scholar]
- 45.Zhou, B., and Zhang, Z. Y. (1999) J. Biol. Chem. 274 35526–35534 [DOI] [PubMed] [Google Scholar]
- 46.Reddy, V. S., Ali, G. S., and Reddy, A. S. (2002) J. Biol. Chem. 277 9840–9852 [DOI] [PubMed] [Google Scholar]
- 47.Kim, M. C., Panstruga, R., Elliott, C., Muller, J., Devoto, A., Yoon, H. W., Park, H. C., Cho, M. J., and Schulze-Lefert, P. (2002) Nature 416 447–451 [DOI] [PubMed] [Google Scholar]
- 48.Liang, L., Lim, K. L., Seow, K. T., Ng, C. H., and Pallen, C. J. (2002) J. Biol. Chem. 275 30075–30081 [DOI] [PubMed] [Google Scholar]
- 49.Wurgler-Murphy, S. M., and Saito, H. (1997) Trends Biochem. Sci. 22 172–176 [DOI] [PubMed] [Google Scholar]
- 50.Rainaldi, M., Yamniuk, A. P., Murase, T., and Vogel, H. J. (2007) J. Biol. Chem. 282 6031–6042 [DOI] [PubMed] [Google Scholar]
- 51.Samuel, M. A., Miles, G. P., and Ellis, B. E. (2000) Plant J. 22 367–376 [DOI] [PubMed] [Google Scholar]