Abstract
CD8+ cytotoxic T lymphocytes (CTL) can recognize and kill target cells expressing only a few cognate major histocompatibility complex (MHC) I-peptide complexes. This high sensitivity requires efficient scanning of a vast number of highly diverse MHC I-peptide complexes by the T cell receptor in the contact site of transient conjugates formed mainly by nonspecific interactions of ICAM-1 and LFA-1. Tracking of single H-2Kd molecules loaded with fluorescent peptides on target cells and nascent conjugates with CTL showed dynamic transitions between states of free diffusion and immobility. The immobilizations were explained by association of MHC I-peptide complexes with ICAM-1 and strongly increased their local concentration in cell adhesion sites and hence their scanning by T cell receptor. In nascent immunological synapses cognate complexes became immobile, whereas noncognate ones diffused out again. Interfering with this mobility modulation-based concentration and sorting of MHC I-peptide complexes strongly impaired the sensitivity of antigen recognition by CTL, demonstrating that it constitutes a new basic aspect of antigen presentation by MHC I molecules.
A hallmark of CD8+ CTL4 is their ability to recognize and kill target cells expressing only a few cognate MHC I-peptide complexes (1, 2). The initial encounter of CTL with target cells is a nonspecific adhesion, mainly based on intercellular interactions between LFA-1 (CD18,CD11a) and ICAM-1 (CD54) (3-6). This nonspecific adhesion starts at an initial contact site, and after about 2.5 min LFA-1-ICAM-1 ring junctions are formed (4). In such antigen-nonspecific conjugates LFA-1 triggers reorganization of the actin cytoskeleton and phosphorylation of molecules involved in cell adhesion on CD8+ T cells, which lowers the threshold of T cell activation (5). At this stage CTL scan target cells for expression of cognate MHC I-peptide complexes. Effective eradication of transformed or virus-infected cells depends heavily on the efficiency of this scanning; to recognize target cells expressing only a few cognate MHC I-peptide complexes, CTL need to be able to recognize these among high numbers (>106) of vastly diverse MHC I-peptide complexes during the brief nonspecific CTL target cell encounter. Interaction of cognate MHC I-peptide complexes with TCR induces elevation of intracellular calcium, diverse phosphorylation events, activation of integrins, rearrangement of the cytoskeleton, translocation of adhesion and auxiliary molecules and their ligands to the contact site, resulting in the formation of an immunological synapse (IS) (2-6). As the IS matures, antigen recognition molecules (e.g. TCR/CD3, MHC molecules, and coreceptor) localize in the central supramolecular activation complex (cSMAC) and adhesion molecules (e.g. LFA-1/ICAM-1) in peripheral SMAC (pSMAC) (6-10). This molecular segregation occurs by differential actin cytoskeleton-mediated translocations and allows avid polyvalent interactions of TCR and coreceptor with MHC-peptide complexes, respectively, and integrins with their ligands, as both sets of molecules are of different size (6, 11).
The mobility of human and murine MHC-I molecules has been examined on different cell types using fluorescence recovery after photobleaching (FRAP), indicating that a fraction of MHC I molecules was immobile within the measured time scale of 50 ms to 30 s (12-14). Studies using single particle tracking (SPT) showed mobile MHC I molecules following anomalous diffusion (i.e. nonlinear time dependence) over time scales of 100 ms to 300 s or transient confinements within domains of various sizes (13-16). The lateral diffusion of MHC-I molecules in plasma membranes was increased upon truncation of their cytoplasmic tail or replacement of their transmembrane and intracellular portions with a glycosylphosphatidylinositol (GPI) anchor (12, 16). There is compelling evidence that the anomalous diffusion of MHC I molecules depends on the actin cytoskeleton. Actin plays a key role in membrane compartmentalization and determines the barriers of a random walk of MHC I molecules (12, 14, 17). An attractive possibility is that MHC I molecules transiently interact with the actin cytoskeleton via ICAM-1, which has been shown by coimmunoprecipitation and fluorescence resonance energy transfer experiments to associate with MHC I molecules (18-20) and to interact with the actin cytoskeleton (21).
It is not clear what role, if any, the anomalous diffusion of MHC I-peptide complexes has for antigen recognition by CTL. Biological studies on GPI-linked MHC I molecules have reached vastly diverging conclusions. Although some studies showed that GPI-linked MHC I molecules exhibit defective thymic selection and alloantigen recognition by CD8 T cells (22-24), others reported that they are well recognized by antigen-specific CTL (25, 26).
The objective of this study was to investigate the relation between the movements of normal and GPI-linked MHC I-peptide complexes on target cells and their recognition by CTL and to elucidate how cognate and noncognate MHC I-peptide complexes diffuse in nascent IS. The dynamics of MHC II-peptide complexes containing fluorescent peptides have been assessed by single molecule microscopy, a technique that provides unprecedented detailed and accurate information on molecular movements on living cells (11, 27-30). For tracking experiments, we used fluorescent derivatives of the Plasmodium berghei circumsporozoite (PbCS) peptide 252-260 (SYIPSAEKI). This peptide, conjugated with photoreactive 4-azidobenzoic acid (ABA) on Lys-259 (PbCS(ABA)), is recognized by S14 and related CTL clones in the context of Kd (31). These clones were derived from mice immunized with PbCS-(ABA) and hence the ABA group is an essential part of the epitope they recognize (31). Fluorescent ATTO647 dye was introduced in PbCS(ABA) by replacing PbCS Ser-252 with N-γ ATTO647 diaminopropionic acid (Dap(ATTO647)-YIPSAEK-(ABA)I). Alternatively, ATTO647 was introduced in PbCS-(252-260) on Lys-259, i.e. SYIPSAEK(ATTO647)I. For simplicity, the former peptide derivative henceforth is referred to as peptide P1 and the latter as peptide P2. This system has the significant advantage that although both modifications preserved the binding of the peptide to Kd, peptide P1 but not peptide P2 was recognized by S14 CTL (31, 32).
We report that Kd-peptide complexes on fibroblast L-Kd and RMA-S-Kd thymoma cells exhibit dynamic transitions between states of free diffusion and immobility. The immobile periods greatly increased in cell adhesion sites, thus increasing the local concentration of complexes. In the nascent synapse with CTL, cognate but not noncognate Kd-peptide complexes became immobile, resulting in their selective local trapping. The mobility of Kd-peptide complexes was substantially increased upon disruption of the cytoskeleton or for GPI-linked complexes. In both cases the recognition by S14 CTL was greatly reduced, demonstrating for the first time that mobility modulation-based sorting and trapping of MHC I-peptide complexes in the immunological synapse increases the sensitivity of antigen recognition by CTL.
EXPERIMENTAL PROCEDURES
Cells and Kd Transfectants—ICAM-1 and Kd-transfected L cells (L-Kd) and S14 CTL were propagated as described (31, 33). DNA of KdGPI was obtained by truncating C-terminal Kd after Thr-280 and fusion with the GPI anchor of CD55 using the insertion sequence (SRPNKGSGTTSGTTRLLSGHTCFTLTGLLGTLVTMGLLT; GPI anchoring is at the S) as described (34). Both Kd and KdGPI DNA were inserted in the pRRLsin. PPT.hPGK lentiviral vector (35) and viruses produced by transfecting HEK293T cells with envelope (pMD2.VSVG), packaging (pCMVΔR8.91), and lentiviral plasmids. RMA-S cells (105/ml) in DMEM supplemented with Polybrene (4 μg/ml) were cultured for 24 h at 37 °C with viruses incubated for 2.5 h at 37 °C with PbCS(ABA) peptide (10-6 m), stained with anti-Kd monoclonal antibody SF1-1.1.1-FITC, and sorted on an Aria sorter (BD Biosciences). After PbCS(ABA) peptide loading, the Kd surface expression, as assessed upon staining with SF1-1.1.1-FITC, was 81 (MFI) for RMA-S-Kd, 171 for RMA-S-Kd GPI, and 279 for L-Kd cells. The ICAM-1 expression, as determined by staining with monoclonal antibody YN1/1.7.4-PE was 58 (MFI) for RMA-S-Kd, 54 for RMA-S-Kd GPI, and 82 for L-Kd cells.
Peptides and Kd-Peptide Complexes—Chemicals were from Novabiochem-Calbiochem, Sigma, or ATTO-TEC GmbH (Siegen, Germany). Peptides were synthesized on solid phase using Fmoc for transient N-terminal protection as described (31, 32). ATTO-PbCS(ABA) (Dap(ATTO)-YIPSAEK(ABA)I; where Dap is diaminopropionic acid), referred to as peptide P1, was obtained by reacting Fmoc-Dap-YIPSAEK(ABA)I with ATTO647 N-hydroxysuccinimide ester in DMSO/N,N-dimethylformamide/di-isopropylethylamine (3/2/0.1) at 20 °C for 1 h. After removal of Fmoc the deprotected peptide was purified on a semi-preparative C4 column (Vydac, Hesperia, CA). The synthesis of PbCS(ATTO) (SYIPSAEK(ATTO)I), referred to as peptide P2, was analogous, using Fmoc-SYIPSAEKI. All peptides were characterized by matrix-assisted laser desorption ionization time-of-flight-mass spectrometry. 1 μm Kd-peptide P1 containing a free cysteine in position 275 of the Kd heavy chain was reacted with 5 μm of 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidophenyl)butyramide] in phosphate-buffered saline containing 20 mm CHAPS for 2 h at 20 °C and purified by gel filtration on a Superdex S75 column (Amersham Biosciences) in 20 mm CHAPS.
Functional Assays—RMA-S-Kd cells were incubated in DMEM containing 2.5% dialyzed fetal calf serum (Invitrogen), 2 μg/ml of 2 m and 10 mm HEPES with graded concentrations of peptide at 32 °C for 90 min. RMA-S-KdGPI cells were incubated without β2-miroglobulin for 5 min at 37 °C. Peptide binding was assessed by using PbCS(biotin) (SYIPSAEK(biotin)I) peptide and flow cytometric detection of bound peptide via phycoerythrin-labeled streptavidin (Caltag, San Francisco, CA) as described (36). Antigen recognition by S14 CTL was assessed by 51Cr release experiments as described (33). Conjugate formation was assessed as described previously (37). In brief, Indo-1-labeled S14 CTL were mixed with carboxyfluorescein succinimidyl ester (CFSE)-labeled L-Kd cells previously sensitized with graded concentrations of peptide 1 (CTL/target = 1: 1), centrifuged for 1 min at 1000 × g. After 1 min incubation at 37 °C CFSE and Indo-1 cell-associated fluorescence were analyzed by flow cytometry on an LSR flow cytometer (BD Biosciences). The percentage of conjugates was calculated as follows: 2 × CFSE+ Indo-1 + counts/(2 × CFSE + Indo-1 + counts + CFSE + Indo-1 - counts + CFSE - Indo-1 + counts) × 100.
Single Molecule Microscopy—L-Kd cell monolayers on glass coverslips (Assistant, Sondheim, Germany) were incubated in measurement buffer (Hanks' buffered salt solution supplemented with HEPES (10 mm) and β2-microglobulin (2 μg/ml)) with 1.5 nm peptide P1 for 1 h at 32 °C. Alternatively, L-Kd cells were pulsed likewise with 1.5 nm peptide P2 followed by addition of 1 μm PbCS(ABA) peptide after 30 min. RMA-S-Kd cells were incubated with 10 nm of peptide P1 in DMEM supplemented with 2.5% dialyzed fetal calf serum and β2-microglobulin (2 μg/ml) for 90 min at 32 °C. RMA-S-KdGPI cells were incubated with 1 nm of the ATTO peptides at 37 °C for 5 min. Single molecule images were recorded at 37 °C on a modified wide field microscope (Axiovert 200 M, Zeiss, Feldbach, Switzerland) as described (29). Samples were illuminated with 632.8 nm light of a He-Ne laser for 10, 20, or 50 ms per image using average illumination intensities of around 1.2, 0.5, and 0.3 kilowatt/cm2, respectively, and imaged with an intensified CCD camera (I-Pentamax 512 EFT, Roper scientific, Trenton, NJ) or with a back-illuminated electron multiplying CCD camera (Ixon DV 887, Andor Technology, Belfast, UK). Using white light illumination and the settings for single molecule microscopy, we focused on the contact site of newly formed conjugates, just exhibiting high and stable calcium flux and recorded series of single molecule images.
Data Analysis of Single Molecule Trajectories—Fluorescence spots of single Kd-peptide complexes were discriminated from cellular fluorescence background by their near-diffraction-limited sizes and intensities. Analysis of single molecule images was performed with a program written in IGOR PRO (30). Trajectories were evaluated from complexes displaying more than six steps and single-step photobleaching. The diffusion coefficients (D) were determined from a linear fit of the first four data points of the plot of the mean square displacement (MSD) versus the time lag, tlag, using MSD = 4 Dtlag. The confinement length (L) was evaluated from a fit of the plot of MSD versus tlag using a confined diffusion model MSD = noise + A2(1 - exp(- 4 Dtlag/A)), where noise is the measurement noise and A the confinement size. To discriminate between immobile Kd-peptide complexes and those diffusing in confined domains, the distribution of L was compared with those of ATTO647 molecules immobilized on a glass surface (30). Average domain sizes 〈S〉 were evaluated from 〈S〉=〈2A〉= ((〈L〉/2)2 -〈noise〉)1/2 using (〈L〉/2)2 measured on glass-immobile ATTO molecules to assess 〈noise〉. Mobility transitions within trajectories were detected by screening for immobile/confined periods (38). The probabilities of transitions from mobile to immobile, Pmob->immob, and from immobile to mobile, Pimmob->mob, were calculated by dividing the number of detected transitions by the total number of steps summed over all mobile or immobile molecules, respectively. The lifetimes of the mobile, τmob, and immobile periods, τimmo, were calculated using τmob = 1/(ν·Pmob->immob) and τimmo = 1/(ν·Pimmob->mob), where ν is the recording frequency of 10 Hz. Values are given as (mean ± 95% confidence intervals). Error bars in graphs and histograms represent standard deviations and standard errors, respectively.
RESULTS
Single Molecule Microscopy of Kd-Peptide P1 Complexes on Living L-Kd Cells—Single molecule images of individual Kd-peptide complexes on Kd-transfected fibroblasts (L-Kd) were acquired using the fluorescent peptide P1 (ATTO647-PbCS-(ABA)), which was recognized by cloned S14 CTL nearly as efficiently as the parental PbCS(ABA) peptide (Fig. 1A). For tracking experiments L-Kd cells were loaded for 1 h at 32 °C with 1.5 nm peptide P1, corresponding to about maximal lysis by S14 CTL. As determined from images, such L-Kd cells displayed 65 ± 35 (n = 8) Kd-peptide P1 complexes per cell. Individual complexes were discernible as discrete, diffraction limited spots, distinct from diffuse cellular autofluorescence (Fig. 1B). As assessed by confocal microscopy, peptide internalization was barely detectable under these conditions, and unspecific binding on untransfected L cells was negligibly small (data not shown). These spots exhibited single step photo-bleaching, proving that they were single Kd-peptide complexes (Fig. 1C). The diffusion of the complexes was assessed by recording sequences of images at the rate of 10 Hz (Video 1). From the trajectories three types of complexes were discernible as follows: (i) immobile (Fig. 1D); (ii) mobile (Fig. 1E); and (iii) those undergoing transitions between i and ii (Fig. 1F).
FIGURE 1.

Single molecule microscopy of Kd-peptide P1 complexes on L-Kd cells. A, 51Cr-labeled L-Kd cells were pulsed with the indicated concentrations of PbCS (circles), PbCS(ABA) (squares), peptide P1 (triangles), or peptide P2 (crosses) and incubated with cloned S14 CTL (CTL/target = 3/1). After 4 h, the specific lysis was determined from released 51Cr. One representative experiment out of three is shown. B, image of Kd-peptide P1 complexes on the lower glass-adhered membrane of two L-Kd cells (contoured by dashed lines) within an illuminated circular region of 20 μm. Single complexes exhibit diffraction-limited spots, like the one encircled. The scale bar is 5 μm. C, single molecule trace displaying typical single-step photobleaching. D-F, typical trajectories of an immobile/confined complex (D), a mobile one (E), and one undergoing a transition from an immobile/confined to a mobile state (F). The pixel size is 225 nm.
Diffusion of Kd-Peptide P1 Complexes on the Lower Plasma Membrane of L-Kd Cells—The diffusion coefficients (D) of Kd-peptide complexes on the lower, support-adhered plasma membrane of L-Kd cells were calculated from the short time regime of the trajectories of individual complexes (supplemental Fig. S1). Populations of mobile and immobile/confined complexes were clearly distinguishable in the histogram of the diffusion coefficients D and distinct from those of ATTO molecules immobilized on a glass slide (Fig. 2A). Because of insufficient time resolution, D poorly discriminated between complexes that were immobile or confined in small nanometersized domains. We therefore calculated confinement lengths from the long time regime of the trajectories, which scale with sizes of domains explored by single complexes. In the histogram of the confinement lengths L the calculated confinements within regions of varying lengths resulted either from real or apparent confinements, caused by the stochastic nature of the diffusion of mobile complexes (Fig. 2B). Confinement lengths of <360 nm resulted from real confinements, as they did not occur in simulated trajectories of complexes following Brownian diffusion. Using 360 nm as threshold showed that 42 ± 10% of complexes were immobile or confined in small domains (Fig. 2C). The distribution of L was shifted compared with immobilized ATTO molecules (Student's t test, p < 0.05), indicating that these complexes were confined in very small domains. By subtracting the noise from the average confinement length, an average domain size of 90 ± 40 nm was obtained (supplemental Fig. S1).
FIGURE 2.
Trajectories of single Kd-peptide P1 complexes on L-Kd cells. A-C, diffusion of Kd-peptide complexes on the lower substrate-adhered cell membrane. A, diffusion coefficients (D) of single Kd-peptide complexes (red bars; 96 molecules on 12 different cells). B, confinement length (L) for single Kd-peptide complexes (red bars). Molecules with L >1 μm are represented in a bar at >1 μm. Blue bars represent L of complexes following simulated Brownian diffusion and were normalized such that the bars at >1 μm are identical. ATTO molecules immobilized on glass were analyzed the same way (black bars). C, using length of 360 nm as threshold shows that 42 ± 10% of the molecules were immobile/confined and 58 ± 10% mobile (mean value ± 95% confidence interval). The errors in the histogram indicate standard errors. D-F, diffusion of complexes on the upper cell membrane. D, diffusion coefficients (D) of single complexes (red bars; 87 molecules on 27 different cells). E, histogram of length (red bars). Complexes with length >1 μm are represented in a bar at >1 μm. Black bars represent glass slide-immobilized ATTO molecules and blue bars complexes following simulated Brownian diffusion. F, using length of 360 nm as threshold shows that 13 ± 7% of the complexes were immobile/confined and 88 ± 7% mobile. G, bars on the left side represent transitions from mobile to immobile normalized (100% referring to the total number of steps for all mobile molecules) and on the right side transitions of immobile to mobile (100% referring to the total number of steps summed over all immobile molecules). H-I, confocal images of L-Kd cells stained with FITC-phalloidin on the lower membrane (H) or on a median section (I). Scale bars are 10 μm.
A hallmark of the diffusion of Kd-peptide complexes was the mobility transitions between mobile and immobile/confined periods. To quantify this, we used Monte Carlo simulations to calculate the number of stochastic mobility transitions occurring during Brownian diffusion. The simulated trajectories were screened for time periods for which the square displacements remained below 0.016 μm2, indicating that none exceeded six frames. Screening of the trajectories of Kd-peptide complexes for immobile periods exceeding seven frames showed real mobility transition for 4 molecules of the 66 analyzed; in three cases, mobile complexes became immobile, and in one case an immobile one became mobile (supplemental Fig. S2A). The probability of occurrence of immobile periods, Pimmob, as calculated by dividing the number of immobile periods (4) by the total number of steps (1174), was 0.34 ± 0.33%, which was significantly higher than predicted by Brownian diffusion (χ2 test, p < 0.05). There was a probability of Pmob->immob = 0.26 ± 0.29% for a mobile molecule to immobilize and of Pimmob->mob = 0.19 ± 0.37% for an immobile molecule to become mobile (Fig. 2G). From these values estimates for the lifetime of the mobile periods, τmob, of 38 s and of the immobile periods,τimmob, of 53 s were calculated. As we underestimated the real number of transitions, these lifetimes represent upper limits.
Diffusion of Kd-Peptide P1 Complexes on the Upper Plasma Membrane of L-Kd Cells—On the upper surface of L-Kd cells the same analysis yielded a significantly (χ2 test, p < 0.001) lower fraction of immobile/confined complexes (13 ± 7%; Fig. 2, D-F). The distribution of L for the confined molecules (L <360 nm) did not differ from the corresponding distribution on the lower surface (Student's t test, p > 0.2) with a small, but significant shift (Student's t test, p < 0.05) compared with the distribution of glass-immobilized ATTO molecules. The average domain size of 130 ± 60 nm was therefore similar to the domain size determined on the lower surface.
Eight immobile periods were detected among the trajectories of 64 mobile molecules (2085 steps), corresponding to a probability of entering immobilization periods of Pimmob periods = 0.38 ± 0.26% (supplemental Fig. S2C). Although the probabilities of entering immobilization periods were statistically similar (χ2 test, p > 0.8) in both membranes, the trajectories on the upper surface showed shorter immobilization periods than those on the lower (supplemental Fig. S2C). This allowed some Kd-peptide complexes to undergo two mobility transitions in the same trajectory (supplemental Fig. S2B). In total six transitions from mobile to immobile and four from immobile to mobile were observed, corresponding to transition probabilities of Pmob->immob = 0.29 ± 0.23% (τmob ≈ 34 s) and Pimmob->mob = 1.4 ± 1.3% (τimmo ≈ 7.1 s), respectively (Fig. 2G). Pmob->immob values were not statistically different (χ2 test, p > 0.8) on the lower and upper surfaces, indicating that Kd-peptide complexes became immobile with the same frequency. By contrast, Pimmob->mob was statistically higher on the upper surface (χ2 test, p < 0.05), resulting in shorter periods of immobilization and hence in a larger average fraction of mobile molecules (Fig. 2G).
The differences in the diffusion of Kd-peptide complexes between the upper and the lower surfaces correlated with differences in the density of the actin cytoskeleton; the lower substrate-adhered membrane displayed a considerably higher density of actin as compared with a median section (Fig. 2, H and I). The finding that Kd coimmunoprecipitated with ICAM-1 and vice versa (supplemental Fig. S3, lanes 1 and 4) argues that the transient immobilizations of Kd-peptide complexes are explained, at least in part, by their interaction with actin cytoskeleton-associated ICAM-1 (21).
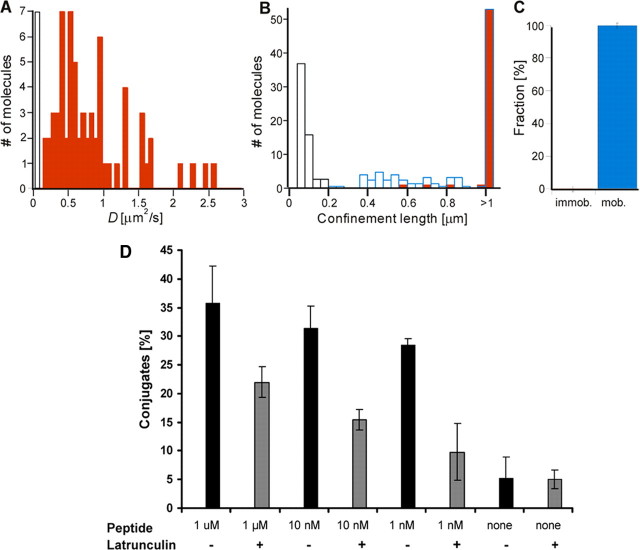
Disruption of the Actin Cytoskeleton Increases the Mobility of Kd-Peptide P1 Complexes and Decreases T Cell Recognition—To find out whether the confinements of the Kd-peptide complexes were related to the cytoskeleton, their diffusion was assessed on L-Kd cells pretreated with latrunculin, to block actin polymerization. Because latrunculin-treated L-Kd cells detached from the substrate, tracking experiments were performed only on the upper cell surface. Remarkably, virtually all complexes were mobile with diffusion coefficients roughly 10 times larger than on untreated cells (Fig. 3, A-C). Also the proportion of molecules exhibiting confinements of less than 1 μm clearly decreased upon latrunculin treatment. These results argue that the transient confinements of Kd-peptide complexes were because of interaction with the cytoskeleton or cytoskeleton-associated entities.
FIGURE 3.
Disruption of the cytoskeleton increases the mobility of Kd-peptide P1 complexes and decreases recognition by S14 CTL. A, diffusion coefficients (D) of complexes on the upper surface of latrunculin-treated L-Kd cells. B, confinement length (L) of the complexes (red bars; 67 molecules on 10 different cells); complexes with length >1 μm are represented in a bar at >1 μm. Black bars represent immobilized ATTO molecules and blue bars molecules following simulated Brownian diffusion. C, all complexes (100 ± 3%) were mobile. D, indo-1-labeled S14 CTL were incubated at 37 °C for 1 min with CFSE labeled L-Kd cells (CTL/target = 1/1) that were sensitized with the indicated concentrations of peptide P1 and pretreated (gray bars) or not (black bars) with latrunculin. The percentage of Indo-1 and CFSE-positive conjugates was analyzed by flow cytometry. Mean values and S.D. were calculated from four experiments.
The proportion of conjugate formation was considerably decreased when target cells were pretreated with latrunculin, especially at low peptide concentrations (Fig. 3D), indicating that the increased mobility of Kd-peptide complexes considerably inhibited the conjugate formation of S14 CTL with L-Kd cells.
GPI-anchored Kd-Peptide P1 Complexes Display Increased Mobility—To generalize these findings we likewise examined Kd-peptide complexes on Kd-transfected RMA-S cells (RMA-S-Kd). Because RMA-S cells are weakly adherent, the diffusion of Kd-peptide P1 complexes was only assessed on the upper cell surface. As on L-Kd cells transient confinements were observed, but the confinement lengths were considerably larger with an average domain size of 240 ± 30 nm (Fig. 4, A and B). 16 ± 8% of the complexes were confined, which is similar as on L-Kd cells (χ2 test, p > 0.3) (Fig. 4C).
FIGURE 4.
Trajectories of single Kd-peptide P1 complexes on RMA-S-Kd cells. A, diffusion coefficients (D) of single complexes (red bars, 94 molecules on 17 different cells). B, histogram of the confinement lengths (L) of complexes (red bars). Complexes with length >1 μm are represented by a bar at >1 μm. Black bars represent immobilized ATTO molecules and blue bars molecules following simulated Brownian diffusion. C, using length of 360 nm as threshold showed that 16 ± 8% of the molecules were confined, and 84 ± 8% were mobile. D-F, Kd-peptide P1 complexes on RMA-S-KdGPI cells. D, diffusion coefficients (D) of single complexes (red bars, 100 molecules on 18 different cells). E, confinement lengths (L)(red bars). Complexes with length of >1 μm are represented in a bar at >1 μm. F, using the length of 360 nm as threshold shows that 1 ± 2% of the molecules were confined and 99 ± 2% mobile.
To find out to what extent the confinements of Kd-peptide P1 complexes were determined by their transmembrane and/or cytoplasmic portion(s), we replaced these by a GPI anchor. KdGPI-peptide complexes displayed diffusion coefficients in average 3-fold larger than Kd-peptide complexes (Fig. 4, A and D). Nearly all complexes (99 ± 2%) were mobile (Fig. 4, E and F). Because GPI-anchored molecules partition into lipid rafts (34), the transient confinements observed for wild type Kd-peptide complexes were not accounted for by partitioning in rafts. Similar findings were obtained for Kd-peptide P1 complexes that were incorporated into the membrane of L cells (H-2k) via the lipid anchor di-palmitoylphosphatidylethanolamine (39). On the lower membrane only one immobile period was detected in the trajectories of 50 complexes, demonstrating that the diffusion of full-length Kd-peptide P1 complexes on the lower membrane was not influenced by interactions of their extracellular portion with the substrate (data not shown). Moreover, Kd,, but considerably less KdGPI, coimmunoprecipitated with ICAM-1 and vice versa (supplemental Fig. S3, lanes 2, 3, 5, and 6), which correlated with the different motilities of Kd and KdGP-peptide complexes (Fig. 4).
GPI-anchored Kd-Peptide P1 Complexes Are Inefficiently Recognized by S14 CTL—RMA-S cells transfected with GPI-linked Kd (RMA-S-KdGPI) expressed considerably higher levels of Kd and bound more peptide than RMA-Kd cells, when tested under identical conditions (Fig. 5A). To conclusively compare the CTL recognition of wild type and GPI-linked Kd-peptide complexes, we determined loading conditions that provided equal peptide binding. To this end we used biotinylated PbCS peptide (PbCS(biotin)), which binds to Kd as well as PbCS(ABA) (31) and flow cytometric detection of cell-associated PbCS(biotin) via phycoerythrin-labeled streptavidin (36). By varying the incubation time and temperature, equal peptide binding was observed when RMA-S-KdGPI cells were incubated at 37 °C for 5 min and RMA-S-Kd cells at 32 °C for 90 min in medium containing dialyzed serum (Fig. 5B). As assessed in a 51Cr release assay, RMA-S-Kd cells were recognized by S14 CTL several hundredfold more efficiently than RMA-S-KdGPI cells loaded with equal amounts of PbCS(ABA) peptide (Fig. 5C).
FIGURE 5.

GPI-linked Kd-peptide P1 complexes are less efficiently recognized. A, RMA-S-Kd cells (squares), RMA-S-KdGPI (circles), or untransfected RMA-S cells (triangles) were incubated with the indicated concentrations of PbCS(biotin) peptide at 37 °C for 1 h, and the peptide binding was assessed by flow cytometry as mean fluorescent intensity (MFI). B, alternatively, RMA-S-Kd cells (open symbols) were incubated at 32 °C with graded concentrations of PbCS(biotin) peptide for 30 min (open squares), 60 min (open circles), or 90 min (open diamonds). For RMA-S-KdGPI cells (closed symbols), the incubations were performed at 37 °C for 1 min (filled triangles), 5 min (filled diamonds), and 10 min (filled squares). One out of four experiments is shown. B, RMA-S-Kd cells (squares) were incubated at 32 °C for 90 min and RMA-S-KdGPI cells (triangles) at 37 °C for 5 min with graded concentrations of PbCS(ABA) peptide and their recognition by cloned S14 CTL assessed in a 51Cr release assay. One out of three experiments is shown.
Cognate Kd-Peptide P1 but Not Noncognate Kd-Peptide P2 Complexes Are Immobilized in the Immunological Synapse—For tracking of Kd-peptide complexes in the immunological synapse formed between peptide P1-pulsed L-Kd cell and S14 CTL, newly formed conjugates were selected that exhibited strong and stable calcium flux in the CTL (Fig. 6A and data not shown). Kd-peptide P1 complexes were exclusively found in the contact site and colocalized with high density of actin (Fig. 6, B and C) and were distributed over most of the extended contact site, typically forming bright clusters (supplemental Fig. S4 and Videos 2 and 3). All complexes were immobile/confined (100 ± 6%) (Fig. 6F). The average confinement length was slightly larger as compared with ATTO dye immobilized on a coverslip, which was explained, at least in part, by the reduced signal-to-noise ratio in the synapse (Fig. 6E). We cannot, however, rule out that some of the Kd-peptide P1 complexes were confined within very small domains.
FIGURE 6.
Kd-peptide P1 complexes in the immunological synapse between a L-Kd cell and a S14 CTL. A, transmission micrograph of an L-Kd cell-S14 CTL conjugate. B and C, actin staining with FITC-phalloidin (B) and ATTO647 fluorescence image of the same conjugate (C). Scale bars are 10 μm. D, diffusion coefficients (D) of single Kd-peptide P1 complexes (red bars, 37 molecules in 19 different synapses). E, confinement length (L) of single Kd-peptide complexes (red bars). Black bars represent glass immobilized ATTO molecules. F, 100 ± 6% of the complexes were immobile.
A strikingly different situation was observed for noncognate Kd-peptide P2 complexes. Because peptide P2 was not recognized by S14 CTL (Fig. 1A), L-Kd cells were pulsed first with a low concentration of peptide P2, followed by a high concentration of unlabeled PbCS(ABA) peptide. As in the previous experiment S14 CTL were selected that just formed stable conjugates exhibiting high calcium flux (Fig. 7A). Importantly, although cognate Kd-peptide P1 complexes exclusively accumulated in the contact site, noncognate Kd-peptide P2 complexes were enriched there but were also found outside (Fig. 7B, supplemental Fig. S5, and Video 4). Whereas in the synapse cognate complexes were immobile (or confined in very small domains), noncognate ones retained mobility (Fig. 7F, and Video 4). As shown in Fig. 7C, about half of the Kd-peptide P2 complexes in the synapse were confined in domains of various sizes (trajectories 4 and 5), others were mobile (trajectory 3), and some exhibited reversible confinements, hopping from one domain to another (trajectories 1 and 2). The diffusion coefficients (D) of Kd-peptide P2 complexes were larger than those of Kd-peptide P1 complexes (Figs. 6D and 7D). Using the confinement length (L) of 360 nm as threshold showed that 56 ± 15% of Kd-peptide P2 complexes were immobile/confined and 44 ± 15% mobile (Fig. 7, E and F). The average size of the domains for Kd-peptide P2 complexes was 130 ± 60 nm.
FIGURE 7.
Single molecule microscopy of Kd-peptide P2 complexes in the synapse between an L-Kd cell and an S14 CTL. A, transmission and overlaid Fluo-3 fluorescence micrograph image. B, corresponding ATTO647 fluorescence image. Scale bars are 5 μm. C, trajectories of Kd-peptide P2 complexes in synapse exhibited reversible confinements followed by free diffusion (1 and 2), free diffusion (3), confinement (4, apparent domain size of 280 nm), or immobility (5, apparent confinement of 140 nm). Scale bar is 500 nm. D, diffusion coefficients (D) of single complexes (red bars, 49 molecules in 12 different synapses). E, confinement lengths (L) of single complexes (red bars). Complexes with length >1 μm are represented in a bar at >1 μm. Blue bars represent complexes following simulated Brownian diffusion, and black bars represent ATTO molecules immobilized on a coverslip. F, using the length of 360 nm as threshold showed that 56 ± 15% of the Kd-peptide complexes were immobile/confined.
Essentially the same findings were obtained on conjugates between RMA-S-Kd cells and S14 CTL. In the synapse formed between RMA-S-Kd cells and S14 CTL cognate Kd-peptide P1 complexes were all localized in the synapse and immobile or confined in small domains (supplemental Fig. S6 and Video 5). By contrast, noncognate Kd-peptide P2 complexes were only enriched in the synapse and rapidly diffused in and out of the synapse (supplemental Fig. S7 and Video 6). In the synapse formed between peptide P2-pulsed RMA-S-KdGPI cells and S14 CTL essentially all complexes were mobile and recruitment into the synapse was scant (supplemental Fig. S8 and data not shown).
DISCUSSION
A key finding of the present study is that MHC I-peptide complexes reversibly immobilize in cell plasma membranes and that the duration of these confinements increased considerably in membranes engaged in adhesion (Figs. 2 and 7 and Videos 4 and 6). Substantially more Kd-peptide P1 complexes were confined in small domains on the lower membrane of L-Kd cells than on the upper membrane (Fig. 2 and Video 1). Moreover, the diffusion of noncognate Kd-peptide P2 complexes in the synapse between L-Kd cells and S14 CTL resembled the one in the lower membrane of L-Kd cells, except that the confinement lengths were smaller (Figs. 2 and 7 and Videos 1 and 4). Transitions between different diffusion modes within the trajectory of single MHC I molecules have been observed previously in single particle tracking experiments (16). However, this study lacks information on the size and frequency of the confinements, making a comparison with our results difficult. The dynamic and reversible confinements of Kd-peptide complexes observed in our study explain why MHC I molecules were found to follow anomalous diffusion in the time scale of minutes (Figs. 2 and 4 and Videos 1-3 and 5) (15). Reversible confinements can give rise to anomalous diffusion by restricting the apparent diffusion of initially mobile molecules over long distances as compared with free Brownian diffusion (40).
Our results on the diffusion of Kd-peptide P1 complexes are consistent with previous studies using FRAP. The diffusion coefficient of Kd-peptide P2 complexes on the upper cell membrane of 0.1 μm2/s (Figs. 2 and 4) lies within the range of 0.06-0.3 μm2/s reported for MHC-I molecules (12-14). Smaller diffusion coefficients (0.004-0.0067 μm2/s) were reported in studies using SPT (14-16), a finding often observed when comparing SPT and FRAP data (13, 41). With a diffusion coefficient of 0.1 μm2/s, it takes Kd-peptide P2 complexes 3.27 min to explore the 78.5-μm2 area of plasma membrane of a cell with a diameter of 5 μm. This is at variance with the observations that nonspecific CTL target cell conjugates are short lived and that cognate MHC I-peptide complexes are rapidly concentrated in nascent CTL target cell contact sites (Fig. 6, supplemental Fig. S4 and Videos 2 and 3) (2). Thus, mechanisms other than purely diffusion controlled transport of MHC I-peptide must exist to make possible efficient TCR scanning.
The observation that MHC I molecules dynamically associate with ICAM-1 explains how they can become transiently linked to the actin cytoskeleton (supplemental Fig. S3) (18-20, 42). Because nonspecific conjugates of CTL and target cells are primarily mediated by LFA-1 and ICAM-1, these molecules are highly enriched in pre-synaptic contact sites (3-5, 18, 42). Dynamic mobility modulation of MHC I-peptide complexes via association with ICAM-1 therefore well explains their efficient diffusional trapping in nascent contact sites (Figs. 2 and 7 and supplemental Fig. S3). The thus greatly increased density of MHC I-peptide complexes in pre-synaptic contact sites in turn permits efficient TCR scanning. This view is consistent with the finding that KdGPI-peptide P1 complexes, which exhibit increased mobility and poorly associated with ICAM-1, were recognized by S14 CTL far less efficiently than wild type complexes (Figs. 4 and 5 and supplemental Fig. S3). It is also consistent with the finding that inhibition of the actin cytoskeleton of L-Kd cells greatly increased the mobility of Kd-peptide P1 complexes and inhibited conjugate formation with S14 CTL (Fig. 3). Stable conjugate and IS formation heavily depend on MHC I-peptide triggered TCR signaling, which activates LFA-1 for high avidity binding to ICAM-1 on target cells (3-5, 42). Moreover, target cell recognition by CTL is independent of costimulation by CD28, but it does rely on costimulation by ICAM-1/LFA-1 (43). ICAM-1 (and possibly other ICAMs) therefore are uniquely suited for the here described mobility modulation-based concentration of MHC I-peptide complexes in newly formed contact sites.
Our assessment of the tempo-spatial dynamics of individual Kd-peptide complexes revealed for the first time that cognate complexes were immobilized in small clusters scattered over most of the extended, nascent synapse with CTL (Fig. 6 and supplemental Figs. S4 and S6 and Videos 2, 3, and 5). Similar microclusters have been observed on CD4+ T cells rapidly after their interaction with APC or ICAM-1 and MHC II-peptide reconstituted planar membranes; they were shown to contain TCR and diverse signaling molecules and to be sites of intense signaling (5, 7-9). Initially these microclusters contain ICAM-1/LFA-1, but as they migrate toward the center to form the cSMAC, adhesion molecules become segregated from antigen recognition molecules (4, 6, 9, 11).
In the nascent synapse between S14 CTL and L-Kd cells, cognate Kd-peptide P1 complexes were immobilized, whereas non-cognate Kd-peptide P2 complexes retained mobility and eventually diffused out again (Figs. 6 and 7 and supplemental Figs. S5-S8 and Videos 2-6). The difference between the two complexes is that one strongly and the other barely detectably interacts with S14 TCR,5 arguing that the retention of MHC I-peptide complexes in the synapse depends on the strength of their interaction with TCR, which upon triggering are cytoskeleton linked (5, 7, 8, 11). Noncognate Kd-peptide P2 complexes were not recognized by S14 CTL (Fig. 1A), yet such ligands can participate in T cell recognition and increase its sensitivity (32, 44). Considering the dimeric recognition modes proposed by these studies and the observation that MHC I-peptide complexes can form compact aggregates, it is conceivable that noncognate complexes partially become immobilized by association with cognate ones (45, 46). This is consistent with the finding that the diffusion coefficients (D) and confinement lengths (L) for cognate and noncognate complexes partially overlap, and both exhibit the same cluster formation (Figs. 6 and 7 and supplemental Figs. S4-S7 and Videos 2-6).
Other factors accounting for the remarkably slow diffusion of noncognate complexes in nascent synapses include the following. 1) the diffusion of noncognate complexes was considerably slower in the synapses of S14 CTL with L-Kd cells than in the synapses with RMA-S-Kd cells and especially with RMA-S-KdGPI cells (supplemental Videos 4 and 6).5 Because L-Kd cells express more ICAM-1 than RMA-S cells (see “Experimental Procedures”) and KdGPI associates poorly with ICAM-1 (supplemental Fig. S3), this argues that the density of cytoskeleton-associated ICAM-1 modulates the mobility of noncognate complexes in the synapse. 2) CD8 in the synapse can bind to and retain noncognate MHC I-peptide complexes (47). These authors proposed that CD8 increases the local density of MHC I-peptide complexes in the synapse and vice versa and that this enhances the sensitivity of antigen recognition. Costimulation by irrelevant peptides was most pronounced on thymocytes, moderate on naive T cells, and mild on effector T cells (48).
The biological significance of dynamic mobility modulation of MHC I-peptide complexes was evidenced by the adverse effects on antigen recognition by measures that interfere with it, such as disruption of the cytoskeleton or replacement of the transmembrane and cytoplasmic portions by a GPI anchor (Figs. 3, 4 and 5). Our finding that KdGPI molecules exhibited greatly increased surface expression and binding of exogenous peptides (Fig. 5A) apparently is accounted for by their inefficient loading with endogenous peptides in the endoplasmic reticulum (49, 50). Because thymic selection of CD8+ T cells and alloantigen recognition rely on presentation of endogenously produced peptides, this explains why GPI-linked MHC I molecules exhibited little or no activity in such studies (12-14). Conversely, in studies where exogenous peptides were presented to CTL of defined specificity, GPI-linked MHC-I molecules were active (25, 26). However, from these studies it is not known with what precise efficiency GPI-linked MHC I molecules present exogenous peptides, because GPI-linked MHC I molecules, lacking avid endogenous peptides, bind exogenous peptides better than wild type molecules. After careful equalization of the peptide binding, we show here for the first time that GPI-linked MHC I-peptide complexes are strikingly less well recognized by CTL than wild type complexes (Fig. 5).
The observation that disruption of the actin cytoskeleton on L-Kd cells rendered Kd-peptide P1 complexes fully mobile and impaired conjugate formation with S14 CTL demonstrates that the dynamic mobility modulation of Kd-peptide P1 complexes improved the initial TCR scanning and TCR triggering (Fig. 3). It also shows that the transient immobilizations of complexes were not accounted for by partitioning in large clusters of MHC I molecules. Such clusters have been observed mainly on activated B and T cells, which express large fractions of free MHC I heavy chains known to aggregate (51). Indeed, we obtained the same results in the absence or presence of excess of β2-microglobulin, which disrupts MHC I clustering.5
In conclusion, this first tracking study of defined, single MHC I-peptide complexes on target cells and their nascent immunological synapse with CTL revealed a new, fundamental aspect of MHC I-restricted antigen presentation. It demonstrates that the anomalous diffusion of complexes is explained by transient association with cytoskeleton-associated ICAM-1. Because ICAM-1 is concentrated in pre-synaptic contact sites formed by CTL and target cells, this results in local diffusional trapping and sorting of complexes. This in turn increases the efficiency of TCR scanning and TCR triggering and hence the sensitivity of antigen recognition.
Supplementary Material
Acknowledgments
We are most grateful to Dr. Y. Sykulev (Thomas Jefferson University) for critical reading of the manuscript and to Dr. E. Uetz-von Allmen (Biotechnology Institute Thurgau) for cloning work on the Kd-GPI construct.
This work was supported in part by Grant 310000-108251 from Swiss National Foundation (to I. F. L.) and internal Ecole Polytechnique Fédérale de Lausanne grants (to H. V.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, Figs. S1-S8, and Videos 1-6.
Footnotes
The abbreviations used are: CTL, cytotoxic T lymphocytes; ABA, 4-azidobenzoic acid; FRAP, fluorescence recovery after photobleaching; GPI, glycosylphosphatidylinositol; IS, immunological synapse; cSMAC, central supramolecular activation cluster; pSMAC, peripheral SMAC; SPT, single particle tracking; TCR, T cell antigen receptor; FITC, fluorescein isothiocyanate; CFSE, carboxyfluorescein succinimidyl ester; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; DMEM, Dulbecco's modified Eagle's medium; MFI, mean fluorescent intensity; MHC, major histocompatibility complex; Fmoc, N-(9-fluorenyl)methoxycarbonyl; Dap, diaminopropionic acid.
J.-M. Segurea and I. F. Luescher, unpublished results.
References
- 1.Sykulev, Y., Joo, M., Vturina, I., Tsomides, T. J., and Eisen, H. N. (1996) Immunity 4 565-571 [DOI] [PubMed] [Google Scholar]
- 2.Purbhoo, M. A., Irvine, D. J., Huppa, J. B., and Davis, M. M. (2004) Nat. Immunol. 5 524-530 [DOI] [PubMed] [Google Scholar]
- 3.Marwali, M. R., MacLeod, M. A., Muzia, D. N., and Takei, T. (2004) J. Immunol. 173 2960-2967 [DOI] [PubMed] [Google Scholar]
- 4.Somersalo, K., Anikeeva, N., Sims, T. N., Thomas, V. K., Strong, R. K., Spies, T., Lebedeva, T., Sykulev, Y., and Dustin, M. L. (2004) J. Clin. Invest. 113 49-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki, J., Yamasaki, S., Wu, J., Koretzky, G. J., and Saito, T. (2007) Blood 109 168-175 [DOI] [PubMed] [Google Scholar]
- 6.Potter, T. A., Grebe, K., Freiberg, K., and Kupfer, A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12624-12629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito, T., and Yokosuka, T. (2006) Curr. Opinion Immunol. 18 305-313 [DOI] [PubMed] [Google Scholar]
- 8.Campi, G., Varma, R., and Dustin, M. L. (2005) J. Exp. Med. 202 1031-1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varma, R., Campi, G., Yokosuka, T., Saito, T., and Dustin, M. L. (2006) Immunity 25 117-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokosuka, T., Sakata-Sogawa, K., Kobayashi, W., Hiroshima, M., Hashimoto-Tane, A., Tokunaga, M., Dustin, M. L., and Saito, T. (2005) Immunology 6 1253-1262 [DOI] [PubMed] [Google Scholar]
- 11.Kaizuka, Y., Douglass, A. D., Varma, R., Dustin, M. L., and Vale, R. D. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 20296-20301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edidin, M., Zuniga, M. C., and Sheetz, M. P. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 3378-3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georgiou, G., Bahra, S. S., Mackie, A. R., Wolfe, C. A. O'Shea, P., Ladha, S., Fernandez, N., and Cherry, R. J. (2002) Biophys. J. 82 1828-1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang, Q., and Edidin, M. (2003) Biophys. J. 84 400-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith, P. R., Morrison, I. E., Wilson, K. M., Fernandez, N., and Cherry, R. J. (1999) Biophys. J. 76 3331-3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capps, G. G., Pine, S., Edidin, M, and Zuniga, M. C. (2004) Biophys. J. 86 2896-2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwik, J., Boyle, S., Fooksman, D., Margolis, L., Sheetz, M. P., and Edidin, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13964-13969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebedeva, T., Anikeeva, N., Kalams, S. A., Walke, B. D., Gaidarov, I., Keen, J. H., and Sykulev, Y. (2004) Immunology 113 460-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bacso, Z., Bene, L., Damjanovich, L., and Damjanovich, S. (2002) Biochem. Biophys. Res. Commun. 290 635-640 [DOI] [PubMed] [Google Scholar]
- 20.Damjanovich, S., Matko, J., Matyus, L., Szabo, G., Szollosi, J., Pieri, J. C., Farkas, T., and Gaspar, R. (1998) Cytometry 33 225-233 [PubMed] [Google Scholar]
- 21.Carpen, O., Pallai, P., Staunton, D. E., and Springer, T. A. (1992) J. Cell Biol. 118 1223-1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zamoyska, R., Ong, T., Kwan-Lim, G., Tomlinson, P., and Robinson, P. J. (1996) Int. Immunol. 8 551-557 [DOI] [PubMed] [Google Scholar]
- 23.Simpson, E., Robinson, P. J., Chandler, P., Millrain, M. M., Pircher, H. P., Brandle, D., Tomlinson, P., Antoniou, J., and Mellor, A. (1994) Immunology 81 132-136 [PMC free article] [PubMed] [Google Scholar]
- 24.Mann, D. W., Stroynowski, I., Hood, L., and Forman, J. (1989) J. Immunol. 142 318-322 [PubMed] [Google Scholar]
- 25.Gao, X. M., Quinn, C. L., Bell, J. I., and McMichael, A. J. (1993) Eur. J. Immunol. 23 653-658 [DOI] [PubMed] [Google Scholar]
- 26.Huang, J. H., Getty, R. R., Chisari, F. V., Fowler, P., Greenspan, N. S., and Tykocinski, M. L. (1994) Immunity 1 607-613 [DOI] [PubMed] [Google Scholar]
- 27.Vrljic, M., Nishimura, S. Y., Brasselet, S., Moerner, W. E., and McConnell, H. M. (2002) Biophys. J. 83 2681-2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umemura, Y. M., Vrljic, M., Nishimura, S. Y., Fujiwara, T. K., Suzuki, K. G., and Kusumi, A. (2008) Biophys. J. doi: 10.1529/biophysj.107.123018 [DOI] [PMC free article] [PubMed]
- 29.Jacquier, V., Prummer, M., Segura, J.-M., Pick, H., and Vogel, H. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 14325-14330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez, J. B., Segura, J.-M., Abankwa, D., Piguet, J., Martinez, K. L., and Vogel, H. (2006) J. Mol. Biol. 363 918-930 [DOI] [PubMed] [Google Scholar]
- 31.Luescher, I. F., Anjuère, F., Peitsch, M. C., Jongeneel, C. V., Cerottini, J.-C., and Romero, P. (1995) Immunity 3 51-63 [DOI] [PubMed] [Google Scholar]
- 32.Cebecauer, M., Guillaume, P., Mark, S., Michielin, O., Boucheron, N., Bezard, M., Meyer, B. H., Segura, J. M., Vogel, H., and Luescher, I. F. (2005) J. Biol. Chem. 280 23820-23825 [DOI] [PubMed] [Google Scholar]
- 33.Jaulin, C., Casanova, J. L., Romero, P., Luescher, I. F., Cordey, A. S., Maryanski, J. L., and Kourilsky, P. (1992) J. Immunol. 149 3990-3994 [PubMed] [Google Scholar]
- 34.Legler, D. F., Doucey, M. A., Schneider, P., Chapatte, L., Bender, F. C., and Bron, C. (2005) FASEB J. 19 73-75 [DOI] [PubMed] [Google Scholar]
- 35.Naldini, L., Blomer, U., Gallay, P., Ory, D., Mulligan, P., Gage, F. H., Verma, I. M., and Trono, D. (1996) Science 272 263-267 [DOI] [PubMed] [Google Scholar]
- 36.López, J. A., Luescher, I. F, and Cerottini, J.-C. (1992) J. Immunol. 149 3827-3835 [PubMed] [Google Scholar]
- 37.Angelov, G. S., Guillaume, P., Cebecauer, M., Bosshard, G., Dojcinovic, D., Baumgaertner, P., and Luescher, I. F. (2006) J. Immunol. 176 3356-3365 [DOI] [PubMed] [Google Scholar]
- 38.Simson, R., Sheets, E. D., and Jacobson, K. (1995) Biophys. J. 69 989-993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Legler, D. F., Doucey, M. D., Cerottini, J.-C., Bron, C., and Luescher, I. F. (2001) FASEB J. 15 1601-1603 [DOI] [PubMed] [Google Scholar]
- 40.Saxton, M. J. (1996) Biophys. J. 70 1250-1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saxton, M., and Jacobson, J. K. (1997) Annu. Rev. Biophys. Biomol. Struct. 26 373-399 [DOI] [PubMed] [Google Scholar]
- 42.Lebedeva, T., Dustin, M. L., and Sykulev, Y. (2005) Curr. Opin. Immunol. 17 251-258 [DOI] [PubMed] [Google Scholar]
- 43.O'Keefe, J. P., and Gajewski, T. F. (2005) J. Immunol. 175 5581-5585 [DOI] [PubMed] [Google Scholar]
- 44.Krogsgaard, M., Li, Q. J., Sumen, C., Huppa, J. B., Huse, M., and Davis, M. M. (2005) Nature 434 238-243 [DOI] [PubMed] [Google Scholar]
- 45.Pentcheva, T., and Edidin, M. (2001) J. Immunol. 66 6625-6632 [DOI] [PubMed] [Google Scholar]
- 46.Gaspar, R., Jr., Bagossi, P., Bene, L., Matko, J., Szollosi, J., Tozser, J., Fesus, L., Waldmann, T. A., and Damjanovich, S. (2001). J. Immunol. 166 5078-5086 [DOI] [PubMed] [Google Scholar]
- 47.Yachi, P. P., Ampudia, J., Gascoigne, N. R., and Zal, T. (2005) Nat. Immunol. 6 785-792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yachi, P. P., Lotz, C., Ampudia, J., and Gascoigne, N. R. (2007) J. Exp. Med. 204 2747-2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diedrich, G., Bangia, N., Pan, M., and Cresswell, P. (2001) J. Immunol. 166 1703-1709 [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Y., and Williams, D. B. (2006) Immunol. Res. 35 151-162 [DOI] [PubMed] [Google Scholar]
- 51.Bodnar, A., Bacso, Z., Jenei, A., Jovin, T. M., Edidin, M., Damjanovich, S., and Matko, J. (2003) Int. Immunol. 15 331-339 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.