Abstract
Genes encoding the peptide deformylase enzyme (def) are present in all eubacteria and are involved in the deformylation of the N-formyl group of newly synthesized polypeptides during protein synthesis. We compared the amino acid sequences of this enzyme in different mycobacterial species and found that they are highly conserved (76% homology with 62% identity); however, when this comparison was extended to other eubacterial homologs, it emerged that the mycobacterial proteins have an insertion region containing three consecutive arginine residues (residues 77–79 in Mycobacterium tuberculosis peptide deformylase (mPDF)). Here, we demonstrate that these three arginines are important for the activity of mPDF. Circular dichroism studies of wild-type mPDF and of mPDF containing individual conservative substitutions (R77K, R78K, or R79K) or combined substitutions incorporated into a triple mutant (R77K/R78K/R79K) indicate that such mutations cause mPDF to undergo structural alterations. Molecular modeling of mPDF suggests that the three arginines are distal to the active site. Molecular dynamics simulations of wild-type and mutant mPDF structures indicate that the arginines may be involved in the stabilization of substrate binding pocket residues for their proper interaction with peptide(s). Treatment with 5′-phosphothiorate-modified antisense oligodeoxyribonucleotides directed against different regions of def from M. tuberculosis inhibits growth of Mycobacterium smegmatis in culture. Taken together, these results hold out the possibility of future design of novel mycobacteria-specific PDF inhibitors.
The first amino acid incorporated during polypeptide biosynthesis in eukaryotes is methionine. On the other hand, in prokaryotes and in eukaryotic organelles (mitochondria and chloroplasts), ribosomal protein biosynthesis is initiated with N-formyl-methionyl-tRNA. Thus, all nascent polypeptides in prokaryotes are formylated at their amino termini (1–3). Since peptidases acting at NH2 termini are unable to utilize formylated NH2 termini as substrates, the removal of the formyl group is mandatory for polypeptide maturation in all cases in which the removal of the NH2-terminal methionine is essential for protein folding or function. The enzyme, peptide deformylase (PDF),3 catalyzes the deformylation of N-formylmethionine in nascent polypeptide chains in prokaryotes (3–5). PDF is a metalloprotease containing iron (as Fe2+) or Zn2+ (6–8). The gene encoding peptide deformylase (def) is present throughout the eubacterial lineage (9). In several bacteria, it has been reported to be an essential gene (10–12), currently considered to be an appropriate target for the development of anti-bacterials (13, 14).
We have earlier cloned, expressed, and characterized the peptide deformylase enzyme from Mycobacterium tuberculosis. We found that M. tuberculosis peptide deformylase (mPDF) is an iron-containing protein that is enzymatically active as a multimer (15, 16). Although conversion of Fe2+ to Fe3+ by environmental oxygen results in inactivation of the PDF of Escherichia coli (17), we found mPDF to be stable at 30 °C with a half-life of ∼4 h. Also, preincubation with an oxidizing agent like H2O2 had no significant effect on activity (15). Sequence analysis of mPDF revealed several features, such as the presence of three conserved motifs (motif I, GXGXAAXQ; motif II, EGCLS; motif III, QHEXXH, in which X represents any hydrophobic residue), one insertion (amino acid residues 74–85) between motifs I and II, and an extended COOH terminus (amino acid residues 182–197). We also showed that the first six amino acids (residues 74–79) of the insertion region mentioned above play a role in stabilizing mPDF and in imbuing it with resistance to oxidizing agents like H2O2 (16). This finding is difficult to reconcile with the finding (7) that deletion of similar insertions in other bacteria fails to affect enzyme activity.
There are three consecutive arginine residues in the insertion region (residues 74–85). Through detailed structure-function analyses, we show here that these three arginines are important for mPDF activity. CD spectroscopic studies of the mutants incorporating conservative substitutions (R77K, R78K, and R79K, individually and as a triple mutant) reveal structural alterations in comparison with wild-type mPDF. Molecular modeling of mPDF based on structural homology with Pseudomonas aeruginosa PDF suggests that these arginines are not in the vicinity of the enzyme active site. Molecular dynamics simulations with wild-type and mutant structures, on the other hand, reveal that these arginines might help in the interaction of substrate with amino acid residues in the substrate binding pocket of mPDF. Thus, all of these lines of evidence indicate the importance of arginines of the insertion region of mPDF for its enzyme activity.
EXPERIMENTAL PROCEDURES
Materials—Restriction/modifying enzymes were obtained from New England Biolabs. ECL Western blotting detection kit, PCR DNA and gel band purification kit, and protein molecular weight markers were from GE Healthcare; the Expand long template PCR system was from Roche Applied Science; Herculase fusion DNA polymerase was from Stratagene; VentR DNA polymerase was from New England Biolabs; and plasmid DNA preparation kits were from Qiagen. All other fine chemicals and reagents, including catalase, N-formyl-Met-Ala, and trinitrobenzene sulfonic acid, were procured from Sigma. Different media for bacterial culture were obtained from Difco. Oligonucleotides used in this study were custom synthesized (Biobasic/IDT/Ocimum Biosolutions).
DNA Manipulation and Generation of mPDF Mutants—Genomic DNA isolated from M. tuberculosis strain H37Ra (18) was used for PCR amplification and subsequent cloning of the def gene (Rv0429c) in expression vector (pET-mPDF) as described elsewhere (15). PCR-based methods were employed for generation of mutations in mPDF. For each mutation, two external (CR26 and CR27) and two internal (designed incorporating mutation) primers (supplemental Table 1) were used. To generate deletion (ΔMTA/ΔIR) or point mutants (R77K, R77A, R77D, R78K, R78A, R78D, R79K, R79A, and R79D), two sets of primary (pET-mPDF as the template) and one set of secondary (mixture of primary reaction products as the template) PCRs were carried out by the overlap extension method (19) using Herculase fusion DNA polymerase. For the triple mutant, R77K/R78K/R79K, a ∼400-bp megaprimer, was PCR-amplified with one internal (CK32; designed incorporating mutations) and one external primer (CR27) using R79K as the template, following a method described previously elsewhere (20). Subsequently, this megaprimer, along with the external primer, CR26, upon PCR amplification using R79K DNA as the template, yielded the triple mutant. PCR products containing desired mutations were digested with SacII/HindIII and incorporated into pET-mPDF. All of these constructs and the wild-type mPDF construct were individually transformed into E. coli strain DH5α to build up the DNA for further processing. All mutations were confirmed by sequencing using an automated DNA sequencer.
Expression and Purification of Recombinant Proteins—The wild-type construct (pET-mPDF) and different mutant constructs were transformed into E. coli strain BL21(DE3) for overexpression and subsequent purification of NH2-terminally histidine-tagged fusion proteins. Overnight cultures of these clones (∼15 h at 37 °C in LB broth containing 50 μg/ml kanamycin) were reinoculated and grown to an A600 of 0.8. Cells were then induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside at 25 °C, harvested after 12 h, suspended in lysis buffer (20 mm phosphate buffer, pH 7.4, containing 5 mm dithiothreitol, 10 μg/ml catalase, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin, and 1 μg/ml leupeptin), and sonicated. The pellet fraction obtained through centrifugation (∼12,000 × g for 30 min at 4 °C) was resuspended in lysis buffer containing 3 m urea and 2% Triton X-100. Following centrifugation, the supernatant was dialyzed (14 h at 4 °C) to remove urea, and the protein was purified on a Ni2+-nitrilotriacetic acid column as described earlier (15). Finally, mPDF and its different mutants were eluted (20 mm phosphate buffer, pH 7.4, containing 300 mm NaCl, 250 mm imidazole, and 10 μg/ml catalase). The concentrations of the stock of the Ni2+-nitrilotriacetic acid-purified mPDF and its different mutants were maintained at 3.5 mg/ml and stored at –80 °C until used in assays. Unless mentioned otherwise, mPDF and its mutants were diluted (20 mm phosphate buffer, pH 7.4, containing 1 mg/ml bovine serum albumin and 10 μg/ml catalase) prior to use in assays.
Enzyme Assay—The ability of mPDF or different mutants to deformylate formyl-Met-Ala was assessed through a spectrophotometric assay, as previously described (15). Briefly, in a total of 50 μl of reaction volume, wild-type (70 ng) or mutant (280 ng for R77K, R77A, R77D, R78K, R78A, R78D, 1 μg for R79K, R79A, R79D, and 1.6 μg for R77K/R78K/R79K) protein in 1× assay buffer (100 mm phosphate buffer, pH 7.4, containing 100 μg/ml catalase) was incubated with the substrate (0–40 mm N-formyl-Met-Ala) at 30 °C for 30 min. The reaction was terminated by the addition of 50 μl of 4% HClO4 and further incubated (30 °C for 2 h) with trinitrobenzene sulfonic acid reagent (0.01% in 0.1 m NaHCO3 buffer, pH 8.4). Following the addition of 10% SDS (250 μl) and 1 m HCl (125 μl), the highly chromogenic derivative generated due to reaction of primary amine with trinitrobenzene sulfonic acid was monitored through measurement of the absorption at 335 nm. The values obtained were corrected by subtracting the blank (all ingredients except enzyme) readings. The specific enzyme activity was calculated from standard curves prepared with methionine (0–42.8 nmol) and expressed as nmol of free amino group produced/min/mg of protein. Km and Vmax values were calculated from Lineweaver-Burk plots, and for calculating kcat values, the molecular mass of recombinant mPDF was taken as 22.6 kDa. Unless mentioned otherwise, data for enzyme activity represent mean ± S.D. of values from three independent experiments.
Western Blotting—The recombinant mPDF protein (∼1 mg/ml) was used to raise polyclonal antibody in rabbit (21), which recognizes native enzymes of M. tuberculosis and Mycobacterium smegmatis. For Western blotting, purified proteins (1 μg of protein/slot) or the soluble fractions of cell lysates (50 μg of protein/slot) were resolved in 12% SDS-PAGE and transferred to nitrocellulose membranes (0.45 μm) using a mini-transblot apparatus (Bio-Rad). Membranes were stained with Ponceau S to ensure transfer, processed using anti-mPDF or anti-His antibody, and detected through an ECL detection system following the manufacturer's (GE Healthcare) recommended protocol.
CD Spectroscopy—CD spectra were recorded in a Jasco J-810 spectropolarimeter. Measurements in the far ultraviolet region (250–187 nm) were performed on protein solutions (0.15–0.3 mg/ml) employing a cell with path length of 0.5 cm at 25 °C. The mean residue ellipticity, [θ], was calculated using a mean residue molecular mass of 110 Da. Each spectrum reported is an average of 10 scans. Blank spectra of aqueous buffer were used to correct the observed spectra.
Confocal Microscopy—M. smegmatis culture (∼105 cells/3 ml of 7H9 Middlebrook broth supplemented with a 10% mixture of albumin, dextrose, and catalase) was grown (37 °C/200 rpm) in the presence and absence of different 5′-phosphothiorate-modified antisense oligodeoxyribonucleotides for 24 h. Cells were harvested, washed, and resuspended in phosphate-buffered saline (100 μl). Following mounting of samples (5–10 μl) on glass slides, fluorescence emitted by cells was visualized on an inverted LSM510 META laser-scanning confocal microscope (Carl Zeiss) fitted with a Plan-Apochromat ×100 1.4 numerical aperture oil immersion objective. For detection of green fluorescence, the 488 line of an argon ion laser was directed over an HFT UV/488 beam splitter, and fluorescence was detected in multitrack configuration using an NFT 490 beam splitter in combination with a BP 505–530 band pass filter. Images were finally processed using Adobe Photoshop 5.5.
Bioinformatic Analysis—Nucleotide-derived amino acid sequence of mPDF has been compared with a nonredundant data base using BLAST (22). The multiple-sequence alignment of the retrieved sequences is carried out using ClustalX (23) with default values for gap opening and extension penalties.
Molecular Modeling—The structure of mPDF was predicted by the ROBETTA server (24) using the structure of P. aeruginosa PDF (Protein Data Bank code 1N5N) (25), since it shows highest sequence homology (38% identity, 49% similarity). Five models of the structure of mPDF were predicted by the server. Superposition of these five structures revealed that the core regions of mPDF are conserved (with an average r.m.s. deviation of 0.3 Å), whereas large deviations were observed at the flexible loops (with average r.m.s. deviation of 6.0 Å). These structures were subjected to energy minimization, and the structure with minimum energy was selected. The convergence criterion used was an r.m.s. gradient of 0.01 Å. The energy-minimized structure of mPDF was stereochemically validated using PROCHECK (26) and subsequently used in molecular dynamics studies. The energy minimization and molecular dynamics were carried out using the Sander module of the AMBER9 package (27). AMBER FF03 all atom force field (28) was used for proteins, ions, and water molecules. Structures for different mutants of mPDF were generated using PYMOL (29). Molecular dynamics simulations were performed on wild-type and mutant (R77K/R78K/R79K) structures. Both proteins were immersed in a truncated octahedron box filled with about 8000 TIP3P water molecules. The expansion and shrinkage of all covalent bonds connecting to a hydrogen atom were constrained using the SHAKE algorithm (30). Periodic boundary conditions were applied to avoid the edge effect in all calculations. The structures of the protein immersed in water were neutralized by the addition of the required amount of counter ions. The energy minimization for this system was achieved in two steps. The solvent and ions were energy-minimized first, keeping the protein fixed with restraint force of 100 kcal/mol/Å2. In the next step, all atoms of solute and solvent were minimized without any restrained energy. In each step, the minimization was executed by 1000 steps of steepest descent and 2000 steps of conjugate gradient methods. The nonbonded interactions were calculated with a 12 Å cut-off. The molecular dynamics simulations were performed on this system at 300 K for 10 ns using an NVT ensemble. A time step of 1 fs was used for integration of Newtonian equations, and the snapshots were saved every 2 ps. Analysis of production run (last 9 ns) of the simulations was done using the ptraj module of AMBER 9.
RESULTS
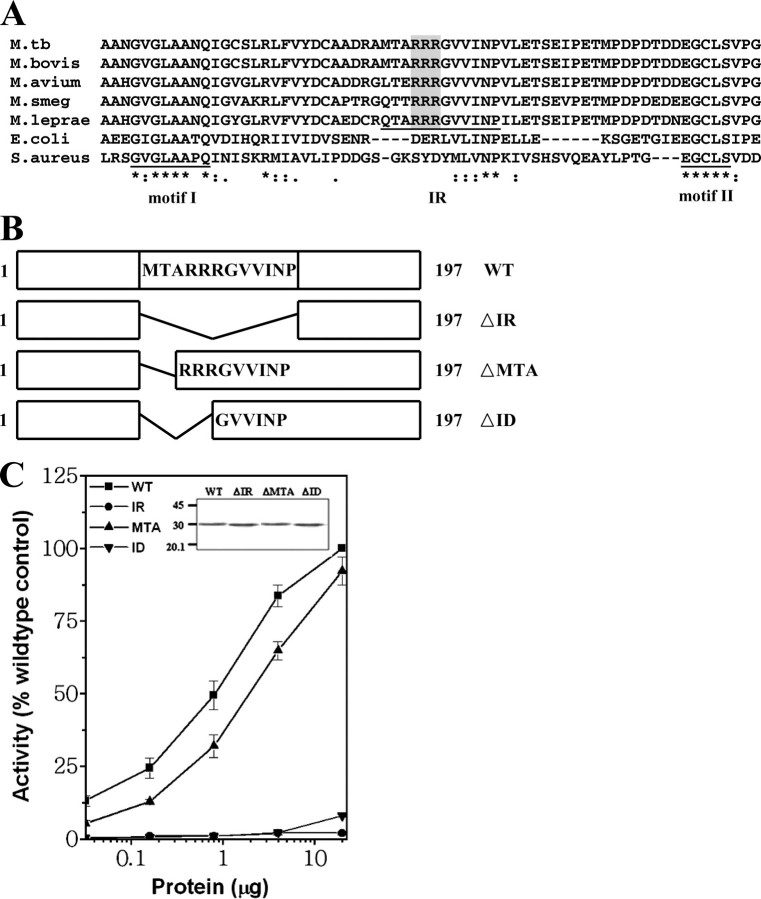
Insertion Region of mPDF Contributes to Peptide Deformylase Activity—The presence of an insertion sequence in PDFs of Gram-positive bacteria is well known (15, 25). The mPDF also possesses insertion between motifs I and II (amino acid residues 74–85; denoted as IR in Fig. 1A). Among different mycobacterial species, this region of PDF exhibits ∼84% identity (Fig. 1A). To identify crucial residues in this region, we constructed three deletion mutants (Fig. 1B), expressed them as His-tagged proteins, and examined their activities. The expression of different mutant proteins was confirmed by Western blot using rabbit polyclonal antiserum against recombinant mPDF (Fig. 1C, inset). In an earlier report (16), we noted that the ΔID mutant (i.e. with residues 74–79 deleted) did not show any activity. Here, we show that the deformylase activity of the ΔMTA mutant (i.e. with only residues 74–76 deleted) is comparable with that of the wild type (Fig. 1C), although deletion of the entire insertion region in the ΔIR mutant (i.e. with residues 74–85 deleted) completely abolishes activity (Fig. 1C). These results establish that the region comprising residues 77–85 in the insertion region is essential for the deformylase activity of mPDF.
FIGURE 1.
Deletion analysis of insertion region of mPDF. A, alignment of amino acid residues between motifs I and II from PDFs of different mycobacterial species (residues 46–111), E. coli (residues 41–96), and Staphylococcus aureus (residues 55–116) showing three consecutive arginines (shaded). The asterisks and dots denote identical and similar amino acids, respectively. M. tb, M. tuberculosis; M. smeg, M. smegmatis. B, deletion scheme. WT, wild-type mPDF; ΔIR, deletion of MTARRRGVVINP residues; ΔMTA, deletion of MTA residues; ΔID, deletion of MTARRR residues. C, assessment of deformylase activity of deletion mutants. Deformylase assays were carried out with the indicated amounts of wild-type or mutant proteins using 5 mm N-formyl-Met-Ala as the substrate. Inset, Western blot of wild-type and mutant proteins (1 μg/slot) using rabbit polyclonal anti-serum against recombinant mPDF.
Three Consecutive Arginines in the Insertion Region Are Important for the mPDF Enzyme Activity—Intriguingly, among the critical residues in the insertion sequence, three consecutive arginines are conserved in the PDF homologs of different mycobacterial species (Fig. 1A). To elucidate the role of these residues, we generated mutants substituting all three arginines with lysine (either individually or together). In comparison with wild-type mPDF, the deformylation ability of all of the mutants was found to be affected (Fig. 2A and Table 1), the effect being most dramatic in the triple mutant in which no activity could be detected, during incubations with increasing concentrations (2.5–40 mm) of substrate (N-formyl-Met-Ala). This was reflected in assays carried out using increasing concentrations of protein (Fig. 2A, inset). The effect was also reflected in the enzyme's turnover rate, since significant decreases in the kcat values were observed (Table 1). Furthermore, as reflected in Km values, mutation of these arginines appears to affect enzyme-substrate affinity (Table 1). The effect of the mutations was not Lys-specific, since substitution of Arg with either Ala or Asp also resulted in significant inhibition in activity compared with wild-type mPDF (Table 1), as would be expected, since nonconservative mutations could be expected to show an effect where conservative mutations also show effects. In subsequent studies, therefore, only arginine-to-lysine (R77K, R78K, R79K) mutants were used.
FIGURE 2.
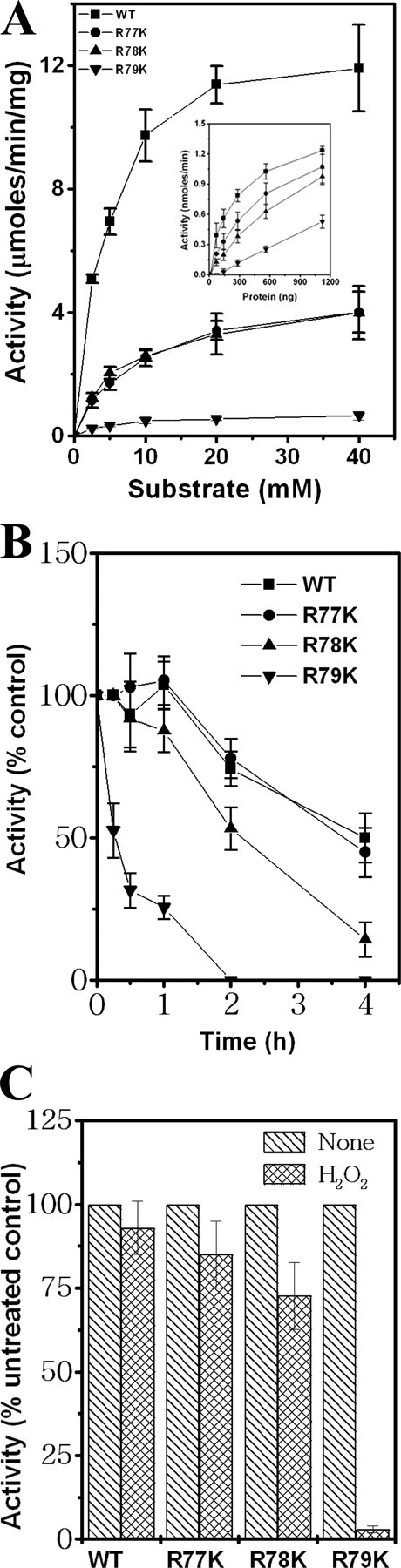
Three consecutive arginines of the mPDF insertion region are crucial for deformylase activity. A, kinetic analyses of deformylase activities of mutant proteins. Inset, enzyme activities as a function of amount of protein using 5 mm substrate. B, comparison of enzymatic stabilities of wild-type (WT) and mutant proteins at 30 °C. C, effect of H2O2 on the deformylase activity of mutants. Wild-type or mutant proteins were preincubated with or without H2O2 (final concentration, 500 mm) at 30 °C for 15 min. This was followed by an enzyme assay using 5 mm substrate.
TABLE 1.
Kinetics of deformylase activity exhibited by mPDF mutants
| Sample | Km | kcat | kcat/Km | Number of experiments |
|---|---|---|---|---|
| mm | s-1 | m-1 s-1 | ||
| Wild type | 4.1 ± 0.2 | 5.0 ± 0.3 | 1220 ± 6 | 3 |
| R77K | 6.9 ± 1.4 | 1.8 ± 0.3 | 266 ± 34 | 4 |
| R77A | 6.5 ± 1.7 | 1.6 ± 0.2 | 252 ± 35 | 4 |
| R77D | 9.3 ± 2.5 | 0.6 ± 0.2 | 67 ± 11 | 3 |
| R78K | 5.6 ± 0.9 | 1.6 ± 0.3 | 293 ± 47 | 3 |
| R78A | 6.4 ± 1.7 | 1.7 ± 0.2 | 270 ± 60 | 5 |
| R78D | 11.7 ± 2.2 | 0.8 ± 0.2 | 57 ± 11 | 3 |
| R79K | 5.0 ± 0.4 | 0.24 ± 0.02 | 52 ± 4 | 3 |
| R79A | 3.4 ± 1.0 | 0.08 ± 0.05 | 22 ± 6 | 3 |
| R79D | NDa | ND | ND | 3 |
| R77K/R78K/R79K | ND | ND | ND | 3 |
ND, not detectable.
The influence of the mutations on the enzymatic stability of mPDF was examined by monitoring the activity as a function of time (Fig. 2B). The activity of R77K exhibited a half-life marginally lower than that of wild-type mPDF (3.7 ± 0.5 h as opposed to 4.1 ± 0.7 h). In comparison, a far more significant decrease in half-life with respect to activity was noticed with R78K (2.2 ± 0.3 h) and R79K (half-life = 0.6 ± 0.1 h). To know the contributions of these arginines in conferring resistance to oxidizing agents, we monitored the effect of H2O2 on the activity of the enzyme. As shown in Fig. 2C, preincubation with H2O2 (500 mm) for 15 min resulted in significant decreases in the deformylase activity of R79K, compared with wild type or R77K or R78K mPDF. Thus, these findings establish the role of three arginines in modulating the activity of mPDF.
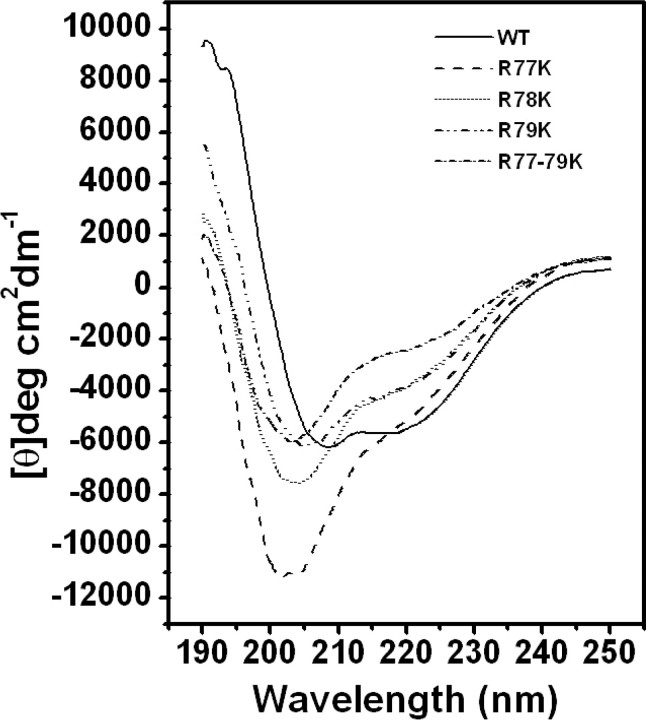
Three Arginines Contribute to the Tertiary Structure of mPDF—The possibility of any structural alterations having occurred in the mutant proteins (R77K, R78K, R79K, or R77K/R78K/R79K) was explored by examining their far-UV CD spectra between 250 and 187 nm. The far-UV CD spectrum of mPDF shows negative minima at 208 and 222 nm (indicative of a predominantly helical structure); however, the low mean residue ellipticity of about –6000 at 222 nm indicates the presence of other (sheet and coil) structures (Fig. 3). Interestingly, the mutant proteins show significant alterations of secondary structures. In all mutants, there was a reduction in the ellipticity at 222 nm and an increase, as well as a blue shifting, of the 208 nm bands, clearly indicating a relative increase in the content of random coil structures in the proteins (Fig. 3). As expected, random coil structures yielded a negative band at 198 nm, and when their content was increased in relation to other structures within the protein, there was a shifting of negative minima in the range of 200–210 nm to lower wavelengths as well as shifting of the crossover point (i.e. the wavelength at which the spectrum crossed the zero line in the ellipticity plot).
FIGURE 3.
Effect of mutations on the secondary structure of mPDF. Far-UV CD spectra of wild-type (WT) and mutant proteins (R77K, R78K, R79K, and R77K/R78K/R79K) in 20 mm phosphate buffer, pH 7.4, were obtained between wavelengths 250 and 187 nm. The Ni2+-nitrilotriacetic acid-purified protein samples (∼0.2 mg/ml) were dialyzed to remove immidazole and were used for spectroscopic analysis.
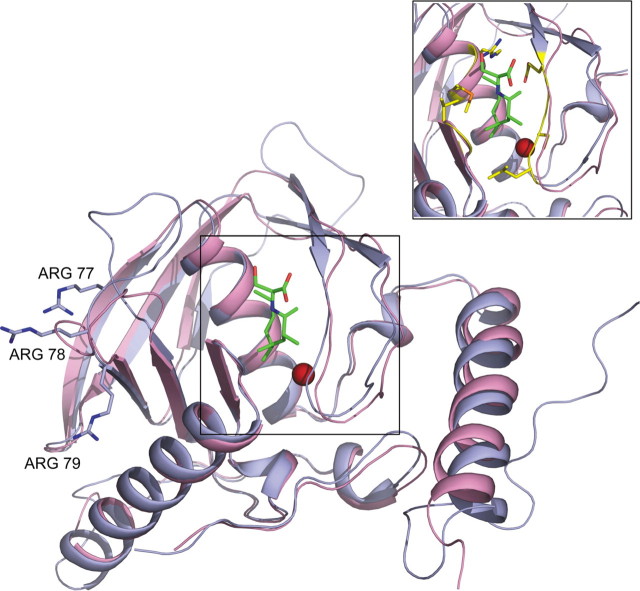
To understand why mutations of the arginines (R77K/R78K/R79K) alter the structure of the protein, molecular modeling and molecular dynamics simulations were carried out on both wild-type and mutant mPDFs. The three-dimensional structure of mPDF was predicted through homology modeling on the ROBETTA server (24) and using P. aeruginosa PDF as template (25), as already mentioned. The modeled structure reveals that all three arginines (residues 77–79) are surface-exposed, and also they are distal to the active site, as shown in Fig. 4. Notably, prediction of disordered regions in this structural model on the SPRITZ server (31) suggests that the three arginine residues, Arg77–Arg79, are in an ordered conformation and not in disordered regions.
FIGURE 4.
Model of mPDF superimposed on ePDF. The superimposition shows that the core regions are conserved in mPDF (blue) and ePDF (pink) along with metal ion (red) and Met-Ala-Ser tripeptide (green). Arginines (R77K/R18K/R79K) are away from the substrate binding pocket. The inset shows the position of residues (yellow) interacting with tripeptide and metal ion in PDF.
To map the substrate binding pocket of mPDF, the predicted structure was superimposed with E. coli PDF, a bacterial structure (ePDF; Protein Data Bank code 1BS8) (32) that is complexed with a tripeptide (Met-Ala-Ser). Furthermore, it has been shown that the interaction between this peptide and PDFs are similar in Gram-positive as well as in Gram-negative bacteria (25, 33). The superposition of mPDF with ePDF, as shown in Fig. 4, reveals that most of the amino acid residues (Gly49, Val50, Gly51, Glu104, Gly105, Cys106, Leu107, Arg144, and Met145) of mPDF lining the cavity within 5 Å of the substrate are identical to that of the ePDF. The amino acid positions Val50, Arg144, and Met145 in mPDF are in correspondence with Ile44, Ile128, and Cys129 of ePDF, respectively. The amino acid residues Cys106, His148, and Glu149, involved in catalytic activity of mPDF (15), are conserved in both the sequences; the orientations of the side chains of these residues are also conserved among various known PDF structures, lending support to the validity of the predicted structure of mPDF.
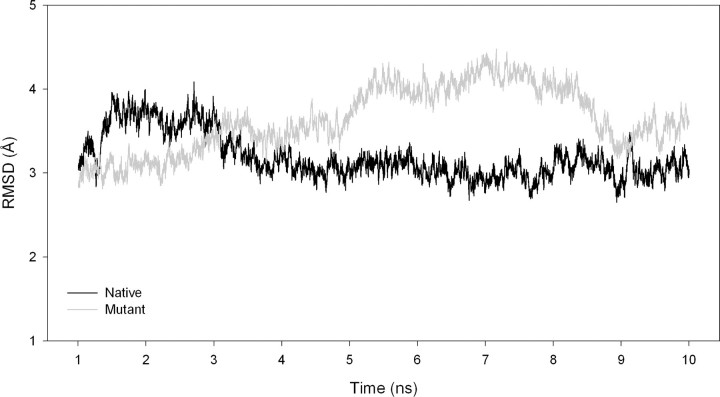
Since the effect of mutations on the activity of the triple mutant was most dramatic (Table 1), it appeared likely that the structural alterations would be most evident in R77K/R78K/R79K, and subsequently in all of the calculations, only this (triple) mutant was used. The wild-type and triple mutant structures of mPDF were subjected to molecular dynamics simulations, and the r.m.s. deviation plots for the same are shown in Fig. 5. During molecular dynamics, the overall structure, nature, and details of interactions of amino acids lining the cavity (within a 5Å distance from the tripeptide mentioned earlier) and that of the metal binding pocket of mPDF were examined. Analysis of the molecular dynamics results suggests that the overall fold of the mutant protein remains the same as that of the wild type. However, we observed few changes in the secondary structure of the mutant, and it became less stable than the wild type. The residue boundaries defining two β strands spanning residues 98–107 and residues 112–120 were found to have shifted to residues 101–106 and residues 112-118, respectively. The propensity of the turn conformation was decreased considerably for residues 107 and 108 near the catalytic site. Turn propensities were increased for residues 146–148 and 153–156. The positions and orientations of the metal binding residues (i.e. Gln-56, Cys-106, His-148, Glu-149, and His-152) in mPDF remained conserved both in terms of sequence and structure among different organisms. During dynamics, no significant changes were detected in the side chain conformations of residues forming the metal binding pocket.
FIGURE 5.
Comparison of r.m.s. deviations (RMSD) as a function of time for wild-type (black) and mutant (gray) mPDF.
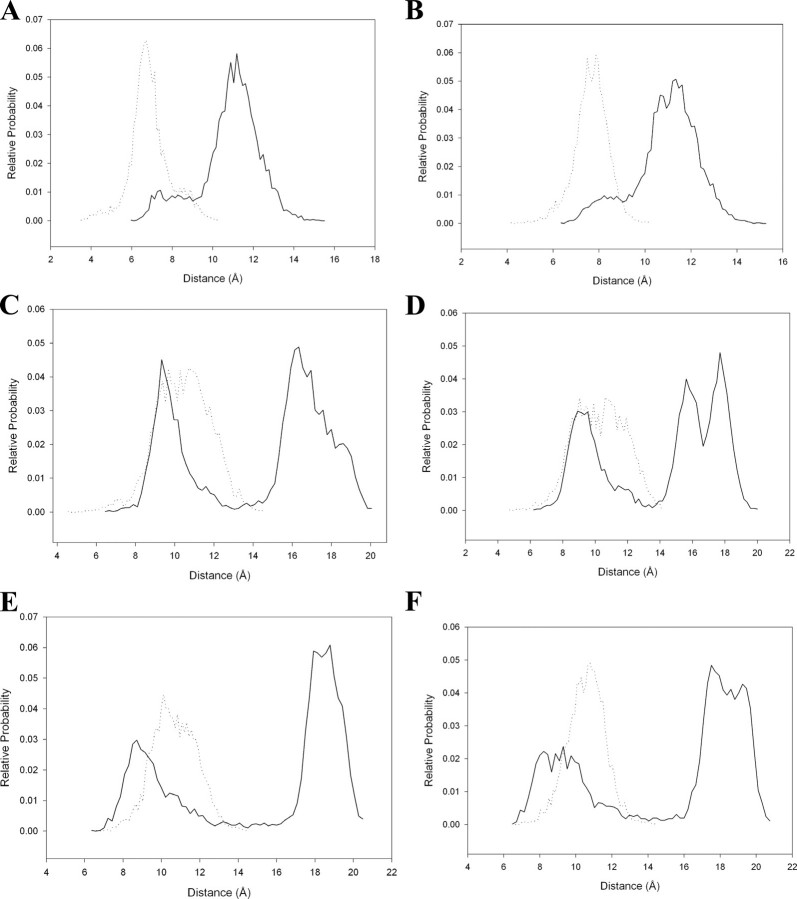
The modeled structure shows that amino acid residues Gly49, Val50, Gly51, Glu104, Gly105, Cys106, Leu107, Arg144, and Met145 lie along the perimeter of the peptide binding cavity. Strikingly, significant changes were observed in the distances between side chains of Arg144 and Leu107. Fig. 6 plots the distribution of distances between side chains of residues Arg144 and Leu107 along the dynamics trajectory. In the triple mutant structure, the average distances between the atoms of side chains of Arg144 and Leu107 were found to have decreased by more than 5 Å. Also, the average distance between side chain atoms of Met145 and Leu107 decreased by nearly 5Å in the mutant. Fig. 7 shows that the decreased distances between the side chain atoms of Arg144 and Leu107 and between Met145 and Leu107 reduce the space available for substrate binding in mutant mPDF (also see supplemental Fig. 2). In addition, the distribution of distances between the Cα atoms of Gly49 and Glu104, Gly105, and Cys106 along the trajectory shows that the average distances were changed by more than 1 Å. However, the distribution of distances shifted by 2 Å and does show more fluctuations in the mutant as compared with the wild type. Furthermore, distribution of distances between the side chains of Glu104 and the backbone oxygen atom (O) of Gly49 shows that the average distance was increased by 2 Å. Similar trends are observed between Val50 and Glu104, Gly105, and Cys106 along the trajectory. The average distance between O of Gly51 and sulfur of Cys106 increased by 1.5 Å, accompanied by a change in the distance of Cα of Gly51 and Cys106 along the dynamics trajectory. On the other hand, minor changes were observed in the distances between Cα of Gly51 and Glu104, Gly105, and Leu107. These results indicate that the nondetectable activity observed in the R77K/R78K/R79K mutant of mPDF is due to structural alterations in the microenvironment of the substrate binding cavity of the mutant, caused by an “action-at-a-distance” mechanism.
FIGURE 6.
Relative probability plots for the distances observed during the course of dynamics between the atoms of wild type (solid line) and mutant (dashed line) mPDF. A, Cδ1 and Cε; B, Cδ2 and Cε of Leu107 and Met145, respectively; C, Cδ1 and NH1; D, Cδ2 and NH1; E, Cδ1 and NH2; F, Cδ2 and NH2 of Leu107 and Arg144, respectively.
FIGURE 7.
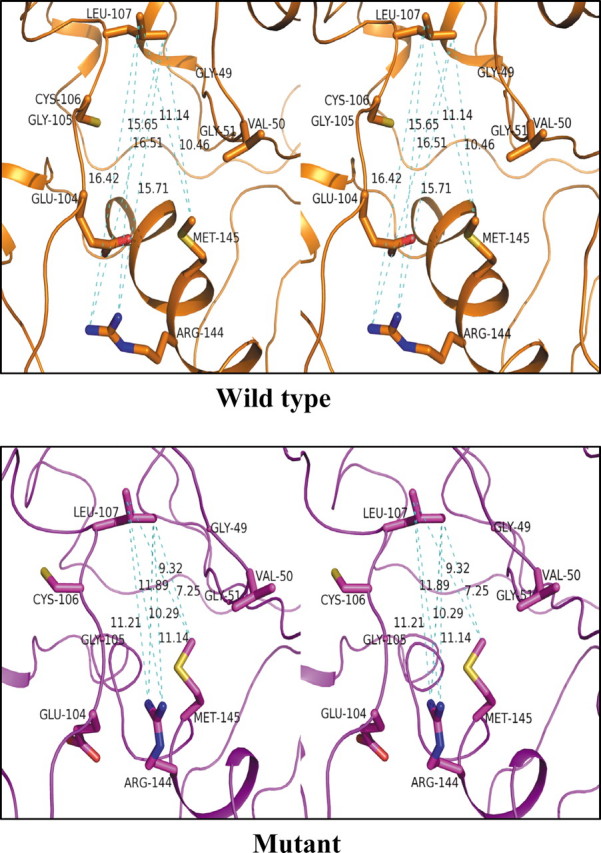
Stereoview of the substrate binding pocket residues of mPDF. The conformation of the substrate binding pocket residues of the averaged structure from simulations for the wild type and the mutant are shown in a stick model and labeled. Dashed lines (cyan) indicate the distance between the atoms in Å units. The figure was generated using PYMOL (29).
Antisense Oligonucleotides against mPDF Affect Mycobacterial Growth—The demonstrated essentiality of def genes in many pathogenic bacteria led to the identification of its protein product, PDF, as a promising drug target (13, 14). To confirm its essentiality in mycobacteria, we used 5′-phosphothiorate-modified antisense oligodeoxyribonucleotides (PS-ODN) against different regions of the gene (Table 2) and monitored their effect on bacterial growth in culture up to 24 h by recording the absorbance at 600 nm as well as by counting colony-forming units. In these experiments, we utilized M. smegmatis strain mc2155, a fast growing saprophyte, which has often been used as a model for genetic studies of M. tuberculosis (34). Comparison of def gene sequences between M. tuberculosis and M. smegmatis exhibited ∼93% homology (∼82% identity; also see supplemental Fig. 1).
TABLE 2.
Effect of PTO modified antisense oligonucleotide on mycobacterial growth
| Treatmenta | Antisense oligonucleotides against region of def gene (5′-3′) | Growth | Number of experiments |
|---|---|---|---|
| % | |||
| None | 100 | 6 | |
| PS-ODN-(34-39) | 100-117b | 101.33 ± 5.36 | 6 |
| PS-ODN-(74-83) | 220-249 | 27.19 ± 1.68 | 6 |
| PS-ODN-(104-111) | 310-333 | 32.70 ± 2.0 | 3 |
| PS-ODN-(147-154) | 439-462 | 31.20 ± 1.87 | 3 |
| PS-ODN-(188-197) | 565-594 | 73.59 ± 7.54 | 6 |
| PS-ODN-(S74-83) | 220-249c | 22.53 ± 1.37 | 3 |
| PS-ODN-(S188-197) | 565-594c | 64.43 ± 2.23 | 3 |
Numbers in parentheses indicate amino acids against antisense oligonucleotides designed.
Antisense oligonucleotides designed to incorporate mismatch between M. tuberculosis (Rv0429c) and M. smegmatis (0832) def gene sequences.
Antisense oligonucleotides designed based on the M. smegmatis def gene.
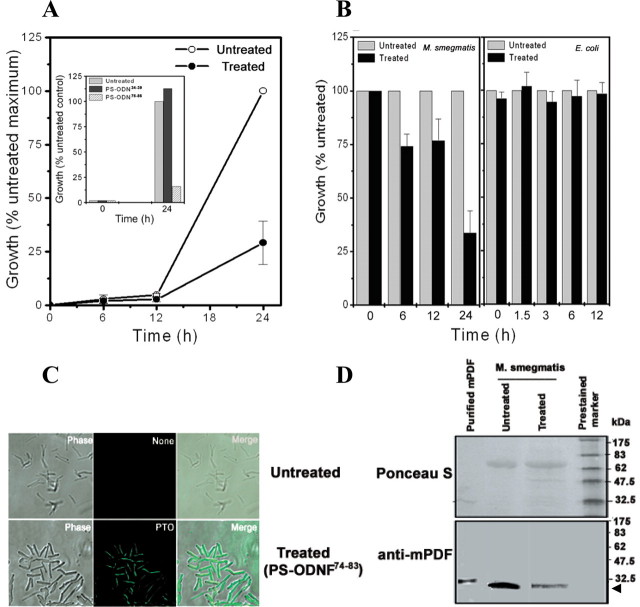
PS-ODNs directed against conserved motifs or insertion region of def inhibited bacterial growth considerably (Fig. 8, A and B (left), and Table 2). Treatment with PS-ODNs directed against the same region of the def gene from M. tuberculosis and M. smegmatis exhibited quite comparable growth reduction (Table 2; compare PS-ODN-(74–83) and PS-ODN-(S74–83) or PS-ODN-(188–197) and PS-ODN-(S188–197)). The results did not significantly differ from each other if only the 5′-end or all bases of PS-ODN were phosphothiorate-modified (compare Fig. 8A and the inset). Since PS-ODN-(74–83) was mycobacteria-specific, it had no effect on the growth profile of E. coli (Fig. 8B, right). Similarly, antisense oligonucleotides against the nonhomologous region of def genes of M. tuberculosis and M. smegmatis hardly affected bacterial growth (Fig. 8A, inset, and Table 2). To ensure that PS-ODN-(74–83) permeated into the M. smegmatis cells, treatment with a fluorescein-conjugated PS-ODN (PS-ODNF-(74–83); see supplemental Table 1) and monitoring of intracellular fluorescence on a confocal microscope were used to confirm entry of the antisense oligonucleotide into cells (Fig. 8C). To monitor the expression of the native PDF protein of M. smegmatis, cultures were grown either in the presence or absence of PS-ODN-(74–83) for 24 h. Following pelleting of cultures, lysates were prepared by resuspending cells (both treated and untreated) in 20 mm phosphate buffer, pH 7.4. The soluble fraction of cell lysates was then subjected to SDS-PAGE and Western blotting using polyclonal antibody against recombinant mPDF. Compared with the untreated control (see the Ponceau S-stained blot, which served as a loading control; Fig. 8D, top), significant reduction in the level of expression of endogenous PDF protein was noticed in M. smegmatis cells treated with PS-ODN-(74–83) (Fig. 8D, bottom). Interestingly, despite the role of the extended carboxyl terminal end in maintaining enzymatic activity (15), PS-ODNs against this region of PDFs from both M. smegmatis and M. tuberculosis affected bacterial growth only marginally (Table 2). However, this may not be surprising, considering that PS-ODNs directed against different regions have variable efficiency in inhibiting bacterial growth (35, 36). Thus, our study confirms that def is essential for survival of mycobacteria.
FIGURE 8.
PTO-modified antisense oligodeoxynucleotides against the insertion region of mPDF affect mycobacterial growth. M. smegmatis or E. coli K12 cultures were grown in the absence or presence of PS-ODNs (10 μm). A, effect of PS-ODN-(74–83) on the growth of M. smegmatis. A, inset, representative experiment showing the effect of PS-ODNAM-(76–85) (all bases PTO-modified) on M. smegmatis growth. B, effect of PS-ODN-(74–83) on the growth of E. coli. C, permeability of flourescein-conjugated PS-ODN-(74–83) within M. smegmatis cells. Culture was grown in the absence (top) or presence (bottom) of PS-ODNF-(74–83) for 24 h and visualized under a confocal microscope. D, expression of M. smegmatis native PDF in response to PS-ODN-(74–83) treatment. Soluble fractions of cell lysates were prepared from treated or untreated cells. Following the protein estimation, samples were resolved by 12% SDS-PAGE, transferred to nitrocellulose membrane, and subjected to Western blotting using anti-mPDF polyclonal antibody. A representative experiment shows Ponceau S staining of nitrocellulose membrane after transfer (top) and following development of Western blot using the ECL detection system (bottom). The reproducibility of the results was checked from three independent experiments. The arrowhead indicates the position of the specific PDF band. The numbers denote the molecular weight (in kDa) of the marker.
DISCUSSION
The PDF enzyme is responsible for the deformylation of N-formylmethionine of the nascent polypeptide chains in the cytoplasm of eubacteria (3–5, 11). Since amino-terminal peptidases are unable to utilize formylated peptides as substrates, the importance of PDF has been realized for a long time (1). In M. tuberculosis, we initiated our studies by characterizing mPDF (15, 16). The enzyme displayed many features similar to other bacterial PDFs. However, being an iron-containing enzyme, its insensitivity to oxidizing agents like H2O2 was notable.
We recently reported the importance of an insertion sequence and a carboxyl-terminal sequence in mPDF (relative to other PDFs) in determining its activity (15, 16), this feature being distinct from the PDFs of other bacteria (7, 37). In this study, we carried out structure-function analyses of mPDF to confirm the identities of crucial amino acids in the insertion region that modulate the enzymatic function of this protein. Studies with different deletion mutants indicated that nine (RRRGVVINP; residues 77–85) of the 12 amino acids (residues 74–85) spanning the insertion region of mPDF are necessary for the deformylase activity (Fig. 1).
Sequence analysis of known iron-containing PDFs revealed that three consecutive arginines in the insertion region of mPDF are typical of different mycobacterial species (Fig. 1A). Our results indicate that alterations in these arginines of mPDF affect the enzyme turnover rates (Table 1). Activity of mPDF is more sensitive to triple mutations (R77K/R78K/R79K) as compared with individual mutations (Fig. 2 and Table 1). Far-UV CD spectra of the individual mutants (R77K, R78K, R79K, and R77K/R78K/R79K) and of the triple mutant showed that distinct structural changes have occurred due to these mutations in relation to the structure of wild-type mPDF (Fig. 3).
We further obtained structural insights of mPDF by predicting its three-dimensional structure. The modeled mPDF shows conserved core and variable loop regions in comparison with other PDF structures. Superposition of mPDF and ePDF structures reveals a conserved pattern of side chain orientation of all residues surrounding the metal ion and the substrate binding pockets. The catalytic site and the three arginines (Arg77– Arg79) of mPDF are distal to each other, and the three arginines are fully exposed to the solvent and not buried within the structure (Fig. 4). Compared with wild-type mPDF, molecular dynamics simulations of the triple mutant (R77K/R78K/R79K) reveal variations in the geometry of residues in the substrate binding pocket (Fig. 6), suggesting that the region containing the arginines has some sort of an action-at-a-distance effect on the active site from which it is physically removed. The details of this effect are as follows. Side chains of the residues Arg144 and Met145 in the binding pocket occupy larger area of binding space available to the substrate in the triple mutant in comparison with wild-type mPDF (Fig. 7; see supplemental Fig. 2). Also, larger fluctuations are observed in the distances between various residues (of segments 49–50 and 104–107) lining the binding pocket of the triple mutant, suggesting that it has become flexible and may not be in an optimal orientation for activity as compared with the pocket of wild-type mPDF. In contrast, the side chains of amino acid residues interacting with the metal atom do not show significant variations during the course of molecular dynamics simulations in the triple mutant, regardless of whether or not the metal atom is included in the simulations. Thus, in the mutant, the reduced space available for substrate, increase in flexibility of the binding pocket residues, and destabilization of the secondary structure may likely hinder the proper interaction between substrate and chemical moieties of the mPDF. This is wholly in consonance with our experimental finding that the triple mutant shows no activity (Table 2). Further examination of the structure of mPDF shows that the loop connected to the segment (containing Arg77– Arg79) interacts with the helix containing residues Arg144 and Met145. Thus, the mutations altering Arg77–Arg79 to Lys77–Lys79 very likely influence the interaction of the loop with the helix with enzyme activity affected in an action-at-a-distance mode.
The protein products of essential genes in pathogens are known to be ideal drug target candidates. The available literature indicates that PDF enzyme activity is essential for the survival of different bacteria (9, 12–14). To explore the essentiality of the gene in mycobacteria, we examined M. smegmatis growth following treatment with antisense PS-ODNs designed against different regions of the M. tuberculosis def gene. Interestingly, PS-ODNs directed against conserved regions of def (PS-ODN-(104–111) and PS-ODN-(147–154)) and also the insertion sequences (PS-ODN-(74–83)) significantly inhibited the bacterial growth (Table 2 and Fig. 8, A and B) while reducing the level of expression of the native PDF protein of M. smegmatis (Fig. 8D) compared with the untreated control. To validate the authenticity of our results, the effects of PS-ODNs (PS-ODN-(74–83) as opposed to PS-ODN-(S74–83) and PS-ODN-(188–197) as opposed to PS-ODN-(S188–197)) directed against the same region of def genes from both M. tuberculosis and M. smegmatis were compared. As expected, the results obtained from this experiment were quite comparable (Table 2). Furthermore, PS-ODN-(34–39) designed against the nonhomologous region of def genes of M. tuberculosis and M. smegmatis did not affect bacterial growth (Fig. 8A, inset, and Table 2). We previously reported the role of the long carboxyl-terminal end of mPDF in maintaining its enzymatic activity (15, 16). However, the PS-ODN-(188–197) or PS-ODN-(S188–197), which were designed against this region of def, marginally affected the mycobacterial growth (Table 2). This does not seem to be unusual, since the efficacy of an antisense construct depends on its targeting efficiency, which could be influenced by a number of factors including the secondary structure of the target sequence (35). Thus, it is apparent that the def is indispensable in mycobacteria, and our findings fit in well with a recent report in which a conditional gene knockout approach showed the essentiality of this gene in M. bovis BCG (38).
A large number of inhibitors of bacterial PDF have been synthesized to date, and a few of them are under clinical trials (14). Several anti-PDF compounds have been shown to inhibit growth of M. tuberculosis in culture (38, 39), and they are reported to be nontoxic to human cell lines (40). However, the presence of PDF enzymes in eukaryotic organelles (plastids/mitochondria), including human mitochondrion (40–43), raises a pertinent question regarding its use as a drug target. Despite the fact that N-formylation is not involved in cytoplasmic protein synthesis in eukaryotes, the presence of this enzyme in organelles raises doubts about its usefulness as a drug target, pending identification of inhibitors of high specificity that would affect only the mycobacterial PDF and not the PDF in the eukaryotic cell organelles. For this, it is necessary to explore a variety of inhibitors, preferably binding to different regions of the PDF fold. The results we present here hold out the potential of designing inhibitors in the future that can bind to the region containing the three arginines and modulate PDF activity, since these three arginines are not present in the organelle PDFs. It is conceivable that synthetic peptides obtained through rational design or screening of a peptide library against the insertion sequence would provide candidate inhibitory agents for mycobacteria, since this region is absent in human def and even in other prokaryotic/eukaryotic PDFs so far characterized. A similar approach involving activity modulation through binding at a distance has already been tried for inhibitors of HIV-1 protease (44). Our results bring mPDF into sharp focus in view of the fact that drug resistance in M. tuberculosis has emerged as a great threat to public health causing millions of deaths (45) worldwide, and to overcome the situation, we need to have new/novel drug intervention strategies.
Supplementary Material
Acknowledgments
We acknowledge the receipt of E. coli strain K12 (MTCC 1302) from the Microbial Type Culture Collection (Chandigarh, India). We are grateful to Dr. P. Guptasarma for CD spectroscopy as well as data interpretation and also for critical reading of the manuscript. We are indebted to Dr. Alok Mondal for confocal microscopic studies. We are thankful to J. Prasad for providing excellent technical assistance.
This work was supported by Council of Scientific and Industrial Research, New Delhi (under network program SMM003) and Department of Biotechnology (New Delhi, India) and financial support in the form of research fellowships (to R. S., P. K., H. H. D., and M. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
Footnotes
The abbreviations used are: PDF, peptide deformylase; ePDF, E. coli peptide deformylase; mPDF, M. tuberculosis peptide deformylase; IR, insertion region; PS-ODN, phosphothiorate-modified antisense oligonucleotide; r.m.s., root mean square.
References
- 1.Kozak, M. (1983) Microbiol. Rev. 471 –45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meinnel, T., and Blanquet, S. (1993) J. Bacteriol. 1757737 –7740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giglione, C., Perrie, M., and Meinnel, T. (2000) Mol. Microbiol. 361197 –1205 [DOI] [PubMed] [Google Scholar]
- 4.Meinnel, T., Blanquet, S., and Mechulam, Y. (1993) Biochimie. (Paris) 751061 –1075 [DOI] [PubMed] [Google Scholar]
- 5.Giglione, C., Vallon, O., and Meinnel, T. (2003) EMBO J. 2213 –23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajagopalan, P. T., Datta, A., and Pei, D. (1997) Biochemistry 3613910 –13918 [DOI] [PubMed] [Google Scholar]
- 7.Li, Y., Chen, Z., and Gong, W. (2002) Biochem. Biophys. Res. Commun. 295884 –889 [DOI] [PubMed] [Google Scholar]
- 8.Zhou, Z., Song, X., Li, Y., and Gong, W. (2004) J. Mol. Biol. 339207 –221 [DOI] [PubMed] [Google Scholar]
- 9.Mazel, D., Coic, E., Blanchard, S., Saurin, W., and Marliere, P. (1997) J. Mol. Biol. 266939 –949 [DOI] [PubMed] [Google Scholar]
- 10.Meinnel, T., and Blanquet, S. (1994) J. Bacteriol. 1767387 –7390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazel, D., Pochet, S., and Marliere, P. (1994) EMBO J. 13914 –923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margolis, P. S., Hackbarth, C. J., Young, D. C., Wang, W., Chen, D., Yuan, Z., White, R., and Trias, J. (2000) Antimicrob. Agents Chemother. 441825 –1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan, Z., and White, R. J. (2006) Biochem. Pharmacol. 71042 –1047 [DOI] [PubMed] [Google Scholar]
- 14.Leeds, J. A., and Dean, C. R. (2006) Curr. Opin. Pharmacol. 6445 –452 [DOI] [PubMed] [Google Scholar]
- 15.Saxena, R., and Chakraborti, P. K. (2005) Biohem. Biophys. Res. Commun. 332418 –425 [DOI] [PubMed] [Google Scholar]
- 16.Saxena, R., and Chakraborti, P. K. (2005) J. Bacteriol. 1878216 –8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajagopalan, P. T., and Pei, D. (1998) J. Biol. Chem. 27322305 –22310 [DOI] [PubMed] [Google Scholar]
- 18.Chaba, R., Raje, M., and Chakraborti, P. K. (2002) Eur. J. Biochem. 2691078 –1085 [DOI] [PubMed] [Google Scholar]
- 19.Shirley, K., Sheng, Y. C., John, J. S., and Alice, W. (1995) in PCR Primer: A Laboratory Manual (Dieffenbach, C. W., Dveksler, G. S., eds.), pp143 –155, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 20.Landt, O., Grunert, H. P., and Hahn, U. (1990) Gene 96125 –128 [DOI] [PubMed] [Google Scholar]
- 21.Sarin, J., Aggarwal, S., Chaba, R., Varshney, G. C., and Chakraborti, P. K. (2001) J. Biol. Chem. 27644590 –44597 [DOI] [PubMed] [Google Scholar]
- 22.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhant, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 253389 –3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997) Nucleic Acids Res. 254876 –4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chivian, D., Kim, D. E., Malmstrom, L., Bradley, P., Robertson, T., Murphy, P., Strauss, C. E., Bonneau, R., Rohl, C. A., and Baker, D. (2003) Proteins 53524 –533 [DOI] [PubMed] [Google Scholar]
- 25.Kreusch, A., Spraggon, G., Lee, C. C., Klock, H., McMullan, D., Ng, K., Shin, T., Vincent, J., Warner, I., Ericson, C., and Lesley, S. A. (2003) J. Mol. Biol. 330309 –321 [DOI] [PubMed] [Google Scholar]
- 26.Laskowski, R. A., MacArthur, M., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Cryst. 26283 –291 [Google Scholar]
- 27.Case, D. A., Cheatham, T. E., 3rd, Darden, T., Gohlke, H., Luo, R., Merz, K. M., Jr., Onufriev, A., Simmerling, C., Wang, B., and Woods, R. J. (2005) J. Comput. Chem. 261668 –1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan, Y., Wu, C., Chowdhury, S., Lee, M. C., Xiong, G., Zhang, W., Yang, R., Cieplak, P., Luo, R., and Lee, T. (2003) J. Comput. Chem. 241999 –2012 [DOI] [PubMed] [Google Scholar]
- 29.DeLano, W. L. (1998) The PyMOL molecular graphics system. DeLano Scientific LLC, San Carlos, CA. (available at: http://www.pymol.org.)
- 30.Ryckaert, J. P., Ciccotti, G., and Berendsen, H. J. C. (1977) J. Comput. Phy. 23327 –341 [Google Scholar]
- 31.Vullo, A., Bortolami, O., Pollastri, G., and Tosatto, S. (2006) Nucleic Acids Res. 34W164 –W168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker, A., Schlichting, I., Kabsch, W., Groche, D., Schultz, S., and Wagner, A. F. (1998) Nat. Struct. Biol. 51053 –1058 [DOI] [PubMed] [Google Scholar]
- 33.Smith, K. J., Petit, C. M., Aubart, K., Smyth, M., McManus, E., Jones, J., Fosberry, A., Lewis, C., Lonetto, M., and Christensen, S. B. (2003) Protein Science 12349 –360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flint, J. L., Kowalski, J. C., Karnati, P. K., and Derbyshire, K. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 10112598 –12603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harth, G., Zamecnik, P. C., Tang, J. Y., Tabatadze, D., and Horwitz, M. A. (2000) Proc. Natl. Acad. Sci. U. S. A. 97418 –423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harth, G., Horwitz, M. A., Tabatadze, D., and Zamecnik, P. C. (2002) Proc. Natl. Acad. Sci. U. S. A. 9915614 –15619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meinnel, T., Lazennec, C., Dardel, F., Schmitter, J. M., and Blanquet, S. (1996) FEBS Lett. 38591 –95 [DOI] [PubMed] [Google Scholar]
- 38.Teo, J. W., Thayalan, P., Beer, D., Yap, A. S., Nanjundappa, M., Ngew, X., Duraiswamy, J., Liung, S., Dartois, V., Schreiber, M., Hasan, S., Cynamon, M., Ryder, N. S., Yang, X., Weidmann, B., Bracken, K., Dick, T., and Mukherjee, K. (2006) Antimicrob. Agents Chemother. 503665 –3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cynamon, M. H., Alvirez-Freites, E., and Yeo, A. E. (2004) J. Antimicrob. Chemother. 53403 –405 [DOI] [PubMed] [Google Scholar]
- 40.Nguyen, K. T., Hu, X., Colton, C., Chakrabarti, R., Zhu, M.X., and Pei, D. (2003) Biochemistry 429952 –9958 [DOI] [PubMed] [Google Scholar]
- 41.Giglione, C., and Meinnel, T. (2001) Trends Plant Sci. 6566 –572 [DOI] [PubMed] [Google Scholar]
- 42.Giglione, C., Boularot, A., and Meinnel, T. (2004) Cell Mol. Life Sci. 611455 –1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fieulaine, S., Juillan-Binard, C., Serero, A., Dardel, F., Giglione, C., Meinnel, T., and Ferrer, J. L. (2005) J. Biol. Chem. 28042315 –42324 [DOI] [PubMed] [Google Scholar]
- 44.Boggetto, N., and Reboud-Ravaux, M. (2002) Biol. Chem. 3831321 –1324 [DOI] [PubMed] [Google Scholar]
- 45.Ginsberg, A. M., and Sigelman, M. (2007) Nat. Med. 13290 –294 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.