Abstract
Halobacterium salinarum sensory rhodopsin I (HsSRI), a dual receptor regulating both negative and positive phototaxis in haloarchaea, transmits light signals through changes in protein-protein interactions with its transducer, halobacterial transducer protein I (HtrI). Haloarchaea also have another sensor pigment, sensory rhodopsin II (SRII), which functions as a receptor regulating negative phototaxis. Compared with HsSRI, the signal relay mechanism of SRII is well characterized because SRII from Natronomonus pharaonis (NpSRII) is much more stable than HsSRI and HsSRII, especially in dilute salt solutions and is much more resistant to detergents. Two genes encoding SRI homologs were identified from the genome sequence of the eubacterium Salinibacter ruber. Those sequences are distantly related to HsSRI (∼40% identity) and contain most of the amino acid residues identified as necessary for its function. To determine whether those genes encode functional protein(s), we cloned and expressed them in Escherichia coli. One of them (SrSRI) was expressed well as a recombinant protein having all-trans retinal as a chromophore. UV-Vis, low-temperature UV-Vis, pH-titration, and flash photolysis experiments revealed that the photochemical properties of SrSRI are similar to those of HsSRI. In addition to the expression system, the high stability of SrSRI makes it possible to prepare large amounts of protein and enables studies of mutant proteins that will allow new approaches to investigate the photosignaling process of SRI-HtrI.
Microorganisms are subjected to a variety of environmental stimuli to which they respond and adapt. Sensory rhodopsin I (SRI),2 a photoactive membrane-embedded retinylidene protein consisting of seven transmembrane helices, functions as a receptor regulating both negative and positive phototaxis in the archaeon Halobacterium salinarum (1, 2). The original state of SRI and its long-lived photointermediate (the M-intermediate) are important for positive and negative phototaxis, respectively, and have absorption maxima at 587 and 373 nm, respectively. Sensory rhodopsin II (SRII, also known as phoborhodopsin) is a negative phototaxis receptor in haloarchaea, including H. salinarum and Natronomonas pharaonis. Those are called HsSRII and NpSRII, respectively, and their absorption maxima are at about 500 nm (3). Thus, haloarchaea are attracted to light with wavelengths longer than 520 nm, and they avoid light with wavelengths shorter than 520 nm because of the functions of SRI and SRII (2). Light >520 nm can activate the ion pumping rhodopsins, bacteriorhodopsin (BR), and halorhodopsin (HR), to obtain light energy, and cells avoid light of shorter wavelengths that contain harmful near-UV radiation (2).
HsSRI and NpSRII form 2:2 signaling complexes with their cognate halobacterial transducer proteins (Htr), HsHtrI and NpHtrII, in membranes, and transmit light signals through changes in protein-protein interactions (4). The excitation light absorbed by HsSRI and NpSRII triggers trans-cis isomerization of the retinal chromophore that is covalently bound to a conserved lysine residue via a protonated Schiff base linkage (5, 6). This photoexcitation results in the sequential appearance of various photointermediates (K, L, M, and O) followed by a return to the unphotolyzed form of the protein (2, 7). The linear cyclic photochemical reaction is referred to as the photocycle.
In the dark (original) state of NpSRII, it has been reported that two specific hydrogen bonds between NpSRII and NpHtrII (8, 9), the deprotonation of Asp-193NpSRII (10) and the HAMP domain of NpHtrII (11), are important for their interactions. Upon formation of the K-intermediate, light activates the photoisomerization of retinal in NpSRII, which leads to steric constraint between Thr-204NpSRII and C14-H in the retinal chromophore (12, 13) and to alteration of the O-H stretching vibration of Thr-204 (14, 15), as revealed by phototaxis and FTIR experiments. Upon formation of the active M-intermediate, helix-F of NpSRII moves toward NpHtrII, as revealed by electron paramagnetic resonance spectroscopy (16) and chemical modification (17), which leads to helix rotation and/or helix movements of NpHtrII (18). At its distal end, activated NpHtrII binds a cytoplasmic histidine kinase, CheA. CheA phosphorylates a phosphoregulator (CheY) that binds to the flagellar motor switch and controls the swimming reversal probability of the cell (19, 20). Thus, compared with HsSRI, the signal relay mechanism of NpSRII is well characterized by various methods (21), because NpSRII is much more stable than H. salinarum SRII (22), especially in dilute salt solutions, and is much more resistant to detergents whereas HsSRI is unstable under those conditions. Therefore, little is known about the molecular mechanism(s) of interactions between SRI and HtrI, about structural changes, or about the signal relay mechanism of positive phototaxis.
New genes encoding the sensory rhodopsin I-like protein have been recently discovered through genome sequencing of S. ruber (23). We present here the first characterization of SRI from the eubacterium S. ruber (SrSRI). SrSRI has all-trans retinal as a chromophore, has a longer absorption maximum (558 nm) than does SRII (500 nm), and has a slower photocycle than the light-driven ion-pumping rhodopsins (BR, HR, and proteorhodopsin), indicating similarities to the prototypic SRI (HsSRI) (1). SrSRI could be expressed in Escherichia coli and shows very high stability, even in detergent micelles, making it possible to prepare large amounts of protein. This also allows preparation of mutant SrSRI, which will allow new approaches to investigate the photosignaling process in the SRI-HtrI system.
EXPERIMENTAL PROCEDURES
Plasmids and Strains—S. ruber (a kind gift from Dr. Dyall-Smith) was grown aerobically at 40 °C at pH 7.0 in medium with the following composition (g/liter): NaCl, 195; MgSO4·7H2O, 25; MgCl2·6H2O, 16.3; CaCl2·2H2O, 1.25; KCl, 5.0; NaHCO3, 0.25; NaBr, 0.625; and yeast extract (Difco), 1.0. Cells were harvested by centrifugation and were stored at –80 °C. Genomic DNA was prepared using the method of Marmur (24). E. coli DH5α was used as a host for DNA manipulation, and BL21(DE3) was used for expressing the genes. For DNA manipulation, the sensory rhodopsin I gene (SRU_2511) was amplified using PCR from the genomic DNA of S. ruber. The forward primer 5′-CATATGGACCCGATTACGATCGTCTA-3′ and the reverse primer 5′-CTCGAGGGCGGCAGACGTGACTGTCGC-3′ were designed (underlining indicates the restriction sites for NdeI and XhoI, respectively). The stop codon was deleted during amplification. The NdeI and XhoI fragment was ligated to NdeI and XhoI sites of pET21c(+) vector (Novagen, Madison, WI). Consequently, the plasmid encodes six histidines at the C terminus, and it was named pTK001. This cloning strategy resulted in the following N- and C-terminal peptide sequences: 1MDPI----TSAA239LEHHHHHH. The constructed plasmid was analyzed using an automated sequencer to confirm the expected nucleotide sequence. Another SRI gene, SRU_2579, was amplified and cloned using almost the same strategy as SRU_2511, except for the use of the restriction site NheI instead of NdeI.
Protein Expression, Purification, and Reconstitution into PG Liposomes—Cells were grown in LB medium supplemented with ampicillin (final concentration of 100 μg/ml). E. coli BL21(DE3) harboring pTK001 was grown to an A660 of 0.3–0.5 in a 30 °C incubator, followed by the addition of 0.5 mm isopropyl-1-thio-β-d-galactopyranoside and 10 μm all-trans retinal. Cells were harvested at 3-h postinduction by centrifugation at 4 °C, resuspended in buffer (50 mm MES, pH 6.5) containing 4 m NaCl, and disrupted by sonication. Cell debris was removed by a low speed centrifugation (5,000 × g, 10 min, 4 °C). For solubilization of the membranes, 1% (w/v) n-dodecyl-β-d-maltoside (DM) was added, and the suspension was incubated overnight at 4 °C. The solubilized membranes were isolated by high speed centrifugation (100,000 × g, 30 min, 4 °C), and the supernatant was incubated with Ni-nitrilotriacetic acid (NTA) superflow (Qiagen, Valencia, CA) for 30 min. Thereafter, the Ni-NTA resin was poured into a chromatography column and was washed extensively with buffer (50 mm MES, pH 6.0) containing 4 m NaCl, 20 mm imidazole, and 0.1% w/v DM to remove unspecifically bound proteins. The histidine-tagged proteins were then eluted with buffer (0.1% DM, 4 m NaCl, 50 mm MES, pH 6.0, 300 mm imidazole). The purified SrSRI sample was then reconstituted into PG liposomes. For reconstitution of l-α-phosphatidylglycerol (Sigma) lipids, the lipids were dissolved in chloroform, and a thin lipid film on the wall of a flask was prepared by careful evaporation of the solvent. SrSRI in buffer (0.1% DM, 1 m NaCl, 10 mm MES, pH 6.0, 100 mm imidazole) was added (at an SrSRI:PG molar ratio of 1:50), and the suspension was gently stirred for 30 min at room temperature. Reconstitution by detergent removal was performed using Bio-Beads SM-2 (Bio-Rad), as previously described (25).
The expression plasmid of HsSRI was constructed essentially as described previously (9). HsSRI, with a tag of six histidines at the C-terminal end, was expressed in E. coli BL21(DE3). Preparation of crude membranes and purification of protein were done as described previously (25).
Room Temperature and Low Temperature UV-Vis Spectroscopy and HPLC Analysis—Room temperature UV-Vis spectra were obtained using a UV2400PC spectrophotometer with an ISR240A integrating sphere (Shimadzu, Kyoto, Japan). The PG-reconstituted sample was washed and resuspended in six buffers (citric acid, MES, HEPES, MOPS, CHES, and CAPS, whose concentrations were 10 mm each) with 1 m NaCl. The buffer composition had the same buffer capacity over a wide range of pH values (2–9). pH values of the sample suspensions were measured using an F-55 pH-meter with a 9669-C10 glass electrode (HORIBA, Kyoto, Japan).
Low-temperature UV-Vis spectra were measured using a V-550DS (JASCO, Tokyo, Japan) spectrophotometer with a cryostat (Optistat DN, Oxford, UK) and a temperature controller (ITC 502, Oxford, UK) with liquid nitrogen as the coolant, as described previously (26). The PG-reconstituted sample with 5 mm NaCl and 2 mm phosphate buffer (pH 7.0) was placed on a BaF2 window, and then was dried to a film under reduced pressure with an aspirator. The films were hydrated with water. SrSRIK minus SrSRI difference spectra were measured at 77 K, as follows: Illumination of the SrSRI film with >450 nm light for 2 min, after which at pH 7.0 and 77 K converted SrSRI to SrSRIK, and subsequent illumination with >640 nm light for 1 min forced SrSRIK to revert to SrSRI. SrSRIM minus SrSRI difference spectra were measured at 200 K and pH 7.0 as follows: To convert SrSRI to SrSRIM, the sample was irradiated with >480 nm light for 2 min, after which a subsequent 1-min illumination with UV light-changed SrSRIM back to SrSRI. The difference spectrum was calculated from the spectra before and after the illumination.
High performance liquid chromatography (HPLC) analysis was performed as described previously (27). The PG-reconstituted sample was analyzed in buffer (1 m NaCl, 50 mm Tris-Cl, pH 7.0). The HPLC system consisted of a PU-2080 pump and a UV-2075 UV/Vis detector (JASCO). The chromatograph was equipped with a silica column (6.0 × 150 mm; YMC-Pack SIL), the solvent was 12% (v/v) ethyl acetate and 0.12% (v/v) ethanol in hexane, and the flow rate was 1.0 ml/min. Extraction of retinal oxime from the sample was carried out by hexane after denaturation by methanol and 500 mm hydroxylamine at 4 °C. The molar composition of retinal isomers was calculated from the areas of the peaks monitored at 360 nm. Assignment of each peak was performed by comparing it with the HPLC pattern from retinal oximes of authentic all-trans- and 13-cis-retinals. For the light-adapted SrSRI, the sample was irradiated with >480 nm light for 1 min. Two independent measurements were averaged.
Time-resolved Laser Spectroscopy—The purified sample in DM detergent micelles was resuspended in 50 mm Tris-HCl, pH 7.0, 0.1% DM, and 1 m NaCl. Flash-induced absorption changes in the 300–700-nm region in the millisecond to second time window with a 10-ms interval were acquired using a commercial flash photolysis system (Hamamatsu Photonics K. K., Hamamatsu, Japan), which consisted of a CCD linear detector (Photonic Multichannel Spectral Analyzer PMA-11 C8808–01), a CW xenon lamp L8004 as a light source and a sample room (flash photolysis optics C9125). Excitation of the SrSRI samples was done by 556-nm nanosecond laser pulses (FWHM; 3.5 nm) from a Nd-YAG laser apparatus (LS-2134UT-10; LOTIS TII: 355 nm, 7 ns, 60 mJ) through an Optical Parametric Oscillator (OPO) (LT-2214-OPO/PM; LOTIS TII). The repetition rate of the laser pulses was 10 Hz and was reduced to 0.5 Hz using a beam shutter unit (BD-10MN-OPO; LOTIS TII) and a frequency divider to wait for completion of the photocycle of SrSRI. The energy of one laser pulse is 3.8 mJ. Synchronization of data acquisition and laser triggering was performed using a delay pulse generator (DG535; Stanford Research Systems, Location Sunnyvale, CA). For signal-to-noise improvement, twenty photoreactions were averaged for one time slice and 3 sets of time-resolved light-induced difference spectra were recorded and averaged for a sample solution (experiment length of ∼6 h). Two experiments were performed and in all, 6 sets of transient spectra were averaged. The bleach of the sample was negligible (∼5%) in each experiment. The temperature of the sample was kept at 20 °C by circulating water through a thermoregulated bath (NCB-1200; EYELA, Tokyo, Japan).
Protein Stability in Low Salt Conditions—Absorption spectra were obtained using a UV-2550 (Shimadzu) spectrometer. To monitor the effects of salt concentration on the absorbance of SRI, PG-reconstituted samples were resuspended in a buffer containing 50 mm Tris-Cl, pH 7.0. Samples were taken at different time intervals, and the absorption spectra were measured.
RESULTS AND DISCUSSION
Two Sensory Rhodopsin I-like Proteins in the Genome of S. ruber—The genome sequence of S. ruber contains two genes coding for sensory rhodopsin I (SRU_2511, which are named SrSRI, and SRU_2579). To date, these are the only examples (other than sensory proteorhodopsin from γ-proteobacteria) of an SRI gene discovered outside the Archaea (28). However, the properties of sensory proteorhodopsin have not been identified yet, because it is not expressed in E. coli, Pichia pastoris, or H. salinarum. The sequences of SRU_2511(SrSRI) and SRU_2579 are distantly related to HsSRI (∼40% identity) (29) and contain most of the amino acid residues (Asn-165, His-166, Asp-201, Arg-215) identified as necessary for function (30–33) (Fig. 1), except for the equivalent of Tyr-163 in SRU_2511. Fig. 1 shows the secondary structure of SRI-like proteins from S. ruber. It is well known that the sensory rhodopsins have seven transmembrane domains and that retinal is the chromophore. A lysine (Lys-205) and an aspartate residue (Asp-72), predicted to be necessary for stabilization of the Schiff base and for the binding between protein and retinal, are completely conserved among SRIs (Fig. 1). Unexpectedly, His-166, one of the most important residues for active site proton transfer and phototaxis signaling (31) in HsSRI is replaced by Tyr in one of the sensory rhodopsin I-like proteins from S. ruber (SRU_2511). These genes, SRU_2511 and SRU_2579, which have a downstream sequence are immediately followed by the same second gene in a probable operon that is under the control of the same promoter as rhodopsin. These putative transducer genes (SRU_2510, Htr for SRU_2511, and SRU_2580, Htr for SRU_2579) encode a ∼650-residue protein with two transmembrane domains at the N-terminal portion followed by an extensive domain with primarily hydrophilic residues. This topology is similar to that of the Htr transducers from the archaeon H. salinarum (34). The eubacterium, S. ruber, also has two light-driven ion pumps, Xanthorhodopsin and a Halorhodopsin-like protein, making an energy source for living cells (23, 35). The sensory rhodopsins would function as a positive phototaxis sensor against longer wavelengths of light (where ion pumping rhodopsins can utilize light energy) and as a negative phototaxis sensor against shorter wavelengths of light (which contains harmful UV).
FIGURE 1.
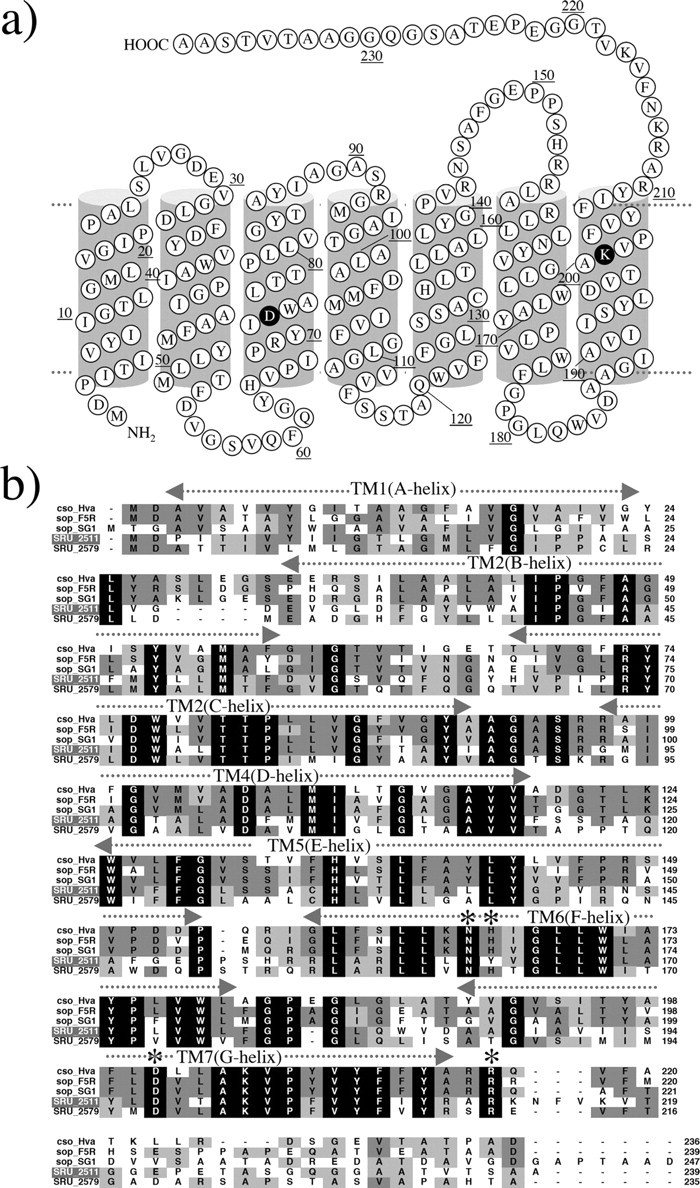
a, secondary structure of S. ruber sensory rhodopsin I (SrSRI, SRU_2511). b, alignment of putative amino acid sequences of SRIs reported so far (29) with SrSRI. Names of archaeal and bacterial sensory rhodopsins and sources from top are: cso_Hva, Haloarcula vallismortis cruxsensory rhodopsin (GenBank™ code: D83748); sop-F5R, H. salinarum strain Flx5R sR (L05603); sop-SGl, Hb. sp. strain SGI sR (X70290); SRU_2511, S. ruber strain DSM 13855 (3852586); SRU_2579, S. ruber strain DSM 13855 (3852268). Amino acid residues marked by an asterisk in helices F and G are important residues for phototaxis in SRI. The amino acid residues identified as necessary for function (31) were conserved among them except for a His residue (His-166 in HsSRI).
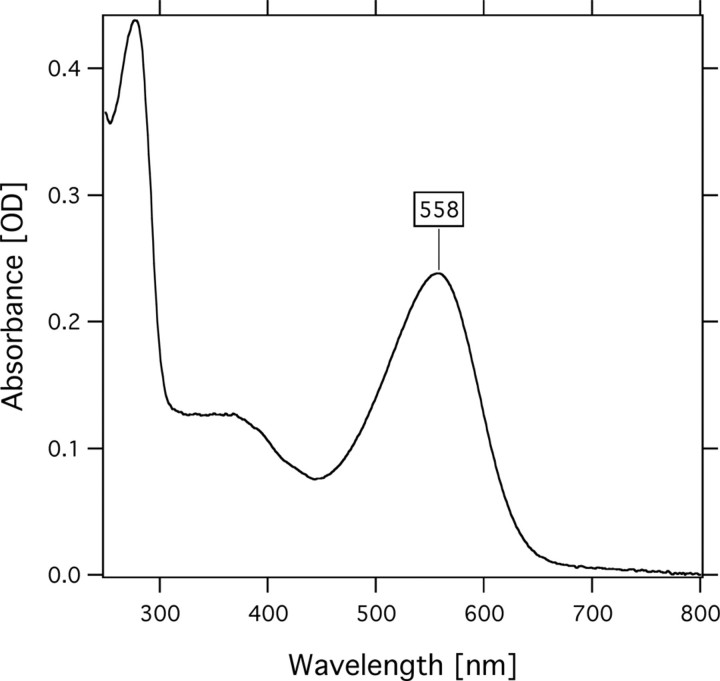
Protein Expression, Purification, and Absorption Spectrum—To determine whether the genes encode functional proteins, we cloned and expressed the two sensory rhodopsins, SRU_2511 and SRU_2579, in E. coli BL21(DE3) cells. Using PCR, we obtained a fragment containing the full-length of the SRU_2511 and SRU_2579 gene coding regions. An NdeI site was created at the translation initiation site in the PCR product using an oligonucleotide primer containing the NdeI site. A XhoI site was created, and the stop codon was deleted during the amplification. The NdeI and XhoI fragment of the PCR product was cloned into the NdeI and XhoI sites of pET21c(+). Induction of E. coli BL21(DE3) with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside caused growth inhibition, suggesting they expressed the desired protein. Actually, upon addition of retinal, a color change of the SRU_2511-transformed cells was recognized, and the color was definitely distinct from cells transformed only by the vector pET21c(+), whereas cells containing SRU_2579 did not show any visible color, implying it was not expressed in the E. coli cells. SRU_2511 is hereafter called SrSRI. Purified SrSRI was reconstituted into PG liposomes, and the absorption spectrum was obtained as shown in Fig. 2. The absorption maximum of SrSRI is located at 558 nm. That maximum wavelength is similar to that of HsSRI (587 nm) (1) and is significantly different from the sensory rhodopsin II whose maximum is located at around 500 nm (36, 37). Thus, we were able to express a sensory rhodopsin I-like protein as a photoactive protein.
FIGURE 2.
The absorption spectrum of SrSRI. The protein was reconstituted into PG liposomes and was resuspended in buffer solution (50 mm Tris-HCl, pH 7.0, 1 m NaCl). The temperature was kept at 20 °C.
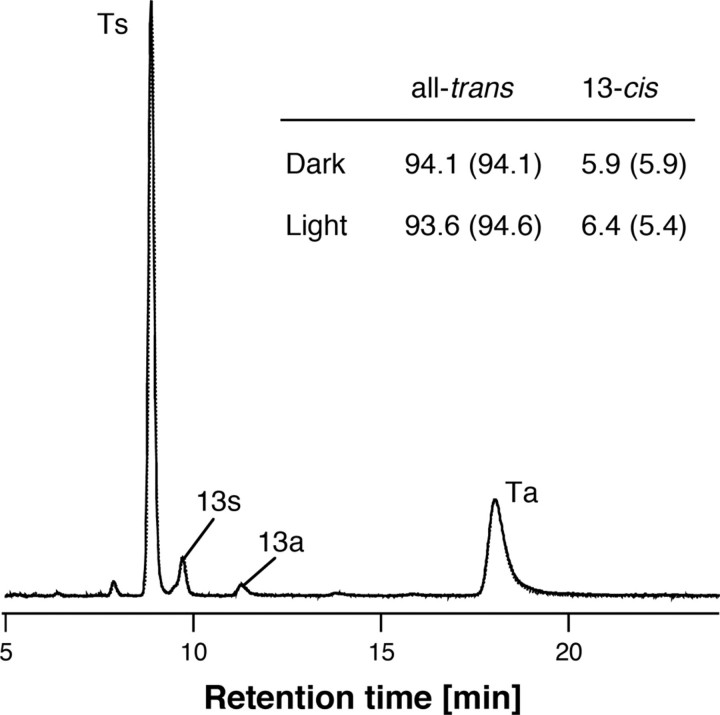
Retinal Configuration and pKa of the Counterion—To investigate the physical properties of SrSRI, HPLC analysis and pH-titration experiments were carried out. In the dark, light-driven ion pumps (BR and HR) use all-trans and 13-cis retinal (38, 39), whereas the photosensors HsSRI, HsSRII, and NpSRII use only all-trans retinal (40, 41). Fig. 3 shows the retinal isomer composition. The isomeric state of retinal is predominantly all-trans, with a small proportion of 13-cis, indicating that the retinal composition is markedly sensory rhodopsin-like. In all retinal proteins (except for Halorhodopsins), the protonated Schiff base is stabilized by an aspartic acid as a counterion. Spectroscopic pH-titration was performed to estimate the pKa value of the counterion of SrSRI (Asp-72) in the unphotolyzed state because protonation of Asp-72 causes a red-shift at the absorption maximum (see Fig. 4a). Fig. 4b shows that difference absorption spectra. Each spectrum was obtained by subtracting that of pH 8.5. The different spectra clearly showed that the acid form has 2-fold larger extinction coefficient than the alkaline form as well as HsSRI (42). The titration curves were analyzed using the Henderson-Hasselbalch equation with a single pKa value (43) (Fig. 4c). The pKa value of Asp-72 for the PG-reconstituted SrSRI was 4.3, which is 3 units lower than that of HsSRI (∼7.2) (44), suggesting a structural difference around the chromophore between SrSRI and HsSRI. Although the pKa value of SrSRI might be affected by interaction with SrHtrI because the value of HsSRI is elevated to 8.5 upon HsHtrI association (44), the lower pKa of SrSRI also indicates that Asp-72 exists in a deprotonated form at neutral pH where the bacteria exist, suggesting the functional importance of the deprotonated state of SrSRI.
FIGURE 3.
Chromophore configuration extracted from SrSRI reconstituted into PG liposomes with 1 m NaCl and 50 mm Tris-Cl (pH 7. 0). The detection beam was set at 360 nm. Ts, Ta, 13s, and 13a stand for all-trans 15-syn retinal oxime, all-trans 15-anti retinal oxime, 13-cis 15-syn retinal oxime, and 13-cis 15-anti retinal oxime, respectively. The molar composition of retinal isomers was calculated from the areas of the peaks in the HPLC patterns. The chromophore configuration extracted from SrSRI suspended in buffer solution containing 0.1% DM is given in parentheses.
FIGURE 4.
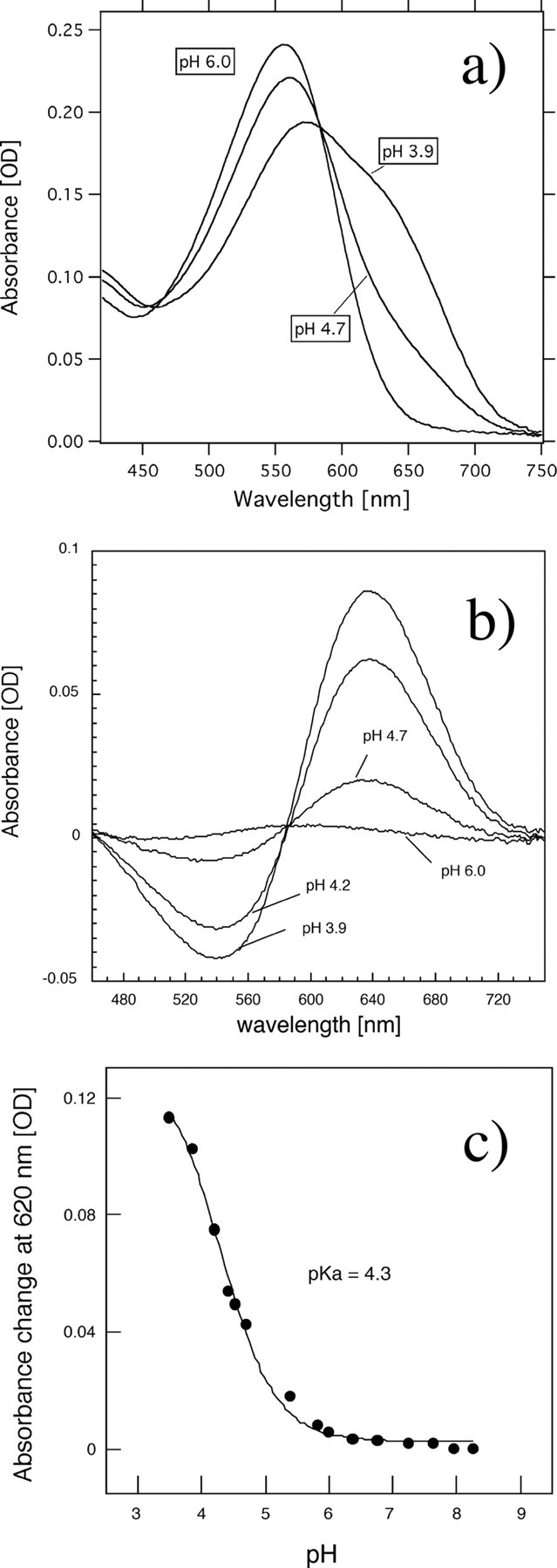
a, absorption spectrum of PG-reconstituted SrSRI at pH 6. 0, 4.7, and 3.9. b, difference absorption spectra. Each spectrum was obtained by subtracting that of pH 8.5. c, pH-titration curves of the counterion, Asp-72, in SrSRI. The SrSRI sample was suspended in a mixture of six buffers with 1 m NaCl. The titration curves were analyzed using the Henderson-Hasselbalch equation with a single pKa value (43). The temperature was kept at 20 °C.
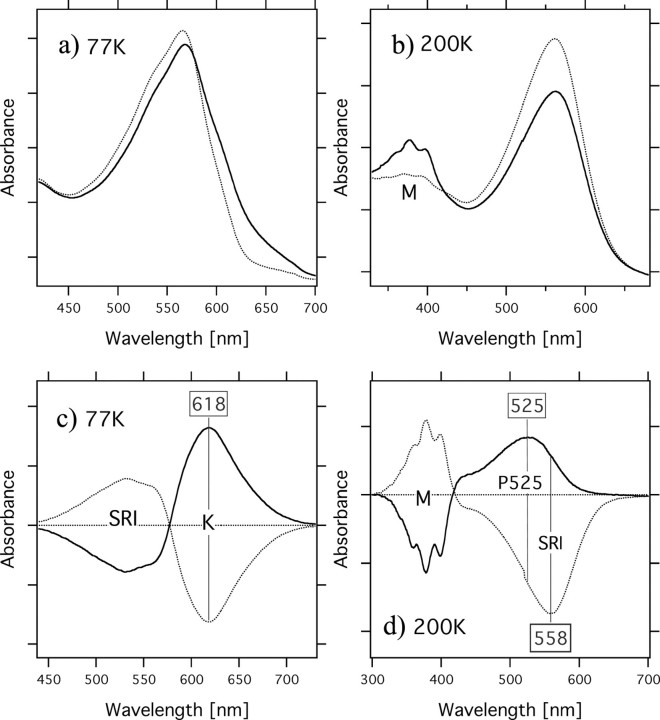
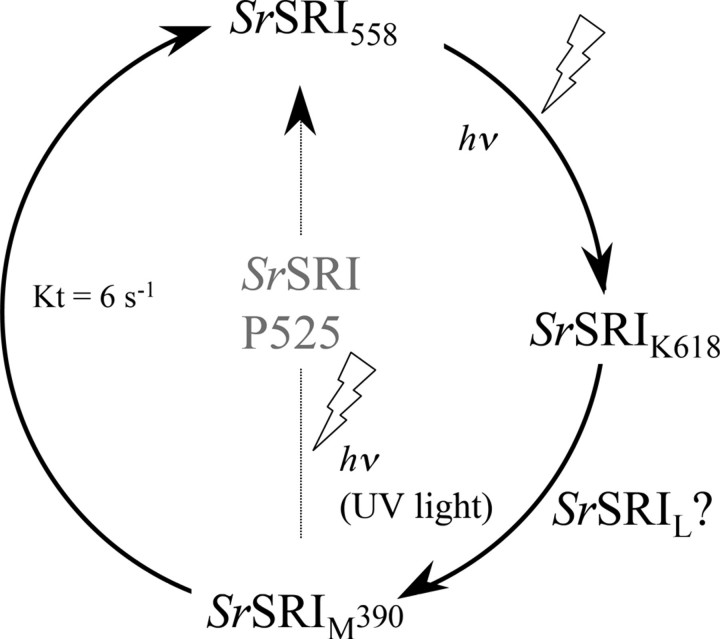
Photocycle of SrSRI as Revealed by Time-resolved Flash Photolysis and Low Temperature UV-Vis Spectroscopy—To confirm the photocycle of SrSRI, flash photolysis and low temperature UV-Vis spectroscopy were used (Figs. 5 and 6), and a schematic drawing of the photocycle based on the results is shown in Fig. 7. The upper panel in Fig. 5 shows the flash-induced difference spectrum of DM-solubilized SrSRI over the spectral range from 300 to 700 nm. In this experiment, the SrSRI sample contained 0.1% DM as a detergent to increase the S/N ratio without inactivation. Depletion by the flash and recovery of the original pigment was observed at 557 nm. The value was almost the same as the absorption maximum of SrSRI shown in Fig. 2 (558 nm). At 390 nm, an increase and a decrease of absorbance were observed, implying the formation and decay of an intermediate SrSRIM similar to HsSRIM (S373) where an absorbance maximum is located at 373 nm. The lower panel in Fig. 5 shows the time courses of the absorbance change at selected wavelengths (390 nm for the M state and 557 nm for the unphotolyzed state). The M-decay rate is estimated as 6 s–1 by single exponential equation, close to that of HsSRI (0.8 s–1) (2) and of NpSRII (0.56 s–1) (45). This slow photocycle is particularly important because a key difference between transport and sensory rhodopsins is the much slower kinetics of the photochemical reaction cycle of the sensors (46). The ion-pumping rhodopsins (BR, HR, and proteorhodopsin) have been optimized for fast photocycling rates to make them efficient pumps (see the gray line in Fig. 5b and Ref. 47). In contrast, the sensory rhodopsins HsSRI and NpSRII have slow photocycles (2, 45), which allows the transient accumulation of long-lived signaling states of the receptors to catalyze a sustained phosphorylation cascade controlling flagellar motor rotation. The slow photocycle of SrSRI therefore is a similar property to that of sensory rhodopsins from other sources.
FIGURE 5.
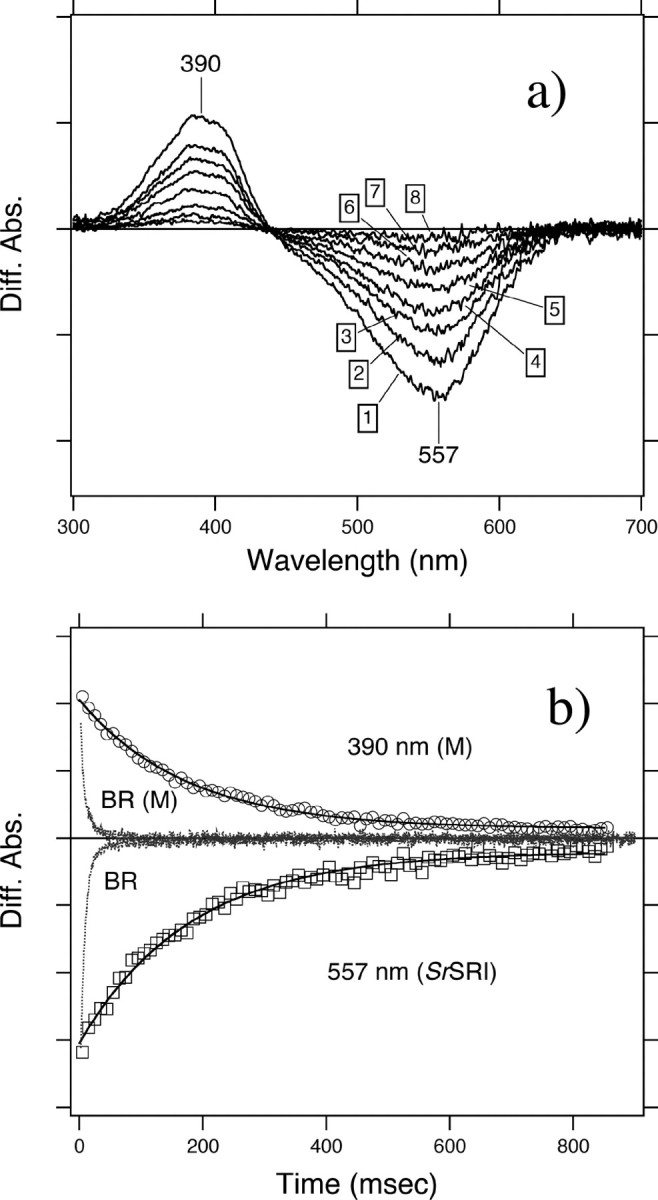
a, flash-induced difference spectra of DM-solubilized SrSRI over a spectral range from 300 to 700 nm. The purified SrSRI was resuspended in 50 mm Tris-HCl, pH 7.0, 0.1% DM, and 1 m NaCl. Curves 1–8 are spectra of 10 ms (1), 50 ms (2), 90 ms (3), 130 ms (4), 210 ms (5), 330 ms (6), 540 ms (7), and 860 ms (8) after the illumination. The temperature was kept at 25 °C. b, flash-induced kinetic data of SrSRI at 390 nm representing the M-decay and at 556 nm representing the recovery of the original SrSRI. The kinetic data of BR were reproduced from Ref. (47) to compare the time range with SrSRI. For BR, the M-intermediate and the unphotolyzed state were monitored at 410 and 570 nm, respectively.
FIGURE 6.
Low temperature UV-Vis absorption spectra of light-activated SrSRI (solid lines) and SrSRI in the dark (dotted lines) measured at 77 K (a) and at 200 K (b). The protein was reconstituted into PG liposomes with NaCl. The difference spectra shown in c and d, were calculated from the spectra recorded before and after irradiation with light. One division of the y-axis of a, b, c, and d corresponds to 0.1, 0.1, 0.05, and 0.05 absorbance unit, respectively.
FIGURE 7.
Schematic of the photochemical reaction cycle of SrSRI identified in this study. hv indicates light-dependent reactions, and the other arrows represent thermal reactions. The decay rate constant of SrSRIm (6 s–1) was obtained by a single exponential equation by the flash photolysis experiments shown in Fig. 5. HsSRI absorbs orange light and forms the long-lived S373 intermediate, which forms the P510 intermediate upon the second photon absorption in the near-UV region (2).
Fig. 6 shows low temperature UV-Vis absorption spectra of light-activated SrSRI (solid lines) and SrSRI in the dark (dotted lines) measured at 77 K (a) and at 200K (b). The difference spectra shown in the panels (c and d) were calculated from the spectra recorded before and after irradiation with light. At 77 K, a light-induced spectral red shift was observed, implying the formation of a K-intermediate of SrSRI. The formation of a K-intermediate was also confirmed by FTIR spectroscopy.3 At 200 K, a spectral blue shift was observed, implying the formation of the M-intermediate of SrSRI. The M-intermediate is one of the most important intermediates for the function of retinal proteins, except for HR. Low temperature FTIR and UV-Vis spectroscopy showed the formation of a small amount of an L-like intermediate of SrSRI as judged by the absorption maximum and by the changes of C=C stretching vibration (data not shown), suggesting the presence of an L-intermediate in SrSRI. In HsSRI as well, K, L, and M intermediates appeared during the photocycle as they do in NpSRII (2, 7). The two difference spectra (dark minus light and light minus subsequent 1-min light illumination) in Fig. 6c are mirror images of each other, indicating that the K intermediates are photoreversible intermediates. However, the two difference spectra (dark minus light and light minus subsequent 1-min light illumination) in Fig. 6d are not mirror images of each other. Upon accumulation of SrSRIM (Fig. 6d, solid line), the SrSRIM forms a P510-like blue-shifted intermediate (P525), which is important for negative phototaxis in HsSRI (1), and the P525 was also trapped at the temperature region between 170 and 243 K. In HsSRI, the M intermediate (S373) becomes the P510-intermediate by illumination with UV light (1). We confirmed that the P510-intermediate of HsSRI is also accumulated in our condition. The identified photocycle scheme is described in Fig. 7.
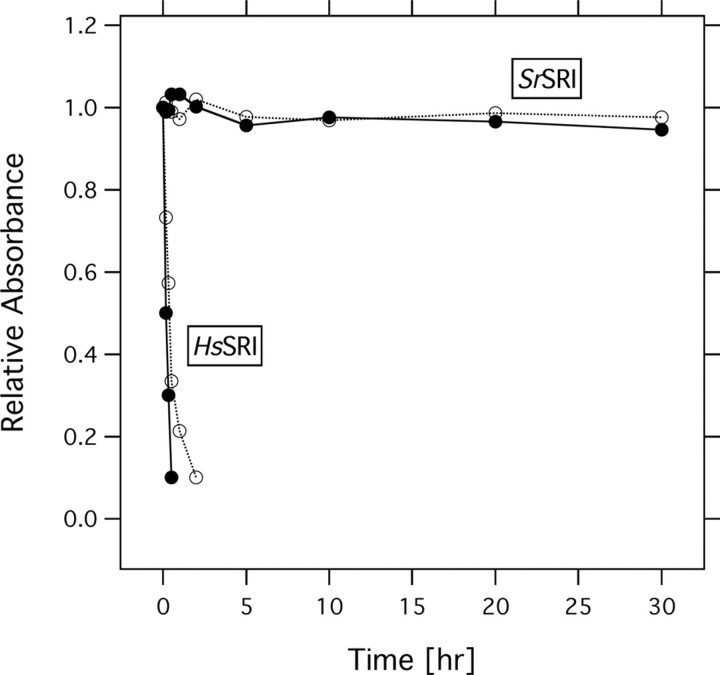
Protein Stability of SrSRI—In 1982, using a BR mutant from the archaeon H. salinarum that was believed to express only HR at that time, Tsuda et al. (48) and Hazemoto et al. (49), found two independent fast and slow photocycles. Bogomolni and Spudich (50) proposed that the pigment having the slow photocycle is the receptor for phototaxis in this bacterium, and they proved their hypothesis by flash photolysis of a mutant lacking both BR and HR, which showed almost the same level of phototaxis as that of the wild type (51). They named this retinal pigment protein sensory rhodopsin I (SRI). Of interest, the ground state of SRI functions as a sensor for positive phototaxis whose action maximum is located at ∼580 nm, while the long-lived photointermediate absorbing maximally at 377 nm works as sensor for negative phototaxis (1). The signal relay mechanism from SR receptors to their cognate Htr transducers has become a focus of interest, in part because of its importance to the general understanding of communication between integral membrane proteins, about which little is known. Important advances in understanding the molecular mechanism of negative phototaxis regulated by the SRII-HtrII complex has been made in recent years because NpSRII is much more stable than HsSRI, especially in dilute salt solutions. Therefore, little is known about the molecular mechanism for positive phototaxis. In fact, the high-resolution crystal structures of BR, HR, and SRII and the SRII-HtrII complex were reported from 1997 through 2006 (52–56), whereas the structure of SRI has not been determined yet. Fig. 8 shows the protein stabilities of HsSRI and SrSRI under low salt conditions (0 and 50 mm NaCl) as judged by the absorbance of the proteins. A time-dependent decrease in the absorbance of HsSRI was observed, implying the degeneration of the protein within 5 h, whereas SrSRI maintained its spectral stability over the 24 h (Fig. 8). As described in the pH-titration experiment, SrSRI is also stable over a wide pH range (pH 3.5–8.5) (Fig. 4). In addition, PG-reconstituted SrSRI is stable even in the absence of NaCl, over a wide pH range (pH 2.5–10) and at high temperature (∼60 °C) (data not shown), while HsSRI is not stable under those conditions. Thus, SrSRI expressed in E. coli shows high stability, making it possible to prepare large amounts of protein and enabling the study of mutant proteins, which will open new avenues to investigate the photosignaling process in the SRI-HtrI system. In the future, we will examine whether SrSRI and SrHtrI are expressed in native cells, and whether they regulate positive and/or negative phototaxis.
FIGURE 8.
Stability of HsSRI and SrSRIin the absence of NaCl (solid lines) and 50 mm NaCl (dotted lines). The time-dependent absorbances of the proteins were plotted. The samples were reconstituted into PG-liposomes and resuspended in a buffer containing 50 mm Tris-Cl (pH 7.0), and the temperature was maintained at 20 °C.
Concluding Remarks—In this study, we present the first characterization of SRI from a eubacterium S. ruber. The SrSRI was expressed well (5 mg/liter culture) as a recombinant protein having all-trans retinal as a chromophore, and it is much more stable than SRI from the archaeon H. salinarum (HsSRI) under various conditions. UV-Vis, low-temperature UV-Vis, pH-titration, and flash photolysis experiments revealed that the photochemical properties of SrSRI are similar to those of SRI from the archaeon H. salinarum. In the future, we will examine whether SrSRI and SrHtrI regulate positive and/or negative phototaxis.
Acknowledgments
We thank Akira Kawanabe for technical assistance in the HPLC experiments, and we are also very grateful to Dr. Mike Dyall-Smith and Dr. David Burns for the gift of the strain S. ruber.
This work was supported by Grants 19042013 and 19045015 (to Y. F.), 19370067 and 20050015 (to H. K.), and 19870010 (to Y. S.) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SRI, sensory rhodopsin I; SrSRI, S. ruber SRI; HsSRI, H. salinarum SRI; NpSRII, N. pharaonis SRII (also known as pharaonis phoborhodopsin, ppR); HtrI, halobacterial transducer protein I; HsHtrI, H. salinarum HtrI; SrHtrI, S. ruber HtrI; NpHtrII, N. pharaonis HtrII; BR, bacteriorhodopsin; SrSRIK, SrSRI K intermediate; SrSRIM, SrSRI M intermediate; HPLC, high performance liquid chromatography; FTIR, Fourier transform infrared spectroscopy; MES, 4-morpholineethanesulfonic acid; MOPS, 4-morpholinepropanesulfonic acid; CHES, 2-(cyclohexylamino)ethanesulfonic acid; CAPS, 3-(cyclohexylamino)propanesulfonic acid.
Y. Sudo, unpublished data.
References
- 1.Spudich, J. L., and Bogomolni, R. A. (1984) Nature 312 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoff, W. D., Jung, K. H., and Spudich, J. L. (1997) Annu. Rev. Biophys. Biomol. Struct. 26 223–258 [DOI] [PubMed] [Google Scholar]
- 3.Sasaki, J., and Spudich, J. L. (2008) Photochem. Photobiol., in press [DOI] [PubMed]
- 4.Sasaki, J., Nara, T., Spudich, E. N., and Spudich, J. L. (2007) Mol. Microbiol. 66 1321–1330 [DOI] [PubMed] [Google Scholar]
- 5.Yan, B., Takahashi, T., Johnson, R., Derguini, F., Nakanishi, K., and Spudich, J. L. (1990) Biophys. J. 57 807–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandori, H., Tomioka, H., and Sasabe, H. (2002) J. Phys. Chem. A 106 2091–2095 [Google Scholar]
- 7.Chizhov, I., Schmies, G., Seidel, R., Sydor, J. R., Luttenberg, B., and Engelhard, M. (1998) Biophys. J. 75 999–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hippler-Mreyen, S., Klare, J. P., Wegener, A. A., Seidel, R., Herrmann, C., Schmies, G., Nagel, G., Bamberg, E., and Engelhard, M. (2003) J. Mol. Biol. 330 1203–1213 [DOI] [PubMed] [Google Scholar]
- 9.Sudo, Y., Yamabi, M., Kato, S., Hasegawa, C., Iwamoto, M., Shimono, K., and Kamo, N. (2006) J. Mol. Biol. 357 1274–1282 [DOI] [PubMed] [Google Scholar]
- 10.Sudo, Y., Iwamoto, M., Shimono, K., and Kamo, N. (2004) Biochemistry 43 13748–13754 [DOI] [PubMed] [Google Scholar]
- 11.Sudo, Y., Okuda, H., Yamabi, M., Fukuzaki, Y., Mishima, M., Kamo, N., and Kojima, C. (2005) Biochemistry 44 6144–6152 [DOI] [PubMed] [Google Scholar]
- 12.Sudo, Y., Furutani, Y., Wada, A., Ito, M., Kamo, N., and Kandori, H. (2005) J. Am. Chem. Soc. 127 16036–16037 [DOI] [PubMed] [Google Scholar]
- 13.Sudo, Y., Furutani, Y., Spudich, J. L., and Kandori, H. (2007) J. Biol. Chem. 282 15550–15558 [DOI] [PubMed] [Google Scholar]
- 14.Sudo, Y., Furutani, Y., Shimono, K., Kamo, N., and Kandori, H. (2003) Biochemistry 42 14166–14172 [DOI] [PubMed] [Google Scholar]
- 15.Sudo, Y., Furutani, Y., Kandori, H., and Spudich, J. L. (2006) J. Biol. Chem. 281 34239–34245 [DOI] [PubMed] [Google Scholar]
- 16.Wegener, A. A., Chizhov, I., Engelhard, M., and Steinhoff, H. J. (2000) J. Mol. Biol. 301 881–891 [DOI] [PubMed] [Google Scholar]
- 17.Yoshida, H., Sudo, Y., Shimono, K., Iwamoto, M., and Kamo, N. (2004) Photochem. Photobiol. Sci. 3 537–542 [DOI] [PubMed] [Google Scholar]
- 18.Wegener, A. A., Klare, J. P., Engelhard, M., and Steinhoff, H. J. (2001) EMBO J. 20 5312–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudolph, J., Tolliday, N., Schmitt, C., Schuster, S. C., and Oesterhelt, D. (1995) EMBO J. 14 4249–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trivedi, V. D., and Spudich, J. L. (2003) Biochemistry 42 13887–13892 [DOI] [PubMed] [Google Scholar]
- 21.Spudich, J. L., and Luecke, H. (2002) Curr. Opin. Struct. Biol. 12 540–546 [DOI] [PubMed] [Google Scholar]
- 22.Sudo, Y., Yamabi, M., Iwamoto, M., Shimono, K., and Kamo, N. (2003) Photochem. Photobiol. 78 511–516 [DOI] [PubMed] [Google Scholar]
- 23.Mongodin, E. F., Nelson, K. E., Daugherty, S., Deboy, R. T., Wister, J., Khouri, H., Weidman, J., Walsh, D. A., Papke, R. T., Sanchez Perez, G., Sharma, A. K., Nesbo, C. L., MacLeod, D., Bapteste, E., Doolittle, W. F., Charlebois, R. L., Legault, B., and Rodriguez-Valera, F. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 18147–18152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marmur, J. (1961) J. Mol. Biol. 3 208–218 [Google Scholar]
- 25.Kandori, H., Shimono, K., Sudo, Y., Iwamoto, M., Shichida, Y., and Kamo, N. (2001) Biochemistry 40 9238–9246 [DOI] [PubMed] [Google Scholar]
- 26.Iwata, T., Nozaki, D., Tokutomi, S., Kagawa, T., Wada, M., and Kandori, H. (2003) Biochemistry 42 8183–8191 [DOI] [PubMed] [Google Scholar]
- 27.Shimono, K., Ikeura, Y., Sudo, Y., Iwamoto, M., and Kamo, N. (2001) Biochim. Biophys. Acta 1515 92–100 [DOI] [PubMed] [Google Scholar]
- 28.Spudich, J. L. (2006) Trends Microbiol. 14 480–487 [DOI] [PubMed] [Google Scholar]
- 29.Yao, V. J., and Spudich, J. L. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 11915–11919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson, K. D., Zhang, X. N., and Spudich, J. L. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 3185–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, X. N., and Spudich, J. L. (1997) Biophys. J. 73 1516–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung, K. H., and Spudich, J. L. (1998) J. Bacteriol. 180 2033–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki, J., Phillips, B. J., Chen, X., Van Eps, N., Tsai, A. L., Hubbell, W. L., and Spudich, J. L. (2007) Biophys. J. 92 4045–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seidel, R., Scharf, B., Gautel, M., Kleine, K., Oesterhelt, D., and Engelhard, M. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 3036–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balashov, S. P., Imasheva, E. S., Boichenko, V. A., Anton, J., Wang, J. M., and Lanyi, J. K. (2005) Science 309 2061–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi, T., Yan, B., Mazur, P., Derguini, F., Nakanishi, K., and Spudich, J. L. (1990) Biochemistry 29 8467–8474 [DOI] [PubMed] [Google Scholar]
- 37.Shimono, K., Hayashi, T., Ikeura, Y., Sudo, Y., Iwamoto, M., and Kamo, N. (2003) J. Biol. Chem. 278 23882–23889 [DOI] [PubMed] [Google Scholar]
- 38.Pettei, M. J., Yudd, A. P., Nakanishi, K., Henselman, R., and Stoeckenius, W. (1977) Biochemistry 16 1955–1959 [DOI] [PubMed] [Google Scholar]
- 39.Kamo, N., Hazemoto, N., Kobatake, Y., and Mukohata, Y. (1985) Arch. Biochem. Biophys. 238 90–96 [DOI] [PubMed] [Google Scholar]
- 40.Imamoto, Y., Shichida, Y., Hirayama, J., Tomioka, H., Kamo, N., and Yoshizawa, T. (1992) Biochemistry 31 2523–2528 [DOI] [PubMed] [Google Scholar]
- 41.Scharf, B., Hess, B., and Engelhard, M. (1992) Biochemistry 31 12486–12492 [DOI] [PubMed] [Google Scholar]
- 42.Olson, K. D., Deval, P., and Spudich, J. L. (1992) Photochem. Photobiol. 56 1181–1187 [DOI] [PubMed] [Google Scholar]
- 43.Sudo, Y., Iwamoto, M., Shimono, K., and Kamo, N. (2002) Biophys. J. 83 427–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spudich, J. L. (1995) Biophys. Chem. 56 165–169 [DOI] [PubMed] [Google Scholar]
- 45.Sudo, Y., Iwamoto, M., Shimono, K., Sumi, M., and Kamo, N. (2001) Biophys. J. 80 916–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spudich, J. L., and Lanyi, J. K. (1996) Curr. Opin. Cell Biol. 8 452–457 [DOI] [PubMed] [Google Scholar]
- 47.Sudo, Y., and Spudich, J. L. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 16129–16134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsuda, M., Hazemoto, N., Kondo, M., Kamo, N., Kobatake, Y., and Terayama, Y. (1982) Biochem. Biophys. Res. Commun. 108 970–976 [DOI] [PubMed] [Google Scholar]
- 49.Hazemoto, N., Kamo, N., Terayama, Y., Kobatake, Y., and Tsuda, M. (1983) Biophys. J. 44 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bogomolni, R. A., and Spudich, J. L. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 6250–6254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spudich, E. N., and Spudich, J. L. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 4308–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pebay-Peyroula, E., Rummel, G., Rosenbusch, J. P., and Landau, E. M. (1997) Science 277 1676–1681 [DOI] [PubMed] [Google Scholar]
- 53.Luecke, H., Schobert, B., Richter, H. T., Cartailler, J. P., and Lanyi, J. K. (1999) J. Mol. Biol. 291 899–911 [DOI] [PubMed] [Google Scholar]
- 54.Luecke, H., Schobert, B., Lanyi, J. K., Spudich, E. N., and Spudich, J. L. (2001) Science 293 1499–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gordeliy, V. I., Labahn, J., Moukhametzianov, R., Efremov, R., Granzin, J., Schlesinger, R., Buldt, G., Savopol, T., Scheidig, A. J., Klare, J. P., and Engelhard, M. (2002) Nature 419 484–487 [DOI] [PubMed] [Google Scholar]
- 56.Moukhametzianov, R., Klare, J. P., Efremov, R., Baeken, C., Goppner, A., Labahn, J., Engelhard, M., Buldt, G., and Gordeliy, V. I. (2006) Nature 440 115–119 [DOI] [PubMed] [Google Scholar]