Abstract
The Tweety proteins comprise a family of chloride ion channels with three members identified in humans (TTYH1-3) and orthologues in fly and murine species. In humans, increased TTYH2 expression is associated with cancer progression, whereas fly Tweety is associated with developmental processes. Structurally, Tweety proteins are characterized by five membrane-spanning domains and N-glycan modifications important for trafficking to the plasma membrane, where these proteins are oriented with the amino terminus located extracellularly and the carboxyl terminus cytoplasmically. In addition to N-glycosylation, ubiquitination mediated by the HECT type E3 ubiquitin ligase Nedd4-2 is a post-translation modification important in regulating membrane proteins. In the present study, we performed a comprehensive analysis of the ability of each of TTYH1-3 to interact with Nedd4-2 and to be ubiquitinated and regulated by this ligase. Our data indicate that Nedd4-2 binds to two family members, TTYH2 and TTYH3, which contain consensus PY ((L/P)PXY) binding sites for HECT type E3 ubiquitin ligases, but not to TTYH1, which lacks this motif. Consistently, Nedd4-2 ubiquitinates both TTYH2 and TTYH3. Importantly, we have shown that endogenous TTYH2 and Nedd4-2 are binding partners and demonstrated that the TTYH2 PY motif is essential for these interactions. We have also shown that Nedd4-2-mediated ubiquitination of TTYH2 is a critical regulator of cell surface and total cellular levels of this protein. These data, indicating that Nedd4-2 differentially interacts with and regulates TTYH1-3, will be important for understanding mechanisms controlling Tweety proteins in physiology and disease.
Ubiquitination is a post-translational modification essential for regulation of many physiological processes, including those as diverse as cell cycle progression (1), transcription (2), immune responses (3), and synapse formation (4). In addition, dysregulated ubiquitination is important in various pathologies, including cancer (5). In these processes, the covalent attachment of ubiquitin, a highly conserved 76-amino acid peptide, serves as a signal for protein localization, facilitating protein degradation, stability, trafficking, interactions, and activity (6).
The addition of ubiquitin to proteins occurs at lysine residues and requires the tightly coordinated action of three enzymes: a ubiquitin-activating enzyme (E1),5 a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3). The E3 enzymes, which are thought to be the predominant mediators of substrate binding (7), can be classified into two broad families. The first of these is the RING (really interesting new gene) domain containing the E3 enzyme family, members of which facilitate ubiquitination by simultaneously binding the substrate and an E2 enzyme. In this case, it is the E2 ubiquitin-conjugating enzyme that transfers the ubiquitin moiety directly to the substrate. In contrast, members of the second E3 family, the HECT (homologous to E6-AP COOH-terminal) domain-containing ubiquitin ligases, transfer ubiquitin from the E2 enzyme to the substrate (8). HECT family E3 ligases are characterized by three structurally conserved domains critical for localization (a C2 module at the amino terminus), substrate binding (up to four WW domains), and ubiquitin transfer (a HECT domain) (9). The WW domains of these ligases belong to the Group I WW domain subfamily, which bind to ligands containing consensus (L/P)PXY motifs, also known as PY motifs (10). In contrast, other WW domain subfamilies bind ligands containing PPLP (Group II), polyproline motifs flanked by arginine or lysine residues (Group III) and phosphoserine or phosphothreonine residues (Group IV) (10).
Recently, members of the HECT E3 family have been shown to regulate trafficking and degradation of membrane proteins, including ion channels (11, 12). In particular, Nedd4-2, a member of the Nedd4-like E3 ligase subfamily (13), has been shown to play a key role in the regulation of several ion channels (14). For example, Nedd4-2 regulates the epithelial Na+ channel (ENaC) by controlling its cell surface stability (15). Also, interactions between the Cl- ion channel ClC-5 and Nedd4-2 regulate cell surface expression of this ion channel as well as its role in albumin uptake (16). In addition, the activities of the voltage-gated Na+ channel 1.2 (Nav1.2), Nav1.7, and Nav1.8 are inhibited by Nedd4-2-mediated ubiquitination (17). Other examples include Nedd4-2-mediated ubiquitin regulation of the voltage-gated K+ channels KCNQ2/3 and KCNQ3/5 (18) and Nav1.5 (19).
The human Tweety proteins, designated TTYH1 (Tweety homologue 1), TTYH2, and TTYH3, comprise a recently identified family of maxichloride ion channels (20) named after the protein encoded by the Drosophila melanogaster gene Tweety (21). TTYH2 and TTYH3 display ionomycin, Ca2+-dependent Cl- channel activity, whereas volume-regulated channel activity is only apparent in a carboxyl-terminal variant isoform of TTYH1 (20). In addition to the human Tweety genes, ortho-logues have been identified for TTYH1 in mice, macaques, and Caenorhabditis elegans (22) and TTYH2 in mice (23), with two additional Tweety genes reported in flies (20, 22). Although studies defining the roles of Tweety family members in patho-physiological processes are lacking, several reports have indicated associations with developmental processes and cancer. For example, in flies, experiments involving simultaneous deletion of the Tweety gene and two adjacent genes, dodo and pendulin, resulted in reduced viability, suggestive of functions for the encoded proteins in development (21, 24). Also, recently increased expression of the second human member of this family, TTYH2, has been linked to renal cell carcinoma (23) and colorectal cancer (25).
Examining mechanisms regulating the cellular localization of Tweety family members, we have recently shown that these proteins contain five membrane-spanning domains with a topology at the cell surface in which the amino terminus is located extracellularly and the carboxyl terminus is located cytoplasmically (26). In the present study, we have further investigated mechanisms regulating Tweety proteins, by examining the role of ubiquitination, mediated by the HECT type E3 ubiquitin ligase Nedd4-2, in the regulation of TTYH1-3. By analyzing the sequence of each of these proteins and orthologous murine Tweety members, we have identified consensus WW domain binding sites and, using immunoprecipitation experiments, shown that TTYH2 and TTYH3, but not TTYH1, are Nedd4-2-binding proteins. We also show that TTYH2 and TTYH3 are efficiently ubiquitinated by Nedd4-2. Identifying endogenous TTYH2 and Nedd4-2 as binding proteins, we have confirmed the PY motif of this Tweety family member as an essential mediator of interactions with Nedd4-2. Consistently, we have shown that Nedd4-2-mediated ubiquitination of TTYH2 regulates cell surface and total cellular levels of this chloride ion channel. Since spatial and temporal protein localization is critical in pathophysiological processes, these data will be important for understanding mechanisms regulating Tweety family proteins in developmental processes and cancer progression.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents—Anti-TTYH2 rabbit polyclonal antibodies were generated against the TTYH2 unique peptides REVTMELTKLSDQTG (T192), PQAWRMATHSPPRGQL (T435), and SVADEHLRHYGNQFPA (T519). The peptides were synthesized by Mimotopes (Clayton, Australia) and conjugated to diphtheria toxoid before injection into rabbits. Immunoglobulins (IgGs) were affinity-purified against the peptide used for immunizations using SulfoLink coupling gel (Pierce) as previously described (27). Monoclonal and rabbit polyclonal antibodies against the Myc (EQKLISEEDL) epitope were purchased from Cell Signaling Technology (Genesearch Pty. Ltd., Arundel, Australia). Monoclonal and polyclonal anti-hemagglutinin (HA; YPYDVPDYA) epitope antibodies were from Roche Applied Sciences. Rabbit polyclonal and mouse monoclonal anti-FLAG epitope (DYKDDDDK) and anti-pancadherin monoclonal antibodies were purchased from Sigma. Monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies were purchased from Chemicon International (Boronia, Australia). A polyclonal anti-ubiquitin antibody was from Sigma. An antibody that recognizes the HECT domains of Nedd4 and Nedd4-2 (16, 28) and a specific anti-Nedd4-2 antibody (29) have been described previously. Horse-radish peroxidase-conjugated and fluorescently conjugated secondary antibodies were from Pierce and Invitrogen, respectively. Control IgGs were from Sigma and Invitrogen, protein A/G-agarose was from Roche Applied Sciences, and glutathi-one-Sepharose 4B beads were from GE Healthcare.
Bioinformatics Analyses—Alignments and searches were performed using algorithms available at the Australian National Genomic Information Service BioManager Web site.
Expression Constructs—TTYH1-HA, TTYH2-Myc, and TTYH3-HA expression constructs were described previously (26). TTYH1 and TTYH3 cDNAs (26) were used as templates to generate the respective expression constructs TTYH1-Myc and TTYH3-Myc in pcDNA3.1 (Invitrogen). To generate a mammalian expression construct encoding the chimera TTYH2-GFP, DNA encoding the complete TTYH2 coding sequence was amplified by PCR and cloned in frame into the EcoRI site of the vector pEGFP-N1 (Clontech, Mountain View, CA). Site-directed mutagenesis, to introduce the TTYH2 mutations S444A, S504A, Y509F, and S510A, was performed using the proofreading polymerase Pfu Ultra (Stratagene, La Jolla, CA). DNA encoding the carboxyl terminus of TTYH2 (residues 409-534) was amplified by PCR and cloned in frame into the vector pGEX-6P-1 (GE Healthcare) to generate a bacterial expression construct encoding the fusion protein glutathione S-transferase (GST)-TTYH2. All constructs and mutations were confirmed by DNA sequencing. The expression vector for Nedd4-2-FLAG has been described previously (29).
Cell Culture and Transfections—Chinese hamster ovary and human embryonic kidney (HEK293) cells were grown in Dulbecco's modified Eagle's medium, and the opossum kidney cell line was maintained in Dulbecco's modified Eagle's medium/Ham's F-12 medium. Cultures were supplemented with 10% fetal bovine serum, 100 units/ml of penicillin, and 100 units/ml of streptomycin unless otherwise specified and incubated at 37 °C in 5% CO2. Transfections were performed using Lipofectamine 2000 (Invitrogen), following the instructions of the manufacturer.
GST Pull-down Assays—Recombinant GST and GST-TTYH2 were generated and purified as previously described (30). Briefly, BL-21 (DE3) cells (Stratagene) transformed with either pGEX-6P-1 or the GST-TTYH2 fusion protein expression construct were induced with isopropyl β-d-1-thiogalactopyranoside (1 mm), harvested by centrifugation, and then lysed by sonication. For pull-down assays, 50 μg of GST or GST-TTYH2 fusion protein was incubated with glutathione-Sepharose 4B beads for 3 h at 4°C. The beads and bound proteins were washed and incubated with ∼1 mg of protein lysate from opossum kidney cells at 4 °C for 18 h. Proteins bound to the beads were eluted into Laemmli sample buffer, separated on SDS-PAGE, and detected by Western blot analysis.
Immunoprecipitation—In experiments to detect interacting proteins, lysates were collected from cells grown in antibiotic-free medium in a buffer consisting of 10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Triton X-100, 5 mm EDTA, and Complete protease inhibitor mixture (Roche Applied Science). In experiments to detect ubiquitination, cells were treated with 20 μm MG-132 (Sigma) for 4 h before collection of lysates in a buffer containing 1% SDS, 2 mm dithiothreitol, and protease inhibitor mixture followed by denaturation at 100 °C for 5 min. This mixture was then diluted 10 fold in buffer containing 0.55% IGE-PAL, 55.55 mm Tris (pH 8.0), 11.1 mm MgCl2, and protease inhibitor mixture. Lysates were precleared with protein A/G-agarose for 1 h at 4°C on a rolling platform. After centrifugation, the supernatants were mixed with appropriate antibodies or isotype IgGs and incubated overnight at 4 °C. Fresh aliquots of protein A/G-agarose beads were then added, and the mixture was incubated for 4 h at 4°C with gentle agitation. The beads were then washed three times in cell lysis buffer. Associated proteins were eluted into Laemmli sample buffer and analyzed by Western blot analysis.
Cell Surface Biotinylation—HEK293 cells transfected with wild type and mutant TTYH2-Myc expression constructs were washed three times with phosphate-buffered saline (PBS), and plasma membrane proteins were biotinylated by incubation with cell-impermeant EZ-link NHS-SSBiotin (1.22 mg/ml; Pierce) at 4 °C for 1 h with gentle agitation. The cells were then washed in PBS prior to lysis in a buffer containing 20 mm HEPES, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100. After centrifugation, ImmunoPure immobilized streptavidin (Pierce) was added to the supernatant and incubated for 15 min on ice to precipitate biotinylated proteins. The streptavidin beads, associated with biotinylated plasma membrane proteins, were then pelleted by centrifugation, and the supernatant, containing cytoplasmic proteins, was transferred to a fresh tube. Plasma membrane and cytoplasmic fractions were examined by Western blot analysis.
Western Blot Analysis—Whole cell lysates were collected in a buffer containing Triton X-100 (1%, v/v), 50 mm Tris-HCl (pH 7.4), NaCl (150 mm) and protease inhibitor mixture (Roche Applied Science). Protein concentrations were determined by a microbicinchoninic acid assay (Pierce). Cell lysates and immunoprecipitated proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes, which were blocked in 5% skim milk in Tris-buffered saline containing 0.1% (v/v) Tween 20 (TBS-T). For detection of ubiquitinated proteins, membranes were autoclaved prior to the blocking step to enhance sensitivity, as previously described (31). Membranes were incubated with appropriate antibodies diluted in blocking buffer for 1 h at room temperature, washed with TBS-T, and then incubated with species-appropriate horseradish peroxidase-conjugated secondary antibodies for 45 min. Following washes, membranes were incubated with SuperSignal West Pico Substrate (Pierce) and then exposed to film. Consistent protein loading and transfer was determined by reprobing membranes stripped in Restore Western blot stripping buffer (Pierce) with either anti-TTYH2, anti-GAPDH, anti-Myc, anti-HA, or anti-FLAG antibodies as appropriate. Where relevant, results are displayed graphically as mean ± S.D., and significance was examined using Student's t test with a p value of <0.05 considered significant.
Confocal Microscopy—HEK293 cells grown on poly-l-lysine-coated coverslips were transfected with expression constructs encoding Nedd4-2-FLAG and either wild type or mutant TTYH2-GFP. Following washes in PBS, cells were fixed with 4% formaldehyde in PBS for 30 min at room temperature, rinsed twice with PBS, and then blocked for 30 min in 0.2% bovine serum albumin in PBS. Cells were incubated with an anti-FLAG monoclonal antibody (dilution 1:1000) in blocking buffer for 1 h at room temperature, washed with PBS, and then incubated with an Alexa Fluor 568-conjugated goat anti-mouse secondary antibody in blocking buffer for 30 min at room temperature. Nuclei were stained with 4′, 6-diamidino-2-phenylindole hydrochloride (Chemicon). Following washes with PBS, cover-slips were mounted on slides, and cells were imaged with a Leica TCS SP5 confocal microscope (Leica Microsystems, Sydney, Australia). Images were processed and displayed using Adobe Photoshop CS3.
RESULTS
Analysis of Tweety Family Members for Consensus Binding Motifs for HECT Type E3 Ubiquitin-Protein Ligases—Interactions between HECT type E3 ubiquitin-protein ligases and substrates are mediated by Group I WW domains present within the ligase (11). Most commonly, these domains bind proline-rich PPXY consensus sequences referred to as PY motifs (11). In addition, examples of WW domain binding to phosphoserine in SP motifs and phosphothreonine in TP motifs have also been reported (32, 33). We analyzed the amino acid sequence of orthologous human, mouse, and rat members of the Tweety family of chloride ion channels for these consensus sequences. Potential binding motifs present within each orthologue are highlighted on the human TTYH1, TTYH2, and TTYH3 in Fig. 1A. TTYH1 contains SP motifs at residues 83 and 198. However, based on the orientation of Tweety proteins at the plasma membrane, described recently by He et al. (26), only 83SP is located intracellularly, within intracellular loop 1. TTYH2 contains within its carboxyl-terminal domain (CTD) SP motifs at residues 504 and 510 flanking the PY motif 506PPTY. An additional intracellular SP motif is present within this domain at residue 444 of human TTYH2 alone. TTYH3 contains within its CTD 489TP and 496SP motifs with the latter immediately adjacent to the PY motif 498PPSY. TTYH2 and TTYH3 also contain extracellularly located SP motifs at residues 35 (amino-terminal domain) and 300 (extracellular loop 2), respectively. As shown in Fig. 1B, the PY motifs of human TTYH2 and TTYH3 closely match PY motifs from mouse and rat ortho-logues and also PY motifs from ENaC subunits and voltage-gated Na+ and K+ channels, which are known to mediate interactions with HECT type E3 ubiquitin-protein ligases.
FIGURE 1.
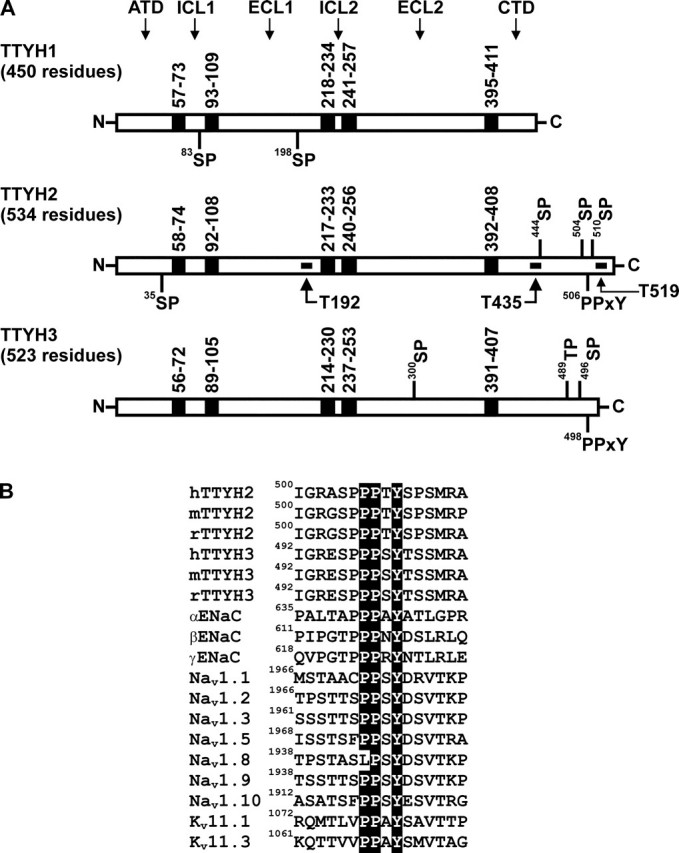
Consensus HECT type E3 ubiquitin-protein ligase binding motifs of human Tweety family members. A, TTYH1, TTYH2, and TTYH3 showing transmembrane domains (black boxes) and consensus SP, TP, and PPXY motifs. The predicted positions of transmembrane domains and the total number of residues are indicated, as are the location of peptides used to generate three anti-TTYH2 polyclonal antibodies (designated T192, T435, and T519). The orientations of the Tweety proteins at the plasma membrane are also shown. ATD, amino-terminal domain; ICL1 and ICL2, intracellular loop 1 and 2; ECL1 and ECL2, extracellular loop 1 and 2; CTD, carboxyl-terminal domain. B, alignment of PY motifs and neighboring sequences (±6 residues) of human, mouse, and rat TTYH2 and TTYH3 against human ENaC subunits α, β, and γ, human PY motif-containing voltage-gated sodium channels (Navs), and mouse voltage-gated potassium channels (Kvs). Each motif is numbered on the left to indicate its position in the amino acid sequence of the respective ion channel.
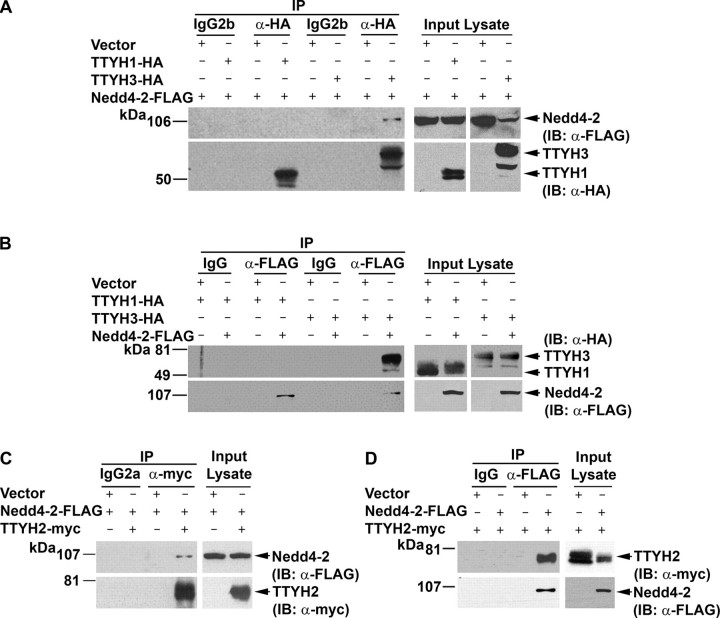
TTYH2 and TTYH3, but Not TTYH1, Interact with the HECT Type E3 Ubiquitin-Protein Ligase Nedd4-2—Experiments with cells transiently co-transfected with constructs encoding TTYH1-HA, TTYH2-Myc, or TTYH3-HA with a Nedd4-2-FLAG expression construct were performed to examine whether interactions occur between Tweety family members and a member of the HECT type E3 ubiquitin-protein ligase family. As shown in Fig. 2A using an anti-HA tag antibody, Nedd4-2-FLAG was immunoprecipitated from lysates from cells co-transfected with TTYH3-HA and Nedd4-2-FLAG expression constructs but not from cells co-transfected with TTYH1-HA and Nedd4-2-FLAG expression constructs. Consistently, performing the reverse immunoprecipitation using an anti-FLAG tag antibody, TTYH3-HA was able to be immunoprecipitated from TTYH3-HA/Nedd4-2-FLAG-transfected cells, whereas TTYH1-HA was not able to be immunoprecipitated from cells co-transfected with TTYH1-HA and Nedd4-2-FLAG expression constructs (Fig. 2B). In addition, using an anti-Myc tag antibody, Nedd4-2-FLAG was immunoprecipitated from cells co-transfected with TTYH2-Myc and Nedd4-2-FLAG expression construct (Fig. 2C). Consistently, performing the reverse immunoprecipitation using an anti-FLAG tag antibody, TTYH2-Myc was able to be immunoprecipitated from these cells (Fig. 2D). Note that use of isotype-matched IgGs in control immunoprecipitations indicated the specificity of the immunoprecipitating antibodies. In summary, the Tweety family proteins TTYH2 and TTYH3, containing CTD SP and PY motifs, immunoprecipitated with the HECT type E3 ubiquitin-protein ligase Nedd4-2. In contrast, TTYH1, which contains an SP motif in intracellular loop 1 but lacks a PY motif, does not immunoprecipitate with Nedd4-2. These data suggest that interactions between Tweety family proteins and Nedd4-2 are mediated by PY and, potentially, SP motifs located within CTDs.
FIGURE 2.
TTYH2 and TTYH3 but not TTYH1 interact with Nedd4-2. HEK293 cells were transfected with Nedd4-2-FLAG and either TTYH1-HA, TTYH2-Myc, or TTYH3-HA expression constructs or vector (pcDNA3.1) control. Proteins were immunoprecipitated (IP) using either monoclonal anti-HA or anti-Myc antibodies or a rabbit anti-FLAG antibody or isotype-matched (IgG2b for anti-HA; IgG2a for anti-Myc) or species-matched (rabbit IgG for anti-FLAG) control IgG. Western blot analysis (IB) was performed using anti-FLAG and anti-HA antibodies (A and B) or anti-FLAG and anti-Myc antibodies (C and D). Lysates were probed with anti-HA and anti-FLAG antibodies (A and B) or anti-FLAG and anti-Myc antibodies (C and D).
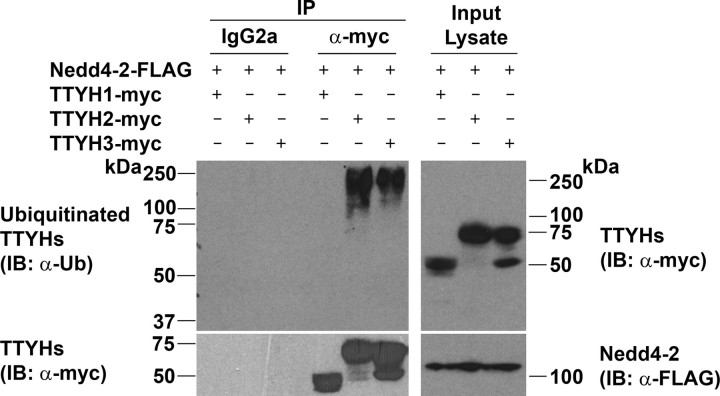
TTYH2 and TTYH3, but Not TTYH1, Are Ubiquitinated by Nedd4-2—We have shown that the PY and SP motif-containing proteins TTYH2 and TTYH3 interact with the E3 ubiquitin-protein ligase Nedd4-2 in transfected cells (Fig. 2). In contrast, TTYH1, which lacks a PY motif, does not interact with Nedd4-2 (Fig. 2). Co-transfection experiments were performed to examine whether the level of ubiquitination of these ion channels correlated with the presence of CTD PY and SP motifs. HEK293 cells were co-transfected with a Nedd4-2-FLAG construct and either a TTYH1-Myc, TTYH2-Myc, or TTYH3-Myc expression construct, lysed in a denaturing buffer, and then subjected to immunoprecipitation using an anti-Myc antibody. Immunocomplexes were then examined by Western blot analysis using an anti-ubiquitin antibody. As shown in Fig. 3 (left), TTYH2 and TTYH3, but not TTYH1, were ubiquitinated by Nedd4-2 (α-Ub panel), although each Tweety family member was efficiently immunoprecipitated (α-Myc panel). Probing whole cell lysates with an anti-Myc antibody indicated that each Tweety protein was efficiently expressed by HEK293 cells, whereas anti-FLAG Western blot analysis indicated that Nedd4-2 was expressed at approximately equal levels in each co-transfection (Fig. 3, right).
FIGURE 3.
Comparison of Nedd4-2-mediated ubiquitination of human Tweety proteins. Lysates from HEK293 cells co-transfected with Nedd4-2-FLAG and either TTYH1-Myc, TTYH2-Myc, or TTYH3-Myc expression constructs were immunoprecipitated (IP) with a mouse anti-Myc antibody or isotype-matched (IgG2a) control IgG. To prevent degradation of ubiquitinated proteins, cells were treated for 4 h with the proteasome inhibitor MG-132 before the collection of lysates. Immunoprecipitated proteins were examined by Western blot analysis (IB) using anti-ubiquitin (Ub) and anti-Myc antibodies. Lysates were examined by Western blot analysis using rabbit anti-Myc and anti-FLAG antibodies. Ubiquitinated and total Tweety proteins (TTYHs) and Nedd4-2-FLAG are indicated.
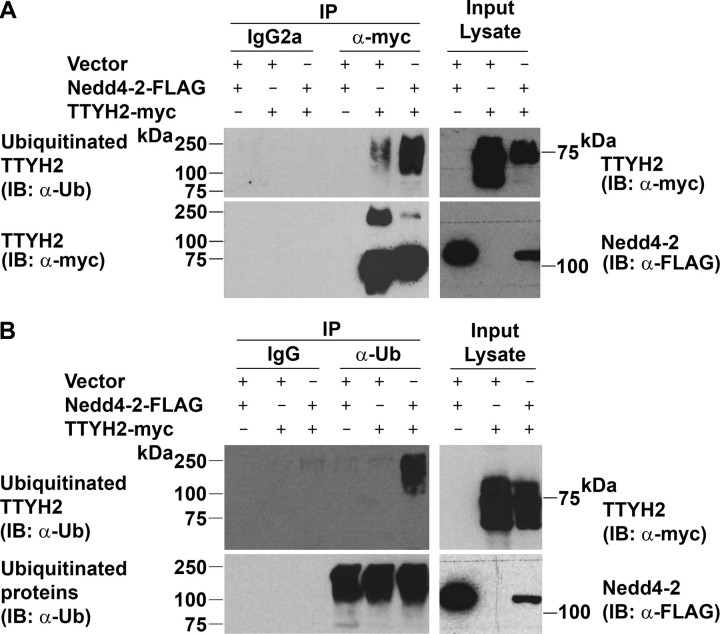
We have shown that the E3 ubiquitin ligase Nedd4-2 interacts with and ubiquitinates the CTD PY and SP motif containing Tweety family members TTYH2 and TTYH3 (Fig. 2). Focusing on TTYH2, we performed further immunoprecipitation experiments to compare the level of ubiquitination of TTYH2-Myc in HEK293 in the presence and absence of Nedd4-2-FLAG. Consistent with the data shown in Fig. 3, anti-ubiquitin Western blot analysis of proteins immunoprecipitated with an anti-Myc antibody detected ubiquitinated TTYH2-Myc from cells co-expressing Nedd4-2-FLAG (Fig. 4A). Interestingly, TTYH2-Myc levels were consistently lower in cells co-transfected with the Nedd4-2-FLAG expression construct (panel showing anti-Myc analysis of lysates), possibly indicating increased degradation of the ion channel in the presence of ubiquitin ligase. Also of note, lower but distinct levels of ubiquitination of TTYH2-Myc were also apparent in cells co-transfected with vector, indicating the action of an endogenous ubiquitin ligation system on TTYH2-Myc. In contrast, anti-Myc Western blot analysis of proteins immunoprecipitated with an anti-ubiquitin antibody detected ubiquitinated TTYH2-Myc only from cells co-expressing Nedd4-2 (Fig. 4B). It is likely that the low level of TTYH2-Myc ubiquitinated by an endogenous ligase was not detected by this Western blot analysis, because the anti-ubiquitin antibody used for immunoprecipitation enriched for all ubiquitinated proteins, whereas when an anti-Myc antibody was used for immunoprecipitation, TTYH2 was specifically enriched.
FIGURE 4.
Ubiquitination of TTYH2 in the presence and absence of Nedd4-2. Lysates from HEK293 cells co-transfected with the indicated constructs were subjected to immunoprecipitation with either anti-Myc or anti-ubiquitin antibodies or control IgGs. To prevent degradation of ubiquitinated proteins, cells were treated for 4 h with the proteasome inhibitor MG-132 before collection of lysates. A, proteins immunoprecipitated (IP) with either an anti-Myc antibody or control IgGs were examined by Western blot analysis (IB) using anti-ubiquitin (Ub) and anti-Myc antibodies. Lysates were examined by Western blot analysis using anti-Myc or anti-FLAG antibodies. B, proteins immunoprecipitated with either an anti-ubiquitin antibody or control IgGs were examined by Western blot analysis using anti-Myc and anti-ubiquitin antibodies. Lysates were examined by Western blot analysis using anti-Myc and anti-FLAG antibodies. Ubiquitinated and total TTYH2-Myc and Nedd4-2-FLAG are indicated.
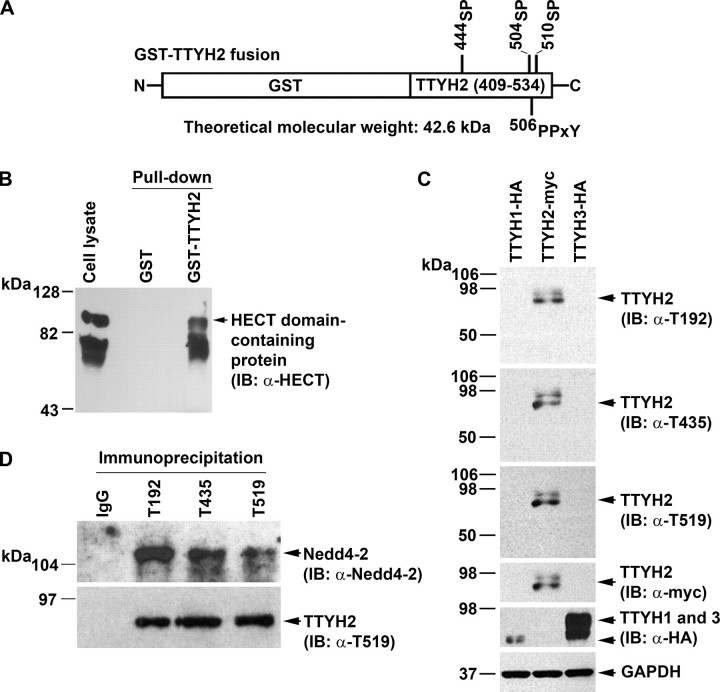
Endogenous TTYH2 and Nedd4-2 Interact—Confirmation of interactions between TTYH2 and endogenous Nedd4-2 was examined initially using a GST pull-down approach. A fusion of GST with the CTD of TTYH2 (residues 409-534) encompassing potential WW domain binding sites at 444SP,504SP, 506PPTY, and 510SP (Fig. 5A) was incubated with lysates from opossum kidney cells. Western blot analysis of proteins interacting with GST-TTYH2, using an antibody that reacts with the HECT domains of the E3 ubiquitin-protein ligases Nedd4 and Nedd4-2 (16, 28), detected prominent bands spanning ∼70-80 and ∼110-120 kDa (Fig. 5B). Since Nedd4 and Nedd4-2 migrate, respectively, as ∼120 and ∼115 kDa bands by Western blot analysis (16), these data are consistent with the proposal that the CTD of TTYH2 interacts with HECT domain-containing proteins, including either Nedd4 or Nedd4-2 and another unknown protein.
FIGURE 5.
Interactions between endogenous TTYH2 and Nedd4-2. A, schematic representation of a fusion of GST and the carboxyl-terminal tail of TTYH2 (residues 409-534). Consensus binding sites for HECT type E3 ubiquitin-protein ligases are indicated. B, lysates from opossum kidney cells were subjected to pull-down using either GST or GST-TTYH2. Input lysates and captured proteins were subjected to Western blot analysis using an anti-HECT domain polyclonal antibody that detects the HECT domain of Nedd4 and Nedd4-2. C, specificity of three anti-TTYH2 polyclonal antibodies (T192, T435, and T519). Chinese hamster ovary cells were transfected with expression constructs encoding either TTYH1-HA, TTYH2-Myc, or TTYH3-HA. Lysates were subjected to Western blot analysis (IB) using anti-TTYH2 T192, T435, and T519 antibodies. GAPDH was used as a loading control, and the expression of TTYH1-HA, TTYH2-Myc, and TTYH3-HA was confirmed by reprobing membranes with anti-HA or anti-Myc antibodies as appropriate. D, Western blot analysis using a specific anti-Nedd4-2 antibody of endogenous proteins immunoprecipitated from HEK293 cells using one of the anti-TTYH2 polyclonal antibodies T192, T435, or T519 or control rabbit IgG. The membrane was stripped and reprobed for immunoprecipitated endogenous TTYH2 using antibody T519 (the same pattern was obtained when the blot was reprobed with either T192 or T435 antibodies; data not shown).
To examine interactions between endogenous TTYH2 and an endogenous HECT type E3 ubiquitin-protein ligase, we developed three rabbit anti-TTYH2 peptide polyclonal antibodies (designated T192, T435, and T519). The selected TTYH2 peptides showed no homology to other proteins and, as shown in Fig. 1A, spanned residues 192-206 (located in ECL1), 435-450 (located in the CTD), and 519-534 (also located in the CTD). These antibodies specifically detected TTYH2 in transfected Chinese hamster ovary cells and did not cross-react with lysates from Chinese hamster ovary cells expressing TTYH1-HA or TTYH3-HA (Fig. 5C). The occurrence of interactions between endogenous TTYH2 and Nedd4-2 was analyzed by performing immunoprecipitations from HEK293 cell lysates using each of these three specific anti-TTYH2 antibodies and control rabbit IgGs. As shown in Fig. 5D, Western blot analysis using a specific anti-Nedd4-2 antibody (29) detected this HECT type E3 ubiquitin-protein ligase at the predicted molecular mass of ∼115 kDa in each of the immunoprecipitations performed using the anti-TTYH2 T192, T435, and T519 antibodies. Reprobing of the membrane with these antibodies specifically detected endogenous TTYH2 (Fig. 5D; only T519 is shown).
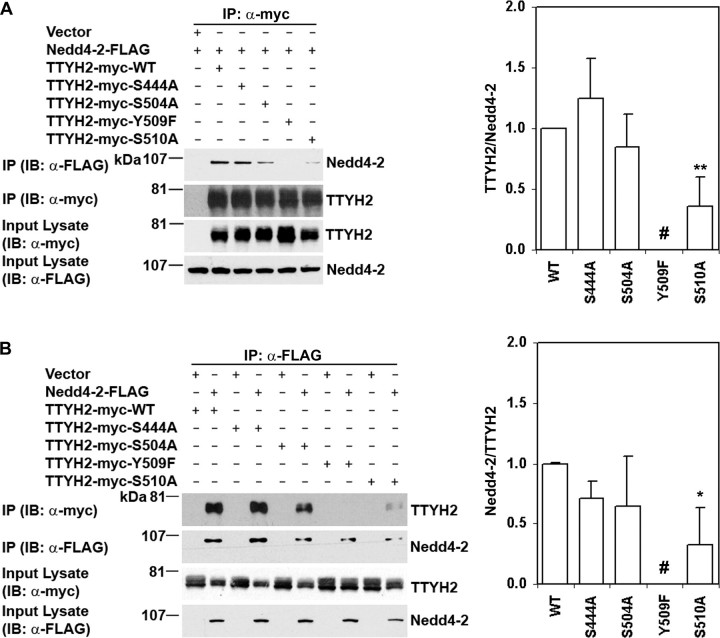
Identification of TTYH2 Motifs Mediating Interaction with Nedd4-2—To identify the residues mediating binding of TTYH2 and Nedd4-2, expression constructs encoding mutant forms of TTYH2 were generated in which consensus WW domain binding sites (SP and PY motifs) had been mutated: 444SP to 444AP (S444A), 504SP to 504AP (S504A), PPT509Y to PPT509F (Y509F), and 510SP to 510AP (S510A). HEK293 cells were co-transfected with expression constructs encoding Nedd4-2-FLAG and either wild type or one of these mutant forms of TTYH2-Myc. Immunoprecipitations were performed with anti-Myc or anti-FLAG antibodies. As shown in Fig. 6A, anti-FLAG Western blot analysis of proteins immunoprecipitated using the anti-Myc antibody indicated that the ability of TTYH2-Myc to bind Nedd4-2-FLAG was completely abolished by mutation of the tyrosine residue of the PY motif (mutant Y509F). In addition, mutation S510A reduced interactions between these proteins by ∼60% (Fig. 6A). Although in some experiments binding between TTYH2 and Nedd4-2 was reduced by the S504A mutation, this was not seen consistently (Fig. 6A). Further, TTYH2-Myc S444A was at least as efficient at forming a complex with Nedd4-2-FLAG as wild type TTYH2-Myc.
FIGURE 6.
The role of carboxyl-terminal domain SP and PY motifs in binding of TTYH2 to Nedd4-2. HEK293 cells were co-transfected with expression constructs encoding Nedd4-2-FLAG and either wild type (WT) or mutant TTYH2-Myc as indicated. Immunoprecipitates from cells co-transfected with vector (pcDNA3.1) and the Nedd4-2-FLAG expression construct were used as controls. Western blot analyses are representative of three experiments. The mean and S.D. of signal intensities, determined by densitometry analysis, from these three experiments, relative to data from TTYH2 wild type (WT), are graphed on the right of each panel. A, proteins immunoprecipitated (IP) with an anti-Myc monoclonal antibody and input lysates were subjected to Western blot analysis (IB) using either anti-FLAG or anti-Myc antibodies (left). The ratio of Nedd4-2-FLAG to TTYH2-Myc in immunoprecipitates is graphed in the right panel. #, binding completely abolished in each of the three experiments; **, p < 0.01. B, proteins immunoprecipitated with an anti-FLAG antibody and input lysates were subjected to Western blot analysis using either anti-Myc or anti-FLAG antibodies (left). The ratio of TTYH2-Myc to Nedd4-2-FLAG in immunoprecipitates is graphed in the right panel. #, binding completely abolished in each of the three experiments; *, p < 0.05.
As shown in Fig. 6B, these data were supported when an anti-FLAG antibody was used as the immunopurifying reagent. Western blot analysis of these immunoprecipitates using an anti-Myc antibody confirmed that the ability of TTYH2-Myc to bind Nedd4-2-FLAG was completely abolished by the Y509F mutation and that binding between these proteins reduced by an average of ∼60% as a result of the S510A mutation (Fig. 6B). In contrast, TTYH2-Myc S444A and S504A were as efficient at binding Nedd4-2-FLAG as wild type TTYH2-Myc (Fig. 6B). These data support the proposal that the TTYH2 PY motif is essential for interactions with the HECT type E3 ubiquitin-protein ligase Nedd4-2 and that 510SP, located immediately downstream of the PY motif, also impacts upon these interactions.
The impact of the TTYH2 SP (S444A, S504A, and S510A) and PY motif (Y509F) mutations on interactions with Nedd4-2 was also examined by confocal microscopy. In these experiments, HEK293 cells were co-transfected with either wild type or mutant TTYH2-GFP constructs and the Nedd4-2-FLAG construct. Cells were counterstained with 4′, 6-diamidino-2-phenylindole hydrochloride to highlight nuclei. As shown in Fig. 7, wild type TTYH2 and Nedd4-2 co-localized to cytoplasmic punctate structures proximal to the nucleus. Overlay of TTYH2-GFP (green) and Nedd4-2-FLAG (red) images indicated that there was considerable overlap (yellow in merged image) in the cellular localization of these proteins (Fig. 7A, merged). In cells co-expressing either of the TTYH2 mutants S444A or S504A and Nedd4-2, the TTYH2 signal was also predominantly localized to punctate cytoplasmic structures (green), and there was considerable overlap (yellow in merged image) with Nedd4-2 (red) expression (Fig. 7, B and C). In contrast, in cells expressing TTYH2-Y509F (green) and Nedd4-2 (red), there was little evidence of co-localization (Fig. 7D, merged), and, even more striking, Nedd4-2 maintained a diffuse cytoplasmic expression pattern (Fig. 7D, Nedd4-2-FLAG panel) similar to the signal apparent in cells transfected only with the Nedd4-2-FLAG expression construct (Fig. 7F). Although in cells co-expressing TTYH2-S510A (green) and Nedd4-2 (red) there was also evidence of co-localization of these proteins (Fig. 7E), overlapping signal (yellow in merged image) was generally less intense compared with cells expressing Nedd4-2 and either wild type, S444A, or S504A TTYH2 (Fig. 7, compare E with A, B, and C).
FIGURE 7.
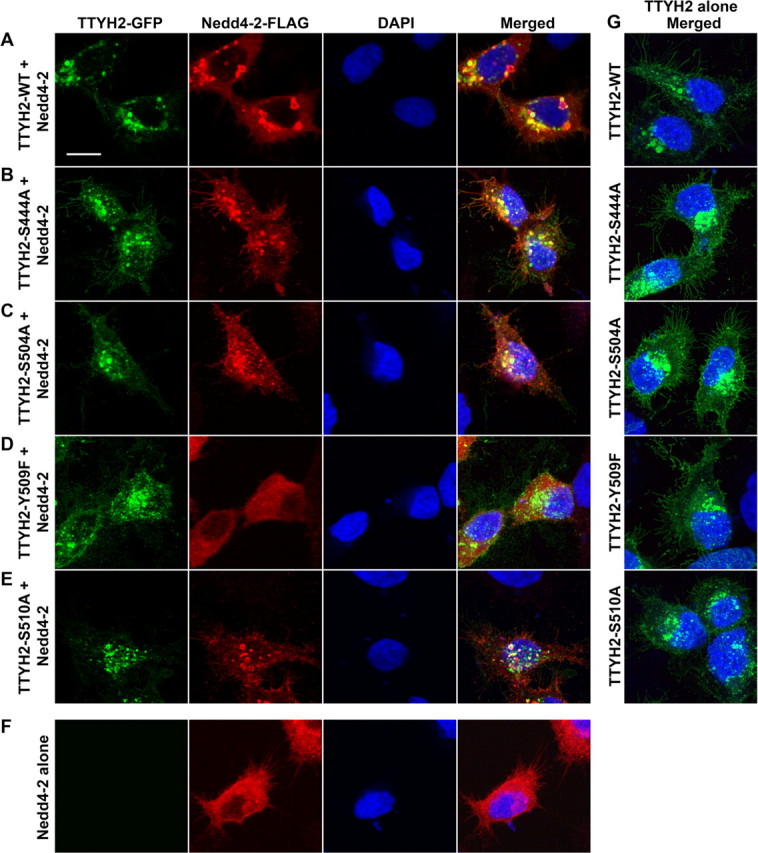
Localization of Nedd4-2 and wild type and mutant TTYH2. HEK293 cells were co-transfected with expression constructs encoding Nedd4-2-FLAG and either wild type (A), S444A (B), S504A (C), Y509F (D), or S510A (E) mutant TTYH2-GFP expression constructs as indicated. In controls, cells were transfected with either Nedd4-2-FLAG (F) or TTYH2-GFP (wild type or mutant) (G) expression constructs alone. Cells were fixed and then incubated with an anti-FLAG antibody, followed by a fluorescently tagged secondary antibody. Nuclei were stained with 4′, 6-diamidino-2-phenylindole hydrochloride. Images were acquired using a Leica TCS SP5 confocal microscope and processed and displayed using Adobe Photoshop CS3.
Comparison of TTYH2 in Nedd4-2-expressing and -nonexpressing cells indicated that the cellular location of TTYH2 in punctate cytoplasmic structures and, to a lesser extent, the cell surface appeared to be unaffected by the presence or absence of the ubiquitin ligase (Fig. 7, compare G with A-E). These data support the role of the PY motif of TTYH2 as an essential mediator of interactions with Nedd4-2 and, to a lesser extent, a role for the 510SP motif in these interactions.
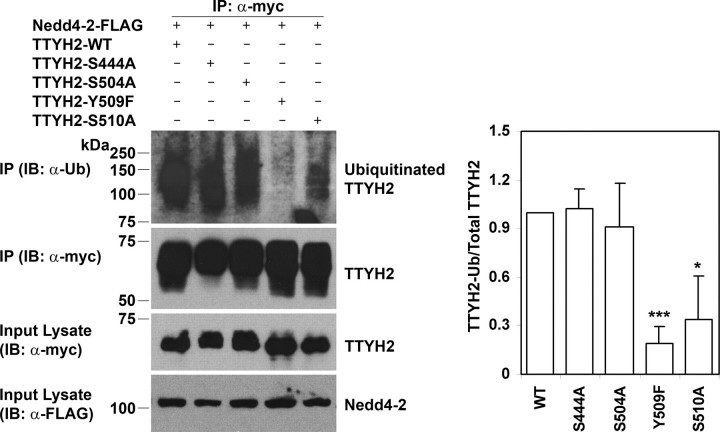
Impact of Mutation of TTYH2 CTD SP and PY Motifs on Nedd4-2-mediated Ubiquitination—The 510SP and PY motifs of TTYH2 impact on the binding of this protein to the E3 ubiquitin-protein ligase Nedd4-2 (Figs. 6 and 7), and TTYH2 is a ubiquitination target of Nedd4-2 (Fig. 3). We next examined whether ubiquitination of TTYH2 would be altered by mutation of the TTYH2 CTD SP and PY motifs, including those with reduced affinity for Nedd4-2. HEK293 cells co-transfected with the Nedd4-2-FLAG construct and either wild type or mutant TTYH2-Myc constructs were treated for 4 h with the proteasome inhibitor MG-132 before collection of lysates to prevent degradation of ubiquitinated proteins. Lysates were fully reduced and denatured, as previously described (34), to disrupt interactions between TTYH2 and other proteins and then subjected to immunoprecipitation using an anti-Myc antibody. As shown in Fig. 8, Western blot analysis of immunoprecipitated complexes, using an anti-ubiquitin antibody, showed that levels of Nedd4-2-mediated ubiquitination of TTYH2-Y509F were markedly reduced relative to wild type TTYH2 (∼80% reduction). Levels of Nedd4-2-mediated ubiquitination of TTYH2-S510A were also reduced relative to wild type TTYH2. Quantitatively, mutation of the 510SP motif reduced ubiquitination of TTYH2 by ∼66% relative to wild type protein. In contrast, TTYH2-S444A and -S504A were as efficiently ubiquitinated as wild type TTYH2 (Fig. 8). These data indicate that in addition to being critical for mediating binding of TTYH2 and Nedd4-2, the TTYH2 PY motif is also essential for Nedd4-2-mediated ubiquitination of this member of the Tweety family of chloride ion channels. In addition, the TTYH2 SP motif flanking the PY motif at position 509, but not SP motifs at positions 444 and 504, also impacts upon Nedd4-2-mediated ubiquitination of TTYH2.
FIGURE 8.
The effect of loss of SP and PY motifs on ubiquitination of TTYH2 by Nedd4-2. Lysates from HEK293 cells co-transfected with Nedd4-2-FLAG and either wild type or mutant TTYH2-Myc expression constructs were subjected to immunoprecipitation using an anti-Myc antibody. To prevent degradation of ubiquitinated proteins, cells were treated for 4 h with the proteasome inhibitor MG-132 before the collection of lysates. Proteins immunoprecipitated (IP) with an anti-Myc antibody were examined by Western blot analysis (IB) using anti-ubiquitin and anti-Myc antibodies. Lysates were also examined by Western blot analysis using anti-Myc and anti-FLAG antibodies. Ubiquitinated and total TTYH2-Myc and Nedd4-2-FLAG are indicated. Western blot analyses are representative of three experiments. Signal intensities, determined by densitometry analysis, from these three experiments were used to calculate the ratio of ubiquitinated to total immunoprecipitated TTYH2-Myc. The mean and S.D. of the ratio for each experimental condition, relative to data from TTYH2 wild type (WT), are graphed in the right panel. *, p < 0.05; ***, p < 0.001.
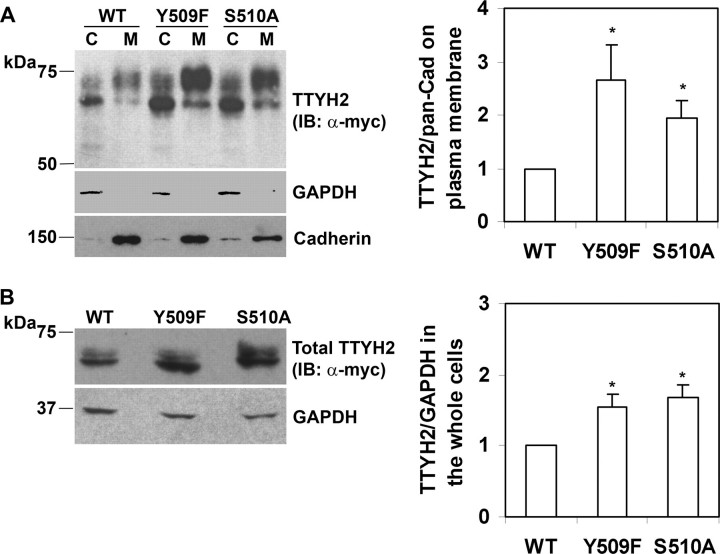
Disruption of TTYH2 Sites for Nedd4-2 Binding Increases the Level of Plasma Membrane and Total Cellular TTYH2—Ubiquitination by HECT domain E3 ubiquitin ligases regulates trafficking and degradation of membrane proteins (11). We have shown that Nedd4-2, a member of this family, binds to and ubiquitinates the membrane-spanning chloride ion channel TTYH2. Accordingly, we examined the impact of modulation of interactions between these two proteins on the levels of plasma membrane and total cellular TTYH2. HEK293 cells were co-transfected with Nedd4-2-FLAG and wild type, Y509F, or S510A TTYH2 expression constructs. TTYH2 levels were assessed by anti-Myc Western blot analysis of plasma membrane, and cytoplasmic fractions were separated by biotinylation of intact cells. The efficiency of fractionation by this approach was confirmed by anti-pancadherin and anti-GAPDH Western blot analysis. The ratio of plasma membrane TTYH2 to pancadherin, determined by densitometric analysis of Western blot analyses from three separate experiments, was used to compare changes in levels of plasma membrane TTYH2. As shown in Fig. 9A, levels of plasma membrane TTYH2 increased markedly when the sites most important in Nedd4-2 binding were mutated; TTYH2-Y509F and TTYH2-S510A cell surface levels were ∼3- and ∼2-fold higher than wild type TTYH2.
FIGURE 9.
Interactions between Nedd4-2 and TTYH2 impact on plasma membrane and total cellular levels of TTYH2. HEK293 cells were co-transfected with Nedd4-2-FLAG and wild type, Y509F, or S510A TTYH2-Myc expression constructs. A, plasma membrane (M) and cytoplasmic (C) fractions, separated by biotinylation of intact cells, were subjected to anti-Myc, anti-GAPDH, and anti-pancadherin Western blot analyses. The displayed blots are representative of three experiments. The mean and S.D. of the ratio of plasma membrane TTYH2 to pancadherin from these three experiments are graphed in the right panel.*, p < 0.05. B, whole cell lysates were subjected to anti-Myc and anti-GAPDH Western blot analyses. The displayed blots are representative of three experiments. The mean and S.D. of the ratio of whole cell TTYH2 to GAPDH from these three experiments, relative to data from TTYH2-WT, are graphed in the right panel.*, p < 0.05. IP, immunoprecipitation; IB, immunoblot; WT, wild type.
The impact of modulation of interactions between TTYH2 and Nedd4-2 on the levels of total cellular TTYH2 were assessed by anti-Myc Western blot analysis of whole cell lysates. The ratio of TTYH2 to GAPDH, determined by densitometric analysis of Western blot analyses from three separate experiments, was used to compare changes in levels of total cellular TTYH2. As shown in Fig. 9B, levels of total cellular TTYH2 were altered less than plasma membrane levels when the sites most important in Nedd4-2 binding were mutated; TTYH2-Y509F and TTYH2-S510A total cellular levels were both ∼1.5-fold higher than wild type TTYH2.
DISCUSSION
The Tweety proteins are a family of chloride ion channels that we have recently shown to contain five membrane spanning domains with a topology at the cell surface in which the amino terminus is located extracellularly and the CTD is located cytoplasmically (26). Three family members have been identified in humans, designated TTYH1, TTYH2, and TTYH3, with orthologous proteins known in murine species (20, 22, 23). Of these proteins, human TTYH2 and TTYH3 have been described as ionomycin, Ca2+-dependent chloride ion channels, whereas a TTYH1 isoform, variant at the carboxyl terminus, is proposed to function as a volume-regulated Cl- channel (20). To date, the mechanisms regulating these proteins are poorly defined.
Since the HECT type E3 ubiquitin ligase Nedd4-2 is known to regulate various ion channels, in this study we have investigated the role of ubiquitination mediated by this E3 ligase in the regulation of the three human members of the Tweety family. Using sequence analysis and immunoprecipitation protocols, we have defined varying levels of interactions between Tweety family proteins and Nedd4-2, which are dependent upon defined structural motifs present within each family member. Significantly, we have demonstrated binding of endogenous TTYH2 and Nedd4-2 and, importantly, shown that, functionally, ubiquitination of TTYH2 by Nedd4-2 regulates cell surface and total cellular levels of this chloride ion channel. In addition, we have shown that in transfected HEK293 cells, another Tweety family member, TTYH3, binds to and is efficiently ubiquitinated by Nedd4-2. We also show that the remaining human member of the Tweety family, TTYH1, does not bind to Nedd4-2. This lack of binding is consistent with the lack of consensus WW domain binding sites within the amino acid sequence of TTYH1.
Interactions between HECT type E3 ubiquitin-protein ligases and substrates are mediated by WW domains present within the ligase (11). Although most commonly, these domains bind substrate PY motifs (11), examples of WW domain binding to phosphorylated SP or TP motifs have also been reported (11). Consistent with these observations, our mutagenesis studies indicated that the CTD PY motif of TTYH2 is essential for Nedd4-2 binding. Using immunoprecipitation and confocal microscopy approaches, we demonstrated that mutation of the TTYH2 PY motif tyrosine residue completely abolished binding between TTYH2 and Nedd4-2, resulting in an almost complete loss of ubiquitination of TTYH2 in the presence of this ligase. Our observation of the essential role of the TTYH2 PY motif in mediating binding to Nedd4-2 is consistent with previous ion channel studies reporting interactions with HECT type E3 ligases. For example, binding of Nedd4-2 and the sodium channel Nav1.5 is abrogated by mutation of the PY motif of the channel (19). In addition, the PY motif of ENaC, which is mutated in a hereditary form of hypertension known as Liddle syndrome, is essential for Nedd4-2-mediated ubiquitination and degradation of this channel (35-37).
Our mutation studies also indicated that an SP motif located immediately downstream of the PY motif impacts upon Nedd4-2 binding and ubiquitination of TTYH2. Elimination of the 510SP motif reduced TTYH2 binding to Nedd4-2 by ∼60% with an accompanying 66% decrease in ubiquitination. Accordingly, it is possible that this motif, like the PY motif, is also directly involved in TTYH2 binding to Nedd4-2. However, based on our observations that the PY motif containing Tweety family member TTYH3 also binds efficiently to Nedd4-2 but lacks a downstream SP motif, it is likely that TTYH2 510SP has an indirect role in binding, potentially supporting the conformation of the adjacent PY motif. Thus, we propose that binding between TTYH2 and Nedd4-2 was reduced by mutation of the 510SP motif because it altered the conformation of the adjacent binding site.
Importantly, our data indicate that Nedd4-2-mediated ubiquitination of TTYH2 regulates cell surface and total cellular levels of this chloride ion channel. In these experiments, levels of cell surface and total cellular TTYH2 increased ∼3- and ∼1.5-fold, respectively, when Nedd4-2 binding and ubiquitination were abolished by mutation of the PY motif. Furthermore, the reduction in Nedd4-2 binding (∼60%) and ubiquitination (∼66%) of TTYH2 induced by mutation of 510SP resulted in ∼2- and ∼1.5-fold increases, respectively, in the levels of cell surface and total cellular TTYH2. These data indicate that signals inducing Nedd4-2-mediated ubiquitination of TTYH2 will impact upon the amount of this Cl- channel located on the cell surface and within cells and also point to an important role for ubiquitination in the regulation of TTYH2 function.
Our observation of regulation of a Tweety family member by the HECT type E3 ubiquitin-protein ligase Nedd4-2 is consistent with reports indicating that membrane proteins and, in particular, ion channels are commonly regulated by Nedd4-2-mediated ubiquitination. For example, Fotia et al. (17) showed that the activities of the voltage-gated Na+ channels 1.2, 1.7, and 1.8 are inhibited by this E3 ubiquitin-protein ligase. Also, Nedd4-2 is the physiological regulator of the activity of ENaC in two epithelial cell lines (38). It has been clearly demonstrated that suppression of ion channel activity by Nedd4-2 is associated with its ubiquitin ligase activity. For example, it has been shown that Nedd4-2-mediated ubiquitination of the sodium channel ENaC (39), the potassium channel KCNQ1 (40), and the amino acid transporter ATA2 (41) leads to internalization of these proteins and, eventually, lysosomal (15) and/or proteosomal (42) degradation.
Interestingly, Hryciw et al. (16) have reported that interactions between ClC-5, which has typically been regarded as an intracellular Cl- channel, and Nedd4-2 regulate both the cell surface expression of ClC-5 and its non-ion channel function in albumin uptake. In fact, it is now recognized that ClC-5 functions as a key component, independent of its role in ion transport, in the assembly of the macromolecular complex involved in protein endocytosis (16, 43, 44). In this respect, our observations here and previously (26) of cell surface-localized TTYH2 and the presence of a consensus integrin-binding RGD motif within ECL1 of human and murine TTYH2 (23) may also indicate that this protein will have functions in addition to ion channel activity, such as mediating cell to cell interactions by binding to integrins. In support of this proposal, other RGD motif-containing cell surface proteins, such as ADAM-15 and the cell adhesion molecule L1, have been shown to bind integrins αvβ3 and α5β1 (45) and αvβ3 (46), respectively.
Rae et al. (23) and Toiyama et al. (25) have previously reported up-regulation of TTYH2 mRNA levels in renal cell carcinoma and colon cancer, respectively. In addition, the latter group demonstrated that small interfering RNA-mediated down-regulation of TTYH2 resulted in reduced proliferation and increased aggregation of colon cancer-derived Caco-2 and DLD-1 cells (25). These cellular changes are consistent with a role for TTYH2 in colon cancer and, potentially, other cancers and suggest that mechanisms increasing TTYH2 protein levels will be important in cancer progression. In the in vivo cancer setting, it is possible that dysregulated Nedd4-2-mediated ubiquitination may promote such an oncogenic transformation. Our data indicate that this could occur through loss or reduction of Nedd4-2-mediated ubiquitination, leading to increased levels of TTYH2. Of relevance, it has recently been reported that mRNA transcript levels of serum and glucocorticoid-regulated kinase-1 (Sgk-1), a negative Nedd4-2 regulator (39), are markedly up-regulated in renal cell carcinoma (47). Thus, a possible mechanism controlling TTYH2 in cancer progression would involve increased phosphorylation of Nedd4-2 by Sgk-1, resulting in reduced Nedd4-2-mediated ubiquitination of TTYH2 and increased levels of this Tweety family member. Sgk-1-mediated phosphorylation of Nedd4-2 may also be relevant in regulating binding of this ligase to TTYH3.
In summary, our data provide the first evidence that the HECT type E3 ubiquitin ligase Nedd4-2 differentially regulates members of the Tweety family of Cl- ion channels. In particular, Nedd4-2 regulates cell surface and total cellular protein levels of TTYH2 by binding and ubiquitination of this second human member of the Tweety family. It is probable, based on studies of other proteins, that this mechanism will regulate the TTYH2 chloride channel activity observed by Suzuki and Mizuno (20) as well as other potential functions of TTYH2. This information will be important for understanding the role of TTYH2 and other Tweety family proteins in normal physiology and disease.
Acknowledgments
We thank Dr. Leonore de Boer for expert technical assistance with confocal microscopy experiments.
This work was supported by National Health and Medical Research Council of Australia grants (to J. D. H., P. P., and S. K.) and a fellowship (to J. D. H.) and Australian Research Council grants (to P. P. and S. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: E1, ubiquitin-activating enzyme; E2, ubiquitin-conjugating enzyme; E3, ubiquitin ligase; CTD, carboxyl-terminal domain; GST, glutathione S-transferase; Kv, voltage-gated potassium channel; Nav, voltage-gated sodium channel; HA, hemagglutinin; GAPDH, glyceralde-hyde-3-phosphate dehydrogenase; PBS, phosphate-buffered saline.
References
- 1.Sullivan, M., and Morgan, D. O. (2007) Nat. Rev. Mol. Cell Biol. 8894 -903 [DOI] [PubMed] [Google Scholar]
- 2.Muratani, M., and Tansey, W. P. (2003) Nat. Rev. Mol. Cell Biol. 4192 -201 [DOI] [PubMed] [Google Scholar]
- 3.Liu, Y. C. (2004) Annu. Rev. Immunol. 2281 -127 [DOI] [PubMed] [Google Scholar]
- 4.DiAntonio, A., and Hicke, L. (2004) Annu. Rev. Neurosci. 27223 -246 [DOI] [PubMed] [Google Scholar]
- 5.Mukhopadhyay, D., and Riezman, H. (2007) Science 315201 -205 [DOI] [PubMed] [Google Scholar]
- 6.Schnell, J. D., and Hicke, L. (2003) J. Biol. Chem. 27835857 -35860 [DOI] [PubMed] [Google Scholar]
- 7.Schwartz, D. C., and Hochstrasser, M. (2003) Trends Biochem. Sci. 28321 -328 [DOI] [PubMed] [Google Scholar]
- 8.Pickart, C. M., and Eddins, M. J. (2004) Biochim. Biophys. Acta 1695 55-72 [DOI] [PubMed] [Google Scholar]
- 9.Kee, Y., and Huibregtse, J. M. (2007) Biochem. Biophys. Res. Commun. 354329 -333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudol, M., and Hunter, T. (2000) Cell 1031001 -1004 [DOI] [PubMed] [Google Scholar]
- 11.d'Azzo, A., Bongiovanni, A., and Nastasi, T. (2005) Traffic 6429 -441 [DOI] [PubMed] [Google Scholar]
- 12.Rotin, D., Staub, O., and Haguenauer-Tsapis, R. (2000) J. Membr. Biol. 1761 -17 [DOI] [PubMed] [Google Scholar]
- 13.Harvey, K. F., and Kumar, S. (1999) Trends Cell Biol. 9166 -169 [DOI] [PubMed] [Google Scholar]
- 14.Staub, O., and Rotin, D. (2006) Physiol. Rev. 86669 -707 [DOI] [PubMed] [Google Scholar]
- 15.Staub, O., Gautschi, I., Ishikawa, T., Breitschopf, K., Ciechanover, A., Schild, L., and Rotin, D. (1997) EMBO J. 166325 -6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hryciw, D. H., Ekberg, J., Lee, A., Lensink, I. L., Kumar, S., Guggino, W. B., Cook, D. I., Pollock, C. A., and Poronnik, P. (2004) J. Biol. Chem. 27954996 -55007 [DOI] [PubMed] [Google Scholar]
- 17.Fotia, A. B., Ekberg, J., Adams, D. J., Cook, D. I., Poronnik, P., and Kumar, S. (2004) J. Biol. Chem. 27928930 -28935 [DOI] [PubMed] [Google Scholar]
- 18.Ekberg, J., Schuetz, F., Boase, N. A., Conroy, S. J., Manning, J., Kumar, S., Poronnik, P., and Adams, D. J. (2007) J. Biol. Chem. 28212135 -12142 [DOI] [PubMed] [Google Scholar]
- 19.van Bemmelen, M. X., Rougier, J. S., Gavillet, B., Apotheloz, F., Daidie, D., Tateyama, M., Rivolta, I., Thomas, M. A., Kass, R. S., Staub, O., and Abriel, H. (2004) Circ. Res. 95 284-291 [DOI] [PubMed] [Google Scholar]
- 20.Suzuki, M., and Mizuno, A. (2004) J. Biol. Chem. 27922461 -22468 [DOI] [PubMed] [Google Scholar]
- 21.Campbell, H. D., Schimansky, T., Claudianos, C., Ozsarac, N., Kasprzak, A. B., Cotsell, J. N., Young, I. G., de Couet, H. G., and Miklos, G. L. (1993) Proc. Natl. Acad. Sci. U. S. A. 9011386 -11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell, H. D., Kamei, M., Claudianos, C., Woollatt, E., Sutherland, G. R., Suzuki, Y., Hida, M., Sugano, S., and Young, I. G. (2000) Genomics 68 89-92 [DOI] [PubMed] [Google Scholar]
- 23.Rae, F. K., Hooper, J. D., Eyre, H. J., Sutherland, G. R., Nicol, D. L., and Clements, J. A. (2001) Genomics 77200 -207 [DOI] [PubMed] [Google Scholar]
- 24.Maleszka, R., Hanes, S. D., Hackett, R. L., de Couet, H. G., and Miklos, G. L. (1996) Proc. Natl. Acad. Sci. U. S. A. 93447 -451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toiyama, Y., Mizoguchi, A., Kimura, K., Hiro, J., Inoue, Y., Tutumi, T., Miki, C., and Kusunoki, M. (2007) World J. Gastroenterol. 132717 -2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He, Y., Ramsay, A. J., Hunt, M. L., Whitbread, A. K., Myers, S. A., and Hooper, J. D. (2008) Biochem. J. 412 45-55 [DOI] [PubMed] [Google Scholar]
- 27.Hooper, J. D., Nicol, D. L., Dickinson, J. L., Eyre, H. J., Scarman, A. L., Normyle, J. F., Stuttgen, M. A., Douglas, M. L., Loveland, K. A., Sutherland, G. R., and Antalis, T. M. (1999) Cancer Res. 593199 -3205 [PubMed] [Google Scholar]
- 28.Kumar, S., Harvey, K. F., Kinoshita, M., Copeland, N. G., Noda, M., and Jenkins, N. A. (1997) Genomics 40 435-443 [DOI] [PubMed] [Google Scholar]
- 29.Konstas, A. A., Shearwin-Whyatt, L. M., Fotia, A. B., Degger, B., Riccardi, D., Cook, D. I., Korbmacher, C., and Kumar, S. (2002) J. Biol. Chem. 27729406 -29416 [DOI] [PubMed] [Google Scholar]
- 30.Hryciw, D. H., Wang, Y., Devuyst, O., Pollock, C. A., Poronnik, P., and Guggino, W. B. (2003) J. Biol. Chem. 27840169 -40176 [DOI] [PubMed] [Google Scholar]
- 31.Swerdlow, P. S., Finley, D., and Varshavsky, A. (1986) Anal. Biochem. 156147 -153 [DOI] [PubMed] [Google Scholar]
- 32.Otte, L., Wiedemann, U., Schlegel, B., Pires, J. R., Beyermann, M., Schmieder, P., Krause, G., Volkmer-Engert, R., Schneider-Mergener, J., and Oschkinat, H. (2003) Protein Sci. 12 491-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verdecia, M. A., Bowman, M. E., Lu, K. P., Hunter, T., and Noel, J. P. (2000) Nat. Struct. Biol. 7 639-643 [DOI] [PubMed] [Google Scholar]
- 34.Miller, J. H., Draper, L. R., and Kempner, E. S. (2003) Biophys. J. 842781 -2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, C., Pribanic, S., Debonneville, A., Jiang, C., and Rotin, D. (2007) Traffic 81246 -1264 [DOI] [PubMed] [Google Scholar]
- 36.Staub, O., Dho, S., Henry, P., Correa, J., Ishikawa, T., McGlade, J., and Rotin, D. (1996) EMBO J. 152371 -2380 [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou, R., Patel, S. V., and Snyder, P. M. (2007) J. Biol. Chem. 28220207 -20212 [DOI] [PubMed] [Google Scholar]
- 38.Snyder, P. M., Steines, J. C., and Olson, D. R. (2004) J. Biol. Chem. 2795042 -5046 [DOI] [PubMed] [Google Scholar]
- 39.Snyder, P. M., Olson, D. R., and Thomas, B. C. (2002) J. Biol. Chem. 2775 -8 [DOI] [PubMed] [Google Scholar]
- 40.Jespersen, T., Membrez, M., Nicolas, C. S., Pitard, B., Staub, O., Olesen, S. P., Baro, I., and Abriel, H. (2007) Cardiovasc. Res. 7464 -74 [DOI] [PubMed] [Google Scholar]
- 41.Hatanaka, T., Hatanaka, Y., and Setou, M. (2006) J. Biol. Chem. 28135922 -35930 [DOI] [PubMed] [Google Scholar]
- 42.Malik, B., Schlanger, L., Al-Khalili, O., Bao, H. F., Yue, G., Price, S. R., Mitch, W. E., and Eaton, D. C. (2001) J. Biol. Chem. 27612903 -12910 [DOI] [PubMed] [Google Scholar]
- 43.Devuyst, O., Jouret, F., Auzanneau, C., and Courtoy, P. J. (2005) Nephron 99 p69-73 [DOI] [PubMed] [Google Scholar]
- 44.Hryciw, D. H., Ekberg, J., Pollock, C. A., and Poronnik, P. (2006) Int. J. Biochem. Cell B 381036 -1042 [DOI] [PubMed] [Google Scholar]
- 45.Nath, D., Slocombe, P. M., Stephens, P. E., Warn, A., Hutchinson, G. R., Yamada, K. M., Docherty, A. J., and Murphy, G. (1999) J. Cell Sci. 112579 -587 [DOI] [PubMed] [Google Scholar]
- 46.Yip, P. M., Zhao, X., Montgomery, A. M., and Siu, C. H. (1998) Mol. Biol. Cell 9 277-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon, P., Schneck, M., Hochstetter, T., Koutsouki, E., Mittelbronn, M., Merseburger, A., Weigert, C., Niess, A., and Lang, F. (2007) Cell Physiol. Biochem. 20 715-728 [DOI] [PubMed] [Google Scholar]