Abstract
Saffron, the processed stigma of Crocus sativus, is characterized by the presence of several apocarotenoids that contribute to the color, flavor, and aroma of the spice. However, little is known about the synthesis of aroma compounds during the development of the C. sativus stigma. The developing stigma is nearly odorless, but before and at anthesis, the aromatic compound β-ionone becomes the principal norisoprenoid volatile in the stigma. In this study, four carotenoid cleavage dioxygenase (CCD) genes, CsCCD1a, CsCCD1b, CsCCD4a, and CsCCD4b, were isolated from C. sativus. Expression analysis showed that CsCCD1a was constitutively expressed, CsCCD1b was unique to the stigma tissue, but only CsCCD4a and -b had expression patterns consistent with the highest levels of β-carotene and emission of β-ionone derived during the stigma development. The CsCCD4 enzymes were localized in plastids and more specifically were present in the plastoglobules. The enzymatic activities of CsCCD1a, CsCCD1b, and CsCCD4 enzymes were determined by Escherichia coli expression, and subsequent analysis of the volatile products was generated by GC/MS. The four CCDs fell in two phylogenetically divergent dioxygenase classes, but all could cleave β-carotene at the 9,10(9′,10′) positions to yield β-ionone. The data obtained suggest that all four C. sativus CCD enzymes may contribute in different ways to the production of β-ionone. In addition, the location and precise timing of β-ionone synthesis, together with its known activity as a fragrance and insect attractant, suggest that this volatile may have a role in Crocus pollination.
Crocus sativus is a triploid sterile plant characterized by its long red stigmas. These stigmas, when desiccated, constitute the spice known as saffron. Saffron is considered a complex mixture of volatile and non-volatile compounds that contribute to its overall aroma and flavor. The main components of saffron responsible for coloring strength are the derived carotenoids cis- and trans-crocins, picrocrocin, and its degradation product, the odor-active safranal that composes up to 70% of total volatiles (1). In addition to safranal, 150 volatile compounds are estimated to be present in the spice, and ∼60 constituents have been identified (2). The structures of several of these reveal an isoprenoid-based origin like safranal, and are assumed to be the products of the oxidative cleavage of carotenoids.
In recent years, a family of enzymes that cleave carotenoid substrates at different double bond positions have been described in plants. This family, the carotenoid cleavage dioxygenases (CCDs),2 are specific for the location of the double bond in the molecule they cleave, but many are promiscuous in their carotenoid substrate choice (3, 4). The founding member of the CCD family was Vp14, a 9-cis-epoxycarotenoid dioxygenase (NCED). This and other closely related dioxygenases catalyze the cleavage at the 11,12 double bond of both 9-cis-violaxanthin and 9-cis-neoxanthin to produce xanthoxin (5–7), which is subsequently converted into the phytohormone abscisic acid (ABA). Homology to VP14 allowed the discovery of other enzymes in different plant species that could be distinguished based on their specific cleavage sites. Among them, two groups have been expanded in the last year, the CCD1 and the CCD4 groups of enzymes. CCD1 enzymes symmetrically cleave the 9,10(9′,10′) double bonds of multiple carotenoid substrates to produce a C14 dialdehyde and two C13 cyclohexenone derivatives (4). More recently, an additional cleavage activity for CCD1 at the 5,6(5′,6′) double bonds of lycopene has been reported (8). CCD1 genes have been identified either genetically or biochemically in Arabidopsis (3), grape (9), nectarine (10), tomato (11), petunia (12), melon (13), and Crocus (14). These CCD1 enzymes seem to be localized in the cytoplasm, in contrast with the CCD4 enzymes that showed a plastid targeting sequence. Among the latter, AtCCD4 has recently been localized in plastoglobules (15), although the cleavage activity and resulting products for the CCD4 group are still to be identified (4).
We describe here the isolation of four CCD genes from C. sativus (CsCCD1a, CsCCD1b, CsCCD4a, and CsCCD4b). All four cDNAs were isolated by homology taking advantage of conservation of carotenoid dioxygenase motifs. Using in vivo bacterial expression assays, we found that all four CsCCD enzymes were able to cleave different carotenoid substrates at their 9,10(9′,10′) double bonds. To study the possible involvement of these CsCCDs in the synthesis of C13-norisoprenoids in C. sativus stigma, the expression patterns of the four CsCCDs were followed during stigma development and compared with the changes in the C13-norisoprenoid levels.
EXPERIMENTAL PROCEDURES
Plant Material and Stress Treatments—Plant tissues and stigmas from C. sativus grown under field conditions in Tarazona de la Mancha, Spain, were used throughout the experiments. Stigmas were collected at developmental stages defined as follows: yellow stigma, closed bud inside the perianth tubes (around 0.3 cm in length); orange stigma, closed bud inside the perianth tubes (around 0.4 mm in length); red stigma, closed bud inside the perianth tubes (0.8 cm in length); -2da, 2 days before anthesis, dark red stigmas in closed bud outside the perianth tubes (3 cm); da, day of anthesis, dark red stigmas (3 cm); +1da, 1 day after anthesis, dark red stigmas; and +3da, 3 days after anthesis, dark red stigmas. Petals, style, and stamens were collected from flowers at the time of anthesis. Young and senescent leaves, roots, and corms were frozen in liquid nitrogen and stored at -80 °C until required. To determine stress-induced gene expression in leaves, whole leaves were collected from plants growing in fields, cut in 1-cm long pieces and transferred to 24-well plates containing 1 ml of water supplemented with ABA (100 μm), 200 mm CaCl2, or distilled water that was used as a control; and incubated under normal conditions (16 h light/8 h dark cycles at 22 °C). Heat stress was applied by raising the temperature in the growth chamber to 38 °C for 6 h. Leaf tissue was collected 24 h after and immediately frozen in liquid nitrogen and stored at -80 °C until required. For the dehydration experiments leaf pieces were subjected to dehydration in Petri dishes on Whatman No. 3MM paper (Whatman International, Maidstone, England) at 60% relative humidity and 22 °C, under dim light (100 lux), and collected at 2, 5, and 10 h.
Seeds from Arabidopsis wild type ecotype Columbia (Col) and transgenic lines were sown in pots containing vermiculite and watered with nutrient solution under a controlled environment with 16-h light/8-h dark cycles at 22 °C. Seeds from transformed Arabidopsis plants were surface sterilized by rinsing them in 70% (v/v) ethanol for 1 min, followed by a 15-min treatment in 10% (v/v) bleach + 0.05% (v/v) Triton X-100 and three rinses in sterile-distilled water.
Cloning of C. sativus CCD cDNAs—Total RNA and mRNA were isolated from developed C. sativus stigma in two different developmental stages by using Ambion PolyAtrack® and following the manufacturer's protocols (Ambion, Inc.). The first-strand cDNAs were synthesized by RT from 2 μg of total RNA using a first-strand cDNA synthesis kit (GE Healthcare) and an oligo(dT) primer. These cDNAs were used as templates for PCR using degenerate primers: 5′-TT(T/C) GA(T/C) GGN GA(T/C) GGN ATG G-3′; and 5′-GC(A/G) TTC CAN AG(A/G) TG(A/G) AA(A/G) CA(A/G) AA(A/G) CA(A/G) TC-3′. The design of these primers was based on the conserved regions of the bean (AAK38744), tomato (Z97215), and maize (U95953) proteins. Conditions for RT were as follows: 65 °C for 5 min, followed by 37 °C for 1 h, followed by 75 °C for 5 min. Thermal cycling parameters were 2 min at 95 °C, 10 times at 30 s at 95 °C, 30 s at 55 °C-0.3 °C/cycles, and 1 min 30 s at 72 °C; 35 times at 30 s at 95 °C, 30 s at 50 °C, and 1 min 30 s at 72 °C; and finally 5 min at 72 °C. The PCR products were separated in an agarose gel, purified, ligated into pGEMTeasy (Promega, Madison, WI), and then introduced into E. coli DH5α. In total three different clones were obtained. The full-length clones were obtained by a reverse transcription-polymerase chain reaction of 3′ and 5′ amplification ends (SMART-RACE cDNA Amplification kit, Clontech) using RNA from stigma tissue, and several primer combinations (Table 1).
TABLE 1.
Primer sequences used for CsCCD gene cloning and analysis
| cDNA | Sequence | Primer | |
|---|---|---|---|
| Primers for CCDs isolation | CsCCDs | CAYGAYTTYGCNATHACNGRACA | Sense |
| CCRTGAAANCCRTANGGNACTCG | Antisense | ||
| CsCCD1a | 5′–ATGGGAGAAGTAGCGAAGAG–3′ | Sense | |
| 5′–TACGTCGGTTTGCTGCCACTGGA–3′ | Antisense | ||
| CsCCD1b | 5′–ATGGCAAATAAGGAGGAGGCA–3′ | Sense | |
| 5′–GTCATGTCTCTGCTTGGTGCT–3′ | Antisense | ||
| CsCCD4 | 5′–ATGCAGGTGGACCCAACCAAG–3′ | Sense | |
| 5′–CTGCTGTGACAGCAGCTCAGC–3′ | Antisense | ||
| Primers for 3′ and 5′ RACE-PCRs | CsCCD1a | 5′–ATGTACAAGGTATCTTTGACT–3′ | Sense |
| 5′–CAAGCATTAGCATTATGGAAT–3′ | Sense | ||
| CsCCD1b | 5′–TGTGGTTCAGAGGCAATATTTGT–3′ | Antisense | |
| 5′–ACAAATATTGCCTCTGAACCACA–3′ | Antisense | ||
| 5′–TGTGTGTTGATTTCACCCCACAA–3′ | Antisense | ||
| 5′–AAGACACACCGATACGAGCTC–3′ | Sense | ||
| 5′–TCAGTAATAGCGAAATCATG–3′ | Antisense | ||
| CsCCD4 | 5′–TTCGGTGTGGGATGTTTCGGA–3′ | Sense | |
| 5′–GCTTCTACACGATCTGGATG–3′ | Antisense | ||
| 5′–CGTCGCCCTCGAAGAGGTGGT–3′ | Sense | ||
| 5′–GATCTTCCTCAGCGTCTTCTC–3′ | Antisense | ||
| Primers for cloning in expression vectors | CsCCD1a | 5′–GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGGGAGAAGTAGCGAAGGA–3′ | Sense |
| 5′–GGGGACCACTTTGTACAAGAAAGCTGGGTCCTACTCTGCTTGGTGCTTCTGAAGTTC–3′ | Antisense | ||
| CsCCD1b | 5′–GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGGCAAATAAGGAGGAGGCAGA–3′ | Sense | |
| 5′–GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAGTCTCTGCTTGGTGCTTCTG–3′ | Antisense | ||
| CsCCD4a/b | 5′–GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGCAGGTGGACCCAACCAAGG–3′ | Sense | |
| 5′–CCGGGGACCACTTTGTACAAGAAAGCTGGGTTACTGCTGTGACAGCAGCTCAGC–3′ | Antisense | ||
| CsCCD4a-211 | 5′–GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGCAGGTGGACCCAACCAAGG–3′ | Sense | |
| 5′GGGGACCACTTTGTACAAGAAAGCTGGGTCCTACTGCTGTGACAGCAGCTCAGCTTC–3′ | Antisense | ||
| Primers for RT-PCR expression analysis | CsCCD1a | 5′–ATGGGAGAAGTAGCGAAGGAGG–3′ | Sense |
| 5′–TACGTCGGTTTGCTGCCACTGGAG–3′ | Antisense | ||
| CsCCD1b | 5′–CATCCGTCGCAGTGGACTTAC–3′ | Sense | |
| 5′–AGGCATGGAATCCATATGGAAC–3′ | Antisense | ||
| CsCCD4a | 5′–CAATCTCAAGTATTAGCATTC–3′ | Sense | |
| CsCCD4b | 5′–CACTACCCATCTCATCAAGA–3′ | Sense | |
| CsCCD4a/b | 5′–CTGCTGTGACAGCAGCTCAGC–3′ | Antisense | |
| Primers for location analysis | CsCCD4 | 5′–GGGGACAAGTTTGTACAAAAAAGCAGGCTGAACCATGGATTATCGGTTGTCATCCTC–3′ | Sense |
| CsCCD4 | 5′–GGGGACCACTTTGTACAAGAAAGCTGGGTCCTGCTGTGACAGCAGCTCAGCT–3′ | Antisense |
DNA Sequencing and Analysis of DNA Sequences—Plasmids were sequenced using an automated DNA sequencer (ABI Prism 310, PerkinElmer Life sciences). Computer analyses of the DNA and amino acid sequences were carried out using the WU-BLAST program at ExPASy. Multisequence alignment of the amino acid sequences was carried out using the Clustal W program at the PBIL (npsa-pbil.ibcp.fr). Relative molecular masses and pI values were calculated from the deduced amino acid compositions with the Compute pI/Mw tool available at the ExPASy Molecular Biology Web server (Geneva, Switzerland). Subcellular sorting was predicted at the PSORT Web server for analyzing and predicting protein-sorting signals at the Institute for Molecular and Cellular Biology (Osaka, Japan). Both the phylogenetic analysis and tree construction were carried out using the Clustal W program and the Phylodendron program.
Expression Analysis—Gene-specific oligos were designed to be used for the expression analysis (Table 1). For RT-PCR, total RNA was isolated from C. sativus anthers, corms, leaves, petals, roots, stigmas, and style, by grinding fresh tissue in liquid nitrogen to a fine powder and extracting in 1 ml of TRIzol reagent (Invitrogen) per 100 mg of fresh tissue weight, according to the protocol of the manufacturer. The RNA was resuspended in 100 μl of RNase-free water and treated with RQ1 RNase-free DNase (Promega, Madison, WI). The DNase was heat inactivated before RT-PCR. The RNA was quantified with a spectro-photometer at OD of 260 and 280 and stored at -80 °C. First-strand cDNAs were synthesized by RT from 2 μg of total RNA using a first-strand cDNA synthesis kit (Amersham Biosciences) and random primers (Promega). As an internal control, the mRNA level of the constitutively expressed ribosomal protein 18 was used (16). For the determination of semi-quantitative RT-PCR conditions, we carried out PCR for a different number of amplification cycles (20, 25, 30, 35, 40, and 45) using cDNA from yellow, red, anthesis, and postanthesis stigmas (1 μl from a 0.5 dilution from the original RT reaction), and specific primers for CsCCD1a, CsCCD1b, CsCCD4a, CsCCD4b, and RPS18. For quantization, the RT-PCR products were imaged using a UV gel documentation system. Densitometry was performed on each band by application of the IP-010-SD photodocumentation system program (Vilber Lourmat). Exponential amplification for these genes occurred during cycles 30–35. Therefore, 35 cycles were performed for each expression analysis.
Confocal Laser Scanning Microscopy and Image Analysis— Confocal laser microscopy analysis was performed with stable transgenic Arabidopsis lines. Arabidopsis leaves from 60-week-old plants were observed using a Leica TCS-SL confocal laser scanning microscope. For green fluorescent protein (GFP) detection, the excitation source was an argon ion laser at 488 nm and emission was observed between 510 and 530 nm. Chloroplast autofluorescence was detected between 660 and 700 nm.
Expression and Enzymatic Activity—For expression of CsCCD1a, CsCCD1b, CsCCD4, and CsCCD4a-211 the cDNAs were amplified with the att-primers (Table 1) and introduced into the vector pDONR™221 (Invitrogen) by a BP recombination reaction, and from this vector to the destination vector pDES14 by a LR recombination reaction, using Gateway Technology (Invitrogen). The recombinant clones were co-transformed in the E. coli BL21-AI strain with plasmids for the production of β-carotene and zeaxanthin. Positive colonies were selected from the Luria-Bertani medium containing ampicillin (100 μg/ml) and chloramphenicol (40 μg/ml) and grown in liquid or solid Luria-Bertani medium before being induced with 2% (w/v) arabinose. Carotenoid-derived metabolites from pelleted or scraped bacteria were extracted and analyzed by HPLC as previously described (17, 18). For volatile analysis, bacteria were grown and analyzed as described (12) with the following modifications. Desorption tubes filled with 200 mg of tenax TA were used instead of SuperQ columns. Volatile organic compounds retained in the tenax tube were thermally desorbed in a Turbomatrix Thermal Desorber (PerkinElmer Life Sciences). Primary desorption was performed at 300 °C for 10 min under an 80 ml/min helium flow, without inlet split, maintaining the cold trap at -30 °C. Secondary desorption was performed at 300 °C for 1 min, with a total flow of 22.5 ml/min and a 20 ml/min outlet split.
Vector Construction and Plant Transformation—To produce transgenic plants in which GFP-tagged CsCCD4a protein was expressed under the control of the 35S promoter, vector pGWB5 was used (19). The strategy followed for cloning CsCCD4a in this vector was based on Gateway Technology, and the oligonucleotides used are indicated in Table 1. The amplified fragment was introduced into the vector pDNOR221 by a BP recombination reaction (Invitrogen), and from this vector to the destination vector pGWB5 by a LR recombination reaction. The recombinant pGWB5 vector was transferred into Agrobacterium tumefaciens strain GV3101 by electroporation and bacteria were selected on YEB agar with 50 μgml-1 kanamycin and hygromicin and 100 μgml-1 rifampicin and 25 μgml-1 gentamicin. Arabidopsis plants were transformed by floral dipping (20) and transformants selected on Murashige and Skoog agar with 50 μgml-1 kanamycin and hygromicin.
Volatile Collection and Analysis from Fresh Stigma Tissue— HS-SPME extraction conditions were used: 10 mg of C. sativus stigma from different stages were introduced into a 22-ml crimp cap vial, together with 10 μl of a 1 ppm methylsalicylate solution as an internal standard. Then 65-μm PDMS/DVB fiber (polydimethylsiloxane/divinylbenzene) (Supelco, Pennsylvania, PA) was exposed to the headspace for 1 h at 30 °C while being shaken at 300 rpm. After sampling, the fiber was immediately thermally desorbed in the GC injector. A desorption time of 1 min at 250 °C was used in the splitless mode. Before sampling, each fiber was reconditioned for 5 min in a GC injector port at 250 °C. Triplicate analyses were performed on each stage.
Volatile organic compounds were analyzed by GC/MS using a Clarus 600 gas chromatograph from PerkinElmer, equipped with a ZB-5ms capillary column (30 m, 0.25 mm, 0.25 μm) (Phenomenex, Torrance, CA). Oven programming conditions were 40 °C for 2 min, 5 °C/min ramp until 180 °C, 15 °C/min ramp until 250 °C, then 5 min at 250 °C. Helium was used as the carrier gas at a constant pressure of 19.5 psi. Detection was performed by a PerkinElmer Clarus 600T mass spectrometer in the EI mode (ionization energy, 70 eV; source temperature, 150 °C). The acquisition was made in scan mode (m/z range 35–250). The identification of peaks was initially determined by mass spectrometry of the collected volatiles and subsequently by co-elution with a known standard (Sigma), except for megastigma-4,6,8-triene, which was identified by comparison with mass spectra in the NIST05 library.
RESULTS
Cloning and Characterization of cDNAs from C. sativus Stigmas Coding CCD Enzymes—To identify dioxygenases involved in apocarotenoid production, we have used a homology-based strategy, taking advantage of specific carotenoid dioxygenase motifs. Two populations of cDNAs were prepared by reverse transcription of poly(A)+ from total RNA isolated from C. sativus stigmas at the yellow and scarlet stages. DNA fragments were amplified by degenerate primers CCD-F and CCD-R. Initially three fragments were obtained, and subsequently labeled CsCCD1a, CsCCD1b, and CsCCD4 (GenBank™ accession numbers AJ416712, AJ416713, and AJ416714). Specific oligonucleotides were designed to obtain the full-length clones by RACE-PCR (Table 1). Complete cDNA clones were obtained for each dioxygenase and 100% identity was found between CsCCD1a and the previously isolated CsCCD (14), and 70% between CsCCD1b and CsCCD. Two different full cDNA clones were obtained for CsCCD4: CsCCD4a and CsCCD4b. A 98% identity was obtained between CsCCD4a and CsCCD4b that can be considered as allelic variants, due to the triploid nature of C. sativus. CsCCD4a (580 amino acids) and CsCCD4b (569 amino acids) showed 100 and 98% similarity in 369 amino acids with the previously isolated CsZCD (369 amino acids) (14), respectively. However, CsCCD4a and -b are more than 200 amino acids longer than CsZCD. A typical cleavable plastid transit peptide was predicted for all three CsCCD4 enzymes (21). The location prediction program PSORT predicted that all the CsCCD4 enzymes would be localized at the chloroplast. Because the stigmas do not have chloroplasts, chromoplasts seem to be the most likely location for these enzymes.
The subcellular localization of CsCCD4 enzymes was experimentally tested using fusions to the GFP. Because CsCCD4a and CsCCD4b proteins are identical in the N-t signal plastid transit peptide and in general very similar, we performed the location experiments only with one of them. Confocal imaging of the 35S-driven CsCCD4a-GFP fusion protein in 5-week-old transgenic Arabidopsis plants confirmed the plastid location of the protein in mature leaves (Fig. 1A). CsCCD4a-GFP accumulates in plastids in concentrated foci, as have been previously observed for other plastoglobule-localized proteins (22–24). Interestingly, no signal was detected in young seedlings, neither in the cotyledons nor in the first young leaves. This fact is in agreement with the variation in the number and size of plastoglobules observed during the development of leaves in Arabidopsis (25). It is probable that the small size and low number of plastoglobules in young leaves did not allow the detection of the GFP signal by confocal imaging. Plastoglobules are ubiquitously found in all types of plastids (26, 27) and recently, chromoplast plastoglobules were shown to contain specific enzymatic functions in carotenoid biosynthesis (15), in addition to their well known function of carotenoid storage and sequestration (24). A dioxygenase from Arabidopsis, At4g19170 (assigned CCD4 for carotenoid cleavage dioxygenase; but also named NCED4 for 9-cis-epoxy-carotenoid dioxygenase) was also localized in chloroplast plastoglobules (15).
FIGURE 1.
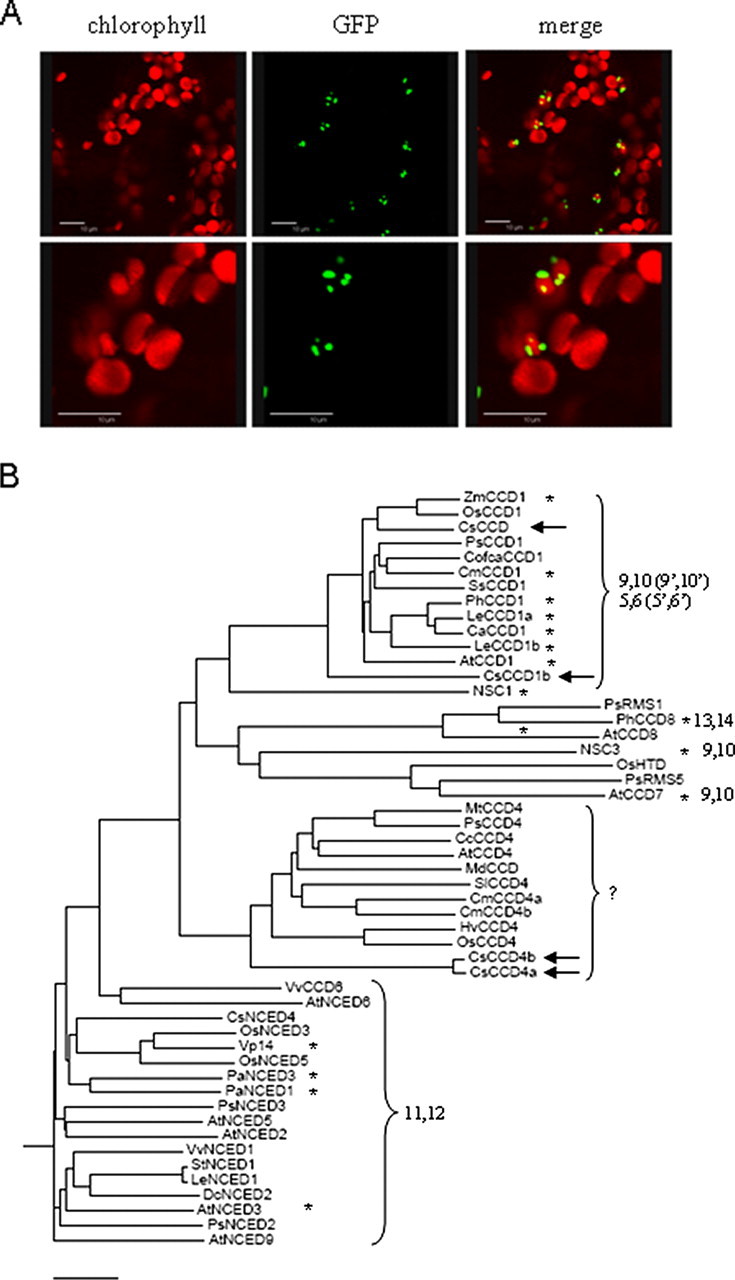
The C. sativus dioxygenases belong to two different subfamilies. A, confocal microscopic detection of CsCCD4a-GFP fluorescence in Arabidopsis leaves at two resolution levels. Merge, overlap of chlorophyll and GFP signals. Scale bars = 10 μm. B, unrooted phylogenetic tree of the CCD proteins based on amino acid sequence similarity. Only full-length members of the family are included. The predicted protein sequences were initially clustered using ClustalW. Accession numbers used are: At, A. thaliana, AtCCD1 (At3g63520), AtNCED2 (At4g18350), AtNCED3 (At3g14440), AtCCD4 (At4g19170), AtNCED5 (At1g30100), AtNCED6 (At3g24220), AtCCD7 (At2g44990), AtCCD8 (At4g32810), and AtNCED9 (At1g78390); Hv, Hordeum vulgare HvCCD4 (ak248229.1); Ca, Capsicum annuum, CaCCD1 (Y14164); Cm, Chrysanthemum × morifolium CmCCD4a (AB247158), CmCCD4b (AB247160); Cc, Citrus clementina CcCCD4 (abc26011); Cofa, Coffea Arabica, CofcaCCD1 (DQ157170); Cs, C. sativus, CsCCD (CsCCD1a) (CAC79592), CsCCD1b (EU523661), CsCCD4a (EU523662), CsCCD4b (EU523663), and CsNCED4 (EU527188); Cm, Cucumis melo, CmCCD1 (DQ269467); Ls, Lycopersicum sculentum LeCCD1a (AB120111), LeCCD1b (AAT68188), LeNCED1 (CAB10168); Ml, Malus × domestica MdCCD (Z93765); Mt, Medicago trunculata MtCCD4 (ac144759); NSC, Nostoc sp NSC1 (NP_485149) NSC3 (NP_488935.1); Os, Oryza sativa OsHTD (XP473418), OsCCD4 (AP005825), OsCCD1(ABG22113), OsNCED3 (NM001057300), OsNCED5 (AY838901); Pa, Persea americana, PaNCED1 (AAK00632), PaNCED3 (AAK00623); Ph, Petunia × hybrida PhCCD1 (AY576003); Ps, Pisum sativum, PsNCED2 (AB080192), PsNCED3 (AB080193), PsRMS1 (Q6Q623), PsRMS5 (ABD67496), PsCCD1(BAC10549), PsCCD4 (BAC10552); Sl, Solanum lycopersicom SlCCD4 (ap009393); Ss, Suaeda salsa, SsCCD1(DQ003599); St, Solanum tuberosum, StNCED1 (AJ276244); Vv, Vitis vinifera, VvNCED1 (AY3337613), VvCCD6 (A7NXT2); Zea mays VP14 (U95953.1), ZmCCD1 (ABF85668.1). The arrows indicate the CsCCD enzymes, and asterisks indicate the tested activity. The numbers refer to the double bond recognized and cleaved. The horizontal scale shows the number of differences per 100 residues derived from the ClustalW alignment.
Sequence alignment of CsCCD1a (CsCCD), CsCCD1b, CsCCD4a, and CsCCD4b with known NCEDs involved in ABA biosynthesis and other plant CCDs was performed using the ClustalW program, thus generating a phylogenetic tree (Fig. 1B). The phylogenetic tree showed four different groups. One included NCED proteins, another included CCD1 enzymes; a third class included the Arabidopsis enzymes CCD7 and CCD8, implicated in synthesis of a novel apocarotenoid hormone that controls lateral shoot growth (28, 29) and a fourth class that included CCD4 enzymes, whose substrates and cleavage position preferences remain unknown. CsCCD1a (CsCCD) is inside the CCD1 cluster, where some of its members are shown to cleave zeaxanthin at the 9,10(9′,10′) positions (14), whereas CsCCD1b is closely related to the CCD1 cluster.
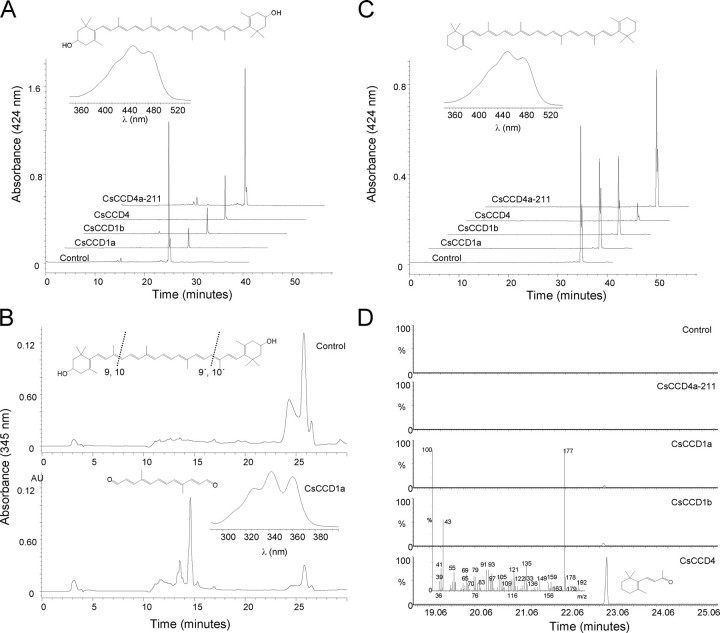
Activity of CsCCD Proteins in E. coli—To determine the potential function of CsCCD1a, CsCCD1b, and CsCCD4a/b, the cDNAs were cloned into the E. coli expression vector pDEST14, and the recombinant proteins were expressed in E. coli strains that accumulate different carotenoid compounds (30, 31). When expression of each of the CsCCD constructs was induced, accumulation of each of those carotenoids was significantly reduced (Fig. 2, A and C). Despite this reduction in carotenoid accumulation, we were unable to identify, in most cases, any cleavage product by HPLC analysis in the cells or in growth media extracts. It is likely that the products were further catabolized in E. coli (32). However, by HPLC, it was possible to detect carotenoid cleavage products from organic extracts of cell pellets from induced E. coli cultures expressing CsCCD1 in zeaxanthin-accumulating E. coli cells. These extracts showed low levels of C14 dialdehyde, indicating that it was the result of a symmetrical cleavage at the 9,10 and 9′,10′ positions of zeaxanthin (Fig. 2B). In addition, only β-ionone, generated by the symmetric 9,10(9′,10′) cleavage of β-carotene, was detected by GC/MS analysis when CsCCD1a and CsCCD1b expression was induced in E. coli cells that accumulate β-carotene (Fig. 2D). Although 5,6(5′,6′) activity has been recently reported for the CCD1 enzymes, we could not detect the presence of 6-methyl-5-hepten-2-one in our assays, probably due to the undetectable levels of lycopene in the induced cultures. The obtained results indicate that, like its homologs in Arabidopsis (3), Crocus (14), tomato (11), petunia (12), Vitis (9), and melon (13), CsCCD1a and CsCCD1b cleave carotenoids at the 9,10 double bond without substrate specificity. In contrast, for CsCCD4a/b no products were detected by HPLC (Fig. 2, A and C) and the β-ionone compound was identified by GC/MS only in the β-carotene producing cells (Fig. 2D). Interestingly, the CsCCD4 enzyme was 15- and 7.5-fold more active than CsCCD1a and CsCCD1b enzymes, respectively, in producing the β-ionone, indicating that perhaps this CCD form has higher activity. Furthermore, due to the similarity between CsCCD4 enzymes and CsZCD, we decided to test whether CsZCD cleaves at 9,10(9′,10′). For CsZCD amplification we used the oligonucleotides previously described (14). Although several clones were sequenced and analyzed, we only obtained partial clones corresponding to CsCCD4a or CsCCD4b, called CsCCD4a-211 and CsCCD4b-200. Nevertheless, we decided to use the truncated clone CsCCD4a-211 in the activity assays. Following the same procedure as used for the other CsCCD enzymes tested, CsCCD4a-211 was expressed in E. coli zeaxanthin and β-carotene producing cells. However, no cleavage product was identified with this truncated protein either by HPLC or GC/MS analysis of collected volatiles (Fig. 2, A, C, and D).
FIGURE 2.
Expression of CsCCD enzymes in carotenoid-producing strains of E. coli. E. coli engineered to accumulate the carotenoids β-carotene and zeaxanthin were transformed with an arabinose-inducible recombinant vector for CsCCD1a, CsCCD1b, CsCCD4a-211, and CsCCD4. A, representative HPLC elution profiles of zeaxanthin levels without (control) and with induction of CsCCD1a, CsCCD1b, CsCCD4a-211, and CsCCD4 expression. Inset, on-line spectrum and structure of zeaxanthin. B, chromatogram at absorbance of 345 nm of the zeaxanthin + CsCCD1a combination with the cleavage product peak at 15 min present in the lower chromatogram. The absorption maximum of the obtained product was consistent with that of C14 dialdehyde reported previously (3). Inset, in the upper part the structure of zeaxanthin is shown and the 9,10 and 9′,10′ double bonds are indicated; in the lower part the on-line spectrum and structure of the obtained product after a 9,10(9′,10′) cleavage. C, representative HPLC elution profiles of β-carotene levels without (control) and with induction of CsCCD1a, CsCCD1b, CsCCD4a-211, and CsCCD4 expression. Inset, on-line spectrum and structure of β-carotene. D, GC/MS analysis showing β-ionone emitted by bacteria following induction of CsCCD1a, CsCCD1b, CsCCD4a-211, and CsCCD4 expression. Inset, in the upper part the structure of β-carotene is shown and the 9,10 and 9′,10′ double bonds are indicated, in the lower part the mass spectrum of the observed of β-ionone product. Controls are representative chromatograms from uninduced cultures.
Expression Patterns of CsCCD Genes in Floral Organs—The expression pattern of the four CsCCD genes was examined in floral and vegetative organs. For each gene, specific primers were used (Table 1). CsCCD1a was detected in all floral organs with high levels in stigma and petals, whereas CsCCD1b expression was restricted to the stigma (Fig. 3A). CsCCD4a and CsCCD4b were detected in all the floral organs but the highest expression levels were present in the stigma. Studies were also performed in mature leaves, corm, and roots, and no expression was found for any CsCCD except for CsCCD1, which was expressed in leaves and roots (data not shown).
FIGURE 3.
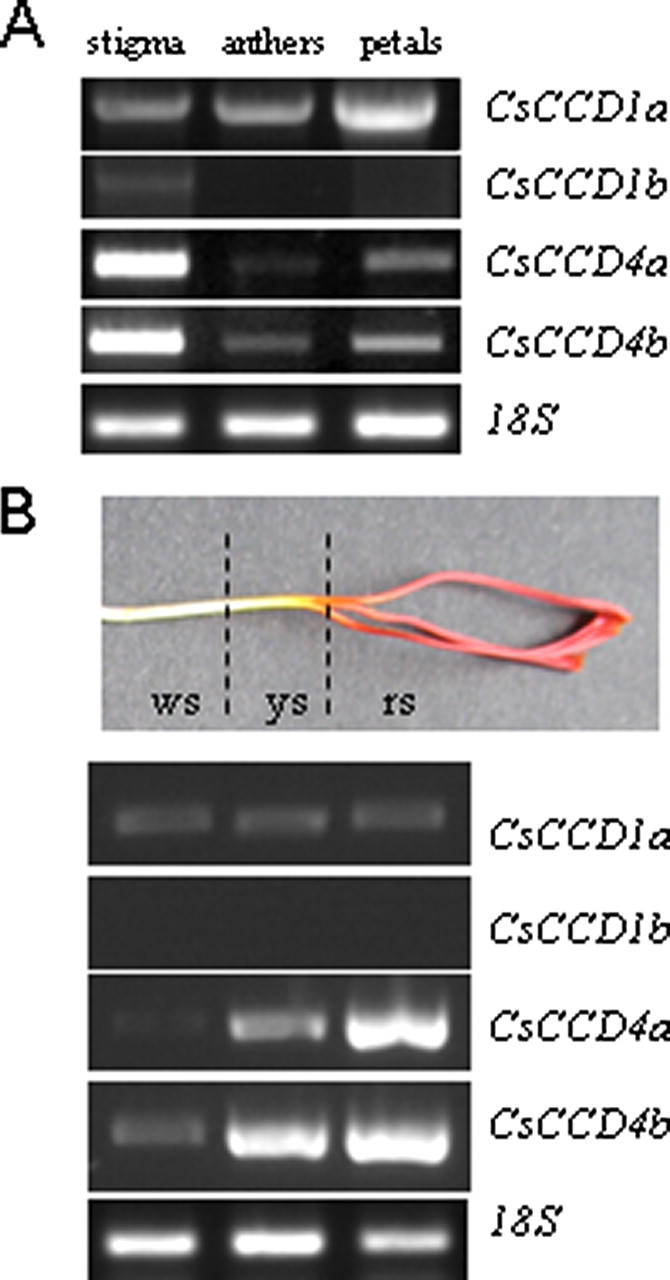
Expression analysis of CsCCDs by RT-PCR in flower organs. A, expression levels in stigma, petals, and stamens. B, expression levels in style and stigma tissues: ws, white style; ys, yellow style; and rs, red stigma. RT-PCR experiments were repeated three times, and representative results are shown. The RPS18 gene for 18 S RNA was amplified as a control. The PCR products were separated by 1% (w/v) agarose gel electrophoresis and visualized by ethidium bromide staining.
The subcellular localization studies revealed the targeting of CsCCD4 enzymes to plastoglobules. In C. sativus stigmas, the presence of plastoglobules decreases from the red stigma to the yellow and drastically from the yellow to the colorless style (33). Therefore, we studied the levels of expression of the CsCCD genes along the style as compared with the stigma (Fig. 3B). The expression pattern suggests that CsCCD4a and -b expressions are associated with the presence of plastoglobules along the style and stigma and that these parallel the increasing concentration of carotenoids, which in turn are responsible for generating the observed color gradient.
CsCCD Expression Patterns during Stigma Development and Their Relationship with C13 Norisoprenoid Volatiles—The expression of the CsCCD genes was followed during stigma development by RT-PCR. Analyses were performed with RNA isolated from different stages of stigma development, i.e. flowers containing yellow, orange, and red stigmas that are characterized by the presence of immature anthers, and small petals that do not show the characteristic purple coloration (Fig. 4A). These immature flowers are contained inside perianth tubes that elongate as flowers develop inside. Only when flowers are completely developed do they emerge from the perianth tubes and open when anthesis (da) occurs a few days later. Upon emerging, all flowers exhibited purple petals and scarlet stigmas (-2da to +3da) (Fig. 4A). The RT-PCR analysis revealed that accumulation of the mRNAs corresponding to the dioxygenase genes studied followed different expression patterns during stigma development (Fig. 4B). The CsCCD1a transcript levels remained relatively constant during the developmental process, and became undetectable at 3 days post-anthesis (Fig. 4B). In contrast, the CsCCD1b transcript levels reached a peak at the orange stage and decreased 2 days before anthesis, becoming undetectable at 1 day postanthesis (Fig. 4B). CsCCD4a transcript was practically undetectable in the yellow and orange stages, and reached a peak at anthesis and then decreased in the final stage (+3da). Both CsCCD4a and -b transcripts reached the highest levels at the scarlet stages (-2da to +3da), although CsCCD4b transcript levels remained constantly high during those stages. The high levels of expression of CsCCD4a and -b suggest a functional role of CsCCD4 in plastoglobules, because plastoglobules increase in number and size as the stigma of C. sativus develops (33).
FIGURE 4.
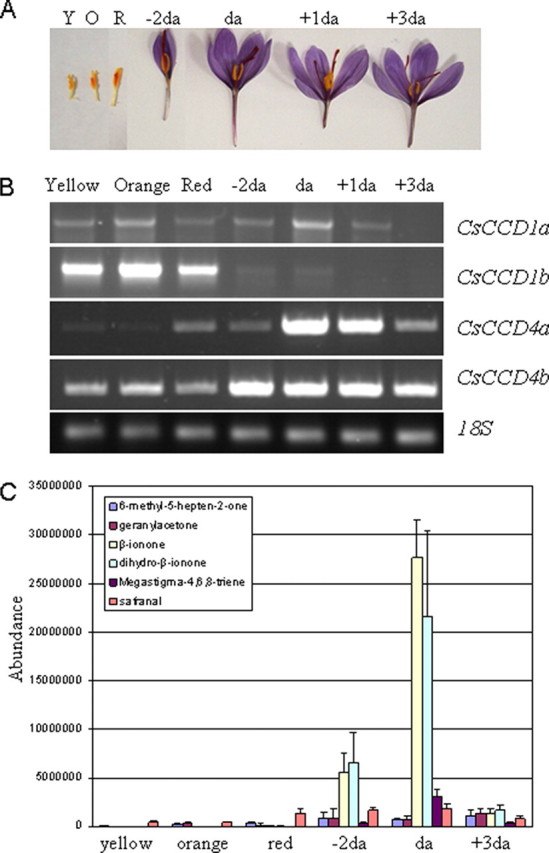
Expression levels of CsCCDs during stigma development and volatile emission. The mRNA levels were determined by RT-PCR amplification using specific oligonucleotides for each CsCCD gene. A, stigma tissue of C. sativus in different developmental stages: yellow (Y), orange (O), red (R), 2 days before anthesis (-2da), anthesis (da), 1 day after anthesis (+1da), and 3 days after anthesis (+3da). B, CsCCD1a, CsCCD1b, CsCCD4a and -b expression in yellow, orange, and red undeveloped stigmas, and in preanthesis, anthesis, and postanthesis developed stigmas as shown in A. Equal amounts of total RNA were used in each reaction. The levels of the constitutively expressed RPS18 coding gene were assayed as controls. C, carotenoid-derived volatile emissions during the stigma development of C. sativus.
Total volatiles from orange, red, and scarlet pre-anthesis (-2da), anthesis (da), and post-anthesis (+3da) stigmas were analyzed using GS/MS. The concentration of volatiles changed with stigma development, with the highest levels in the latest stages (up to 13% by fresh weight). Nine individual volatiles were studied: 9-apo-β-caroten-9-one (β-ionone), 7,8-dihydro-β-ionone, 6,10-dimethyl-3,5,9-undecatrien-2-one (pseudoionone), 9-apo-α-caroten-9-one (α-ionone), 6,10-dimethyl-5,9-undecadien-2-one (geranylacetone), megastigma-4,6,8-triene, 4-(2,6,6-trimethyl-cyclohexa-1,3-dienyl)but-2-en-4-one (damascenone), 6-methyl-5-hepten-2-one (6-methyl-5-hepten-2-one), and 2,6,6-trimethyl-1,3-cyclohexadiene-1-carboxaldehyde (safranal). With the exception of safranal, the other eight compounds were selected because they are the known 9,10(9′,10′)- and 5,6(5′,6′)-carotenoid dioxygenases products that may potentially be generated depending on the availability of specific carotenoids in the stigma. In the yellow, orange, and red stigma stages, apocarotenoid C13 volatiles were practically undetectable (Fig. 4C). Similarly, geranylacetone and 6-methyl-5-hepten-2-one were detected at low levels even in the more advanced developmental stigma stages (Fig. 4C). β-Ionone, 7,8-dihydro-β-ionone, and megastigma-4,6,8-triene were detected prior to anthesis and their levels increased up to the time of anthesis, quickly decreasing thereafter (Fig. 4C). The presence of these volatiles indicate that β-carotene is the principal precursor of the monoterpene volatiles in the developed stigma.
Stress Regulation of CsCCDs—C. sativus is well adapted to stress conditions, actively growing from autumn to spring with very little rainfall, surviving the summer drought below ground, and with a flowering time characterized by hot days and cold nights, shortly after summer. To test whether the transcript levels of the CCDs were up-regulated under stress, RT-PCR were performed on stressed detached leaf discs. CsCCD4a and CsCCD4b expression was significantly induced by dehydration stress (Fig. 5A), accumulating within 5 or 10 h from the start of dehydration. Both heat treatment and senescence also induced the expression of CsCCD4a and -b (Fig. 5B). In contrast, CsCCD1a was detected in all treatments. However, CsCCD1 transcript levels decreased at 10 h post-treatment in dehydrated leaf discs (Fig. 5A), and was up-regulated by senescence (Fig. 5B). The mRNA corresponding to CsCCD1b was undetectable in leaf discs under any of the conditions tested (Fig. 5, A and B).
FIGURE 5.
Transcript levels of CsCCDs in response to stress conditions. A, transcript levels of CsCCD1a, CsCCD1b, CsCCD4a and -b analyzed by RT-PCR from total RNA extracted from control (0 h) and leaf disks dehydrated for 2, 5, and 10 h. B, RT-PCR analysis on control (Ct), ABA treated, CaCl2 treated, heat treated, or senescent leaf disks. RPS18 was used as an internal control.
DISCUSSION
This study describes the isolation of four carotenoid dioxygenase genes from C. sativus stigmas. They belong to two different families, CCD1 or CCD4. The CCD1 family assembles cytosolic enzymes that specifically cleave the 9,10(9′,10′) double bonds of a variety of carotenoid substrates, and this is the case of CsCCD1a and CsCCD1b. The enzymes belonging to the CCD4 family have an N-terminal domain for plastid targeting, as we have shown that the CsCCD4a and CsCCD4b proteins are targeted to plastid plastoglobules in plants. We also determined that these C. sativus enzymes have a 9,10(9′,10′) cleavage activity on β-carotene. Therefore, our studies identified the first proteins from the CCD4 cluster whose enzymatic activity has been characterized. CsCCD4a and CsCCD4b showed high similarity with CsZCD (14), but the predicted CsCCD4a and -b proteins are much larger proteins in the N-terminal region. CsZCD was identified as the enzyme responsible for the initial step in the synthesis of crocin in C. sativus stigmas by a zeaxanthin 7,8(7′,8′)-cleavage activity (14). However, we have been unable to isolate the CsZCD gene and to assay its activity; instead, the isolated partial clone CsCCD4a-211 was assayed and it showed no activity under our experimental conditions. CsCCD4a-211 and CsZCD, lack apparently important residues and domains for the dioxygenase activity (34). Therefore, we suggest that the described structure and catalytic activity of CsZCD should be reconsidered.
Until this report, the CCD1 enzymes were the only 9,10(9′,10′) cleavage enzymes reported in aroma production in many different plants (4). The CCD7 enzyme has broad activity, cleaving carotenoids asymmetrically at the 9,10 or 9′,10′ positions (17, 35). CCD7 works in sequence with CCD8 to release a yet unidentified branching-inhibitor signal (36). However, CCD7 cannot be dismissed as a source of volatile compounds that potentially contribute to flavor (35). In the last couple of years, more dioxygenases genes have been identified as part of several ESTs and genome projects. Among the dioxygenases identified, the CCD4 group has notably increased, suggesting its members perform a general function in all the plant species where it has been identified. We have shown that CCD4 enzymes cleave double bonds at 9,10(9′,10′) positions, and seem to be more active than the CCD1 enzymes, at least for the substrate β-carotene. In addition, CCD4 seems to be targeted to the plastids, whereas CCD1 enzymes are cytosolic. The plastid, or more exactly, the plastoglobule location of the CCD4 enzymes, as is the case of the C. sativus and the Arabidopsis enzymes, allows these enzymes to obtain access to plastid carotenoids, whereas the CCD1 activity is limited to carotenoids out of these organelles or once these organelles have lost homeostasis or are targeted for degradation. Furthermore, the observed 9,10(9′,10′) cleavage activity of the CCD4 enzymes could explain why transgenic tomato, with a significant LeCCD1 decrease expression (∼90%), was accompanied by only a 40–60% reduction in β-ionone (11); and also why in transgenic petunia co-suppressed for CCD1 expression, the plants still produced β-ionone (12). Additionally, chrysanthemum plants expressing a petal specific enzyme, CmCCD4a, showed white petals, whereas plants lacking it showed yellow petals, due to β-carotene accumulation (37). Altogether, this constitutes additional evidence of CCD4 activity.
The presence of gene families with the same enzymatic activity appears to be common in secondary metabolism and might confound the assignment of gene function based on sequence information alone (38). The biosynthesis of secondary metabolites is often restricted to a particular tissue and may occur at a specific stage of development. Several carotenoid biosynthesis enzymes are encoded by at least two genes. In each of these enzymes, one isoform is constitutively expressed, whereas the other is specific for chromoplasts in flowers and/or fruits (39, 40). Perhaps CCD1 and CCD4 are also examples of this type of specialization. From the phylogenetic relationships, it could be speculated that CCD1 and CCD4 enzymes evolved from a common ancestor to give rise to two forms that evolved to different specific activities, and were targeted to different compartments where the access to substrates in a cell or plastid compartment generated new possibilities for reaction and eventually imparted different benefits to the plant. CsCCD4a and -b expression is restricted to flower tissues and the RT-PCR results showed a clear relationship between CsCCD4a and -b mRNA abundance and the carotenoid content: when the level of CsCCD4a and -b was high, the carotenoid content was low. Furthermore, the same behavior was observed when CsCCD4 expression (referred to as CsZCD) was analyzed in the stigma tissue of different Crocus species (18). Crocus goulimyi, with the lowest carotenoid content, showed the highest CsCCD4 levels. The results of the expression analysis support a role for CsCCD4a and -b in β-ionone biosynthesis in the stigma tissue at the time of anthesis, which is most likely associated with a role of attracting insects for pollination (41), a process essential for reproductive success in self-incompatible species such as Crocus (42), whose pollen grains have dimensions too large to be airborne (43). Therefore, it is possible that β-ionone plays a role in attracting insect pollinators, and it has previously been identified as an effective attractant for some insects (44–47). Several insects such as Bombus silvestris (48, 49), overwintering bees (50), or Apis mellifica (51) have been reported as active pollinators in C. sativus.
CsCCD1a and CsCCD1b expression does not precisely mirror the emission of apocarotenoid volatiles in the stigma tissue. The lag between CsCCD1b gene expression and a putative intervention in apocarotenoid production could be due to differences in subcellular localization of the enzymes and the consequent availability of substrates. Carotenoid synthesis and accumulation is localized to plastids, but CsCCD1a and CsCCD1b proteins do not seem to be imported into plastids. Immunohistochemistry have been used to show a cytoplasmic location for CsCCD1a (14), and due to the high homology of CsCCD1a and CsCCD1b, and the lack of an obvious plastid transit peptide, we also expect CsCCD1b to be cytoplasmic. Alternatively, the lag in volatile emission might be due to the sequestration of the apocarotenoid products, whose release could be regulated temporally or spatially. There are numerous glycosides in saffron (2) that may undergo hydrolysis to yield different volatiles that could be released later during stigma development, and this could be what happens with the products of CsCCD1a and CsCCD1b. Furthermore, CsCCD1a, as well as other CCD1 genes, was constitutively expressed and is the only gene detected in non-stressed leaves. Thus, CsCCD1a could be responsible for the rapid production of a variety of C13 cyclohexone apocarotenoids, depending on the substrate, either after local damage or loss of homeostasis that occurs in processes such as plant defense (with β-ionone having known antifungal and antimicrobial activity) (52) and in other processes where the C13 product of zeaxanthin or lutein cleavage, 3-hydroxy-β-ionone, may control plant growth (53).
Mature fruit and flower tissues are characterized by the presence of chromoplasts, which are capable of storing large amounts of carotenoids (54) in the hydrophobic core of chromoplast plastoglobules and fibrils (55–57). The expression pattern for the CsCCD4a and -b genes is correlated with the increase in plastoglobule size and number detected in C. sativus stigmas during development (33), and suggests that the expression of CsCCD4a and -b, and most likely their protein levels, are finely tuned with the availability of substrates in the plastoglobules. Furthermore, CsCCD4a and -b expression was found to be up-regulated in stressed and senescent leaves, which showed an enlargement of plastoglobules as part of the plant stress response (58–62). Carotenoids are known to accumulate in green tissues experiencing stress conditions, and a number of studies indicate that they provide efficient protection against oxidative stress. However, much less is known about the roles of apocarotenoids in higher plants under stress conditions or during senescence, with the exception of ABA. It has been shown that extreme temperatures, high irradiance, or ultraviolet (UV) stress induced the synthesis of β-ionone and β-cyclocitral among other apocarotenoid volatiles (63). Our finding that CsCCD4a and -b are induced under those conditions fits with their participation in the synthesis of stress response compounds in leaves.
β-Ionone, pseudoionone, and geranylacetone have been previously detected in saffron spice, the dried and toasted stigmas of C. sativus, at very low levels (64, 65). However, the presence of these volatiles has never been investigated during natural stigma development in plants. Comparison between volatiles from the spice and from fresh tissue pointed out that the volatile profile present in the fresh tissue is completely different from that obtained from the processed material, where safranal is the major volatile (66). The presence of β-ionone in fresh tissue is directly correlated with the accumulation of β-carotene in the latest stages of stigma development (18). In contrast, the low levels of geranylacetone, 6-methyl-5-hepten-2-one or the absence of pseudoionone, are in accordance with the low levels of δ-carotene and lycopene previously detected (18). These findings further corroborate previous observations in tomato, watermelon, and melon fruits that indicate that apocarotenoid composition is related with the carotenoid content (13, 67, 68).
In conclusion, we have shown that C. sativus has two classes of CCDs with the same enzymatic activity even though they are expressed and localized differently. Our data strongly suggest that each class of enzymes may be responsible for the metabolism of carotenoids at different and specific subcellular sites, during both normal development and in response to stress conditions.
Acknowledgments
We thank Dr. F. X. Cunningham (University of Maryland College Park, MD) for donating bacterial plasmids for carotenoid production; Dr. Nakagawa (Shimane University, Japan) for providing plasmid pGWB5 and Dr. D. Bradley (IBMCP, Spain) and K. A. Walsh Costello (ETSIA; UCLM) for critical reading of the manuscript and for English grammar correction.
This work was supported by Ministerio de Educación y Ciencia Grants BIO2003-05259 and BIO2006-00841. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CCD, carotenoid cleavage dioxygenase; NCED, 9-cis-epoxycarotenoid dioxygenase; ABA, abscisic acid; RT, reverse transcriptase; GFP, green fluorescent protein; HPLC, high pressure liquid chromatography; RACE, rapid amplification of cDNA ends.
References
- 1.Tarantilis, P. A., Polissiou, M., and Manfait, M. (1994) J. Chromatogr. 664 55-61 [DOI] [PubMed] [Google Scholar]
- 2.Winterhalter, P., and Straubinger, M. (2000) Food Rev. Int. 16 39-59 [Google Scholar]
- 3.Schwartz, S. H., Qin, X., and Zeevaart, J. A. D. (2001) J. Biol. Chem. 276 25208-25211 [DOI] [PubMed] [Google Scholar]
- 4.Auldridge, M. E., McCarty, D. R., and Klee, H. J. (2006) Curr. Opin. Plant Biol. 9 315-321 [DOI] [PubMed] [Google Scholar]
- 5.Schwartz, S. H., Tan, B. C., Gage, D., Zeevaart, J. A. D., and McCarty, D. R. (1997) Science 276 1872-1874 [DOI] [PubMed] [Google Scholar]
- 6.Tan, B. C., Schwartz, S. H., Zeevaart, J. A. D., and McCarty, D. R. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 12235-12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chernys, J., and Zeevaart, J. A. D. (2000) Plant Physiol. 124 343-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogel, J. T., Tan, B. C., McCarty, D. R., and Klee, H. J. (2008) J. Biol. Chem. 283 11364-11373 [DOI] [PubMed] [Google Scholar]
- 9.Mathieu, S., Terrier, N., Procureur, J., Bigey, F., and Gunata, Z. (2005) J. Exp. Bot. 56 2721-2731 [DOI] [PubMed] [Google Scholar]
- 10.Baldermann, S., Naim, M., and Fleischmann, P. (2005) Food Res. Int. 38 833-836 [Google Scholar]
- 11.Simkin, A. J., Schwartz, S. H., Auldridge, M., Taylor, M. G., and Klee, H. J. (2004) Plant J. 40 882-892 [DOI] [PubMed] [Google Scholar]
- 12.Simkin, A. J., Underwood, B. A., Auldridge, M., Loucas, H. M., Shibuya, K., Schmelz, E., Clark, D. G., and Klee, H. J. (2004b) Plant Physiol. 136 3504-3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibdah, M., Azulay, Y., Portnoy, V., Wasserman, B., Bar, E., Meir, A., Burger, Y., Hirschberg, J., Schaffer, A. A., Katzir, N., Tadmor, Y., and Lewinsohn, E. (2006) Phytochemistry 67 1579-1589 [DOI] [PubMed] [Google Scholar]
- 14.Bouvier, F., Suire, C., Mutterer, J., and Camara, B. (2003) Plant Cell 15 47-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ytterberg, A. J., Peltier, J. B., and van Wijk, K. J. (2006) Plant Physiol. 140 984-997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubio, A., Fernández, P., Fernández, J. A., and Gomez-Gomez, L. (2004) Planta 219 955-96615605174 [Google Scholar]
- 17.Schwartz, S. H., Qin, X., and Loewen, M. C. (2004) J. Biol. Chem. 279 46940-46945 [DOI] [PubMed] [Google Scholar]
- 18.Castillo, R., Fernandez, J. A., and Gomez-Gomez, L. (2005) Plant Physiol. 139 674-689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., Toyooka, K., Matsuoka, K., Jinbo, T., and Kimura, T. (2007) Biosci. Bioeng. 104 34-41 [DOI] [PubMed] [Google Scholar]
- 20.Clough, S. J., and Bent, A. F. (1998) Plant J. 16 735-743 [DOI] [PubMed] [Google Scholar]
- 21.Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997) Protein Eng. 10 1-6 [DOI] [PubMed] [Google Scholar]
- 22.Naested, H., Holm, A., Jenkins, T., Nielsen, H. B., Harris, C. A., Beale, M. H., Andersen, M., Mant, A., Scheller, H., Camara, B., Mattsson, O., and Mundy, J. (2004) J. Cell Sci. 117 4807-4818 [DOI] [PubMed] [Google Scholar]
- 23.Lohmann, A., Schöttler, M. A., Bréhélin, C., Kessler, F., Bock, R., Cahoon, E. B., and Dörmann, P. (2006) J. Biol. Chem. 281 40461-40472 [DOI] [PubMed] [Google Scholar]
- 24.Vidi, P. A., Kanwischer, M., Baginsky, S., Austin, J. R., Csucs, G., Dörmann, P., Kessler, F., and Bréhélin, C. (2006) J. Biol. Chem. 281 11225-11234 [DOI] [PubMed] [Google Scholar]
- 25.Kaup, M. T., Froese, C. D., and Thompson, J. E. (2002) Plant Physiol. 129 1616-1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lichtenthaler, H. K. (1968) Endeavor 27 144-149 [Google Scholar]
- 27.Vishnevetsky, M., Ovadis, M., and Vainstein, A. (1999) Trends Plant Sci. 4 232-235 [DOI] [PubMed] [Google Scholar]
- 28.Booker, J., Auldridge, M., Wills, S., McCarty, D., Klee, H., and Leyser, O. (2004) Curr. Biol. 14 1232-1238 [DOI] [PubMed] [Google Scholar]
- 29.Sorefan, K., Booker, J., Haurogne, K., Goussot, M., Bainbridge, K., Foo, E., Chatfield, S., Ward, S., Beveridge, C., Rameau, C., and Leyser, O. (2003) Genes Dev. 17 1469-1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cunningham, F. X., Pogson, B., Sun, Z., McDonald, K. A., and DellaPenna, D. (1996) Plant Cell 8 1613-1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun, Z., Gantt, E., and Cunningham, F. X., Jr. (1996) J. Biol. Chem. 271 24349-24352 [DOI] [PubMed] [Google Scholar]
- 32.von Lintig, J., and Vogt, K. (2000) Biol. Chem. 275 11915-11920 [DOI] [PubMed] [Google Scholar]
- 33.Grilli, M. C., and Canini, A. (2004) Plant Biosys. 138 43-52 [Google Scholar]
- 34.Kloer, D. P., and Schulz, G. E. (2006) Cell Mol. Life Sci. 63 2291-2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auldridge, M. E., Block, A., Vogel, J. T., Dabney-Smith, C., Mila, I., Bouzayen, M., Magallanes-Lundback, M., DellaPenna, D., McCarty, D. R., and Klee, H. J. (2006b) Plant J. 45 982-993 [DOI] [PubMed] [Google Scholar]
- 36.Beveridge, C. A. (2006) Curr. Opin. Plant Biol. 9 35-40 [DOI] [PubMed] [Google Scholar]
- 37.Ohmiya, A., Kishimoto, S., Ainda, R., Yoshioka, S., and Sumitomo, K. (2006) Plant Physiol. 142 1193-1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pichersky, E., and Gang, D. R. (2000) Trends Plant Sci. 5 439-445 [DOI] [PubMed] [Google Scholar]
- 39.Galpaz, N., Ronen, G., Khalfa, Z., Zamir, D., and Hirschberg, J. (2006) Plant Cell 18 1947-1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siddique, M. A., Grossmann, J., Gruissem, W., and Baginsky, S. (2006) Plant Cell Physiol. 47 1663-1673 [DOI] [PubMed] [Google Scholar]
- 41.Moise, A. R., von Lintig, J., and Palczewski, K. (2005) Trends Plant Sci. 10 178-186 [DOI] [PubMed] [Google Scholar]
- 42.Heslop-Harrison, Y. (1977) Ann. Bot. 41 913-922 [Google Scholar]
- 43.Chichiricco, G. (1999) Grana 38 31-41 [Google Scholar]
- 44.Del Mazo Cancino, A., and Damon, A. (2007) Plant Species Biol. 22 129-134 [Google Scholar]
- 45.Nemesio, A., and Silveira, F. A. (2006) Neotrop. Entomol. 35 313-323 [DOI] [PubMed] [Google Scholar]
- 46.Milet-Pinheiro, P., and Schlindwein, C. (2005) Rev. Bras. Zool. 22 853-858 [Google Scholar]
- 47.Hammack, L. (2001) J. Chem. Ecol. 27 1373-1390 [DOI] [PubMed] [Google Scholar]
- 48.Mathew, B. (1999) in Reproduction Biology in Saffron and Its Allies (Negbi, M., ed) pp. 19-30, Harwood Academic Publishers, Amsterdam, The Netherlands
- 49.Grilli-Caiola, M. (1999) in Saffron (Negbi, M., ed) pp. 31-44, Harwood Academic, Amsterdam, The Netherlands
- 50.McKee, J., and Richards, A. J. (1998) Bot. J. Linnean Soc. 128 369-384 [Google Scholar]
- 51.Negbi, M. (1999) in Saffron (Negbi, M., ed) pp. 1-18, Harwood Academic, Amsterdam, The Netherlands
- 52.Mikhlin, E. D., Radina, V. P., Dmitrossky, A. A., Blinkova, L. P., and Button, L. G. (1983) Prikl. Biokhim Mikrobiol. 19 795-803 [PubMed] [Google Scholar]
- 53.Kato-Noguchi, H. (1993) Physiol. Plantarum 89 423-426 [Google Scholar]
- 54.Kirk, J. T., and Tiliney-Bassett, R. A. (1978) in The Plastids: Their Chemistry, Structure, Growth and Inheritance (Kirck, S. T., and Tiliney-Bassett, R. A., eds) pp. 217-239, Elsevier/North Holland Biomedical Press, Amsterdam, The Netherlands
- 55.Deruere, J., Romer, S., d'Harlingue, A., Backhaus, R. A., Kuntz, M., and Camara, B. (1994) Plant Cell 6 119-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sitte, P., Falk, H., and Liedvpgel, B. (1980) in Pigments in Plants (Czygan, F.-C., ed) pp. 117-148, G. Fischer, Stuttgart
- 57.Wrischer, M., Prebeg, T., Magnus, V., and Ljube, N. (2007) Acta Bot. Croat. 66 81-87 [Google Scholar]
- 58.Tevini, M., and Steinmüller, D. (1985) Planta 163 91-96 [DOI] [PubMed] [Google Scholar]
- 59.Locy, R. D., Chang, C. C., Nielsen, B. L., and Singh, N. K. (1996) Plant Physiol. 110 321-328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rey, P., Gillet, B., Römer, S., Eymery, F., Massimino, J., Peltier, G., and Kuntz, M. (2000) Plant J. 21 483-494 [DOI] [PubMed] [Google Scholar]
- 61.Bondada, B. R., and Syvertsen, J. P. (2003) Tree Physiol. 23 553-559 [DOI] [PubMed] [Google Scholar]
- 62.Munne-Bosch, S., and Alegre, L. (2004) Funct. Plant Biol. 31 203-216 [DOI] [PubMed] [Google Scholar]
- 63.Bouvier, F., Isner, J. C., Dogbo, O., and Camara, B. (2005) Trends Plant Sci. 10 187-194 [DOI] [PubMed] [Google Scholar]
- 64.Kanakis, C. D., Daferera, D. J., Tarantilis, P. A., and Polissiou, M. G. (2004) J. Agric. Food Chem. 52 4515-4521 [DOI] [PubMed] [Google Scholar]
- 65.D'Auria, M., Mauriello, G., Racioppi, R., and Rana, G. L. (2006) J. Chromatogr. Sci. 44 18-21 [DOI] [PubMed] [Google Scholar]
- 66.Tarantilis, P. A., and Polissiou, M. (1997) J. Agric. Food Chem. 45 459-462 [Google Scholar]
- 67.Lewinsohn, E., Sitrit, Y., Bar, E., Azulay, Y., Meir, A., Zamir, D., and Tadmor, Y. (2005) J. Agric. Food Chem. 53 3142-3148 [DOI] [PubMed] [Google Scholar]
- 68.Tieman, D. M., Zeigler, M., Schmelz, E. A., Taylor, M. G., Bliss, P., Kirst, M., and Klee, H. J. (2006) J. Exp. Bot. 57 887-896 [DOI] [PubMed] [Google Scholar]