Abstract
cgi-58 (comparative gene identification-58) is a member of α/β-hydrolase family of proteins. Mutations in CGI-58 are shown to be responsible for a rare genetic disorder known as Chanarin-Dorfman syndrome, characterized by an excessive accumulation of triacylglycerol in several tissues and ichthyosis. We have earlier reported that YLR099c encoding Ict1p in Saccharomyces cerevisiae can acylate lysophosphatidic acid to phosphatidic acid. Here we report that human CGI-58 is closely related to ICT1. To understand the biochemical function of cgi-58, the gene was overexpressed in Escherichia coli, and the purified recombinant protein was found to specifically acylate lysophosphatidic acid in an acyl-CoA-dependent manner. Overexpression of CGI-58 in S. cerevisiae showed an increase in the formation of phosphatidic acid resulting in an overall increase in the total phospholipids. However, the triacylglycerol level was found to be significantly reduced. In addition, the physiological significance of cgi-58 in mice white adipose tissue was studied. We found soluble lysophosphatidic acid acyltransferase activity in mouse white adipose tissue. Immunoblot analysis using anti-Ict1p antibodies followed by mass spectrometry of the immunocross-reactive protein in lipid droplets revealed its identity as cgi-58. These observations suggest the existence of an alternate cytosolic phosphatidic acid biosynthetic pathway in the white adipose tissue. Collectively, these results reveal the role of cgi-58 as an acyltransferase.
The most abundant form of energy is stored as triacylglycerol (TAG)3 in the lipid droplets (LDs) or adiposomes. LDs also have a high concentration of cholesterol esters, enclosed within the coat of proteins bound to phospholipids (1). Adiposomes are specialized subcellular structures in adipocytes and are ubiquitously present in most of the cell types of vertebrates. LDs are implicated in several pathological conditions including obesity, inflammation, ichthyosis, myopathy, atherosclerosis, and neurological abnormalities (2). It is known that various proteins associated with LDs regulate their dynamics. The most prominent proteins are perilipin, adipophilipin, S3-12, and TIP-47 (3). All of these proteins share a conserved N-terminal region. Another interesting protein annotated as cgi-58 or abhd5 (α/β hydrolase domain-containing protein 5) was shown to interact with perilipin and adipocyte triglyceride lipase (ATGL) on the LD surface (3, 4). Mutations in CGI-58 are associated with Chanarin-Dorfman syndrome, an autosomal recessive disease characterized by TAG accumulation in several tissues (5). Clinical manifestations include ichthiosys, hepatic steatosis, cardiomyopathy, ataxia, and mental retardation (6). It is known that cgi-58 in association with ATGL maintains TAG homeostasis in adipocytes. The mutations that affect this association hinder both TAG hydrolysis (4) and recycling of neutral lipids to phospholipids (7). cgi-58 is localized in LDs, endoplasmic reticulum, and Golgi bodies, all of which are involved in the assembly of lipoprotein particles and VLDL-TAG secretion (3). It is likely that cgi-58 is involved in the secretion of apolipoprotein B-containing lipoproteins (8). cgi-58 belongs to esterase/lipase/thioesterase family of proteins, but the presence of an asparagine in place of serine in GXSXG motif abolishes the lipase activity (9). Although the role of cgi-58 in TAG hydrolysis remains unequivocal, the association of cgi-58 with metabolically active LDs signifies its role in phospholipid and TAG biosyntheses. However, the biochemical role of cgi-58 still remains unknown.
Recently, we have shown that YLR099c encoding Ict1p, in Saccharomyces cerevisiae, mediates acylation of lysophosphatidic acid (LPA) to phosphatidic acid (PA), thereby enhancing phospholipid biosynthesis under cellular stress (10). In this study, we show that cgi-58 is a homologue of Ict1p and has an acyl-CoA-dependent LPA-specific acyltransferase activity. In addition, we also demonstrate that cgi-58 is a primary source for the soluble LPA acyltransferase activity observed in white adipose tissue of mice.
EXPERIMENTAL PROCEDURES
Materials—Human CGI-58 clone was obtained from Open Biosystems. S. cerevisiae (BY4741: MATa; his 3Δ1; leu2Δ0; met15Δ0; ura3Δ0) and deletion mutant strain of ICT1 (BY4741 background) were obtained from Invitrogen. [1-14C]Oleoyl-CoA (54 mCi/mmol) and [3H]1-oleoyl LPA (47 Ci/mmol) were purchased from PerkinElmer Life Sciences. Silica gel 60F254 TLC plates were from Merck. Oligonucleotide primers, chemicals, and solvents were purchased from Sigma. Polyclonal antibodies were raised against the nickel-nitrilotriacetic acid affinity-purified recombinant Ict1p as described (10).
White Adipose Tissue—White adipose tissues surrounding the reproductive tract of mice were dissected, frozen in liquid nitrogen, and stored at –80 °C for further use. Frozen tissue (1 g) was thawed and homogenized in phosphate-buffered saline (PBS, pH 7.4) containing 1 mm 2-mercaptoethanol, 1 mm EDTA, and a mixture of protease inhibitors (1 mm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 50 μg/ml leupeptin, 10 μg/ml pepstatin, and 10 μg/ml aprotinin). The components were homogenized with four to five strokes in a Potter-Elvehjem glass-Teflon homogenizer, and the homogenate was centrifuged at 300 × g for 15 min. Supernatant was subjected to centrifugation at 10,000 × g for 15 min. The floating layer was washed twice with PBS followed by ultracentrifugation at 100,000 × g. The washed floating layer was used as lipid droplets. The 10,000 × g supernatant thus obtained was further centrifuged at 240,000 × g for 60 min to obtain cytosol (soluble fraction). The pellet was washed with PBS and centrifuged again at 240,000 × g for 60 min to obtain total membranes. All of the operations were carried out at 4 °C (11).
Size Exclusion Chromatography— The cytosol from mice white adipose tissue (4 mg of protein) was applied to a Superdex-200 FPLC column fitted to AKTA-FPLC system (GE Healthcare). The column was pre-equilibrated with 50 mm Tris-HCl (pH 7.5) containing 0.1 m NaCl. The fractions (1 ml each) were collected at a flow rate of 0.5 ml/min and assayed for LPA acyltransferase activity. The column was calibrated with standard molecular mass markers: thyroglobulin (669 kDa), aldolase (232 kDa), bovine serum albumin (66 kDa), ovalbumin (43 kDa), and chymotrypsin (25 kDa).
Immunoblotting—Proteins were separated by 12% SDS-PAGE and transferred onto a nitrocellulose membrane. Rabbit anti-Ict1p antibodies were used at a dilution of 1:1000 in 0.05% gelatin in PBS.
Purification of Recombinant cgi-58—pCMV-SPORT6 vector containing CGI-58 open reading frame was used as a template for amplification of the gene. Forward primer (5′-CCCGGATCCATGGCGGCGGAGGAGGAG-3′) and reverse primer (5′-CCCCTCGAGTCAGTCCACAGTGTCGCAGATCTCC-3′) were used. PCR (1 min of denaturation at 94 °C, 1 min of annealing at 55 °C, and 1 min of elongation at 72 °C) was performed using Pfu polymerase for 30 cycles with 10 pmol concentration of each primer. The purified PCR product and pRSET A vector (N-terminal histidine tag) were digested with BamHI and XhoI and ligated directionally. The construct was transformed into Escherichia coli BL21 (DE3) cells and induced with 1 mm isopropyl β-d-thiogalactopyranoside for 4 h at 37 °C. The cell pellet was resuspended in lysis buffer containing 50 mm Tris-HCl (pH 8.0) and 300 mm NaCl. The cells were disrupted by sonication. The 10,000 × g supernatant was allowed to bind to the nickel-nitrilotriacetic acid matrix. The column was washed with lysis buffer containing 25 mm imidazole. The bound protein was eluted with 250 mm imidazole in lysis buffer. Fractions (1 ml each) were collected and analyzed on 12% SDS-PAGE followed by Coomassie Brilliant Blue staining.
LPA Acyltransferase Assay—The reaction mixture contained 10 μm [1-14C]oleoyl-CoA (110,000 dpm/assay), 1–15 μg of enzyme source, and 50 μm LPA (1-oleoyl) in lysis buffer with a total volume of 100 μl. The reaction was carried out at 30 °C for 10 min, and the reaction was terminated by extracting the lipids (10). In addition, acyltransferase activity was also assayed using 50 μm [1-oleoyl-9,10-3H]LPA (220,000 dpm/assay) and 20 μm acyl-CoA. The lipids were separated on a TLC plate using chloroform: methanol:acetone:acetic acid:water (50:10:20:15:5, v/v/v/v/v) as the solvent system.
The lipids were also analyzed by two-dimensional TLC using chloroform:methanol:ammonia (65:25:5, v/v/v) as the first dimensional solvent system and chloroform:methanol:acetic acid:water (40:20:5:0.5, v/v/v/v) as the second dimensional solvent system (10). Individual phospholipids were identified by comparing the Rf values of the unknown with the Rf values of the standards. The TLC plates were subjected to autoradiography, and the PA spots were scraped and counted with toluene-based scintillation mixture.
Site-directed Mutagenesis—Mutations (Q130P and E260K) in cgi-58 were introduced using the following primers: forward primer for Q130P, 5′-AGAAGAAGTGGAGAATCCGTTTGTGGAATCCATTGA-3′ and reverse primer for Q130P, 5′-TCAATGGATTCCACAAACGGATTCTCCACTTCTTCT-3′; forward primer for E260K, 5′-TGCAGACTCCAAGTGGTAAGACAGCTTTCAAGAAT-3′; and reverse primer for E260K, 5′-ATTCTTGAAAGCTGTCTTACCACTTGGAGTCTGCA-3′. Wild type CGI-58 template (80 ng) and primers (25 pmol each) were added to PCR tubes containing 0.2 mm dNTPs, 1 mm MgSO4, 2.5 units of Pfu polymerase, and 1× reaction buffer. Amplification was done using the following conditions: denaturation of the template at 94 °C for 4 min followed by 20 cycles at 94 °C for 45 s (denaturation), 52 °C for 1 min (annealing) and 72 °C for 6 min (extension). The reaction was continued for another 20 min at 72 °C to complete the extension. The product was treated with DpnI at 37 °C for 6 h to digest the methylated template and transformed into E. coli DH5α strain, and the mutants were confirmed by sequencing.
Overexpression of cgi-58 in S. cerevisiae—Full-length CGI-58 cDNA was subcloned from pRSET A to pYES2 vector at the BamHI-XhoI site and transformed into yeast cells by the lithium chloride method (12). Transformants were selected on synthetic minimal medium devoid of uracil (SM-U) containing 2% glucose and were grown to the late log phase. The cells were harvested by centrifugation and inoculated at an A600 level of 0.4 in SM-U medium containing 2% galactose and grown for 24 h. To confirm the expression, cells (A600 = 5) were resuspended in 50 mm Tris-HCl (pH 7.5), 2% SDS and then lysed using glass beads. The proteins were separated by 12% SDS-PAGE and transferred onto a nitrocellulose membrane. The overexpression of cgi-58 was confirmed using anti-Ict1p antibodies at a dilution of 1:1000 (v/v).
Preparation of Yeast Cell Lysate—To measure the enzymatic activity of S. cerevisiae overexpressing cgi-58 and vector control, the cell extracts were prepared by lysing with glass beads followed by centrifugation at 1000 × g to remove glass beads and unbroken cells. Cell-free extract was used as the enzyme source.
[32P]Orthophosphate Incorporation into Phospholipids— pYES2-CGI-58 and pYES2 transformants were grown to late log phase in 5 ml of SM-U containing 2% glucose and then transferred to 50 ml of the same medium, such that the absorbance is 0.1. The cells were grown till the absorbance reached 3. For labeling of total cellular phospholipids, A600 = 0.4 of the cells were inoculated in a fresh medium containing 2% galactose and 200 μCi of [32P]orthophosphate and grown for 24 h. The cells (A600 = 25) were harvested by centrifugation, and the lipids were extracted and analyzed by two-dimensional TLC. The solvents for the first dimension were chloroform:methano-l:ammonia (65:25:5, v/v/v); solvents for the second dimension were chloroform:methanol:acetone:acetic acid:water (50:10:20: 15:5, v/v/v/v/v) (10).
[14C]Acetate Incorporation into Neutral Lipids—The transformants (pYES2-CGI-58 and pYES2) were grown to late log phase in 5 ml of SM-U containing 2% glucose and then transferred to 10 ml of the fresh medium with an absorbance of 0.1. The cells were grown till the absorbance reached 3. For neutral lipid labeling, A600 = 0.4 of the cells were inoculated in a fresh medium containing 2% galactose and 4 μCi/ml of [14C]acetate and grown for 24 h. The cells (A600 = 10) were harvested, and the lipids were extracted and separated on a silica TLC using chloroform:methanol:acetic acid (98:2:0.5, v/v/v) as the solvent system.
Identification of cgi-58 by Matrix-assisted Laser Desorption Ionization (MALDI) Mass Spectrometry—Protein from LDs were separated on 12% SDS-PAGE and stained by Coomassie Brilliant Blue. The band of interest was excised, rehydrated with water, washed thrice for 15 min each with 50% acetonitrile in 100 mm ammonium bicarbonate (pH 8.0), and dried. The sample was digested with 0.80 ng/ml proteomics grade trypsin (Promega) in 100 mm ammonium bicarbonate (pH 8.0) for 12 h at 37 °C. The tryptic fragments were then extracted with 50% acetonitrile and 0.1% trifluoroacetic acid, dried, resuspended in saturated 4-hydroxy-cyanocinnamic acid in 50% acetonitrile and 0.1% trifluoroacetic acid, and applied to a MALDI sample plate that was dried and washed with water to remove excess buffer salts. MALDI mass spectrometry analysis was performed on the ULTRA FLEX TOF-TOF, Bruker Daltonics instrument equipped with a pulsed N2 laser and analyzed in the reflectron mode using a time delay of 90 ns, accelerating voltage of 25 kV in the positive ion mode. Initially spectra of 200 laser shots were acquired, and the spectra were calibrated externally to a spectrum of peptide mixture of known masses ranging from 1046 to 2465 Da. The intense peaks in the spectrum were selected for fragmentation by laser-induced dissociation using the LIFT program of ULTRA FLEX TOF-TOF instrument. cgi-58 was identified by subjecting the peptide masses obtained to Mascot version 2.1 (Matrix Science) search engine.
RESULTS
CGI-58 Is Homologous to ICT1—BLAST analysis of Ict1p in the nonredundant data base of NCBI revealed that Ict1p is a close homologue of cgi-58. Clustal W analysis of Ict1p with human cgi-58 and its homologues in Mus musculus and Xenopus suggests a wide phyletic distribution of cgi-58, thereby indicating an evolutionary conserved function (Fig. 1). However, most of the human orthologues have not been characterized. A close homologue of cgi-58 is abhd4, which has been found to be important in endocannabinoid biosynthesis (13). cgi-58, a 350-amino acid protein, is a member of α/β-hydrolase family of proteins. In addition to the hydrolase domain, cgi-58 possesses an esterase (pfam 00756), hydrolase/acyltransferase domain (COG 0596), and lysophospholipase domain (COG 2267). The GXSXG motif of cgi-58 is highly conserved in majority of the known lipases, phospholipases, lysophospholipases, esterases, and serine proteases; however, the conserved Ser is replaced by an Asn in this case. A distinct structural motif HX4D is also present in cgi-58 at the C-terminal region. Hydropathy plot of cgi-58 predicted the absence of any transmembrane domain (data not shown).
FIGURE 1.
cgi-58 is a close homologue of yeast Ict1p. BLAST analysis of Ict1p with nonredundant protein data base available at NCBI was performed. Multiple sequence alignment of proteins homologous to human cgi-58 was carried out using sequences from representative organisms such as Xenopus, mice, and yeast. The Clustal W analysis suggests the conserved nature of the sequence with a wide phyletic distribution. The accession numbers were NP_057090.2 for Homo sapiens, NP_080455.1 for M. musculus, NP_001086565.1 for Xenopus laevis, and NP_013200.1 for S. cerevisiae. The conserved HX4D and GXNXG motifs are indicated in the boxes.
cgi-58 Encodes LPA Acyltransferase—Human CGI-58 was cloned in pRSET A and expressed in BL21 (DE3) cells. Immunoblot analysis using anti-His6 antibody was done to confirm the expression. Recombinant protein was purified by nickel-nitrilotriacetic acid column chromatography (Fig. 2A). To confirm whether anti-Ict1p antibodies cross-react with human cgi-58, immunoblot analysis was performed. Anti-Ict1p antibody did cross-react with human cgi-58 (Fig. 2B). LPA acyltransferase activity of cgi-58 was analyzed, and the enzymatic product was confirmed by two-dimensional TLC as PA (Fig. 2C). To assess acyl acceptor specificity, lysophosphatidylcholine, lysophosphatidylethanolamine, lysophosphatidylserine, and lysophosphatidylinositol were used. The recombinant cgi-58 showed no significant activity toward these substrates. These data suggest that cgi-58 is an acyl-CoA-dependent LPA-specific acyltransferase (Fig. 2D). The recombinant cgi-58 showed no lipase activity with 3H-labeled triolein and phospholipase D activity toward 14C-labeled PC and 14C-labeled PE (data not shown).
FIGURE 2.

Purification of recombinant human cgi-58. A, purification of cgi-58 from E. coli BL21(DE3) using nickel-nitrilotriacetic acid affinity column chromatography. The proteins were resolved on 12% SDS-PAGE and stained with Coomassie Brilliant Blue. Lane 1, uninduced lysate; lane 2, induced lysate; lane 3, the purified cgi-58. B, immunoblot analysis with anti-Ict1p antibodies was carried out. Lane 1, vector transformed; lane 2, CGI-58 transformed. C, LPA acyltransferase reaction product was resolved on a two-dimensional TLC using chloroform:methanol:ammonia (65:25:5, v/v/v) as the first dimension (1D) and chloroform:methanol:acetic acid:water (40:20:5:0.5, v/v/v/v) as the second dimension (2D) solvent systems. SF, solvent front. D, to study the acyl acceptor preference, the purified cgi-58 was assayed with various lysophospholipids under standard assay conditions. LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; LPI, lysophosphatidylinositol; LPS, lysophosphatidylserine; G3P, glycerol-3-phosphate.
Characterization of the Recombinant cgi-58—The enzyme showed a protein-dependent (Fig. 3, A and B) and time-dependent (Fig. 3C) increase in the incorporation of [14C]oleoyl-CoA into LPA to form PA. To study the effect of other phospholipids on the LPA acyltransferase activity, the assay was performed in the presence of other lysophospholipids and phospholipids. The activity was inhibited by the high concentrations of all phospholipids studied. However, the effect of lysophosphatidylcholine was most prominent. Lysophosphatidylcholine inhibited 68% of the enzyme activity at 30 μm (Fig. 3D). To assess the preference of acyl-CoA by the enzyme, the assays were performed using palmitoyl-, oleoyl-, and stearoyl-CoAs as acyl donors and [3H]LPA as an acyl acceptor. A slightly higher activity was observed with oleoyl-CoA (14.75 nmol of PA formed per min/mg of protein) as compared with palmitoyl-CoA (12.20 nmol of PA formed per min/mg of protein). When stearoyl-CoA was used as an acyl donor, only 33% of the activity was found under the standard assay conditions (Fig. 3E). Based on the above observations, we suggest that cgi-58 is a soluble LPA acyltransferase, which plays an important role in PA biosynthesis.
FIGURE 3.
Characterization of the purified recombinant human cgi-58. A and B, LPA acyltransferase activity was monitored under the standard assay conditions with increasing amounts of protein (0, 0.25, 0.5, 0.75, and 1 μg). C, time-dependent formation of PA. The assay was performed with 50 μm LPA, 10 μm [14C]oleoyl-CoA and 5 μg of enzyme in a final volume of 0.5 ml. An aliquot of 100 μl was withdrawn at different time intervals. The activity was assessed as described under “Experimental Procedures.” D, effect of various (lyso)phospholipids on the acyltransferase activity. E, LPA acyltransferase activity was monitored using 50 μm [3H]LPA and 10 μm of various acyl-CoAs. LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; LPI, lysophosphatidylinositol; LPS, lysophosphatidylserine; PS, phosphatidylserine; PI, phosphatidylinositol.
Acylation and Coactivation Functions Are Segregated in cgi-58—Q130P and E260K mutations in cgi-58 (Fig. 4A) were shown earlier to have a debilitating effect on the activation of ATGL (4). These are also two of the several mutations found in Chanarin-Dorfman syndrome patients (9). To assess the role of these mutations on LPA acyltransferase activity, mutants and wild type proteins were purified. Western blotting with anti-His6 antibody confirmed the purity (Fig. 4B), and acyltransferase activity was analyzed. Q130P and W260K did not have a significant effect on the acyltransferase activity (Fig. 4C), suggesting that LPA acyltransferase activity is functionally segregated from its role as a coactivator.
FIGURE 4.

Effect of point mutations in cgi-58 on LPA acyltransferase activity. A, the schematic representation of the domains retrieved from conserved domain data base at NCBI. α/β-Hydrolase fold (pfam 0059; COG 0596) and PldB, lysophospholipase (COG 2267). B, Coomassie Brilliant Blue-stained gel showing the purity of human and mutant cgi-58 proteins. Lane 1, wild type (WT); lane 2, Q130P; lane 3, E260K. Immunoblot analysis of wild type and mutants of cgi-58 was performed using anti-His monoclonal antibody using 20 μg of protein of cell extract. C, the purified recombinant cgi-58 and the mutants (1.0 μg of protein) were used for the assay.
cgi-58 Overexpression Enhances Phospholipids in S. cerevisiae— To study the effect of cgi-58 overexpression on cellular phospholipids, S. cerevisiae was transformed with pYES2-CGI-58. Immunoblotting with anti-Ict1p antibodies confirmed the overexpression of the protein (Fig. 5A). Overexpression of human cgi-58 in yeast led to an increase in the major phospholipids (Fig. 5B). Total radioactivity in the chloroform fraction of pYES2-CGI-58 cells showed a 5-fold increase in the incorporation, when compared with the pYES2 vector control yeast cells (Fig. 5C). When we analyzed the chloroform extract in a two-dimensional TLC (excluding origin), the overall increase in phospholipids alone was found to be 3.6-fold (Fig. 5D). PA showed a ∼2-fold increase as compared with the vector control (Fig. 5D). To our surprise, the amount of labeled PC and PE was found to be ∼3.3- and ∼4.7-fold higher than the vector control, respectively (Fig. 5D). It is worth noting that overexpression of cgi-58 led to an increased biosynthesis of all the major phospholipids. To test whether human cgi-58 can complement the PA biosynthetic defect in Δict1 cells (10), cells were transformed with pYES2-CGI-58, and [32P]orthophosphate incorporation into lipids, in particular to PA, was studied. CGI-58 overexpression in Δict1 cells was found to rescue the metabolic defect of the mutant (supplemental Fig. S1).
FIGURE 5.
Heterologous expression of human cgi-58 in S. cerevisiae enhances overall phospholipids. A, pYES2 and pYES2-CGI-58 transformed yeast were grown overnight in the presence of galactose, and cells (A600 = 5) were lysed using glass beads. The proteins were separated on a 12% SDS-PAGE, and immunoblotting was performed using anti-Ict1p antibodies at a dilution of 1:1000 (v/v). A segment of the Coomassie Brilliant Blue-stained gel was used as a loading control. B, yeast cells overexpressing cgi-58 and the vector control were grown for 24 h in galactose medium in the presence of 200 μCi of [32P]orthophosphate. The lipids were extracted from cells (A600 = 25) and resolved on two-dimensional silica TLC using chloroform:methanol:ammonia (65:25:5, v/v/v) as first dimension and chloroform:methanol:acetone:acetic acid:water (50:10:20:15:5, v/v/v/v/v) as second dimension solvent systems. The lipids are indicated as 1, phosphatidylinositol (PI); 2, phosphatidylserine (PS); 3, PC; 4, PE; 5-8, unknown (UK); 9, PA; and 10, cardiolipin (CL). O represents the origin. C, [32P]orthophosphate incorporated in total lipids (chloroform extract) in pYES2 and pYES2-CGI-58 is represented as the cpm/A600 of cells of labeling. D, the amount of [32P]orthophosphate incorporated into individual phospholipids is represented as the cpm/A600 of cells/24 h of labeling. Each data point represents the mean of three independent experiments ± S.D. E, LPA acyltransferase assay was done using cell-free extract of pYES2 and pYES2-CGI-58 transformed yeast. F, yeast cells overexpressing cgi-58 and the vector control were grown for 24 h in galactose medium in the presence of 4 μCi of [14C]acetate/ml of medium. Lipids were extracted from cells (A600 = 10) and resolved on silica TLC using chloroform:methanol:acetic acid (98:2:0.5, v/v/v) followed by autoradiography. G, [14C]acetate incorporation into TAG, DAG, and free fatty acid (FFA) of pYES2 and pYES2-CGI-58 transformed yeast cells is represented as the counts in dpm/10 units of A600 of cells/24 h of labeling. Each point represents the average of two independent experiments.
Cell-free extracts of transformants (pYES2-CGI-58) were prepared and assayed for the LPA acyltransferase activity, which showed nearly a 1.8-fold increase in acylation when compared with the vector control (Fig. 5E), suggesting a possible role for cgi-58 in phosphatidic acid biosynthesis.
To understand the fate of neutral lipid biosynthesis, [14C]acetate labeling study was performed in pYES2-CGI-58 transformed yeast cells. Overexpression of cgi-58 in S. cerevisiae decreased the TAG levels by 2-fold and showed a corresponding increase in the DAG level (Fig. 5, F and G).
PA Formation in WAT Cytosol—PA can be synthesized by three different routes (i) phosphorylation of DAG by DAG kinase, (ii) acylation of LPA by LPA acyltransferase, and (iii) hydrolysis of phosphoalcohol head group from phospholipids by phospholipase D. To determine the contribution of each of these routes to the total PA pool, cytosol of mice WAT was used. DAG kinase was assayed in the presence of DAG, [γ-32P]ATP, MgCl2, phosphatidylserine, octyl-β-glucopyranoside, 500 μm [γ-32P]ATP, and 20 μg of cellular protein (14). Commercially available DAG kinase (CalBiochem) was used as the positive control. There was no formation of labeled PA, suggesting that the DAG kinase is not involved in the formation of PA in the cytosol of mice WAT. Hydrolysis of 14C-labeled PC and 14C-labeled PE were monitored using cytosol as the enzyme source. There was no significant formation of labeled PA (data not shown). LPA acylation by the cytosol showed a time-dependent incorporation of [14C]oleoyl-CoA (data not shown). These results suggest that mice WAT cytosol has PA biosynthesizing activity, which is attributed to LPA acyltransferase.
Distribution of PA Biosynthetic Activity—We investigated the PA formation in WAT and found that the cytosol (240,000 × g supernatant), lipid droplets and membrane fractions were capable of synthesizing PA. Table 1 summarizes the LPA acylation in these fractions. Interestingly, cytosol had 28% of the total LPA acyltransferase activity, whereas LDs and membrane fraction had 15 and 57% activity, respectively (Table 1). These results indicate that there is a PA biosynthetic pathway in the cytosol. To demonstrate that this activity is not due to the nonsedimentable membrane fragments generated during fractionation, a gel exclusion FPLC column (Superdex-200) was used. Most of the LPA acyltransferase activity eluted around 232 kDa (Fig. 6A), and this could be due to specific/nonspecific association of cgi-58 with other proteins. However, the identity of these proteins is not clear. The association of cgi-58 with other proteins was confirmed by immunoblotting with anti-ict1p antibodies (data not shown). These results indicate that the PA forming activity is also present in the cytosol of white adipose tissue.
TABLE 1.
Distribution of LPA acyltransferase activity in white adipose tissue
White adipose tissue (1 g of wet weight) cell lysate was subjected to differential centrifugation. Enzyme was assayed under the standard assay conditions. The values are the means ± S.D. of three separate experiments, each performed in duplicate.
| Fraction | Protein | Total activity | Specific activity | Total activity |
|---|---|---|---|---|
| mg | pmol/min | pmol/min/mg | % | |
| Cytosol | 8.03 | 603.61 | 75.16 ± 11.09 | 28.35 ± 2.38 |
| Lipid droplets | 0.81 | 328.30 | 471.21 ± 1.98 | 15.42 ± 1.41 |
| Membranes | 1.76 | 1196.64 | 648.80 ± 1.11 | 56.23 ± 3.65 |
FIGURE 6.
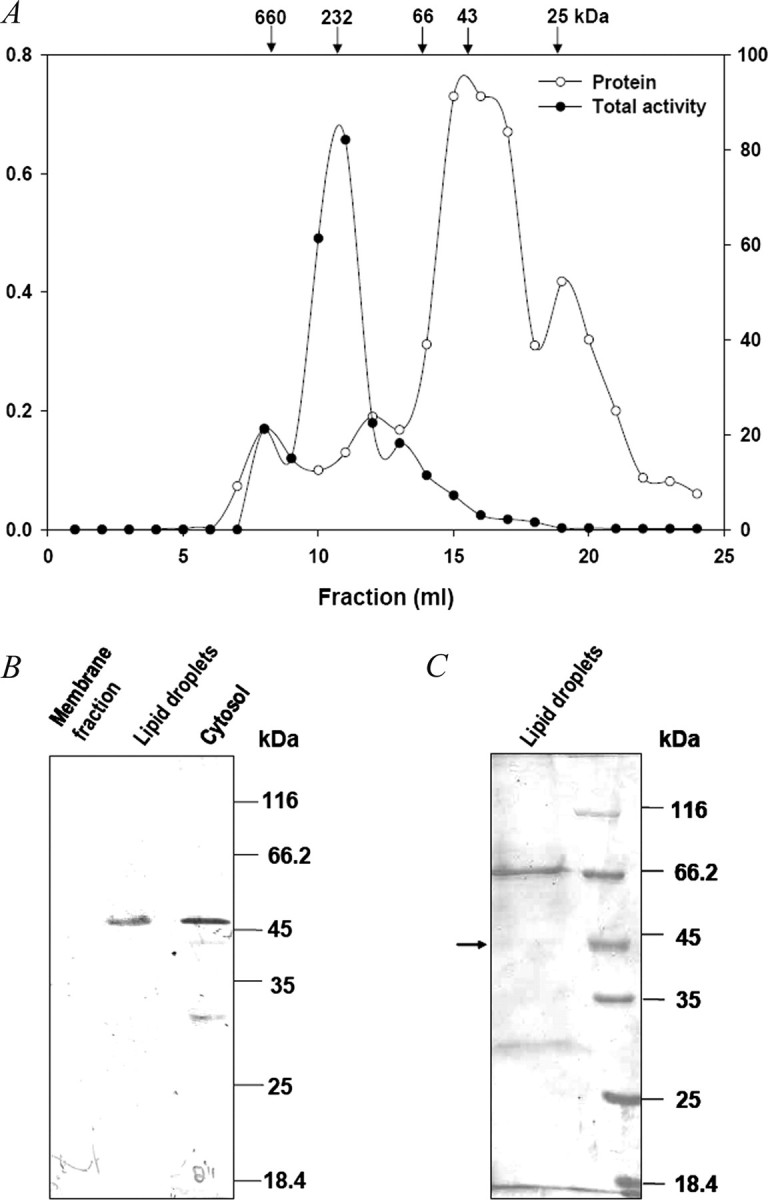
Detection of cgi-58 in cytosol and lipid droplets of mice white adipose tissue using anti-Ict1p antibodies. A, cytosol was loaded onto a Superdex-200 analytical FPLC gel filtration column to obtain the soluble fraction free of membranes contamination, and the elution profile of protein and the LPA acyltransferase activity were monitored. B, lipid droplets, cytosol, and membranes from mice white adipose tissue were separated by differential centrifugation. Proteins from cytosol (60 μg), membranes (60 μg), and lipid droplets (10 μg) were resolved by 12% SDS-PAGE and transferred onto a nitrocellulose membrane. Immunoblotting was performed with anti-Ict1p polyclonal antibodies. C, proteins (10 μg) of lipid droplets were separated by 12% SDS-PAGE and stained with Coomassie Blue. The indicated band was processed by MALDI mass spectrometry.
Identification of cgi-58 in Mice WAT—Our studies on S. cerevisiae showed that Ict1p, a cytosolic protein, could acylate LPA to PA (10). To identify Ict1p-like proteins, we resorted to immunoblotting of WAT subcellular fractions. Immunoreactive bands were observed in the LDs and cytosol when probed with anti-Ict1p antibodies (Fig. 6B). The band (Fig. 6C) corresponding to ∼45 kDa in LDs was found to be cgi-58 by MALDI mass spectrometry analysis. These findings suggest that the cytosolic LPA acyltransferase activity is contributed by cgi-58.
DISCUSSION
The present study is based on our earlier report on Ict1p, a protein that is highly expressed upon organic solvent stress in S. cerevisiae. Ict1p was found to acylate specifically LPA to PA, and this function was found to be important for membrane lipid biosynthesis and repair on organic solvent stress. BLAST analysis of Ict1p revealed that cgi-58 is a close relative of Ict1p. cgi-58 is a member of the α/β-hydrolase family of proteins containing a highly conserved tertiary fold of alternating α-helices and β-sheets. It is found to be conserved from bacteria to humans. Human cgi-58 is a soluble protein with α/β-hydrolase domain-containing hydrolase/acyltransferase/esterase/lipase and phospholipase motifs (5, 15, 17). Mutations in cgi-58 are known to be associated with Chanarin-Dorfman syndrome, a rare autosomal recessive disorder, characterized by the accumulation of TAG in various tissues and ichthyosis (5, 6, 9). The exact biochemical function of cgi-58 and its role in lipid metabolism still remains elusive.
Amino acids 69–87 in cgi-58 form a highly hydrophobic region corresponding to the lipid-binding motif of the protein (5). The presence of this domain suggests that the enzyme could utilize lysophospholipids. However, phospholipase and lysophospholipase activities in the purified recombinant cgi-58 were not observed, probably because of the absence of Ser residue in the active site (18). In addition, it has a distinct structural motif, HX4D, observed in some acyltransferases. Histidine in the HX4D motif abstracts a proton from the hydroxyl group of an acyl acceptor, thereby facilitating the nucleophilic attack on thioester of an acyl donor (19).
The bacterially expressed, purified recombinant human cgi-58 was examined for the possible acyltransferase activities. It was found to specifically acylate LPA in an acyl-CoA-dependent manner to form PA. It showed a preference for both palmitoyl- and oleoyl-CoAs.
It is known that Gln130 and Glu260 in cgi-58 are important residues involved in the interaction with ATGL, and these interactions are important for normal physiology of the cell (4). The two point mutations (Q130P and E260K) did not have any effect on the acyltransferase activity, thereby suggesting that the ATGL coactivation and LPA acylation by cgi-58 are unrelated.
Our results provide evidence for the presence of a soluble acyl-CoA:LPA acyltransferase in the eukaryotic system. Overexpression of cgi-58 in yeast enhances the PA level and thereby overall phospholipid biosynthesis. Our study on the fate of neutral lipids indicates a significant reduction in the TAG level on cgi-58 overexpression in yeast. Similar studies in COS-7, Hep-G2, or McA-RH7777 cells suggest a decrease in TAG level on cgi-58 overexpression (4, 8). The decrease in TAG could be because of the coactivation of ATGL-like lipase by cgi-58 in yeast (20).
PA is the major intermediate in membrane and storage lipid biosyntheses. It is possible that cgi-58 could be involved in providing PA for glycerolipid biosynthesis (7, 10, 21). CGI-58 overexpression studies in Hep-G2 or McA-RH7777 cells suggest a minor change in the levels of phospholipids and cholesteryl esters (8). However, studies involving fibroblasts from Chanarin-Dorfman syndrome patients indicate that although there was no apparent lipolysis defect in those cell lines, accumulations of some major phospholipids and sphingomyelins were decreased when compared with normal fibroblast cells (7).
It was shown earlier that β-adrenergic stimulation releases cgi-58 from its interacting protein (perilipin), which thereafter traverses into the cytoplasm. However, the precise role of cgi-58 in the cytoplasm remains unknown (2). It is possible that cgi-58 can associate with ATGL and facilitate lipolysis (4, 20), or it can facilitate recycling of TAG-hydrolyzed products into phospholipids (7), thereby maintaining the TAG homeostasis.
To understand the physiological significance of cgi-58 in vivo, we resorted to mice white adipose tissue. We provide a direct evidence for the presence of PA biosynthetic activity in cytosol of mammalian system based on the following observations. First, subcellular distribution studies indicate that more than 60% of the total activity is associated with the membranes (Table 1). Second, the enzyme involved in PA synthesis is permeable in the gel filtration column. The presence of soluble enzymes that provide important precursors for glycerolipid biosynthesis is well documented. Cytosolic monoacylglycerol acyltransferase (22), diacylglycerol acyltransferase (23), and LPA phosphatase (24) were shown to be present in the immature seeds of Arachis hypogaea. Recently, cytosolic LPA phosphatase (25) and PA phosphatase have also been reported in S. cerevisiae (16). In addition, we have earlier demonstrated in Rhodotorula glutinis the presence of a cytosolic multienzyme complex for the synthesis of TAG (11). The present study highlights the existence of a cytosolic acyltransferase in mammals and provides an insight into the molecular and biochemical function of cgi-58.
Supplementary Material
Acknowledgments
We thank Pooja Kumari for the technical assistance and Venky Sreedhar Reddy for helpful discussion and comments on the manuscript.
This work was supported by a grant from the Department of Biotechnology (New Delhi, India). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: TAG, triacylglycerol; LPA, lysophosphatidic acid; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; WAT, white adipose tissue; LD, lipid droplet; ATGL, adipocyte triglyceride lipase; PBS, phosphate-buffered saline; FPLC, fast protein liquid chromatography; MALDI, matrix-assisted laser desorption ionization; DAG, diacylglycerol.
References
- 1.Brasaemle, D. L., Rubin, B., Harten, I. A., Gruia-Gray, J., Kimmel, A. R., and Londos, C. (2000) J. Biol. Chem. 27538486 –38493 [DOI] [PubMed] [Google Scholar]
- 2.Brasaemle, D. L. (2007) J. Lipid Res. 482547 –2559 [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi, T., Omatsu, N., Matsushita, S., and Osumi, T. (2004) J. Biol. Chem. 27930490 –30497 [DOI] [PubMed] [Google Scholar]
- 4.Lass, A., Zimmermann, R., Haemmerle, G., Riederer, M., Schoiswohl, G., Schweiger, M., Kienesberger, P., Strauss, J. G., Gorkiewicz, G., and Zechner, R. (2006) Cell Metabolism 3309 –319 [DOI] [PubMed] [Google Scholar]
- 5.Akiyama, M., Sawamura, D., Nomura, Y., Sugawara, M., and Shimuzu, H. (2003) J. Investig. Dermatol. 1211029 –1034 [DOI] [PubMed] [Google Scholar]
- 6.Musumeci, S., Agata, A., Romano, C., Patane, R., and Cutrona, D. (1988) Am. J. Med. Genet. 29377 –382 [DOI] [PubMed] [Google Scholar]
- 7.Igal, R. A., and Coleman, R. A. (1998) J. Lipid Res. 3931 – 43 [PubMed] [Google Scholar]
- 8.Brown, J. M., Chung, S., Das, A., Shelness, G., Rudel, L. L., and Yu, L. (2007) J. Lipid Res. 482295 –2305 [DOI] [PubMed] [Google Scholar]
- 9.Lefevre, C., Jobard, F., Caux, F., Bouadjar, B., Karaduman, A., Heilig, R., Lakhdar, H., Wollenberg, A., Verret, J. L., Weissenbach, J., Ozguc, M., Lathrop, M., Prud'homme, J. F., and Fischer, J. (2001) Am. J. Hum. Genet. 691002 –1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh, A. K., Ramakrishnan, G., and Rajasekharan, R. (2008) J. Biol. Chem. 2839768 –9775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gangar, A., Karande, A. A., and Rajasekharan, R. (2001) J. Biol. Chem. 27610290 –10298 [DOI] [PubMed] [Google Scholar]
- 12.Schiestl, R. H., and Gietz, R. D. (1989) Curr. Genet. 16339 –346 [DOI] [PubMed] [Google Scholar]
- 13.Simon, G. M., and Cravatt, B. F. (2006) J. Biol. Chem. 28126465 –26472 [DOI] [PubMed] [Google Scholar]
- 14.Bunting, M., Tang, W., Zimmerman, G. A., McIntyre, T. M., and Prescott, S. M. (1996) J. Biol. Chem. 27110230 –10236 [PubMed] [Google Scholar]
- 15.Ollis, D. L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S. M., Harel, M., Remington, S. J., Silman, I., and Schrag, J. (1992) Protein. Eng. 5197 –211 [DOI] [PubMed] [Google Scholar]
- 16.Han, G.-S., Wu, W.-I., and Carman, G. M. (2006) J. Biol. Chem. 2819210 –9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nardini, M., and Dijkstra, B. W. (1999) Curr. Opin. Struct. Biol. 9732 –737 [DOI] [PubMed] [Google Scholar]
- 18.Hilton, S., and Buckley, J. T. (1991) J. Biol. Chem. 266997 –1000 [PubMed] [Google Scholar]
- 19.Heath, R. J., and Rock, C. O. (1998) J. Bacteriol. 1801425 –1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurat, C. F., Natter, K., Petschnigg, J., Wolinski, H., Scheuringer, K., Scholz, H., Zimmermann, R., Leber, R., Zechner, R., and Kohlwein, S. D. (2006) J. Biol. Chem. 281491 –500 [DOI] [PubMed] [Google Scholar]
- 21.Liu, P., Ying, Y., Zhao, Y., Mundy, D. I., Zhu, M., and Anderson, R. G. W. (2004) J. Biol. Chem. 2793787 –3792 [DOI] [PubMed] [Google Scholar]
- 22.Tumaney, A. W., Sekhar, S., and Rajasekharan, R. (2001) J. Biol. Chem. (2001) 27610847 –10852 [DOI] [PubMed] [Google Scholar]
- 23.Shekar, S., Tumaney, A. W., Rao, T. J. V. S., and Rajasekharan, R. (2002) Plant Physiol. 128988 –996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha, S., Enugutti, B., Rajakumari, S., and Rajasekharan, R. (2006) Plant Phys. 1411533 –1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy, V. S., Singh, A. K., and Rajasekharan, R. (2008) J. Biol. Chem. 2838846 –8854 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





