Abstract
KCNE1 (minK), found in the human heart and, cochlea is a transmembrane protein that modulates the voltage-gated potassium KCNQ1 channel. While KCNE1 has previously been the subject of extensive structural studies in lyso-phospholipid detergent micelles, key observations have yet to be confirmed and refined in lipid bilayers. In this study a reliable method for reconstituting KCNE1 into lipid bilayer vesicles composed of POPC and POPG was developed. Microinjection of the proteoliposomes into Xenopus oocytes expressing the human KCNQ1 (KV7.1) voltage-gated potassium channel led to native-like modulation of the channel. CD spectroscopy demonstrated that the %-helicity of KCNE1 is significantly higher for the protein reconstituted in lipid vesicles relative to the previously described structure in 1.0% LMPG micelles. SDSL EPR spectroscopic techniques were used to probe the local structure and environment of Ser28, Phe54, Phe57, Leu 59, and Ser64 KCNE1 in both POPC/POPG vesicles and in LMPG micelles. Spin-labeled KCNE1 cysteine mutants at Phe54, Phe57, Leu 59, and Ser64 were found to be located inside POPC/POPG vesicles, whereas Ser28 was found to be located outside the membrane. Ser64 was shown to be water-inaccessible in vesicles, but found to be water-accessible in LMPG micelle solutions. These results suggest that key components of the micelle-derived structure of KCNE1 extend to the structure of this protein in lipid bilayers, but also demonstrates the need to refine this structure using data derived from bilayer-reconstituted protein in order to more accurately define its native structure. This work establishes the basis for such future studies.
Introduction
KCNE1 (minK) is a member of the KCNE family of transmembrane proteins that modulate the activity of certain voltage-gated K+ channels, including KCNQ1 (KV7.1). KCNE1 complexes with KCNQ1 to both slow its voltage-stimulated activation and to enhance its open state conductance, resulting in the IKs current of the cardiac action potential (1-3). Hereditary mutations in KCNE1 have been linked to long QT syndrome and deafness (1, 4-8). The structure of KCNE1 was recently determined in LMPG detergent micelles, where its transmembrane domain was observed to be a curved α-helix, with both N- and C-termini consisting of flexibly-linked α-helices, some of which have an affinity for the micellar surface (9). Based on this structure and experimentally-restrained docking into KCNQ1 channel models, it was hypothesized that KCNE1 interacts with S4-S5 linker domain of KCNQ1 to restrict the opening of the gate in S6 which generates the delayed channel opening observed in functional studies of KCNQ1 co-expressed with KCNE1 (9-14).
The LMPG micelle environment used in determination of KCNE1's structure provided a suitable environment for solution NMR experiments due to their isotropic nature. Moreover, the lyso-phospholipid LMPG represents a mild class of detergents that is especially phospholipid-like. However, even the most optimal detergent micelles do not provide a completely natural environment for studies of membrane proteins. This caveat can lead to structural distortions and changes in the stability of both local and global conformations. This means that structures determined in micelles should be validated by structural measurements made under conditions in which the protein of interest is reconstituted into vesicles (Figure 1). Moreover, even in cases where the micellar structure captures the key features of the bilayer-associated structure, the micellar structure may require refinement to correct for minor differences such as the exact beginnings and ends of secondary structural elements. In this paper we describe methods for reconstituting KCNE1 in lipid vesicles and present data that demonstrates that direct structural study of KCNE1 in bilayer conditions is indeed merited in order to refine the micellar structure so as to better reflect the actual bilayer structure.
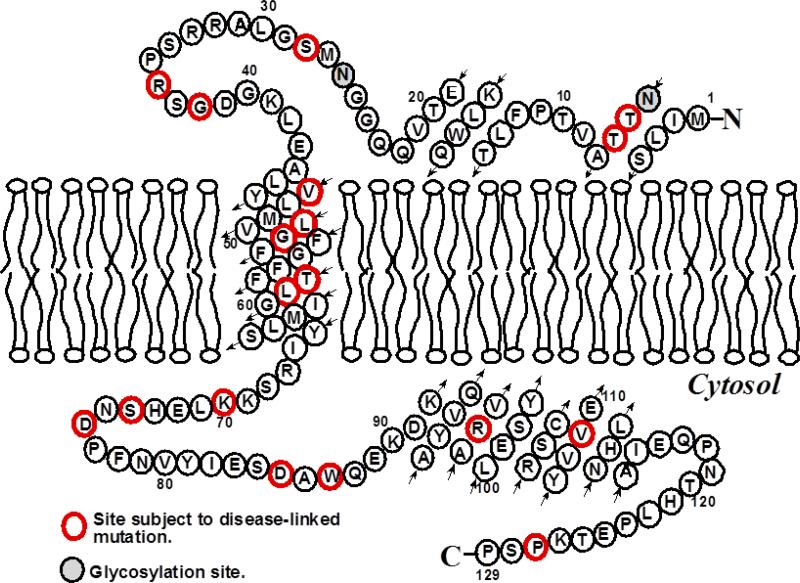
Figure 1.
Topological illustration of KCNE1 with respect to a lipid bilayer assuming that the TMD spans residues 46-71 as previously determined for the protein in 7.5% LMPG micelles at 40ºC and pH 6.0 (9-14). The small arrows show the tracing of the peptide chain.
MATERIALS AND METHODS
Site-Directed Mutagenesis
The His-tag expression vectors (pET-16b) containing wild type and a cysteine-less mutant form of KCNE1 (in which its single cysteine site (105) was mutated to Ala) have previous been described (9). These vectors were transformed into XL1-Blue E. coli cells (Stratagene). Plasmid extracts from these cells were obtained using the QIAprep Spin Miniprep Kit (Qiagen). Site-direct mutagenesis to generate single cysteine mutants from the cysless vector was performed using the QuikChange Lightning Site-Directed Mutagenesis Kit (Strategene). The KCNE1 mutations were confirmed by DNA sequencing from XL10-Gold E. coli (Stratagene) transformants using the T7 primer (Integrated DNA Technologies). Successfully mutated vectors were transformed into BL21(DE3) CodonPlus-RP E. coli cells (Stratagene) for protein overexpression. Mutations S28C and S64C were chosen for EPR measurements to avoid structural perturbations because of the close relationship in structure between Serine and Cysteine. F54C, F57C, and L59C mutations were chosen as additional sites on the TMD to confirm that the TMD was embedded inside the membrane.
Expression and Purification
A 5mL pre-culture of Luria Broth containing 50μg/mL Ampicillin was inoculated with the BL21(DE3) KCNE1 transformant of choice. After overnight growth, the pre-culture was added to 1 L sterile M9 minimal media (12.8 g Na2HPO4·7H2O, 3.0 g KH2PO4, 0.5 g NaCl, 1.0 g NH4Cl, 0.24 g MgSO4, 11 mg CaCl2, 4 g Dextrose, 50 μg/mL Ampicillin, pH 7.0) and grown in a shaker-incubator at 37°C at 220 rpm. Upon reaching an OD600 of 0.8 - 0.9, protein expression was induced by addition of IPTG to a final concentration of 1 mM, and cultures were allowed to grow overnight at 37°C.
Cells were collected by centrifugation of cultures. The cell pellets were resuspended in 40 mL lysis buffer (70 mM Tris-HCL, 300 mM NaCl, pH 8.0). After the addition of 0.5 mg DNase I (Bovine Pancrease, Fischersci), 1 mg lysozyme (Egg White, Fischer Scientific), 40 μL RNase A (20 mg/mL, Thermo Scientific), and magnesium acetate to a concentration of 5 mM, the cell lysates were allowed to rotate at 4°C for 2 hours. After rotating, the samples were subjected to 20 minutes of sonication (5 s on 5 s off, 10 minutes total on time, Fisher Scientific Sonic Dismembrator Model 500, amplitude 40%). The lysates were centrifuged at 40,000 g for 20 minutes. The resulting pellet was resuspended in Urea Buffer (8 M urea, 20 mM Tris-HCl, 150 mM NaCl, 0.2% SDS, 2 mM β-ME, pH 8.0) and rotated overnight at room temperature to dissolve the inclusion bodies.
Overnight suspensions were again centrifuged at 40,000 g for 20 minutes. The resulting supernatant was mixed with 4 mL Ni-NTA resin (Thermo Scientific) which had been previously equilibrated with urea buffer. Samples were then rotated at room temperature for 2 hours. Excess protein was washed away by pouring the resin suspension into a 20 mL gravity flow column and rinsing with 6 volumes of TS/SDS Buffer (20 mM Tris-HCl, 200 mM NaCl, 0.2% SDS, 2 mM β-ME, pH 8.0). To produce LMPG micelle samples, 0.1% LMPG was substituted for 0.2% SDS in the TS Buffer washes. Purified His-tagged KCNE1 was eluted into five 2 mL fractions using elution buffer (250 mM imidazole, 0.2% SDS, 2 mM β-ME, pH 7.0). 0.1% LMPG was substituted for 0.2% SDS for LMPG micelle samples. The fractions were pooled together and concentrated to a volume of 1.5 mL using a 3.5 kD MW cutoff spin column (Millipore). The total concentration was obtained by measuring the OD280 and using an extinction coefficient of 1.2mg/mL per 1.0 absorbance in a 1 cm cell (15). Purity of the KCNE1 protein was confirmed by SDS PAGE.
MTSL Labeling
MTSL (Toronto Research Chemicals) stock (250 mM in methanol) was added directly to the concentrated protein in elution buffer at a 10:1 MTSL:protein molar ratio. The solution was allowed to react overnight at room temperature in the dark with vigorous stirring. The MTSL reaction mixture was loaded into a 3.5 kDa MW cutoff regenerated cellulose dialysis bag (Fisher Scientific) and dialyzed two times against 4 L phosphate buffer (100 mM NaH2PO4, 0.2% SDS, pH 7.2) for 24 hours each time. 0.1% LMPG was substituted for 0.2% SDS in the case of LMPG micelle samples. After dialysis, the samples were centrifuged at 6,000 g for 5 minutes to remove any precipitates. The resulting supernatant was used for EPR and CD spectroscopy. This method was found to be most effective for removing excess MTSL while preserving the detergent environment and minimizing protein aggregation.
Reconstitution into Lipid Bilayers
POPC and POPG powdered lipids (Avanti, Alabaster, AL) were dissolved to a final concentration of 5 mM (9:1 POPC/POPG molar ratio) in 100 mM NaH2PO4, 0.5% DOM, pH 7.2 by repeating at least 10 freeze/thaw/sonication cycles until clear. SM2 Bio Beads (Bio-Rad) were washed with eight 15 mL volumes of methanol followed by eight 15 mL volumes of water prior to use. Reconstitution was carried out by mixing 1 mL of the lipid slurry with 157 g of dialyzed MTSL-labeled protein in 0.2% SDS (500:1 lipid:protein molar ratio) and freeze/thawing twice to equilibrate. The lipid/protein mixture was then added to 300 mg wet Bio Beads and rotated at room temperature for 2 hours. Previous studies showed that this amount of Bio Beads combined with this concentration of detergent effectively removes all detergent and forms MLVs (16). The resulting sample was centrifuged at 2,000 g for 5 minutes to remove Bio Beads. The supernatant was concentrated by ultracentrifugation at 300,000 g for 30 minutes. The proteoliposome pellet was thoroughly resuspended in 50 L of 100 mM NaH2PO4 pH 7.2 and immediately used in spectroscopic studies. The resulting sample was a white, opaque, and highly viscous.
CW-EPR Spectroscopy
CW EPR spectroscopy was carried out on a Bruker EMX X-Band spectrometer. Spectra were recorded over a scan width of 100 G with a field modulation of 1 G at a frequency of 100 kHz. Samples were placed in a glass capillary tube and were 50 μL in volume with a total protein concentration of approximately 200 μM for micelle samples and 50 M for proteoliposomes. Each sample was scanned 10 - 20 times with a microwave intensity of 10 mW at 298 K.
CW-EPR Power Saturation Studies
Insertion of KCNE1 into POPC/POPG bilayers was verified using CW EPR Power Saturation at X-Band. Power saturation experiments were performed on a Bruker EMX X-band CW-EPR spectrometer consisting of an ER 041XG microwave bridge coupled with an ER 4123D CW-Resonator (Bruker BioSpin). Samples were loaded into gas permeable TPX capillary tubes with a total volume of 3 - 4 μl at a concentration of 30 - 50 μM. EPR data collection was carried out using a modulation amplitude of 1 G and varying microwave power of 0.4 - 100 mW. The scan range of all spectra was 100 G, and the final spectra were obtained by signal averaging 10 scans. The power saturation method works on the principle that under non-saturating conditions, the height of the spectral lines is linearly proportional to the square root of the incident microwave power, P1/2 (17). If the microwave power is subsequently increased, the increase in signal amplitude becomes less linear with P1/2, and signal height starts to decrease as the sample saturates.
Nitrogen is used as a control to purge the sample of oxygen and other paramagnetic relaxing agents. The power saturation curves were obtained for the S28C (probe outside of model membrane) and F54C, F57C, L59C, and S64C (probe inside) sites on KCNE1 under three conditions: 1) equilibrated with nitrogen as a control; 2) equilibrated with lipid-soluble paramagnetic reagent 20% oxygen (air); and 3) equilibrated with nitrogen in the presence of a water-soluble paramagnetic reagent NiEDDA (1 mM) as previously synthesized (18). The samples were purged with gas for at least 60 minutes at a rate of 10 mL per minute before performing each measurement. High purity nitrogen and house supply compressed air lines were used during the experiment. The resonator remained connected to the gas during all measurements and the sample temperature was held at 298 K. The peak-to-peak amplitude of the first derivative mI =0 resonance line (A) was measured and plotted against the square root of the incident microwave power. The data points were then fit using a Matlab software script according to Equation 1:
| (1) |
where I is a scaling factor, P½ is the power where the first derivative amplitude is reduced to half of its unsaturated value, and ε is a measure of the homogeneity of saturation of the resonance line. In equation 1, I, ε, and P½ are adjustable parameters and yield a characteristic P½ value. The corresponding Φ depth parameters were calculated (17, 19-21). The membrane depth (Å) of each spin-labeled KCNE1 mutant was calculated according to Equation 2:
| (2) |
The above bilayer depth calibration equation was determined using vesicles incorporating standard n-doxyl labeled POPCs (Avanti Polar Lipids) containing unlabeled WT KCNE1. Depth parameters (Φ) were measured and plotted against known depths for each labeled POPC (17, 22, 23). A linear dependence of Φ was found for the PCSLs, and equation 2 was derived via linear regression analysis.
Functional analysis of KCNQ1 and KCNE1
Complementary DNAs (cDNAs) encoding KCNQ1 and KCNE1 were constructed in plasmid vectors pSP64T and pRc/CMV, respectively, as previously described (24). All constructs were verified by sequencing. cRNA was transcribed in vitro from EcoRI (pSP64T-KCNQ1) or XbaI (pRc/CMV-KCNE1) digested linear DNA templates using Sp6 or T7 RNA polymerase and the mMessage mMachine transcription system (Ambion Inc, Austin, TX). The size and integrity of cRNA preparations were evaluated by formaldehyde-agarose gel electrophoresis, and full-length cRNA concentrations were estimated by comparison with a 0.24-9.5-kb RNA ladder (SIGMA, St. Louis, MO).
Female Xenopus laevis were anesthetized by immersion in 0.2% tricaine for 15 - 30 minutes. Ovarian lobes were removed and oocytes were manually defolliculated. Stage V–VI oocytes were injected with 25 nl of cRNA (KCNQ1, 6 ng/oocyte; KCNE1 constructs, 3 ng/oocyte) and incubated at 18°C for 48 - 96 hours in L-15, (Leibovitz's media, Invitrogen, Carlsbad, CA) diluted 1:1 with water and supplemented with penicillin (150 units/mL) and streptomycin (150 g/mL). Some oocytes were injected with 25 nl of water as a control for endogenous currents. Because previous studies revealed that Xenopus oocytes express endogenous KCNE genes(25), currents from oocytes injected with KCNQ1 cRNA alone were always recorded from representative oocytes in each batch to test for endogenous KCNEs.
Incorporation of recombinant His-KCNE1 protein into Xenopus oocytes
Our previous studies demonstrated that, following microinjection into oocytes, purified KCNE1 protein solubilized in LMPG micelles incorporates into the plasma membrane, where it modulates exogenous human KCNQ1 expressed therein (9-14). In this study, a similar protocol was employed, but used KCNE1 in POPC/POPG vesicles at a 2.5 mg/mL protein concentration. Vesicle samples were successfully prepared by removing detergent through either dialysis or Bio Beads. The oocytes were first injected with KCNQ1 cRNA, followed by injection of 25 nl of protein-containing or protein-free liposomes 24 hours later.
Electrophysiological analysis
Oocyte membrane currents were recorded at room temperature 2 days after injection using a two-microelectrode voltage-clamp technique with an OC-725B amplifier (Warner Instruments Corp., Hamden, CT). Oocytes were bathed at room temperature in a modified ND96 solution containing 96 mM NaCl, 4 mM KCl, 2 mM MgCl2, 0.1 mM CaCl2, 5 mM HEPES, pH 7.6, ~200 mosmol/kg. Data were recorded using Clampex 7 (Molecular Devices Corp., Sunnyvale, CA), filtered at 500 Hz and digitized at 2 kHz. Data were analyzed and plotted using a combination of Clampex, SigmaPlot 2000 (SPSS Science, Chicago, IL), and Origin 7.0 (OriginLab, Northampton, MA). Current-voltage and normalized isochronal voltage– activation relationships were obtained by measuring current at 2 s during depolarizing pulses between -50 and +60 mV from a holding potential of -80 mV. The normalized isochronal data were fit with a Boltzmann function of the form: 1/{1 + exp[(V – V1/2)/kv]}, where V1/2 is the half-maximal activation voltage and kv is the slope factor. Oocytes with currents at -80 mV (holding potential) larger than currents measured for water injected oocytes (-0.15 A) were considered leaky and not used for analysis.
CD Spectroscopy
CD experiments were performed on a Jasco model J-810 Spectropolarimeter over a wavelength range from 190 - 250 nm. Each sample was prepared in triplicate. Samples were subjected to 10 scans with a step size of 1 nm. The cuvette path length was 1.00 mm, and spectra were recorded at 298 K. Instrument calibration was performed with a standard solution of d(+)-10-camphorsulfonic acid. Protein concentrations were held at 15 M as confirmed by BCA assay (Thermo Scientific). When necessary, samples were diluted in 100 mM NaH2PO4 pH 7.2. Scattering effects produced by the sample matrix were found to be reduced when passing the vesicle samples through a 100 nm pore size extruder.
CD Spectral Simulations
CD spectral simulations were performed using the DICHROWEB website located at http://dichroweb.cryst.bbk.ac.uk which was supported by grants to the BBSRC Centre for Protein and Membrane Structure and Dynamics (CPMSD) (26). The Contin-LL algorithm with reference set 4 and a spectral width of 190 - 240 nm was used for all simulations (27-29). All CD and EPR data were plotted using Igor software (Wavemetrics Tigard, OR).
RESULTS
Homogeneous Reconstitution of KCNE1 in Bilayered Lipid Vesicles
POPC/POPG vesicles were chosen as the bilayer model membrane system in which to solubilize KCNE1. These two lipids are fully miscible, are akin with the most common phospholipids found in mammalian cell membranes, and have phase transition temperatures to the L αliquid crystalline phase below 0º C. The POPC:POPG mol:mol ratio was chosen to be 9:1 so as to mimic the 10-20 mol% of anionic phospholipids typically found in mammalian membranes (30).
Several reconstitution methods were tried including dialysis from aqueous SDS detergent into POPC/POPG vesicles and detergent removal by size exclusion chromatography. The dialysis method led to extensive sample aggregation while size exclusion chromatography proved inconsistent in sample preparation. Reconstitution using SM2 Bio Beads proved the most efficient and consistent for reliable sample reproduction. Interestingly, samples produced using dialysis still showed proper channel modulation upon injection into oocytes.
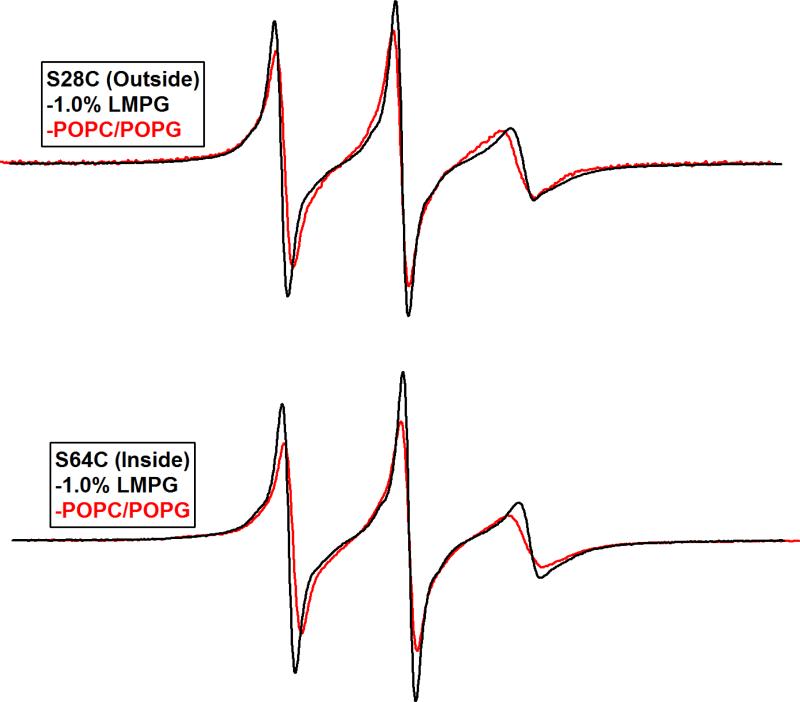
Figure 2 shows X-Band CW-EPR spectra of spin-labeled S28C and S64C KCNE1 in 1% LMPG micelles and in POPC/POPG liposomes at pH 7.2. Inspection of the data reveals a broadening in the spectral linewidths when moving from the micelles to the lipid bilayer and that residue S28C is more mobile than residue S64C in a lipid bilayer. This indicates that the dynamic motion of the nitroxide spin label is slower in the membrane environment when compared to the micelles and that resides within the membrane are less mobile than those outside. Similar line broadening has previously been reported (22, 31-33).
Figure 2.
CW EPR (X-Band) spectra of KCNE1 mutants S28C (Top) and S64C (Bottom). Black, 1.0% LMPG micelles. Red, POPC/POPG 9:1 Proteoliposomes (500:1 Lipid:Protein). The pH was 7.2 and the temperature was 298 °K for all samples. Spectra were normalized by spin concentration to show the difference in dynamic properties between S28C and S64C in a bilayer as compared to LMPG micelles.
Microinjection of Bilayer-Reconstituted KCNE1 into KCNQ1 Channel-Expression Oocytes
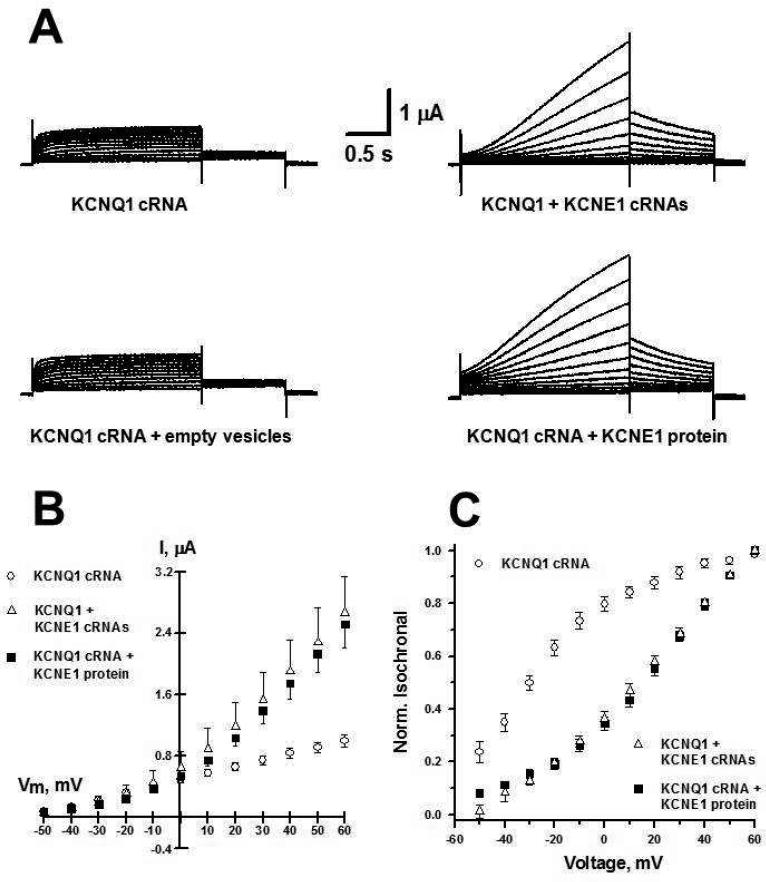
The ability of KCNE1 to associate with and modulate human KCNQ1 that was exogenously expressed in Xenopus oocytes was preserved when KCNE1 in POPC/POPG vesicles was microinjected into oocytes. The characteristic delayed channel opening resulting from KCNQ1-KCNE1 co-assembly was observed and was similar for both microinjected recombinant protein and for KCNE1 that was expressed in the oocytes following KCNE1 cRNA injection (Figure 4) (15). A negative control experiment in which protein-free POPC/POPG vesicles were injected did not result in modulation of KCNQ1 channel function, as expected. This indicates that KCNE1 in POPC/POPG vesicles somehow transferred into the oocyte membranes, perhaps by fusion of the vesicles with the native membranes, to yield a properly functioning channel complex at the plasma membrane.
Figure 4.
Functional expression of KCNE1 protein in Xenopus oocytes. A, Representative current traces recorded from oocytes injected with KCNQ1 cRNA, KCNQ1 + KCNE1 cRNAs, KCNQ1 cRNA + empty vesicles and KCNQ1 cRNA + KCNE1 protein in lipid vesicles; B, Current-voltage relationship for currents measured after 2 s test pulses from a holding potential of -80 mV for oocytes injected with KCNQ1 cRNA (○, n = 10), KCNQ1 + KCNE1 cRNA (8, n = 12) and KCNQ1 cRNA + KCNE1 protein in lipid vesicles (!, n =12); C, Normalized isochronal activation curves for oocytes, data were fit with a Boltzmann function and resulted in the following fit parameters; KCNQ1 cRNA (○): V½ = -32.5 ± 3.0 and kV = 20.4 ± 2.4; KCNQ1 + KCNE1 cRNAs (8): V½ = 28.8 ± 0.6 and kV = 20.6 ± 0.7; and for KCNQ1 cRNA + KCNE1 protein in lipid vesicles (!): V½ = 34.1 ± 1.3 and kV = 23.9 ± 1.1.
Probing the Topology of KCNE1 using Power Saturation EPR
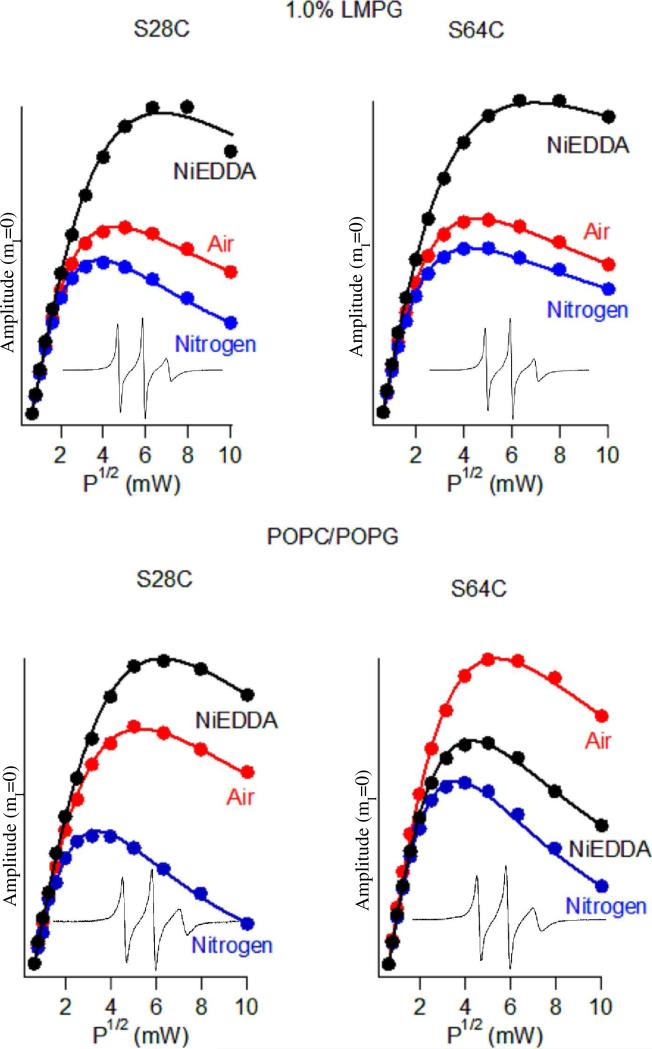
Power Saturation EPR experiments were conducted on KCNE1 to examine the location of and insertion of different residues with respect to the membrane (Figure 3). KCNE1 samples in LMPG micelles and POPC/POPG vesicles were analyzed under conditions where samples were exposed to NiEDDA, N2, or air (which contains O2). NiEDDA is highly polar and resides mostly in the aqueous phase. Conversely, paramagnetic O2 is non-polar and resides mostly within the hydrophobic lipid bilayer. Since NiEDDA and O2 function as paramagnetic-relaxing agents that decrease the T1 of the unpaired electron in the nitroxide spin label, the relative location of an MTSL spin label with respect to the aqueous phase and the membrane can be determined by examining signal intensity as a function of microwave power.
Figure 3.
EPR Power saturation curves for KCNE1 in 1% LMPG micelles and POPC/POPG vesicles pH 7.2 298 °K. Mutation S28C is outside the lipid bilayer and mutation S64C is part of the trans-membrane domain.
Figure 3 reveals that the addition of NiEDDA to S28C KCNE1 has a dramatic effect on the power saturation curve in both LMPG micelles and in POPC/POPG vesicles. This indicates that S28C is water-accessible in both matrices. Inspection of the power saturation profile of KCNE1 S64C (Figure 3(B)) indicates that O2 in air alters the relaxation profile much greater than both N2 and NiEDDA in POPC/POPG vesicles but not in LMPG micelles (9), demonstrating that site 64 is buried inside the hydrophobic vesicle membrane but water accessible in LMPG micelles. The N2 control experiments showed similar saturation results for both mutants. Both S28C and S64C are water-exposed in LMPG micelles, but their water access diverges upon reconstitution into lipid vesicles where S28C remains water exposed but S64C appears to be buried in a water-inaccessible part of the bilayer.
Additional CW-EPR power saturation experiments (See Supporting Information) and depth parameter calculations were conducted on additional mutants (F54C, F57C, and L59C) to confirm that KCNE1 has a transmembrane domain. The power saturation results for these mutants are similar to S64C. Table 1 shows the calculated depth parameter (Φ) values from the power saturation experiments. The mutants F54C, F57C, L59C, and S64C follow a trend as the depth parameter (Φ) increases from 0.8 to 1.9 then decreases again to 1.1 indicating that the predicted TMD is correctly spanning the membrane (Figure 1). Mutation L59C has the largest Φ value and is located near the center of the membrane while mutants F54C, F57C, and S64C have lower Φ values indicating that they are within the membrane, but not as deep. S28C has a negative Φ value indicating it resides outside of the membrane in the aqueous phase (17, 20, 21).
Table 1.
Depth parameter values (Φ) and membrane depth for different spin-labeled KCNE1 residues in POPC/POPG vesicles.
| Label Position | Depth Parameter (Φ) | Depth (Å) |
|---|---|---|
| S28C | -1.0 | |
| F54C | 0.8 | 6.8 |
| F57C | 1.4 | 10.1 |
| L59C | 1.9 | 12.9 |
| S64C | 1.1 | 8.5 |
An increase in Φ indicates that a residue is buried more deeply in the membrane. Negative Φ values indicate that a residue is not associated with the membrane.
A calibration curve based on known locations of n-doxyl POPCs was established for our membrane protein samples. Wild-type (unlabeled) KCNE1 was incorporated into each of the spin-labeled lipid samples in the same manner as labeled protein (17, 22). The Φ values for n = 5, 7, and 10 doxyl-POPC, along with published depths for each PCSL, were used to calculate distances into the membrane for each KCNE1 mutant (23). Based upon the solution NMR structure of KCNE1 in LMPG micelles, residue G60 should be at or near the center of the membrane bilayer (15). Taking into account that each residue is approximately 1.5 Å in depth from the next residue, this calibration of distance indicates that the center of our membrane system (POPC/POPG) is approximately 15 Å (17). The errors in these measurements are approximately +/- 2 Å due to the uncertainty introduced in the measurement of the depth parameter (Φ). This value is consistent with the length of the TMD of KCNE1 from the published solution NMR structure (15). Similar trends in Φ values and overall membrane thickness have been observed in the literature for transmembrane proteins (22, 34).
Secondary Structure Content in Phospholipid Vesicles Versus Micelles
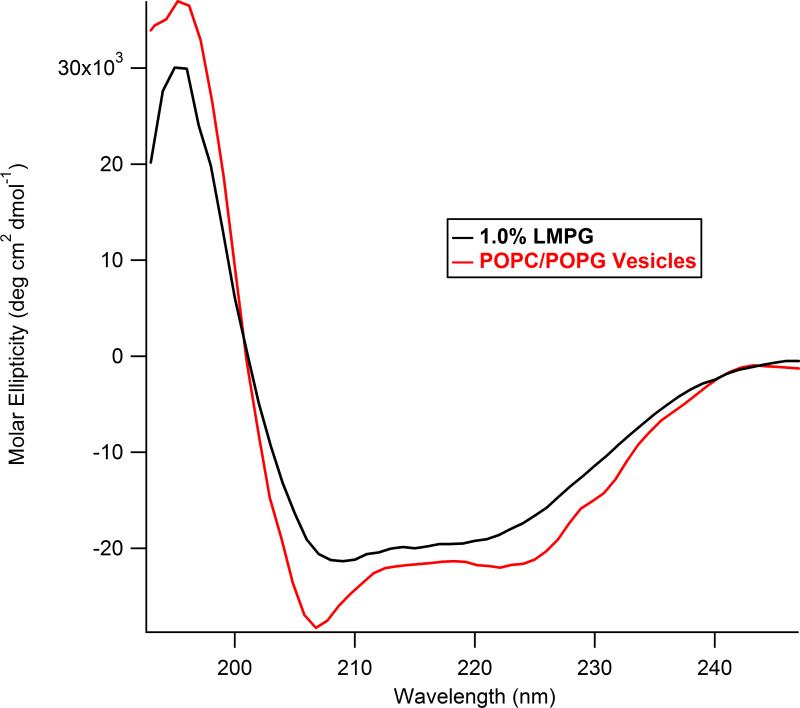
Figure 5 shows CD spectra of WT KCNE1 in 1.0% LMPG micelles and in POPC/POPG vesicles at pH 7.2 and 298 K. CD Spectroscopy of WT KCNE1 showed a predominately α-helical pattern in both micelle and lipid bilayer environments. Spectra collected in LMPG and POPC/POPG show similar α-helical contents as indicated by the peak at 195nm and troughs at 208 and 222nm. KCNE1 in 1.0% LMPG was found to have 44% helical content, in good agreement with the a value of 43% as determined in the NMR-based structural analysis which was conducted at pH 6.0 and 313 K (9). An increase in α-helical content to 60% was observed when moving from a micelle environment to POPC/POPG vesicles as indicated by an increase in the height of the peak at 195nm and depth of the troughs at 208nm and 222nm. The CD data clearly indicates that KCNE1 forms a more ordered structure upon insertion into a lipid bilayer with a higher α-helical content.
Figure 5.
CD spectra of WT KCNE1 in micelle and vesicle environments. Black, 1.0% LMPG micelles, 15 μM WT KCNE1, pH 7.2 298 °K; Red, POPC/POPG MLVs, 500:1 Lipid:Protein, WT KCNE1, pH 7.2 298 °K.
DISCUSSION
Previous structural studies of KCNE1 were carried out in 7.5% LMPG micelles at pH 6.0 and 313 K, a medium that is believed to be relatively native-like in terms of its ability to support the native stability, structure, and functionality of membrane proteins. However, even the best micelles are an imperfect mimic of actual lipid bilayers, such that it is imperative to test key features of the micelle-derived structure of KCNE1 for extrapolation to bilayered membrane conditions. For example, the transmembrane helix of KCNE1 was found to be curved in LMPG micelles, an observation that figures prominently in a structural detailed working model for KCNE1 modulation of the KCNQ1 (9). Is that curvature truly present in actual lipid bilayers or it is a micellar artifact? To cite a second example, the transmembrane helix of KCNE1 in micelles extends from residues 46-71 (9). It is important to test whether or not in more ordered matrix of lipid bilayers the transmembrane domain helix extends beyond the limits observed in micelles. The reconstitution in this work of KCNE1 in POPC/POPG vesicles combined with data that demonstrates the feasibility of conducting site-directed spin labeling EPR spectroscopic measurements set the stage for a rigorous testing/refinement of the KCNE1 structure in a bilayer environment.
That POPG/POPC vesicles represent a suitable model membrane composition for studying the structure of KCNE1 is at least partially validated by the fact that injection of recombinant vesicle-reconstituted KCNE1 into oocytes led to delivery of KCNE1 to the oocyte membranes where KCNE1 was able to co-assemble with the KCNQ1 potassium channel and modulate its function in a native-like fashion. Similar results were previously obtained when KCNE1 in LMPG micelles was injected into oocytes (Tian, Vanoye, et al., 2007). Given that we have confirmed that KCNE1 is largely helical in the vesicles it is very unlikely that its secondary structure would be dramatically altered during the process of protein transfer from the vesicles to the oocyte membranes. This suggests that KCNQ1 associates with the mostly-helical form of KCNE1, not a beta sheet-dominated structure as was previously seen for a peptide corresponding to the TMD of KCNE1 following reconstitution of this peptide into vesicles by co-solubilizing the peptide with lipid in an organic solvent and then dialyzing out the solvent in an aqueous reservoir (35).
Previous work suggests that the method used when incorporating KCNE1 into a lipid bilayer is crucial to determining what type of secondary structure the protein adopts (36, 37). The TMD of KCNE1 is very hydrophobic and has several polar residues on either end. It was hypothesized that these polar residues are responsible for anchoring the protein to the polar membrane surface while the inner non-polar residues interact with the lipid carbon chains. This amphipathic nature makes KCNE1 very insoluble and prone to aggregation in aqueous solution where it adopts a primarily β-sheet conformation. Membrane reconstitution based on selective removal of detergent from mixed detergent-lipid micelle samples using SM Bio Beads, as employed in the present paper, keeps KCNE1 in contact with high concentrations of either detergent or lipids at all times to avoid contact with other proteins and prevent aggregation. The lipid:protein molar ration is also significantly higher (500:1) as compared to previous studies where a lipid:protein ratios as low as 7.8:1 led to the formation of β-sheets in a peptide version of the TMD (35).
Having established methods for reconstituting both wild type and spin-labeled mutant forms of KCNE1 and verifying structural and functional integrity, we also carried out preliminary EPR and CD spectroscopic experiments that not only set the stage for future more extensive studies of KCNE1's bilayer-associated structure and dynamics, but also provide some interesting comparisons of it properties in bilayered vesicles vs. mild detergent (LMPG) micelles. In particular we observed that LMPG micelles do not completely mimic the natural environment of a lipid bilayer as indicated by CW-EPR power saturation data. Residue S64C, previously shown to reside inside an LMPG micelle, is still water accessible in the micelle but not in a vesicle bilayer, likely due to the water permeable nature of the micelle. S28C and S64C are thus indistinguishable in LMPG micelles with respect to their water accessibility. It may prove that S64C still sits at a similar depth in the membrane compared to the micelles but is simply more water accessible in micelles.
As mentioned previously, the LMPG structure of KCNE1 shows several peripheral α-helices that are docked into the micelle at specific residues (9). CW Power Saturation EPR will prove to be an invaluable technique in confirming these docking sites in lipid bilayers. EPR spectroscopy will also be used to fully explore the TMD of KCNE1 and determine which residues span this region in POPC/POPG vesicles as compared to LMPG micelles. Recent structure-function studies suggest that an amphipathic helix located just after the TMD may sit on the membrane surface (38, 39). Considering this helix, the ability of the bilayer to stabilize α-helical structures, and the many helical segments that exist in the previous LMPG micelle structure, it is easy to see how a bilayer could function to stabilize and extend these helical segments, giving rise to a higher helical content than previously reported in micelles (9). Our future work will also seek to determine where the additional α-helical content arises as discovered through CD spectroscopy using the wide array of techniques EPR spectroscopy offers for studying membrane protein structure.
Supplementary Material
Acknowledgments
We greatly appreciate the EPR expertise of Dr. Robert M. McCarrick of the Miami University Department of Chemistry and Biochemistry, and we would like to thank Dr. Amanda C. Dabney-Smith of the Miami University Department of Chemistry and Biochemistry for her extensive knowledge and advice in molecular biology.
Abbreviations
- β-ME
2-Mercaptoethanol
- CD
Circular Dichroism
- CMC
Critical Micelle Concentration
- CW
Continuous Wave
- DEER
Double Electron-Electron Resonance
- DOM
n-Dodecyl-β-D-Maltoside
- DPC
Dodecyl Phosphatidylcholine
- EPR
Electron Paramagnetic Resonance
- IPTG
Isopropyl β-D-thiogalactopyranoside
- LMPG
1-Myristoyl-2-Hydroxy-sn-Glycero-3-Phospho-(1'-rac-Glycerol) (Sodium Salt)
- LUV
Large Unilamellar Vesicle
- MLV
Multilamellar Vesicle
- MTSL
(S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate)
- NiEDDA
Nickel(II) ethylenediaminediacetate
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (sodium salt)
- SDS
Sodium Dodecyl Sulfate
- TMD
Transmembrane Domain
- WT
Wild Type
Footnotes
This work was generously supported by the National Institute of Health Grants RO1 GM60259-01 (to GAL), RO1 DC007416 (to CRS). Funding was also provided by the National Science Foundation CHE-1011909 (to GAL) and the NSF REU (CHE1004875) program.
Supporting Information
Supporting information available. Supporting information includes CW EPR and Power Saturation data for KCNE1 residues F54C, F57C, and L59C in POPC/POPG vesicles. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 2.Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, Keating MT. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 3.McCrossan ZA, Abbott GW. The MinK-related peptides. Neuropharmacology. 2004;47:787–821. doi: 10.1016/j.neuropharm.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 4.Abbott GW, Goldstein SA. Disease-associated mutations in KCNE potassium channel subunits (MiRPs) reveal promiscuous disruption of multiple currents and conservation of mechanism. FASEB J. 2002;16:390–400. doi: 10.1096/fj.01-0520hyp. [DOI] [PubMed] [Google Scholar]
- 5.Chen YH, Xu SJ, Bendahhou S, Wang XL, Wang Y, Xu WY, Jin HW, Sun H, Su XY, Zhuang QN, Yang YQ, Li YB, Liu Y, Xu HJ, Li XF, Ma N, Mou CP, Chen Z, Barhanin J, Huang W. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 6.Jespersen T, Grunnet M, Olesen SP. The KCNQ1 potassium channel: from gene to physiological function. Physiology (Bethesda) 2005;20:408–416. doi: 10.1152/physiol.00031.2005. [DOI] [PubMed] [Google Scholar]
- 7.Peters TA, Monnens LA, Cremers CW, Curfs JH. Genetic disorders of transporters/channels in the inner ear and their relation to the kidney. Pediatr Nephrol. 2004;19:1194–1201. doi: 10.1007/s00467-004-1626-6. [DOI] [PubMed] [Google Scholar]
- 8.Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT. Mutations in the hminK gene cause long QT syndrome and suppress IKs function. Nat Genet. 1997;17:338–340. doi: 10.1038/ng1197-338. [DOI] [PubMed] [Google Scholar]
- 9.Kang C, Tian C, Sonnichsen FD, Smith JA, Meiler J, George AL, Jr., Vanoye CG, Kim HJ, Sanders CR. Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel. Biochemistry. 2008;47:7999–8006. doi: 10.1021/bi800875q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panaghie G, Tai KK, Abbott GW. Interaction of KCNE subunits with the KCNQ1 K+ channel pore. J Physiol. 2006;570:455–467. doi: 10.1113/jphysiol.2005.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melman YF, Krumerman A, McDonald TV. A single transmembrane site in the KCNE-encoded proteins controls the specificity of KvLQT1 channel gating. J Biol Chem. 2002;277:25187–25194. doi: 10.1074/jbc.M200564200. [DOI] [PubMed] [Google Scholar]
- 12.Melman YF, Um SY, Krumerman A, Kagan A, McDonald TV. KCNE1 binds to the KCNQ1 pore to regulate potassium channel activity. Neuron. 2004;42:927–937. doi: 10.1016/j.neuron.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Yarov-Yarovoy V, Baker D, Catterall WA. Voltage sensor conformations in the open and closed states in ROSETTA structural models of K(+) channels. Proc Natl Acad Sci U S A. 2006;103:7292–7297. doi: 10.1073/pnas.0602350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panaghie G, Abbott GW. The role of S4 charges in voltage-dependent and voltage-independent KCNQ1 potassium channel complexes. J Gen Physiol. 2007;129:121–133. doi: 10.1085/jgp.200609612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian C, Vanoye CG, Kang C, Welch RC, Kim HJ, George AL, Jr., Sanders CR. Preparation, functional characterization, and NMR studies of human KCNE1, a voltage-gated potassium channel accessory subunit associated with deafness and long QT syndrome. Biochemistry. 2007;46:11459–11472. doi: 10.1021/bi700705j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambert O, Levy D, Ranck JL, Leblanc G, Rigaud JL. A new “gel-like” phase in dodecyl maltoside-lipid mixtures: implications in solubilization and reconstitution studies. Biophys J. 1998;74:918–930. doi: 10.1016/S0006-3495(98)74015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Averill DF, Smith DL, Legg JI. 5-Coordinate, Square-Pyramidal Chelate Complexes of a Novel Tetradentate Amino-Acid Like Ligand. Inorg Chem. 1972;11:2344–&. [Google Scholar]
- 19.Rauch ME, Ferguson CG, Prestwich GD, Cafiso DS. Myristoylated alanine-rich C kinase substrate (MARCKS) sequesters spin-labeled phosphatidylinositol 4,5-bisphosphate in lipid bilayers. J Biol Chem. 2002;277:14068–14076. doi: 10.1074/jbc.M109572200. [DOI] [PubMed] [Google Scholar]
- 20.Frazier AA, Wisner MA, Malmberg NJ, Victor KG, Fanucci GE, Nalefski EA, Falke JJ, Cafiso DS. Membrane orientation and position of the C2 domain from cPLA2 by site-directed spin labeling. Biochemistry. 2002;41:6282–6292. doi: 10.1021/bi0160821. [DOI] [PubMed] [Google Scholar]
- 21.Kohout SC, Corbalan-Garcia S, Gomez-Fernandez JC, Falke JJ. C2 domain of protein kinase C alpha: elucidation of the membrane docking surface by site-directed fluorescence and spin labeling. Biochemistry. 2003;42:1254–1265. doi: 10.1021/bi026596f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klug CS, Su W, Feix JB. Mapping of the residues involved in a proposed beta-strand located in the ferric enterobactin receptor FepA using site-directed spin-labeling. Biochemistry. 1997;36:13027–13033. doi: 10.1021/bi971232m. [DOI] [PubMed] [Google Scholar]
- 23.Dalton LA, Mcintyre JO, Fleischer S. Distance Estimate of the Active-Center of D-Beta-Hydroxybutyrate Dehydrogenase from the Membrane-Surface. Biochemistry. 1987;26:2117–2130. doi: 10.1021/bi00382a009. [DOI] [PubMed] [Google Scholar]
- 24.Tapper AR, George AL., Jr. MinK subdomains that mediate modulation of and association with KvLQT1. J Gen Physiol. 2000;116:379–390. doi: 10.1085/jgp.116.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anantharam A, Lewis A, Panaghie G, Gordon E, McCrossan ZA, Lerner DJ, Abbott GW. RNA interference reveals that endogenous Xenopus MinK-related peptides govern mammalian K+ channel function in oocyte expression studies. J Biol Chem. 2003;278:11739–11745. doi: 10.1074/jbc.M212751200. [DOI] [PubMed] [Google Scholar]
- 26.Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 27.Sreerama N, Venyaminov SY, Woody RW. Estimation of the number of alpha-helical and beta-strand segments in proteins using CD spectroscopy. Biophys J. 1999;76:A381–A381. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Stokkum IH, Spoelder HJ, Bloemendal M, van Grondelle R, Groen FC. Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal Biochem. 1990;191:110–118. doi: 10.1016/0003-2697(90)90396-q. [DOI] [PubMed] [Google Scholar]
- 29.Provencher SW, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 30.Gennis RB. Biomembranes: Molecular Structure and Function. Springe-Verlag; New York: 1989. [Google Scholar]
- 31.Pohl T, Spatzal T, Aksoyoglu M, Schleicher E, Rostas AM, Lay H, Glessner U, Boudon C, Hellwig P, Weber S, Friedrich T. Spin labeling of the Escherichia coli NADH ubiquinone oxidoreductase (complex I) Biochim Biophys Acta. 2010;1797:1894–1900. doi: 10.1016/j.bbabio.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 32.Naber N, Rice S, Matuska M, Vale RD, Cooke R, Pate E. EPR spectroscopy shows a microtubule-dependent conformational change in the kinesin switch 1 domain. Biophys J. 2003;84:3190–3196. doi: 10.1016/S0006-3495(03)70043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glasgow BJ, Gasymov OK, Abduragimov AR, Yusifov TN, Altenbach C, Hubbell WL. Side chain mobility and ligand interactions of the G strand of tear lipocalins by site-directed spin labeling. Biochemistry. 1999;38:13707–13716. doi: 10.1021/bi9913449. [DOI] [PubMed] [Google Scholar]
- 34.Fanucci GE, Cadieux N, Piedmont CA, Kadner RJ, Cafiso DS. Structure and dynamics of the beta-barrel of the membrane transporter BtuB by site-directed spin labeling. Biochemistry. 2002;41:11543–11551. doi: 10.1021/bi0259397. [DOI] [PubMed] [Google Scholar]
- 35.Horvath LI, Knowles PF, Kovachev P, Findlay JBC, Marsh D. A single-residue deletion alters the lipid selectivity of a K+ channel-associated peptide in the beta-conformation: Spin label electron spin resonance studies. Biophys J. 1997;73:2588–2594. doi: 10.1016/S0006-3495(97)78288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aggeli A, Bannister ML, Bell M, Boden N, Findlay JBC, Hunter M, Knowles PF, Yang JC. Conformation and ion-channeling activity of a 27-residue peptide modeled on the single-transmembrane segment of the IsK (minK) protein. Biochemistry. 1998;37:8121–8131. doi: 10.1021/bi972112h. [DOI] [PubMed] [Google Scholar]
- 37.Mercer EAJ, Abbott GW, Brazier SP, Ramesh B, Haris PI, Srai SKS. Synthetic putative transmembrane region of minimal potassium channel protein (minK) adopts an alpha-helical conformation in phospholipid membranes. Biochem J. 1997;325:475–479. doi: 10.1042/bj3250475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rocheleau JM, Gage SD, Kobertz WR. Secondary structure of a KCNE cytoplasmic domain. Journal of General Physiology. 2006;128:721–729. doi: 10.1085/jgp.200609657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lvov A, Gage SD, Berrios VM, Kobertz WR. Identification of a protein-protein interaction between KCNE1 and the activation gate machinery of KCNQ1. Journal of General Physiology. 2010;135:607–618. doi: 10.1085/jgp.200910386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.