Abstract
Formins are key regulators of actin nucleation and polymerization. They contain formin homology 1 (FH1) and 2 (FH2) domains as the catalytic machinery for the formation of linear actin cables. A subclass of formins constitutes the Diaphanous-related formins, members of which are regulated by the binding of a small GTP-binding protein of the Rho subfamily. Binding of these molecular switch proteins to the regulatory N-terminal mDiaN, including the GTPase-binding domain, leads to the release of auto-inhibition. From the three mDia isoforms, mDia1 is activated only by Rho (RhoA, -B, and -C), in contrast to mDia2 and -3, which is also activated by Rac and Cdc42. Little is known about the determinants of specificity. Here we report on the interactions of RhoA, Rac1, and Cdc42 with mDia1 and an mDia1 mutant (mDiaN-Thr-Ser-His (TSH)), which based on structural information should mimic mDia2 and -3. Specificity is analyzed by biochemical studies and a structural analysis of a complex between Cdc42·Gpp(NH)p and mDiaN-TSH. A triple NNN motif in mDia1 (amino acids 164-166), corresponding to the TSH motif in mDia2/3 (amino acids 183-185 and 190-192), and the epitope interacting with the Rho insert helix are essential for high affinity binding. The triple N motif of mDia1 allows tight interaction with Rho because of the presence of Phe-106, whereas the corresponding His-104 in Rac and Cdc42 forms a complementary interface with the TSH motif in mDia2/3. We also show that the F106H and H104F mutations drastically alter the affinities and thermodynamics of mDia interactions.
The dynamics of the actin cytoskeleton play an important role in many physiological processes, e.g. cytokinesis, cell migration, cell morphology, and phagocytosis (1-3). Although the Arp2/3 complex leads to the formation of a branched actin network and is regulated by WASP and VASP and others, the formins and the protein Spire catalyze the nucleation and polymerization of unbranched actin filaments (3-8). Formins are multidomain proteins characterized by formin homology domains. All formins contain the FH22 domain, the actual catalytic machinery for actin polymerization, which is often in tandem with FH1, a polyproline domain mediating profilin·actin recruitment (9-13). The FH3 domain is believed to play a role in the subcellular localization of the protein and is the least conserved FH domain (12, 14-16).
The formins are subdivided into seven groups as follows: Dia (diaphanous), DAAM (dishevelled-associated activator of morphogenesis), FRL (formin-related gene in leukocytes), FHOD (formin homology domain-containing protein), INF (inverted formin), FMN (formin), and delphilin (24). The structure of the FH2 domain from yeast and mouse showed a doughnut-shaped all-helical molecule, which forms a dimer binding with high affinity to actin (13, 17-21). From the structure of the FH2 alone and in complex with tetramethylrhodamine-labeled actin and from biochemical experiments, the mechanism of actin polymerization by the formin was described as a stair-stepping mechanism, where the FH2 dimer binds to the barbed ends of actin filaments as a leaky capper that processively catalyzes barbed-end actin assembly (11, 13, 17, 22-28). The processive movement could be shown in vitro as well as in vivo (11, 22, 23).
For most formins, the tandem arrangement of FH1-FH2 domains is necessary for function. FH1 contains several stretches of poly-l-prolines that are targets for Src homology 3 and WW domain-containing proteins and for profilin (19, 29, 30). By binding to G-actin as well as to FH1, profilin delivers actin monomers to the barbed end of the filament and increases the concentration of actin near the FH2 domain (13, 28). Therefore, FH1-FH2 accelerates the association rate for ATP-G-actin monomers at the barbed end, but it also accelerates the hydrolysis rate of actin-bound ATP (11, 22). This leads to the release of profilin from the barbed-end making this site accessible for the next round of G-actin incorporation (11).
N-terminal to the FH1/FH2 domains, the subclass of Diaphanous-related formins contains a regulatory unit consisting of the GBD and FH3 domains (mDiaN in short), which interact with the C-terminal Diaphanous-autoregulatory domain (DAD) (31-36). The deletion of either GBD or DAD leads to a constitutively active protein that induces actin stress fibers and serum-response factor-dependent nuclear transcription (25, 37-39). The Drosophila protein Diaphanous was isolated in Drosophila as a gene involved in cytokinesis. There are three mammalian homologues mDia1-3 and two Diaphanous-related formin proteins Bni1p and Bnr1p in Saccharomyces cerevisiae (12). Binding of activated small GTP-binding proteins from the Rho family leads to the release of the inhibitory interaction between DAD and mDiaN and activation of the FH2 that is not yet completely understood (31-33, 35, 40).
It has been shown by us and others that mDiaN consists of three subdomains as follows: GBDN, Armadillo repeat region (ARR) also called DID (Diaphanous-inhibitory domain), and dimerization domain (Dim) (33, 35, 40). The structure of the mDiaN in complex with activated RhoC shows that Rho interacts via switch I and II with the GBD but also with the ARR via the Rho insert helix. The DAD-binding site on mDiaN (DID) has also been described biochemically and structurally (32). The structure shows that the 16 highly conserved residues of the DAD core region (DCR) form an α-helix, which binds to the ARR in a site not directly overlapping with the Rho-binding site (32, 41). Nevertheless, Rho actively dissociates DAD from its binding site via long range electrostatic repulsion and short range steric clashes (32).
Small guanine nucleotide-binding proteins (small GNBP, G proteins) cycle between a GDP-bound inactive form and a GTP-bound active form (42-44). They interact with their effector proteins preferentially in the GTP-bound form (44, 45). This discrimination is because of the conformation of two loop regions, switch I and II, that adopt a defined conformation when connected to the γ-phosphate. Rho proteins interact with a number of different effectors, which in turn regulate the actin cytoskeleton either directly, as with mDia, or indirectly by modifying the activity of actin-binding proteins (44, 46-53). Binding motifs in Rho effectors are divergent, with a Cdc42/Rac-interactive binding motif in Cdc42/Rac effectors WASP, WAVE, ACK, and PAK (47, 54). The Rho effectors Rhophilin, Rhotekin, and protein kinase N, and ROCK bind via an anti-parallel coiled-coil motif (47, 55, 56), whereas mDia binding to Rho is again via a different structural motif (35). The common feature of these interactions is that Rho proteins not only recruit their effectors to a particular membrane site but that they also allosterically activate the effector (5, 47, 57). This involves the release of an intramolecular autoinhibited state that can be found in the cases of kinases, e.g. ROCK, protein kinase N, or PAK, but also in the case of WASP, WAVE, and mDia1 (47). For mDia1 the release of autoinhibition by Rho binding does not lead to full activation of the actin polymerization activity catalyzed by FH2 (25, 40).
The activation of Rho proteins plays an important role in spatio-temporal regulation of the actin cytoskeleton inside the cell. Very often, different signal transduction pathways via Rho proteins lead to similar or overlapping cellular responses (58). Effectors of Rho proteins are nevertheless not promiscuous in responding to either Rho and its homologues, i.e. RhoA, -B, -C, or to Cdc42 and Rac and its homologues such as TC10. Because a particular Rho protein interacts with a number of structurally different effectors via the highly conserved switch I and II regions, a high plasticity in the interface is required (59). Numerous Rho family effectors have been described so far, defined as proteins that only bind to the GTP-bound state. ROCK I and II and Rhotekin specifically interact with and are activated by binding of Rho but not Rac or Cdc42 (39, 60). It has been postulated that for ROCK II only residue Glu-40 of RhoA is important for discriminating between RhoA, Cdc42, and Rac1 (61). The Cdc42/Rac-interactive binding motif containing proteins PAK, WASP, and WAVE and others are specific effectors for members for either Rac or Cdc42 but do not interact with Rho.
In case of mDia, the situation is different in that mDia1 specifically binds to and is activated by RhoA, -B, and -C (39), whereas mDia2 and -3 seem to be highly promiscuous and activated by RhoA-C, Cdc42, and RhoD (36, 39, 62, 63). The question to ask is whether, like in protein kinase C-related kinase, the effector binds to multiple Rho family members at different binding sites or whether different G proteins use the same binding site. Here we analyzed the specificity of interaction between mDia isoforms and Rho proteins. We show that Rho, Rac, and Cdc42 use a similar binding site and that most of the specificity is determined by a Asn-Asn-Asn (triple N) motif of mDia1 that is replaced by Thr-Ser-His (TSH) in mDia2 and -3. On the Rho site, specificity is determined by a Phe-His substitution of Phe-106. We also observe for the first time that the insert helix, a specific hallmark of Rho proteins, is involved in effector binding and specificity.
EXPERIMENTAL PROCEDURES
Recombinant Proteins—Proteins were expressed as glutathione S-transferase fusions using the vector pGEX-4T1 (Amersham Biosciences). All proteins were expressed in Escherichia coli BL21(DE3) cells. Cells were grown to an A600 of 0.7 (37 °C, 160 rpm) and induced with 100 μm isopropyl β-d-thiogalactopyranoside overnight at 20 °C for mDiaN and mDiaN-TSH and at 18 °C for RhoA, Rac1, and Cdc42. Cells were harvested and resuspended in buffer A (20 mm Tris/HCl, pH 7.5, 100 mm NaCl, 2 mm β-mercaptoethanol, and 5 mm MgCl2 in cases of RhoA, Rac1, and Cdc42) containing 0.1 mm phenylmethylsulfonyl fluoride and 2 mm EDTA. Cells were lysed by microfluidizing; the extract was applied to a GSH-Sepharose column (Amersham Biosciences), and the column was washed with buffer B (20 mm Tris/HCl, pH 7.5, 100 mm NaCl, 2 mm β-mercaptoethanol, 5 mm MgCl2). The fusion protein was digested with 5 units of thrombin (Serva) per mg of glutathione S-transferase fusion protein on the column overnight at 4 °C. Final purification was done with size exclusion chromatography in buffer B using a Superdex S200 for mDiaN and mDiaN-TSH or S75 for RhoA, Rac1, and Cdc42.
Point mutations were made using the QuikChange method of Stratagene. Mutant proteins were purified like the wild type proteins. Concentrations of mDiaN and mDiaN-TSH were determined using the absorption at 280 nm under denaturing conditions with the extinction coefficient deduced from the sequence, and RhoA/Cdc42/Rac1 concentrations were determined by the Bradford assay. Rho was purified and loaded with Gpp(NH)p and mant-Gpp(NH)p as described elsewhere (35). Rac1 and Cdc42 were purified using the same strategy.
Stopped-flow Kinetics—The experiments were done at 20 °C using a SX18 MW Applied Photophysics apparatus (Leather-head, UK). The mant fluorophore (Molecular Probes) was excited at 366 nm with a bandpass of 6.4 nm. Emission (at 450 nm) was recorded using a 408-nm cutoff filter. All measurements were done with a final concentration of 100 nm mant-Gpp(NH)p-loaded GNBP/mutated GNBP. Increasing concentrations of wild type or mutant mDiaN (1-15 μm) were used in the experiments using pseudo first-order conditions. The observed fluorescence transients were fitted to a single exponential function to obtain kobs. The association rate constant kon was derived from the slope of kobs versus protein concentration. The dissociation rate constants were obtained by mixing a preformed 100 nm mDiaN·RhoA·mant-Gpp(NH)p (or mutated proteins) complex with a 100-fold excess of unlabeled Rho protein. All measurements were done in buffer C (300 mm NaCl, 50 mm Tris/HCl, pH 7.5, 2 mm β-mercaptoethanol, 5 mm MgCl2), which in experiments with RhoA additionally contained 5 mm MgCl2. GraFit 3.0-5.0 was used for data analysis.
ITC Measurements—The interaction of mDiaN/mDiaN-TSH and RhoA/Cdc42/Rac1 and DAD-(1145-1200) and mutants was done by ITC based on Wiseman et al. (64) (MicroCal™; VP-ITC MicroCalorimeter). All measurements were carried out in buffer B at 20 °C. DAD, RhoA, Cdc42, and Rac1 (or mutants) (syringe) were stepwise-injected to the mDiaN, mDiaN-TSH solution (cell). The heating power per injection was observed over the reaction time until equilibrium was reached. The data were analyzed using the software provided by the manufacturer.
Crystallization, Data Collection, and Structure Solution—Crystals of the mDiaN[TSH] domain (69-451) in complex with Cdc42-(1-178)·Gpp(NH)p were grown in buffer D containing Bistris propane, pH 8.8 (pH adjusted with citric acid), 26% PEG3350, 250 mm sodium tartrate to a size of ∼100 × 200 × 100 μm using the hanging drop/vapor diffusion method. The protein was solved in buffer B containing 100 mm NaCl, 20 mm Tris/HCl, pH 7.5, 2 mm β-mercaptoethanol, and 5 mm MgCl2.1 μl of protein solution of various concentrations (2.5, 5, and 10 μg/μl) was mixed with the reservoir solution. After 1 day crystals were grown to their final size and flash-frozen in liquid nitrogen using reservoir solution with 35% PEG3350 as cryoprotectant.
Data set collection has been performed at the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland (Table 4). The native dataset was collected at 100 K on beam line PXII at a wavelength of 0.95 Å using a Mar225 CCD detector. Data were indexed, integrated, and scaled with the XDS package (65).
TABLE 4.
Statistics of data collection and structure refinement
Values in parentheses correspond to those in the outer resolution shell.
| Data collection | |
| Space group | P32 |
| X-ray wavelength | 0.9763 Å |
| Unit cell dimension | a = b = 71.27 Å, c = 244.96 Å |
| Resolution (Å) | 43.5 to 2.7 Å (2.8-2.7) |
| Rsyma | 7.4 (38.1) |
| <I>/σ<I> | 13.5 (3.7) |
| Completeness (%) | 99.5 (99.8) |
| No. of reflections unique | 38,038 (3,957) |
|
Redundancy
|
3.2 (3.2)
|
| Refinement | |
| Rcrystb / Rfreec | 20.6%/24.3% |
| No. of atoms | 8,341 |
| Proteins | 8,228 |
| Gpp(NH)p/Mg2+ | 66 |
| Water | 47 |
| r.m.s.d.d bond lengths (weight) | 0.007 Å (0.022 Å) |
|
r.m.s.d.d
bond angles (weight)
|
1.104° (1.998°)
|
| Ramachandrane | |
| Favored | 98.9% |
| Allowed | 100.0% |
Rsym = (∑|(I - 〈I〉)/(∑I), where 〈I〉 is the average intensity of multiple measurements.
Rcryst = (∑|Fobs - F|)/(∑|Fobs|).
Rfree is the R-factor based on 5% of the data excluded from refinement.
r.m.s.d. is root mean square deviation.
Data were analyzed by MolProbity (70).
Initial phases were obtained by molecular replacement with the program Molrep (66) using the mDiaN·RhoC (PDB code 1Z2C) as a search model. The initial solution containing two Cdc42·Gpp(NH)p·mDiaN-TSH complexes per asymmetric unit was subjected to crystallographic simulated annealing as implemented in CNS (67). The program Coot (68) was used to build the model into the 2Fo - Fc and Fo - Fc maps in iterative rounds of NCS refinement, including translation, liberation, and screw-rotation parameters with REFMAC (69). The final model has a good geometry with 100% of all residues in the allowed regions of the Ramachandran plot as judged by the Molprobity server (70). A summary of data collection and refinement statistics is given in Table 4. Figures of molecular structures were prepared with PyMOL (71).
Coordinates—The structure factors and coordinates have been deposited at the PDB with PDB code 3EG5.
RESULTS
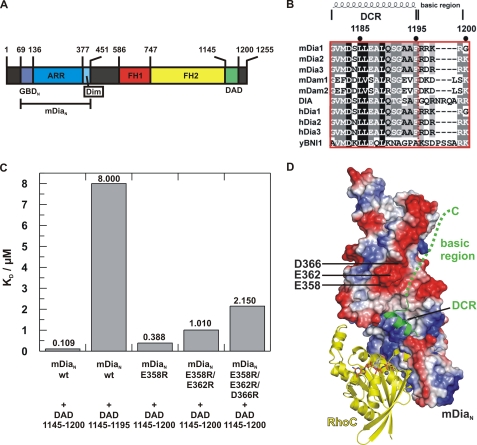
Specificity of the DAD-N-terminal Auto-inhibitory Interaction—Activation of mDia1 involves binding of Rho·GTP, which actively dissociates the interaction between the regulatory N terminus and the C-terminal DAD of mDia1 (32) (see Fig. 1A for domain organization of mDia1). The crystal structure of the [mDiaN·DAD] complex showed that the DCR (see Fig. 1B for an alignment of DCRs) forms an α-helix that interacts mainly via hydrophobic interactions with the ARR of mDiaN and that the binding sites of DAD and RhoA·Gpp(NH)p on mDiaN are partially overlapping such that a ternary complex cannot be formed (32). Although the C-terminal basic region 1196RRKRG1200 of DCR is not visible in the crystal structure (32), its deletion in DAD-(1145-1200) reduces the affinity toward mDiaN 73-fold to 8 μm (Fig. 1C), as determined by ITC. To exclude that this extra part of DAD interferes with the Rho-binding site and thereby contributes to specificity, we tried to locate the binding site for the basic region of DAD in mDiaN. We reasoned that a negatively charged patch near the third helix of ARM5 and the Dim domain could represent the binding site of the basic motif. Single, double, and triple mutants of acidic residues were introduced (E358R, E358R/E362R, and E358R/E362R/E366R, respectively), and binding of the mutant constructs toward DAD-(1145-1200) was followed by ITC. Although only marginal effects on affinity could be measured for the single point mutation (E358R), an additive effect on affinities is observed for the single, double, and triple mutant (388 nm, 1.01 μm, and 2.15 μm) (Fig. 1C) with the affinities converging to the value measured for the DAD construct lacking the C-terminal basic region. These experiments clearly establish that the C-terminal basic region of DAD interacts with the negatively charged patch located on mDiaN remote from the Rho-binding sites (Fig. 1D). Thus, an involvement of this part of DAD in generating specificity toward activated Rho proteins appears unlikely.
FIGURE 1.
Localization of the DAD on mDia. A, schematic representation of the domain organization of mDia1, with GBDN, GTPase-binding domain; ARR, Armadillo-repeat region; Dim, dimerization domain; FH1-2, Formin homology domains 1-2; DAD, Diaphanous autoregulatory domain; mDiaN, the N-terminal regulatory region. B, sequence alignment of DAD-core region (DCR) and basic region from different genes and organisms (SwissProt accession numbers in parentheses): mDia1-3, mouse (O08808, Q9Z207, and O70566); mDam1-2, mouse (Q8BPM0 and Q80U19); Dia, Drosophila (P48608); hDia1-3, human (O60610, O60879, and Q9NSV4); yBNI1, S. cerevisiae (P41832). 100% conserved, black-shaded; 80% conserved, dark gray; 60% conserved, light gray. C, equilibrium dissociation constants (KD) (from ITC) for WT and mutant DAD constructs as indicated. D, model of the ternary mDiaN·RhoC·DAD complex obtained by superimposition of the mDiaN·DAD complex (PDB 2BAP) with the RhoC·mDiaN complex structure (1Z2C), with mDiaN shown in electrostatic surface representation (blue for positively charged surfaces, red for negatively charged surfaces, and white for neutral surfaces). Residues Glu-358, Asp-362, and Glu-366 of mDiaN are indicated. RhoC and the DAD core are displayed as yellow and green ribbons, respectively, Gpp(NH)p as stick model, and Mg2+ as sphere. The green dashed line represents the course of the C-terminal basic region of DAD as suggested from data described in the text.
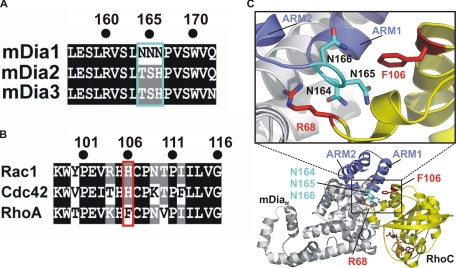
Major Specificity Determinants—By analyzing the RhoC·mDiaN complex structure and comparing them to the sequence of mDia2 and -3, we identified a triple Asn motif (Asn-164-166) of mDia1 in the central portion of the interface, which is replaced by TSH in mDia2/3 (Fig. 2, A and C). Asn-165 and Asn-166 are located in the loop connecting ARM1 (ARM is Armadillo repeat motif) and ARM2 of the ARR of mDiaN and are apparently required for the correct positioning of the Arg-68R (throughout the whole text, superscripts R, C, NNN, and TSH indicate residues of Rho, Cdc42, wild type mDiaN-NNN, and mutated mDiaN-TSH, respectively), a residue being highly crucial for the binding because it wedges deeply into the space between the GBDN and ARR subdomains of mDiaN (35) (Fig. 2C). From the residues of Rho that are in interaction distance with the triple Asn motif only, Phe-106 is variant among Rho, Rac, and Cdc42, and in the latter two it is replaced by His (Fig. 2, B and C). This together with our previous finding that mutation of F106H in Rho drastically reduces the affinity to mDia1 (35) argue for Phe-106 of Rho and His-104 of Cdc42/Rac being major determinants of affinity and probably specificity on the G protein site. Because constructs of mDia2 and -3 were not available in soluble form, we mutated the mDia triple Asn motif to Thr-Ser-His (termed mDiaN-TSH) to produce a mimic of mDia2 and mDia3 for studying its influence on specificity of Rho family proteins to mDia isoforms.
FIGURE 2.
A, sequence alignment of mDia isoforms 1-3 (SwissProt accession numbers O08808, Q9Z207, and O70566, respectively) in the region of the triple NNN- and TSH motifs of mDia1, -2, and -3. B, sequence alignment of mouse Rac1, Cdc42, and RhoA (SwissProt accession numbers P63001, P60766, and Q9QUI0, respectively) in the region around Phe-106 of RhoA. C, ribbon diagram (lower part) of the structure of the RhoC·mDiaN complex (PDB 1Z2C) with a blowup (upper part) highlighting the residues crucial for specificity, with Arg-68 and Phe-106 of RhoC and the triple NNN motif Asn-164-166 of mDiaN located in a loop between ARM1 and ARM2, with mDiaN as gray ribbons, ARM1 and ARM2 highlighted in blue, Asn-164-166 of as cyan sticks, RhoC as yellow ribbons, Arg-68 and Phe-106 as red sticks, and Gpp(NH)p·Mg2+ as orange sticks.
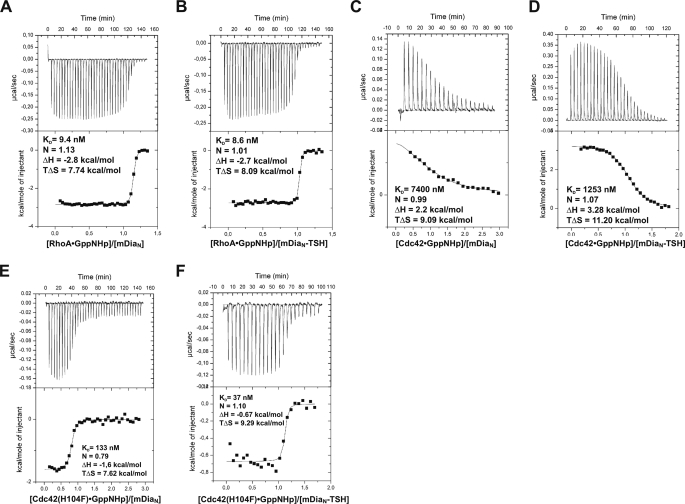
Although the interaction between RhoA (bound to Gpp(NH)p) and mDiaN could previously not be detected by isothermal titration calorimetry (ITC) under high salt conditions (300 mm NaCl, see “Experimental Procedures”), a KD of 9 nm could conveniently be determined at low salt, with favorable values for enthalpy and entropy (Fig. 3A and Table 2). This is very similar to what was previously found by stopped-flow fluorescence measurements under high (35) or low salt,3 indicating that the affinity is not salt-sensitive in contrast to the enthalpy of the reaction. Furthermore, insertion of the TSH motif into mDiaN did not affect its binding affinities to DADs of mDia1, mDia2, and mDia3, as confirmed by ITC (Table 1). We then investigated binding of RhoA, Rac1, or Cdc42 to mDiaN and mDiaN-TSH using ITC (Fig. 3 and Table 2). Whereas mutation of the triple N motif to TSH does not change the affinity toward RhoA (9 nm) (Fig. 3, A and B), it increases affinity toward Cdc42 and Rac1 such that it becomes easily measurable (Fig. 3, C and D). Although the interaction of RhoA toward either mDiaN or mDiaN-TSH is highly exothermic, driven by both enthalpy and entropy, the interactions of Cdc42 and Rac1 toward mDiaN or mDiaN-TSH, are endothermic, with a compensating favorable entropy. As a control we also measured binding of DAD-(1145-1200) to mDiaN-TSH, which was basically unperturbed as compared with WT, an endothermic enthalpy, and a KD of 115 nm (Table 1).
FIGURE 3.
ITC analysis of the interaction between WT and mutant Rho and mDia proteins, with RhoA·Gpp(NH)p and mDiaN (A), RhoA·Gpp(NH)p and mDiaN-TSH (B), Cdc42·Gpp(NH)p and mDiaN (C), or mDiaN-TSH (D) and Cdc42(H104F)·Gpp(NH)p and mDiaN (E) or mDiaN-TSH (F).
TABLE 2.
ITC analysis of binding of WT and mutant G proteins to WT and mutant mDiaN
N is the stoichiometry of binding.
| Construct | KD, nm | ΔH/kcal·mol−1 | TΔS/kcal·mol−1 | N |
|---|---|---|---|---|
| mDiaN | ||||
| RhoA | 9.35 ± 2.7 | −2.8 ± 0.015 | 7.74 | 1.13 |
| RhoA F106H | 180 ± 30 | −1.4 ± 0.014 | 7.68 | 1.02 |
| Cdc42 | 7400 ± 1207 | 2.2 ± 0.232 | 9.09 | 0.99 |
| Cdc42 H104F | 133 ± 32 | −1.6 ± 0.027 | 7.62 | 0.79 |
| Rac1 | 1600 ± 4885 | 1.9 ± 0.237 | 8.30 | 0.84 |
|
Rac1 H104F
|
57 ± 15
|
−2.68 ± 0.037
|
7.04
|
0.86
|
| mDiaN-TSH | ||||
| RhoA | 8.6 ± 2.6 | −2.73 ± 0.015 | 8.09 | 1.01 |
| RhoA F106H | 1418 ± 342 | −0.69 ± 0.020 | 7.15 | 1.14 |
| Cdc42 | 1253 ± 86 | 3.28 ± 0.026 | 11.20 | 1.07 |
| Cdc42 H104F | 37 ± 27 | −0.67 ± 0.015 | 9.29 | 1.10 |
| Rac1 | 758 ± 138 | 1.65 ± 0.031 | 9.85 | 0.76 |
| ac1 H104F | 51 ± 22 | −2.11 ± 0.037 | 7.65 | 0.96 |
TABLE 1.
Thermodynamic analysis by ITC of binding of DAD from mDia1, -2, and -3 to WT and TSH mDiaN
N is the stoichiometry of binding.
| Construct | KD, nm | ΔH/kcal·mol−1 | TΔS/kcal·mol−1 | N |
|---|---|---|---|---|
| mDiaN | ||||
| DADmDia1a | 109 ± 10 | 10.16 ± 0.053 | 19.49 | 0.92 |
| DADmDia2a | 325 ± 20 | 6.87 ± 0.040 | 15.57 | 0.99 |
|
DADmDia3a |
277 ± 23
|
6.65 ± 0.045
|
15.45
|
1.10
|
| mDiaN-TSH | ||||
| DADmDia1a | 115 ± 5 | 6.45 ± 0.016 | 15.74 | 0.88 |
| DADmDia2a | 322 ± 25 | 5.08 ± 0.031 | 13.78 | 0.96 |
| DADmDia3a | 164 ± 12 | 5.97 ± 0.027 | 15.07 | 0.89 |
DADmDia1 encompasses residues 1145-1200, DADmDia2 residues 1004-1060, and DADmDia3 residues 1116-1170.
We further assayed via ITC the influence of the Phe-106 to His substitution in Rho and of His-104 to Phe in Rac1 and Cdc42 on binding to mDiaN and mDiaN-TSH. Mutation of Phe-106 to His in RhoA leads to 20-fold reduced affinity to mDiaN and to a drastically reduced affinity to mDiaN-TSH (KD = 1418 nm), whereas wild type RhoA has an equal affinity to both mDiaN and mDiaN-TSH (KD = 9 nm in both cases). For Rac1 and Cdc42 the exchange of His-104 with the more hydrophobic Phe in turn increases the affinities of Cdc42 and Rac toward mDia1N by 55- and 28-fold, respectively, and is similar with either the triple N or TSH motif (Fig. 3, E and F, and Table 2).
A notable feature of the His-Phe substitutions is that introduction of Phe dramatically alters the enthalpy of the binding reaction (Fig. 3 and Table 2). For Rac1 and Cdc42, the H104F mutation shifts the reaction enthalpy of binding from endothermic to exothermic, with a difference in ΔH of between 3.7 and 4.6 kcal/mol, irrespective of whether wild type or mDiaN-TSH is used. Although this leads to an increase in affinity, it is still partly diminished by a decrease in favorable (positive) entropy of ΔG of 1.5-2.2 kcal/mol, an effect often observed in protein-protein interactions as the so-called enthalpy-entropy compensation (72). For RhoA the opposite F106H exchange is less dramatic, with a decrease in favorable enthalpy of 1.4 (for WT) and 2 kcal/mol (for TSH), and does not significantly modify the entropy term of the binding reaction. This, at least in part, explains why RhoA shows the highest affinity to both mDiaN and mDiaN-TSH compared with Rac1(H104F) and Cdc42(H104F). Rho binding is tighter because of favorable enthalpy and entropy contributions.
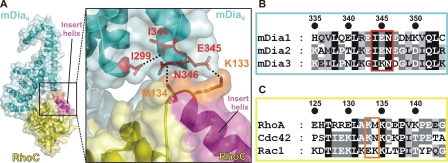
Rho Insert Helix and Specificity—From the biochemical experiments presented above, it is obvious that the specificity and affinity of the interactions between mDia isoforms and Rho subfamily members is not entirely due to the central contact point located between the GBD and ARR subdomains of mDia. A particular structural feature of Rho proteins is an extra helix located between the fifth β-strand and the fourth α-helix of the canonical Ras fold, the so-called Rho insert helix (Fig. 4). In the crystal structure of the Rho·mDia complex, the Rho insert helix is close enough to the interface to assume a contribution to the interaction (Fig. 4A). Residues in direct contact distance (cutoff level 3.6 Å) include Lys-133R and Met-134R from one partner and Glu-345NNN and Asn-346NNN from the other partner. The interaction is mediated via a salt bridge between Lys-133R and Glu-345NNN, which then enables Met-134R and Asn-346NNN to come into van der Waals interaction distance. Although not directly involved in the interaction with Rho, Ile-299NNN appears to play a crucial role in stabilizing the spatial integrity of the insert helix-binding site via a long range hydrophobic interaction with Ile-344NNN, which then can correctly position Glu-345NNN and Asn-346NNN. Strikingly, there is considerable sequence variability for the residues involved in this interaction among Rho proteins and mDia isoforms (Fig. 4, B and C). Although Asn-346NNN is completely conserved among mDia1, -2, and -3, Glu-345 present in both mDia1 and -2 (Glu-368) is replaced by Lys (Lys-360) (Fig. 4B) in mDia3. Lys-133 of Rho is also present in Cdc42 (Lys-131), whereas Rac1 has a Glu-131 at this position. Met-134 of Rho in turn is replaced by Asn-132 in Cdc42 and by Lys-132 in Rac1 (Fig. 4C).
FIGURE 4.
Interaction of the Rho insert helix with mDiaN. A, ribbon and surface representation of the RhoC·mDiaN complex (PDB 1Z2C) with RhoC and mDiaN in yellow and cyan, respectively, in an overview (left section) and as a close-up view (right section). The Rho insert helix is colored in magenta and particular residues (Lys-133 and Met-134 of RhoC and Ile-299, Ile-344, Glu-345, and Asn-346 of mDiaN) are highlighted as sticks and interactions as dashed lines. B and C, sequence alignment of mDia isoforms and Rho proteins (accession numbers as in Fig. 2). Residues Lys-133 and Met-134 of RhoC and Ile-299, Ile-344, Glu-345, and Asn-346 of mDiaN, described in the text and depicted as sticks in A, are highlighted.
We thus studied the influence of residues involved in this interaction via stopped-flow binding kinetics (results are summarized in Table 3). Replacing the positively charged Lys-133R by a negatively charged Glu to disrupt the salt bridge with Glu-345D results in an 8-fold decreased affinity caused by an 8-fold decrease in the association rate constant ka. Surprisingly, mutation of Glu-345D to Lys shows only a very small decrease in affinity to RhoA. Mutation of Met-134NNN to Glu decreases the affinity of Rho to mDiaN by a factor of 2, whereas mutation of Met-134NNN to Ala has no effect implying that electrostatic repulsion between the negative charges of M134ER and Glu-345NNN has more impact on the affinity than the decreased hydrophobic interactions resulting from the M134AR exchange. Mutation of Lys-135R, conserved among Rho proteins but not directly involved in the interaction, to Ala and to Glu does not show an effect on binding. Interestingly, mutation of Ile-299NNN, which does not directly interact with RhoC in the crystal structure, to either Ala or Glu drastically reduces the affinity to Rho by 3000- and 7000-fold, respectively, which is mainly due to a 2250- and 3600-fold increase in the dissociation rate constant, respectively. Because binding of DAD is not affected by the Ile-299NNN mutations as confirmed by ITC measurements (data not shown), we conclude that the Ile-299NNN mutations disturb the local structural integrity of the interface to the Rho insert helix.
TABLE 3.
Stopped-flow kinetic analysis of interaction of WT and mutant Rho with mDiaN
The following abbreviations are used: kd, dissociation rate constant; ka, association rate constant; KD, kd/ka where KD is dissociation equilibrium constant.
| Interaction partners | kd/s−1 | ka/μm−1·s−1 | KD, nM |
|---|---|---|---|
| RhoA/mDiaN | 0.003 | 0.497 | 6 |
| RhoA K133E/mDiaN | 0.003 | 0.064 | 46 |
| RhoA M134A/mDiaN | 0.002 | 0.337 | 6 |
| RhoA M134E/mDiaN | 0.004 | 0.329 | 12 |
| RhoA K135A/mDiaN | 0.002 | 0.458 | 4 |
| RhoA K135E/mDiaN | 0.002 | 0.482 | 4 |
| RhoA/mDiaN 1299A | 6.76 | 0.333 | 20,300 |
| RhoA/mDiaN 1299E | 10.78 | 0.257 | 42,000 |
| RhoA/mDiaN E345K | 0.004 | 0.473 | 8 |
| RhoA/mDiaN N346A | 0.008 | 0.404 | 20 |
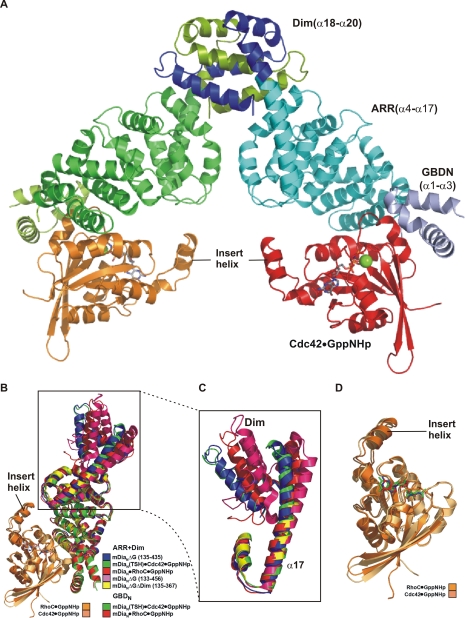
Structure of the Cdc42·Gpp(NH)p·mDiaN-TSH Complex—To analyze the structural basis of the determinants of specificity, we solved the x-ray structure of the complex between Cdc42·Gpp(NH)p and the mDiaN-TSH mutant. Due to the only micromolar affinity of the complex, Cdc42·Gpp(NH)p and mDiaN-TSH were purified separately and mixed with 1:1 stoichiometry prior to crystallization. Initial crystals of the space group P321 were optimized to a final size of ∼100 × 200 × 100 μm suitable for data collection. Molecular replacement with the RhoC·mDiaN complex (PDB 1Z2C) structure as search model revealed a solution with two Cdc42·mDiaN-TSH complexes per asymmetric unit, which eventually was refined to 2.7 Å resolution (see Table 4 for data collection and refinement statistics). The overall structure is very similar to that of the RhoC·mDiaN complex, with Cdc42 occupying the same binding site as RhoC and no appreciable differences in the GBD and the ARR subdomains (Fig. 5, A and B) reflected in the low root mean square deviation value of 1.65 Å. Superimposition of Cdc42·mDiaN-TSH-with the three available structures of mDiaN, in complex with DAD (32, 41) and with RhoC·Gpp(NH)p (35), reveals that the dimerization domain is oriented differently in each of these structures (Fig. 5B) due to different bending of the C-terminal half of the long helix (α17) connecting ARR and Dim (Fig. 5C). The orientation of Cdc42·Gpp(NH)p in the mDiaN-TSH complex is essentially the same as Rho·Gpp(NH)p in the mDiaN complex (Fig. 5B), and the structures of both G proteins are basically identical (Fig. 5D).
FIGURE 5.
Structure of the Cdc42·mDiaN-TSH complex. A, ribbon and surface representation of the Cdc42·mDiaN-TSH dimer, with color coding as indicated, Gpp(NH)p as stick model and Mg2+ ions as green spheres. B, superimposition of the Cdc42·mDiaN-TSH complex structure (mDiaN-TSH colored in green and Cdc42 in pale orange) with RhoC·mDiaN complex (PDB 1Z2C, mDiaN colored in red and RhoC colored in orange), mDiaN of mDiaN·DAD complex (PDB 2BAP, mDiaN colored in blue), mDia-(131-516) (PDB 2BNX (46), colored in magenta), and mDia1-(135-367) (PDB 2F31 (44), colored in yellow). C, close-up view of the superimposition from B, highlighting the long helix α17, helix 3 of the of the fifth ARM of ARR, and the dimerization domain (Dim). D, close up view of the superimposition from B showing Cdc42 and RhoC (color coding as in B) with the Gpp(NH)p molecules as sticks.
It should be noted that the numbering of the Armadillo repeat motifs has been changed compared with our earlier report (35). Because of sequence comparison with the consensus Armadillo repeat motifs, we now consider the first Armadillo repeat of mDiaN to be incomplete rather then the ARM5. ARM1 contains only the second and the third α-helices. α17 is thus the third α-helix of ARM5. The differences in α17 and ARM5 positions indicate a conformational flexibility of the dimerization domain relative to the rest of the molecule.
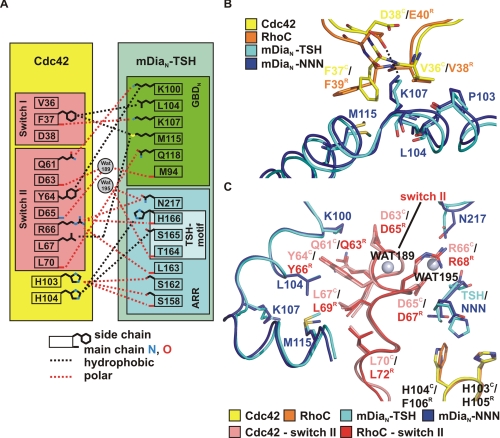
Structural Basis of Specificity—A schematic summary of the interactions found in the Cdc42·mDiaN-TSH complex structure is given in Fig. 6A. The superimposition of the two complex structures reveals that the regions involved in complex formation overall align quite well. Cdc42 binds via switch I and switch II regions in a basically analogous manner as described previously for the Rho complex (34, 35). The switch I region, as already observed for the Rho complex, exclusively interacts with the GBD of mDia (Fig. 6B). Residue Phe-37C in switch I undergoes hydrophobic interactions with the aliphatic side chains of both Met-115TSH and Leu-104TSH, although Phe-37C adapts a different rotameric conformation as compared with Phe-39R in the Rho complex. Although in the Rho complex a salt bridge is formed between Glu-40R and Lys-107NNN, no such interaction is found for the corresponding residue Asp-38C probably due to its smaller side chain impeding a closer distance to the ε-amino group of Lys-107TSH (Fig. 6B). In line with this, the electron density for the side chain of Lys-107TSH is less well defined arguing for more flexibility because of a weaker interaction. The absence of this salt bridge may thus contribute to the weaker affinity of Cdc42 to mDiaN-TSH as compared with Rho.
FIGURE 6.
Structural analysis of the Cdc42 mDiaN-TSH interaction. A, schematic drawing of interacting residues in Cdc42 and mDiaN-TSH (cutoff distance 3.6 Å). B, superimposition of the Cdc42 and the RhoC complexes in the contact area of switch I and mDiaN (color legend in figure), highlighting particular residues, including the salt bridge between Glu40R and Lys-107NNN. C, superimposition of the Cdc42 and the RhoC complexes in the contact area of switch II (color legend in figure), with water molecules displayed as gray spheres and with selected residues highlighted as sticks. For clarity, only important parts of mDiaN-TSH and Cdc42 are displayed. The switch II region of RhoC and Cdc42 is highlighted in red and salmon, respectively.
A notable feature of the switch II region is that it is invariant among Rho isoforms A-C, Cdc42, and Rac isoforms 1-3. As in the Rho complex, switch II of Cdc42 interacts with both the GBDN and the ARR subdomains. In the superimposition of the two complexes, the switch II regions almost perfectly align (Fig. 6C). The side chain of Asp-67R displays a polar contact to the side chain of Asn-164NNN in the triple NNN motif. No such interaction can be formed with Asp-65C because of the N165T mutation in mDiaN-TSH. Here the γ-methyl group of Thr-164TSH points toward the carboxyl side chain of Asp-65C. The orientation of Arg-66C is almost identical to that of Arg-68R, which we have previously proven to be one of the major contributors of the affinity of Rho to mDia binding (35).
The higher resolution of the Cdc42·mDiaN-TSH complex structure allowed us to localize protein-bound solvent. Two water molecules are coordinated in the polar interface surrounding switch II of Cdc42 and the GBDN and the ARR subdomain (Fig. 6C). Wat195 is coordinated by the main chain carbonyl of Thr-164TSH and the side chain amino group of Asn-217TSH and closely located to the guanidinium side chain of Arg-66C. A second water molecule, Wat189, is caged by hydrogen bonds with the carbonyl oxygen of Met-94TSH and Asp-63C and the main chain nitrogen atom of Asp-65C. Although the lower resolution of the Rho·mDiaN complex structure did not reveal such details, enough space in combination with an equivalent protein anatomy is present for a similar water coordination network.
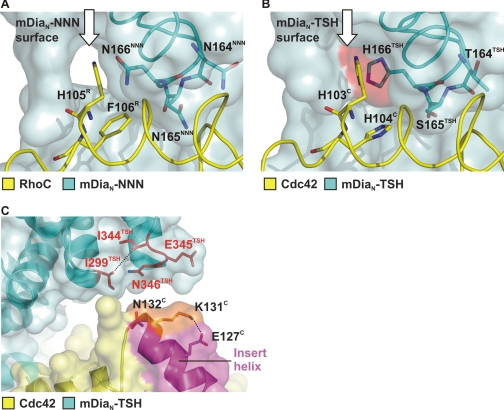
The major difference between mDia1 and mDia2/3 in the G protein binding interface is the NNN-TSH substitution. In the superimposition of the two complex structures, the NNN motif and the THS motif adapt the same backbone conformation (Fig. 6C). The γ-amido group of the Asn-165NNN side chain is involved in a polar contact to the carboxyl group of Glu-102R. Furthermore, Asn-165NNN forms a hydrophobic interaction to Phe-106R via its β-methyl group. In contrast, Ser-165TSH is only involved in the formation of a nonpolar interaction with its β-methyl group and His-104C. Both Asn-166NNN and His-166TSH interact via their main chain carbonyl with the side chain of Arg-68R/Arg-66C. The side chain of His-166TSH presents a more extended hydrophobic surface for interaction with His-104C compared with the smaller polar side chain of Asn-166NNN. A space-filling representation in this area of the interface shows a better complementation with the more bulky hydrophobic side chain of His-166TSH compared with the smaller polar side chain of Asn-166NNN (Fig. 7, A and B). In the case of mDiaN-NNN, a tunnel large enough to harbor water molecules is located close to the hydrophobic surface created by His-105R and Phe-106R (Fig. 7A), whereas this tunnel is completely occluded in the Cdc42·mDiaN-TSH complex by the bulkier side chain of H166C, which creates a continuous hydrophobic surface (Fig. 7B). We thus believe that the more extended hydrophobic surface offered to His-103C and His-104C, the better space complementation and the occlusion of a solvent-filled tunnel in the Cdc42-binding surface on mDia are responsible for the increased affinity of Cdc42 toward mDiaN-TSH.
FIGURE 7.
Structural basis of specificity. A and B, superimposition the Cdc42 and RhoC complexes with WT and mDiaN-TSH around the central portion around the NNN/TSH-motifs, with color coding as indicated. The G proteins are displayed as ribbons on a transparent surface of WT (A) or TSH-mutant (B) mDiaN, with the NNN and TSH motifs (in blue sticks) shining through. The white arrow points to a solvent-accessible tunnel present in the RhoC·mDiaN-NNN structure (A), which is closed by the larger side chain of His-166TSH in the Cdc42·mDiaN-TSH structure (B). The additional van der Waals surface provided by the larger His-166TSH side chain is highlighted in red. (B). C, ribbon and surface representation of the Cdc42·mDiaN-TSH complex around the Cdc42 insert helix, with color coding as indicated and special residues highlighted in stick representation. The Cdc42 insert helix is highlighted in magenta, and residues Lys-131C and Asn-132C are colored in orange (compare Fig. 4).
Although the Rho·mDiaN complex shows an additional contact between the C-terminal end of the Rho insert helix and the ARR (Fig. 4A), no such interaction is present in the Cdc42·mDiaN-TSH complex structure, and the distance between the corresponding elements is longer (Fig. 5, B and D, and Fig. 7C). Instead, the ε-amino group of Lys-131C forms an intramolecular salt bridge with Glu-127C thereby neutralizing the positive charge of Lys-131C. In contrast, the residue corresponding to Glu-127C is Arg-129R, which might even amplify the positive charge on Lys-133R thus strengthening the ionic interaction with Glu-345NNN (compare Fig. 4C). Notably, in Rac1 Arg-129R is replaced by Glu-127Rac, and Lys-133R is replaced by the negatively charged Glu-131Rac and could thus be responsible for the differences in binding affinity toward mDia isoforms.
DISCUSSION
Here we have extended our analysis of the interaction of the N-terminal part of mDia with their cognate G proteins and whether this affects the auto-inhibitory regulation of these formins. We have furthermore dissected the molecular details of the interface between Rho proteins and mDia to derive explanations for the specificity of their interactions.
First, we show that the C-terminal polybasic region of the C-terminal auto-inhibitory region does not overlap with the G protein-binding site. In previous structures of the mDiaN·DAD complexes (32, 41), only 16 residues comprising the DCR forming an amphipathic helix were visible. Mutational analysis shows that the effect of mutating the negative charges C-terminal to the DCR has an additive effect on the affinity of a DAD peptide for the N terminus. Although the release of autoinhibition by the binding of Rho is mediated by the mutual exclusive binding of the core DAD region and G proteins, as analyzed previously by biochemistry and structure (32, 40, 41), our studies here further confirm and extend previous assumptions that the binding sites of G proteins and DAD relevant for affinity are only partially overlapping. The mutations in the polybasic DAD-binding region on mDiaN did not influence Rho binding (RhoA-mDiaN E358R/E362R, 9 nm; RhoA-mDiaN E358R/E362R/E366R, 16 nm; data not shown). This is strengthened by our biochemical data that show that any of the mutations that influence specificity and affinity of Rho binding have almost no effect on DAD binding (Table 1) (32, 35). It should be added that the DAD regions of mDia1, -2, and -3 are sufficiently conserved, such that they bind with almost equal affinity to the N-terminal part of mDia1, such that inhibition in trans between mDia isoforms cannot be excluded (Table 1).
We also show that Cdc42 binds in exactly the same position as Rho and that the specificity of interaction between Rho, Rac, Cdc42 versus mDia1, -2, and -3 isoforms is mediated mostly by two surface epitopes in the complex interface. Surprisingly, the specificity is not dictated by sequence differences in the switch regions even though both of them are involved in the interaction but is in line with the high sequence conservation of the switches. A minor contribution to specificity comes from the switch I residue Glu-40 in Rho, which in Rac and Cdc42 is Asp-38. Switch II is completely conserved between Rho, Rac, and Cdc42 and is involved in a large number of interactions.
The first specificity epitope occurs in the central portion of the interface where the mDia1 NNN motif is replaced by the TSH motif of mDia2 and -3. The crucial Arg-68R residue in switch II, which inserts like a wedge into the interface formed between GBD and ARR subdomains, is one of the most important mediators of the interaction and is conserved as Arg-66 in Rac/Cdc42. The substitution relevant for affinity and specificity is the replacement of Phe-106 by His-104 in Cdc42/Rac. The structural analysis shows that the structures of the NNN and TSH motifs, which form a loop connecting the third helix of ARM1 and the first helix of the ARM2 of the mDiaN ARR, superimpose very well. The major effect of the replacements seems to be a more complementary interface that is formed when His-104C points toward the TSH rather than the NNN module. In the latter case a solvent-accessible tunnel remains in the interface and would thus trap water molecules in the Cdc42-mDia1 interface which would be negatively affecting the affinity. Our data also show that the Phe-106 residue in this part of the interface is more important for high affinity than the NNN motif, because its introduction into Rac increases affinity drastically, whereas the effect is less pronounced in Cdc42.
The second epitope involved in specificity is formed by the Rho insert helix that contacts the ARR subdomain at the loop connecting α16 and α17 of ARM5 of mDiaN. The insert helix is a particular feature unique to the Rho subfamily of the Ras superfamily. Although the Rho insert helix has been implicated previously to be involved in a number of biological effects, i.e. the interaction of Rac with the NADPH oxidase (73) or for Rho kinase activation by RhoA (74) and to be essential for membrane ruffling (75), no structural analysis has yet shown such an interaction. The evidence for the involvement of the insert helix has mostly come from deletion experiments that may or may not produce a proper folding protein.4 The analysis here shows for the first time that the insert helix makes a substantial contribution to the affinity but may be even more important for the specificity. Although the interface between Rho and mDia1 is very tight and complementary, the interface between Cdc42 and the mDia-TSH mutant leaves much space between the corresponding epitopes. Although the difference in complementarity cannot directly be assigned to particular residues, the sequence differences in the area are sufficiently different between the three mDia isoforms to assume a significantly different surface. Our mutational analysis shows that residues apparently not directly involved in this interface have nevertheless a large effect on binding affinity. We can thus speculate that the replacement of the contact area by the respective sequences from mDia2 and -3 would, in conjunction with the TSH motif, create a high affinity protein for Rac and/or Cdc42.
Previously, we have shown that Rho actively displaces DAD from binding to the N-terminal region of mDia (34). Due to the partial overlap of Rho·GTP and DAD, we have postulated a two-step binding mechanism in which the G protein makes a preliminary contact for a loosely bound complex in a first step. In a second step, steric interference and charge-charge repulsion lead to dissociation of DAD and formation of a tight complex. In such a scenario, the Rho insert helix might thus make a first weak interaction which, however, already differentiates between the Rho isoforms RhoA, -B, and -C and Cdc42/Rac. The second step involves the central portion of the interface, where the interaction with the NNN or TSH motif leads to a tight binding complex where DAD is released and the FH2 domain is ready to polymerize actin. Whether or not the FH2 domains, once released from auto-inhibited mDia, have the same activity toward actin polymerization remains to be determined.
Acknowledgments
We are grateful to the machine and beamline groups whose outstanding efforts have made these experiments possible. We thank Andrea Scrima and Wulf Blankenfeldt for data collection.
The atomic coordinates and structure factors (code 3EG5) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: FH, formin homology; Bistris propane, 1, 3-bis[tris(hydroxymethyl)methylamino]propane; GBD, GTPase-binding domain; Dia, Diaphanous; mant, N-methylanthraniloyl; Gpp(NH)p, guanosine 5′-(β,γ-imido)triphosphate; ARR, Armadillo repeat region; DID, Diaphanous-inhibitory domain; DAD, Diaphanous-autoregulatory domain; ARM, Armadillo repeat motif; DCR, DAD core region; GNBP, guanine nucleotide-binding protein; PDB, Protein Data Bank; ITC, isothermal titration calorimetry; WT, wild type; ROCK, Rho-associated coiled-coil kinase.
M. Lammers, unpublished data.
R. Ahmadian, unpublished observations.
References
- 1.Castrillon, D. H., and Wasserman, S. A. (1994) Development (Camb.) 120 3367-3377 [DOI] [PubMed] [Google Scholar]
- 2.Pantaloni, D., Le Clainche, C., and Carlier, M. F. (2001) Science 292 1502-1506 [DOI] [PubMed] [Google Scholar]
- 3.Pollard, T. D., and Borisy, G. G. (2003) Cell 112 453-465 [DOI] [PubMed] [Google Scholar]
- 4.Bompard, G., and Caron, E. (2004) J. Cell Biol. 166 957-962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buck, M., Xu, W., and Rosen, M. K. (2004) J. Mol. Biol. 338 271-285 [DOI] [PubMed] [Google Scholar]
- 6.Eden, S. R., Rohatgi, A. V., Podtelejnikov Mann, M., and Kirschner, M. W. (2002) Nature 418 790-793 [DOI] [PubMed] [Google Scholar]
- 7.Pruyne, D., Evangelista, M., Yang, C., Bi, E., Zigmond, S. H., Bretscher, A., and Boone, C. (2002) Science 297 612-615 [DOI] [PubMed] [Google Scholar]
- 8.Quinlan, M. E., Heuser, J. E., Kerkhoff, E., and Mullins, R. D. (2005) Nature 433 382-388 [DOI] [PubMed] [Google Scholar]
- 9.Evangelista, M., Zigmond, S., and Boone, C. (2003) J. Cell Sci. 116 2603-2611 [DOI] [PubMed] [Google Scholar]
- 10.Higgs, H. N. (2005) Trends Biochem. Sci. 30 342-353 [DOI] [PubMed] [Google Scholar]
- 11.Romero, S., Le Clainche, C., Didry, D., Egile, C., Pantaloni, D., and Carlier, M. F. (2004) Cell 119 419-429 [DOI] [PubMed] [Google Scholar]
- 12.Wallar, B. J., and Alberts, A. S. (2003) Trends Cell Biol. 13 435-446 [DOI] [PubMed] [Google Scholar]
- 13.Zigmond, S. H., Evangelista, M., Boone, C., Yang, C., Dar, A. C., Sicheri, F., Forkey, J., and Pring, M. (2003) Curr. Biol. 13 1820-1823 [DOI] [PubMed] [Google Scholar]
- 14.Kato, T., Watanabe, N., Morishima, Y., Fujita, A., Ishizaki, T., and Narumiya, S. (2001) J. Cell Sci. 114 775-784 [DOI] [PubMed] [Google Scholar]
- 15.Ozaki-Kuroda, K., Yamamoto, Y., Nohara, H., Kinoshita, M., Fujiwara, T., Irie, K., and Takai, Y. (2001) Mol. Cell. Biol. 21 827-839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zigmond, S. H. (2004) Curr. Opin. Cell Biol. 16 99-105 [DOI] [PubMed] [Google Scholar]
- 17.Harris, E. S., Li, F., and Higgs, H. N. (2004) J. Biol. Chem. 279 20076-20087 [DOI] [PubMed] [Google Scholar]
- 18.Higgs, H. N., and Peterson, K. J. (2005) Mol. Biol. Cell 16 1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pring, M., Evangelista, M., Boone, C., Yang, C., and Zigmond, S. H. (2003) Biochemistry 42 486-496 [DOI] [PubMed] [Google Scholar]
- 20.Shimada, A., Nyitrai, M., Vetter, I. R., Kuhlmann, D., Bugyi, B., Narumiya, S., Geeves, M. A., and Wittinghofer, A. (2004) Mol. Cell 13 511-522 [DOI] [PubMed] [Google Scholar]
- 21.Xu, Y., Moseley, J. B., Sagot, I., Poy, F., Pellman, D., Goode, B. L., and Eck, M. J. (2004) Cell 116 711-723 [DOI] [PubMed] [Google Scholar]
- 22.Higashida, C., Miyoshi, T., Fujita, A., Oceguera-Yanez, F., Monypenny, J., Andou, Y., Narumiya, S., and Watanabe, N. (2004) Science 303 2007-2010 [DOI] [PubMed] [Google Scholar]
- 23.Kovar, D. R., and Pollard, T. D. (2004) Nat. Cell Biol. 6 1158-1159 [DOI] [PubMed] [Google Scholar]
- 24.Kovar, D. R., Wu, J. Q., and Pollard, T. D. (2005) Mol. Biol. Cell 16 2313-2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, F., and Higgs, H. N. (2003) Curr. Biol. 13 1335-1340 [DOI] [PubMed] [Google Scholar]
- 26.Shemesh, T., Otomo, T., Rosen, M. K., Bershadsky, A., and Kozlov, M. M. (2005) J. Cell Biol. 170 889-893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wear, M. A., Yamashita, A., Kim, K., Maéda, Y., and Cooper, J. A. (2003) Curr. Biol. 13 1531-1537 [DOI] [PubMed] [Google Scholar]
- 28.Otomo, T., Tomchick, D. R., Otomo, C., Panchal, S. C., Machius, M., and Rosen, M. K. (2005) Nature 433 488-494 [DOI] [PubMed] [Google Scholar]
- 29.Bedford, M. T., Chan, D. C., and Leder, P. (1997) EMBO J. 16 2376-2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macias, M. J., Wiesner, S., and Sudol, M. (2002) FEBS Lett. 513 30-37 [DOI] [PubMed] [Google Scholar]
- 31.Alberts, A. S. (2001) J. Biol. Chem. 276 2824-2830 [DOI] [PubMed] [Google Scholar]
- 32.Lammers, M., Rose, R., Scrima, A., and Wittinghofer, A. (2005) EMBO J. 24 4176-4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otomo, T., Otomo, C., Tomchick, D. R., Machius, M., and Rosen, M. K. (2005) Mol. Cell. 18 273-281 [DOI] [PubMed] [Google Scholar]
- 34.Rose, R., Wittinghofer, A., and Weyand, M. (2005) Acta Crystallogr. F Struct. Biol. Crystalliz. Comm. 61 225-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose, R., Weyand, M., Lammers, M., Ishizaki, T., Ahmadian, M. R., and Wittinghofer, A. (2005) Nature 435 513-518 [DOI] [PubMed] [Google Scholar]
- 36.Watanabe, N., Madaule, P., Reid, T., Ishizaki, T., Watanabe, G., Kakizuka, A., Saito, Y., Nakao, K., Jockusch, B. M., and Narumiya, S. (1997) EMBO J. 16 3044-3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Copeland, J. W., and Treisman, R. (2002) Mol. Biol. Cell 13 4088-4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishizaki, T., Morishima, Y., Okamoto, M., Furuyashiki, T., Kato, T., and Narumiya, S., (2001) Nat. Cell Biol. 3 8-14 [DOI] [PubMed] [Google Scholar]
- 39.Watanabe, N., Kato, T., Fujita, A., Ishizaki, T., and Narumiya, S. (1999) Nat. Cell Biol. 1 136-143 [DOI] [PubMed] [Google Scholar]
- 40.Li, F., and Higgs, H. N. (2005) J. Biol. Chem. 280 6986-6992 [DOI] [PubMed] [Google Scholar]
- 41.Nezami, A., Poy, F., and Eck, M. (2006) Structure (Lond.) 14 257-263 [DOI] [PubMed] [Google Scholar]
- 42.Ihara, K., Muraguchi, S., Kato, M., Shimizu, T., Shirakawa, M., Kuroda, S., Kaibuchi, K., and Hakoshima, T. (1998) J. Biol. Chem. 273 9656-9966 [DOI] [PubMed] [Google Scholar]
- 43.Pai, E. F., Kabsch, W., Krengel, U., Holmes, K. C., John, J., and Wittinghofer, A. (1989) Nature 341 209-214 [DOI] [PubMed] [Google Scholar]
- 44.Vetter, I. R., and Wittinghofer, A. (2001) Science 294 1299-1304 [DOI] [PubMed] [Google Scholar]
- 45.Wittinghofer, A., and Pai, E. F. (1991) Trends Biochem. Sci. 16 382-387 [DOI] [PubMed] [Google Scholar]
- 46.Arai, R., and Mabuchi, I. (2002) J. Cell Sci. 115 887-898 [DOI] [PubMed] [Google Scholar]
- 47.Bishop, A. L., and Hall, A. (2000) Biochem. J. 348 241-255 [PMC free article] [PubMed] [Google Scholar]
- 48.Etienne-Manneville, S., and Hall, A. (2002) Nature 420 629-635 [DOI] [PubMed] [Google Scholar]
- 49.Ho, H. Y., Rohatgi, R., Lebensohn, A. M., Ma, L., Li, J., Gygi, S. P., and Kirschner, M. W. (2004) Cell 118 203-216 [DOI] [PubMed] [Google Scholar]
- 50.Raftopoulou, M., and Hall, A. (2004) Dev. Biol. 265 23-32 [DOI] [PubMed] [Google Scholar]
- 51.Ridley, A. J., and Hall, A. (1992) Cell 70 389-399 [DOI] [PubMed] [Google Scholar]
- 52.Takai, Y., Sasaki, T., Tanaka, K., and Nakanishi, H. (1995) Trends Biochem. Sci. 20 227-231 [DOI] [PubMed] [Google Scholar]
- 53.Wennerberg, K., and Der, C. J. (2004) J. Cell Sci. 117 1301-1312 [DOI] [PubMed] [Google Scholar]
- 54.Burbelo, P. D., Drechsel, D., and Hall, A. (1995) J. Biol. Chem. 270 29071-29074 [DOI] [PubMed] [Google Scholar]
- 55.Jacobs, M., Hayakawa, K., Swenson, L., Bellon, S., Fleming, M., Taslimi, P., and Doran, J. (2006) J. Biol. Chem. 281 260-268 [DOI] [PubMed] [Google Scholar]
- 56.Maesaki, R., Ihara, K., Shimizu, T., Kuroda, S., Kaibuchi, K., and Hakoshima, T. (1999) Mol. Cell 4 793-803 [DOI] [PubMed] [Google Scholar]
- 57.Leung, D. W., and Rosen, M. K. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 5685-5690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Aelst, L., and D'Souza-Schorey, C. (1997) Gen. Dev. 11 2295-2322 [DOI] [PubMed] [Google Scholar]
- 59.Dvorsky, R., and Ahmadian, M. R. (2004) EMBO Rep. 12 1130-1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsui, T., Amano, M., Yamamoto, T., Chihara, K., Nakafuku, M., Ito, M., Nakano, T., Okawa, K., Iwamatsu, A., and Kaibuchi, K. (1996) EMBO J. 15 2208-2216 [PMC free article] [PubMed] [Google Scholar]
- 61.Leung, T., Chen, X. Q., Manser, E., and Lim, L. (1996) Mol. Cell. Biol. 16 5313-5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gasman, S., Kalaidzidis, Y., and Zerial, M. (2003) Nat. Cell Biol. 5 195-204 [DOI] [PubMed] [Google Scholar]
- 63.Yasuda, S., Oceguera-Yanez, F., Kato, T., Okamoto, M., Yonemura, S., Terada, Y., Ishizaki, T., and Narumiya, S. (2004) Nature 428 767-771 [DOI] [PubMed] [Google Scholar]
- 64.Wiseman, T., Williston, S., Brandts, J. F., and Lin, L. N. (1989) Anal. Biochem. 179 131-137 [DOI] [PubMed] [Google Scholar]
- 65.Kabsch, W. (1993) J. Appl. Crystallogr. 26 795-800 [Google Scholar]
- 66.Vagin, A., and Teplyakov, A. (1997) J. Appl. Crystallogr. 20 1022-1025 [Google Scholar]
- 67.Brünger, A. T., Krukowski, A., and Erickson, J. W. (1990) Acta Crystallogr. 46 585-593 [DOI] [PubMed] [Google Scholar]
- 68.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 69.Murshudov, G. N., Lebedev, A., Vagin, A. A., Wilson, K. S., and Dodson, E. J. (1999) Acta Crystallogr. 55 247-255 [DOI] [PubMed] [Google Scholar]
- 70.Lovell, S. C., Davis, I. W., Arendall, W. B., III, de Bakker, P. I., Word, J. M., Prisant, M. G., Richardson, J. S., and Richardson, D. C. (2003) Proteins Struct. Funct. Genet. 50 437-450 [DOI] [PubMed] [Google Scholar]
- 71.DeLano, W. L. (2002) The PyMOL User's Manual, DeLano Scientific, San Carlos, CA
- 72.Frisch, C., Schreiber, G., Johnson, C. M., and Fersht, A. R. (1997) Mol. Biol. 267 696-706 [DOI] [PubMed] [Google Scholar]
- 73.Freeman, J. L., and Lambeth, J. D. (1996) J. Biol. Chem. 271 22578-22582 [DOI] [PubMed] [Google Scholar]
- 74.Zong, H., Kaibuchi, K., and Quilliam, L. A. (2001) Mol. Cell. Biol. 21 5287-5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karnoub, A. E., Der, C. J., and Campbell, S. L. (2001) Mol. Cell. Biol. 21 2847-2857 [DOI] [PMC free article] [PubMed] [Google Scholar]