Abstract
Duchenne muscular dystrophy is caused by a genetic defect in the dystrophin gene. The absence of dystrophin results in muscle fiber necrosis and regeneration, leading to progressive muscle fiber loss. Utrophin is a close analogue of dystrophin. A substantial, ectopic expression of utrophin in the extrasynaptic sarcolemma of dystrophin-deficient muscle fibers can prevent deleterious effects of dystrophin deficiency. An alternative approach for the extrasynaptic up-regulation of utrophin involves the augmentation of utrophin transcription via the endogenous utrophin A promoter using custom-designed transcriptional activator proteins with zinc finger (ZFP) motifs. We tested a panel of custom-designed ZFP for their ability to activate the utrophin A promoter. Expression of one such ZFP efficiently increased, in a time-dependent manner, utrophin transcript and protein levels both in vitro and in vivo. In dystrophic mouse (mdx) muscles, administration of adenoviral vectors expressing this ZFP led to significant enhancement of muscle function with decreased necrosis, restoration of the dystrophin-associated proteins, and improved resistance to eccentric contractions. These studies provide evidence that specifically designed ZFPs can act as strong transcriptional activators of the utrophin A promoter. These may thus serve as attractive therapeutic agents for dystrophin deficiency states such as Duchenne muscular dystrophy.
Utrophin is a large cytoskeletal protein that is closely related to dystrophin. Although utrophin is present throughout the sarcolemma in muscle fibers during development (1), in normal adult muscle, utrophin is only present at the neuromuscular and myotendinous junctions (2–5). In dystrophin deficiency, there is a spontaneous overexpression of extrasynaptic utrophin (2, 6, 7), but it is not sufficient to compensate for dystrophin deficiency (8). The therapeutic effect of up-regulating extrasynaptic utrophin has been tested experimentally through transgenesis (9) or gene transfer (10, 11). These studies have revealed that, if utrophin expression is augmented sufficiently, the dystrophic phenotype can be negated. In this regard, compared with dystrophin replacement, up-regulation of utrophin may have an advantage because utrophin is a self protein and not recognized as a neoantigen as is dystrophin.
Utrophin is expressed from two distinct promoters, utrophin A and utrophin B (12, 13). The two promoters are regulated independently. The utrophin A promoter lies within a CpG island (12), whereas the utrophin B promoter is found within a large intron 50 kb downstream of exon 2 and generates a transcript with a unique exon 1 that splices into exon 3 of the utrophin gene (13). Synapse-specific expression of utrophin A is mediated, in part, via the binding of the ets-factors GABP alpha and beta to the N-box (14–16), which is a characteristic of the promoter of other neuromuscular junction proteins such as the acetylcholine receptor ε subunit (17). In addition, the GC-elements near the N-box are bound by Sp1 and Sp3 factors, which interact with GABP alpha and synergize in the transcriptional activation effected by GABP (16, 18). The nerve-derived trophic factor heregulin (14, 19) and PGC-1 alpha (20, 21) cooperate to increase the activity of GABP to regulate gene transcription of utrophin A at the neuromuscular synapse. In contrast, the N-box is absent in the utrophin B promoter (22, 23), which is expressed ubiquitously in non-muscle tissues.
In this study, we have evaluated the ability of artificial transcriptional factors (24) to activate the endogenous utrophin A promoter in extrasynaptic myonuclei. These custom-designed factors are comprised of proteins with zinc finger (ZF)3 domains arranged in tandem to recognize and bind to specific DNA sequences (25). ZF proteins (ZFP) target G-rich sequences. The utrophin A promoter is a TATA-less promoter with a GC-rich 5′-upstream area, located within a CpG island (12). Thus, we have chosen to target the utrophin A promoter with ZFPs to activate it in a constitutive manner (i.e. without need for the sustained presence of neural factors). We have evaluated the ZFPs in vitro and selected the best one for in vivo testing in the mdx mouse.
EXPERIMENTAL PROCEDURES
Design and Synthesis ZFP Gene—DNase I-accessible chromatin regions of the utrophin promoter A were mapped in nuclei isolated from mouse primary hepatocytes (26). DNase I digestion followed by Southern blot analysis was performed as described previously (27). The amino acid code of the three-finger proteins designed to bind utrophin-A-specific sequences was selected as described (28, 29). The Zif268 wild-type sequence was used as a scaffold to assemble the designed fingers (30). A PCR-based assembly procedure was used to create the synthetic gene encoding the α-helix and β-sheet regions of each three-finger protein as described (27). Briefly, six overlapping oligonucleotides, with an overlap region of 15 bp were used in the presence of Pfu thermostable DNA polymerase (Promega, Madison, WI), in a four-cycle PCR reaction at an annealing temperature of 45 °C. Each fragment was directly cloned into pUC19 for sequencing.
Purification of ZFP Fusion Protein and Electrophoretic Mobility Shift Assay—Each resulting ZFP gene was cloned into the pMal-c2 vector (New England Biolabs) as a fusion with DNA sequences encoding maltose-binding protein. Maltose-binding protein-ZFP fusions were then expressed and affinity-purified using an amylose resin (New England Biolabs). Binding experiments were performed as described before (30). In competitive experiments, unlabeled oligonucleotides were added at 50- or 100-fold molar excess of the amount of labeled DNA. The oligonucleotides used as probes were 20 bp in length and contained 9-bp core sequences for ZFP to target. Mutant probes were designed to substitute 9-bp core sequences. Signal detection was preformed by using a PhosphorImager (Amersham Biosciences) using the ImageQuaNT program.
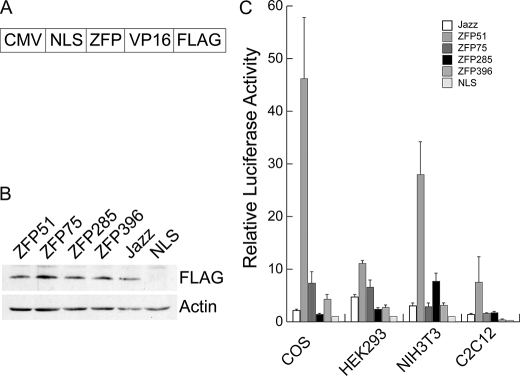
Construct of Expression Vector and Preparation of Recombinant Adenoviruses—The engineered ZFP gene fragment was subcloned into p190E8, which contained a 7-amino acid nuclear localization sequence (Pro-Lys-Lys-Lys-Arg-Lys-Val) from simian virus 40 large T-antigen, a VP16 activator domain from human herpes simplex virus amino acids 413–490, and a FLAG peptide (Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys). A HindIII-EcoRI fragment, which contained all elements, was then cloned into pcDNA3 mammalian expression vector (Invitrogen) (Fig. 2). Control vector pNLS modified from pcDNA3 only contains nuclear localization sequence and FLAG peptide.
FIGURE 2.
Analysis of activation of mouse utrophin A promoter in vitro. A, a schematic illustration of ZFP/VP16 transcription factor construct. The expression cassette was driven by the CMV promoter and contained a nuclear localization signal (NLS) and was tagged with the FLAG epitope. B, expression levels of various ZFPs after transfection. Western blotting with an anti-FLAG antibody was used to monitor the levels of the FLAG-tagged ZFP after transfection into the different cell lines. The examples shown are the results obtained in COS-1 cells. Sample loading was verified by an anti-β-actin antibody. C, ability of various ZFP transcription factors to activate a luciferase reporter linked to 2.98 kb of utrophin A upstream sequences. Results are expressed relative to the control NLS plasmid and are normalized to transfection efficiency as described under “Experimental Procedures.” Error bar: mean ± S.E., n = 3.
To generate the adenoviral vector, ZFP fragment containing the elements NLS, VP16, and FLAG was obtained by PCR and cloned into vector pIRES2-EGFP (Clontech) XhoI-EcoRI site. The 1.7-kb CB promoter (CMV enhancer and chicken β-actin promoter) was released with SalI and XhoI from plasmid pCAGGS and cloned into pIRES2-EGFP NheI-XhoI site; a poly(A) signal fragment was obtained by PCR from pCAGGS and cloned into NotI site of pIRES2-EGFP. A 4.2-kb expression cassette containing all elements was cloned into the adenovirus vector pC4SHU (Microbix Biosystems Inc., Ontario, Canada) NotI site to construct vector pH6. For preparation of recombinant Ad-ZFP, plasmid pH6 was transfected into Cre-293 cells by Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations, and infection rate was confirmed by observing the GFP signal. Adenovirus CMV-GFP was produced as a control. Adenoviruses were amplified using the 293Cre-loxP system and purified by two consecutive continuous CsCl gradient centrifugations as described previously (31). The titer (virus particles/ml) was determined by measuring the optical density at 260 nm. Infectious titers were determined by counting GFP green cells after infection of 293A cells.
Luciferase Reporter Gene Assays—For reporter gene analysis, a 2.98-kb HindIII-KpnI fragment of the utrophin promoter A was subcloned from a BAC clone, which contains the mouse utrophin genomic DNA 5′-region (Clone ID: RP23–111d4, BACPAC Resources Center) and inserted into pGL3 basic vector (Promega) to generate the luciferase reporter plasmid pUA-luc. All cell lines (COS-1, HEK293, NIH3T3, and C2C12) were obtained from American Type Culture Collection. The pUA-luc reporter construct, ZFP plasmid, and pSV-β-galactosidase control plasmid (Promega) were co-transfected into the cells by Lipofectamine 2000 (Invitrogen) or SuperFect reagent (Qiagen) according to the manufacturer's recommendations. Transfection efficiency was assessed in each independent experiment by the transfection of a pCMV-LacZ expression plasmid (Clontech) as control. Cells were collected 48 h after transfection, and luciferase and β-galactosidase activity assays were performed according to the manufacturer's protocol (Promega). Luciferase activity was normalized to β-galactosidase activity.
Activation of Endogenous Utrophin A Promoter in Vitro—Mouse myoblast C2C12 cells were plated at a density of 1 × 106 cells/10-cm plate. One day later, plasmids encoding ZFP were transfected into the cells using SuperFect reagent (Qiagen) according to the manufacturer's recommendations. After selection for 2 weeks in the presence of G418 (800 μg/ml) (Invitrogen), the pools of cells were harvested for assaying protein expression. For Ad-ZFP infections, myoblasts were plated at 90% confluency, Ad-ZFP was added at a multiplicity of infection of 500, and cells were harvested after 24 h. To produce myotubes, medium containing 10% fetal bovine serum was substituted by adding 2% horse serum. After 4 days, myoblast cultures were completely differentiated into myotubes, Ad-ZFP was added into the medium, and cells were cultured further for 4 days prior to harvest for Western blot analysis.
Activation of Endogenous Utrophin A Promoter in Vivo—All aspects of the study were approved by the institutional animal ethics committee according to McGill University guidelines for animal care. Dystrophin-deficient mdx neonatal mice (4-day-old, Jackson Laboratory) were directly injected with 5 μl of Ad-ZFP at a titer of 5 × 1011 virus particles/ml into the right tibialis anterior muscle, whereas the contralateral side was injected with Ad-GFP to serve as a within-animal control. The mice were euthanized at either 20 or 80 days following which the TA muscles were removed for further analysis.
Analysis of mRNA and Protein Levels—For utrophin mRNA analysis, total RNA was extracted from the muscle using RNeasy Mini Kit (Qiagen) according to the manufacturer's recommendations. Quantitative reverse transcriptase (RT)-Real-Time PCR was performed on a SmartCycleR (Cepheid Inc.) using QuantiTect Probe RT-PCR Kit (Qiagen). The primer and probe set (AAACAGGGAGGCACATTGTC, GCCCAGGTCATTGTAGAGGA, and FAM-ACGGACAACCCGAAAAGCATGGACGA-TAMRA) were used to measure utrophin expression. The glyceraldehyde-3-phosphate dehydrogenase primer and probe set (GTGTTCCTACCCCAATGTG, AGGAGACAATGGTCCTCA, and FAM-AGGCATCTGAGGGCCCACTGAAGGGCAT-TAMRA) were used to monitor the internal control glyceraldehyde-3-phosphate dehydrogenase mRNA. For each RNA sample, triplicates were amplified in a one-step reaction, and cycle threshold values were obtained. Utrophin mRNA expression levels were normalized by glyceraldehyde-3-phosphate dehydrogenase. For Western blot analysis, cells were lysed in sample buffer (50 mm Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 100 mm dithiothreitol). Muscle sections were lysed in buffer (50 mm Tris-HCl, pH 6.8, 3.8% SDS, 20% glycerol, 5% β-mercaptoethanol) by boiling at 95 °C for 5 min. Total protein extracts (25 μg) were resolved by SDS-PAGE on 5.5 or 12% polyacrylamide gels (Bio-Rad) and transferred onto nitrocellulose membranes. After blocking, membranes were probed with the primary antibodies anti-utrophin monoclonal antibody (Vector Laboratories) at a dilution of 1:300, anti-FLAG M2 monoclonal antibody (Sigma) at dilution 1:100, VP16 monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at dilution 1:100, and EGFP polyclonal antibody (Abcam) at a dilution of 1:2500. Immunocomplexes were detected with horseradish peroxidase-conjugated species-specific secondary antibodies (Bio-Rad) followed by enhanced chemiluminescence. The blots were also stained with a monoclonal antibody against β-actin or vinculin (Sigma) as a loading control.
Immunocytochemical Analysis—For in vitro analysis, C2C12 cells transfected with plasmid encoding ZFP and cultured in 8-well chamber slides were washed with PBS and then fixed with 4% formaldehyde for 10 min at room temperature. After washing with PBS, the cells were incubated for 30 min in PBS-4% bovine serum albumin with 0.5% Triton X-100, then probed with anti-FLAG-M2 monoclonal antibody-Cy3 conjugate at a dilution of 1:500 in PBS-4% bovine serum albumin or VP16 monoclonal antibody at a dilution of 1:100 overnight at 4 °C. The cells were rinsed in PBS overnight mounted in Fluoromount-G (Southern Biotech) and visualized by fluorescent microscopy. For utrophin or EGFP immunostaining and histopathology analysis, TA muscles of mice of 20 or 80 days of age were embedded in mounting medium and snap-frozen in isopentane precooled with liquid N2. Transverse sections (6 μm thick) were obtained in a cryostat and then fixed on slides in 1% acetone. Immunostaining procedures were carried out to detect utrophin expression using a primary monoclonal antibody diluted 1:300 (Novocastra Laboratories) followed by reaction with a Cy3-conjugated affinity-purified goat anti-mouse IgG. Some sections were immunostained with both utrophin and EGFP antibody. Additional sections were also stained using antibody against β-dystroglycan (NCL-43DAG, Novocastra), and α-sarcoglycan (NCL-50DAG, Novocastra), as described previously (9, 10, 32). Muscle sections were also counterstained with hematoxylin-eosin to allow determination of the number of central nuclei. The number of utrophin-positive myofibers on the entire muscle cross-section was counted as previously described (10, 32) and expressed as the percentage of the entire muscle fiber number.
Analysis of Muscle Physiological Function—These studies were performed as described previously in detail (33). At 80 days postinjection, mice were anesthetized (130 mg/kg intramuscular ketamine and 20 mg/kg intramuscular xylazine) to achieve a loss of deep pain reflexes. Ad-ZFP and control Ad-GFP treated TA muscles were subjected to eccentric contractions. Each contraction involved supramaximal stimulation at 100 Hz for a total of 300 ms; the muscle was held at Lo during the initial 100 ms (isometric component) and then lengthened through a distance of 15% of Lo during the last 200 ms (eccentric component). Peak muscle length was maintained for an additional 100 ms after the cessation of electrical stimulation, followed by a return to Lo during the next 100 ms. A total of five such contractions were imposed on the muscle strip, each being separated by a 30-s recovery period. The resulting force deficit was defined as the percent decline in isometric force from the first to the last eccentric contraction. Because the damage and isometric force deficit associated with eccentric contractions are directly correlated with the peak mechanical stress placed on the muscle, the force deficit was normalized to peak muscle stress.
Statistical Analysis—Differences between Ad-ZFP-injected and contralateral control Ad-GFP-injected muscles were determined using two-tailed t test. All values are means ± S.E. Statistical significance was defined as p < 0.05.
RESULTS
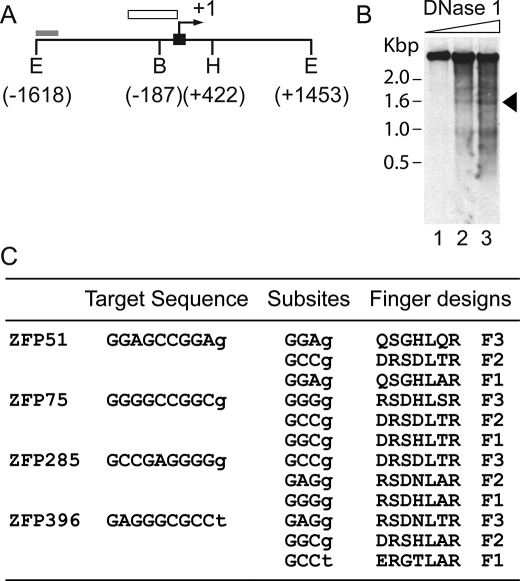
Design and Characterization of ZFPs Targeting the Proximal Utrophin A Promoter—It has been estimated that only 1% of genomic DNA is available for ZFP binding (34), because the vast majority of the binding sites will not be accessible due to nucleosome structure and chromatin condensation. We performed DNase I mapping of the proximal 5′-sequences of the utrophin A promoter, which revealed several DNase I-accessible sites, including one region within 400 bp of the transcription start site (Fig. 1, A and B), suggesting that these regions might provide good targets for ZFPs. We selected 9-bp target sequences within the DNase I-hypersensitive region to design three-finger artificial ZFP transcription factors. Four GC-rich sequences were chosen, and the appropriate ZFPs were designed based on the recognition modules described by Liu et al. (28) (ZFP51, ZFP75, ZFP285, and ZFP396) (Fig. 1C). The modules were linked by the five residue linkers (TGEKP) that are conserved in many naturally occurring multifinger domain proteins.
FIGURE 1.
Design and synthesis of ZFP genes. A, schematic depiction of the 5′-upstream region of mouse utrophin A promoter indicating the location of the probe used for Southern blot analysis in B (gray box), exon A1 (black box), transcription start site (arrow), and general DNase I sensitivity region (open box). The numbering is relative to the start of transcription. (E = EcoR I; B = BamHI; H = HindIII). B, mapping of DNase I-sensitive regions in the 5′-upstream sequences of the mouse utrophin A promoter in the presence of increasing concentrations of DNase I. The arrow indicates location of accessible regions. C, the selected ZFP target sequences and the amino acids chosen for design of each finger. The amino acid residues are listed from position –1 to +6 of the α helix. The ZFP name stems from the location of the target site relative to the transcription start.
To test the transcriptional activity of the artificial ZFPs, these DNA-binding domains were linked to an activation domain provided by the VP16 protein of herpes simplex virus, tagged with the FLAG epitope for monitoring ZFP expression and a nuclear localization signal (NLS) to ensure nuclear trafficking (Fig. 2A). The control plasmid NLS consisted of the NLS linked to FLAG epitope. In subsequent experiments these ZFPs were compared with the previously described ZFP that activated utrophin transcription (labeled Jazz) (30). The ability of each ZFP to activate a reporter gene (luciferase) driven by the utrophin A promoter was assessed by transient transfection assays in four different cell lines (COS-1, NIH 3T3 fibroblasts, human embryonic kidney 293 cells, and mouse C2C12 myoblasts) (Fig. 2, B and C). In each case, one of the designed ZFPs (ZFP51) performed better than all the other ones (Fig. 2C), including the published one (Jazz).
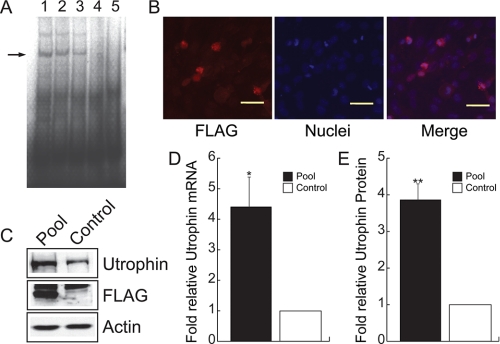
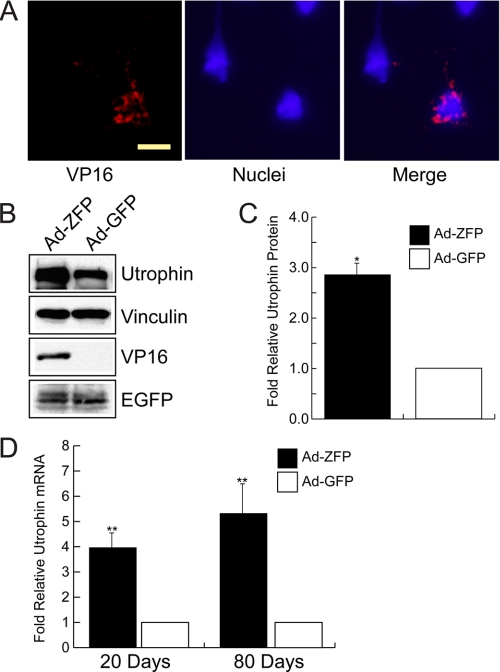
Characterization of ZFP51 in Vitro—The biological activity of ZFP51 was further characterized by a series of in vitro experiments (Figs. 3 and 4). ZFP51 bound its recognition sequence with good specificity as determined by electrophoretic mobility shift assay (Fig. 3A). Transfection of the C2C12 myoblast cell line with ZFP51 resulted in nuclear localization of ZFP51 as revealed by immunostaining of the pooled stable clones (Fig. 3B). Expression of ZFP51 led to a 4.4-fold increase in utrophin transcripts measured by quantitative real-time RT-PCR, and produced higher levels of utrophin protein accumulation (on average ∼3.8-fold increase as normalized to actin) (Fig. 3, C–E). To evaluate the potential of ZFP51 to transactivate the utrophin promoter in vivo, we generated a helper-dependent adenoviral vector (Ad-ZFP) in which the ZFP51 sequence was linked to an IRES-EGFP to allow monitoring of gene transfer in skeletal muscle (Fig. 4A). The biological activity of Ad-ZFP was first verified in C2C12 myoblasts and myotubes in culture (Fig. 4). Transduction with Ad-ZFP resulted in increased utrophin transcript levels (Fig. 4D) and utrophin protein accumulation in both myoblasts and myotubes (Fig. 4, C and E). Taken together, these data show that ZFP51 bound to its cognate sequence, localized to the nucleus after gene transfer, and that it transactivated the endogenous utrophin A promoter with robust transcriptional activity in cultured myoblasts and myotubes.
FIGURE 3.
Characterization of ZFP51 activity. A, gel-shift assay of ZFP51-Mal fusion protein DNA binding specificity. Binding of ZFP-Mal fusion protein to ZFP51 DNA target probe is shown in lane 1. Competition with mutant ZFP51 target probe (lane 2, 50-fold excess; lane 3, 100-fold excess) did not abolish binding, which could be competed with excess unlabeled ZFP51 DNA target probe (lane 4), with migration of labeled probe alone shown in lane 5. B, immunocytochemical analysis with the FLAG monoclonal antibody M2 (red, left panel) was used to detect the expression of the ZFP51 fusion protein in transfected C2C12 cells (blue Hoechst dye, middle panel). ZFP51 localizes to nuclei (right panel, merge of signals). Scale bar = 15 μm. C, ZFP51 activates expression of the endogenous utrophin locus in mouse myoblast C2C12 cells as determined by Western blot analysis on pooled, stably transfected ZFP51 clones as compared with control C2C12 cells. ZFP51 fusion protein expression was visualized using anti-FLAG antibody (arrow); β-actin served as loading control. D, quantification of utrophin transcripts was performed by quantitative real-time RT-PCR of RNA isolated from pooled ZFP51 transfectants. The values reported in the graph are the mean ± S.E., n = 3 independent experiments; *, p < 0.05. E, quantitation of endogenous utrophin protein levels in pooled ZFP51 transfectants. The values reported in the graph are the mean ± S.E., n = 3 independent experiments; **, p < 0.01.
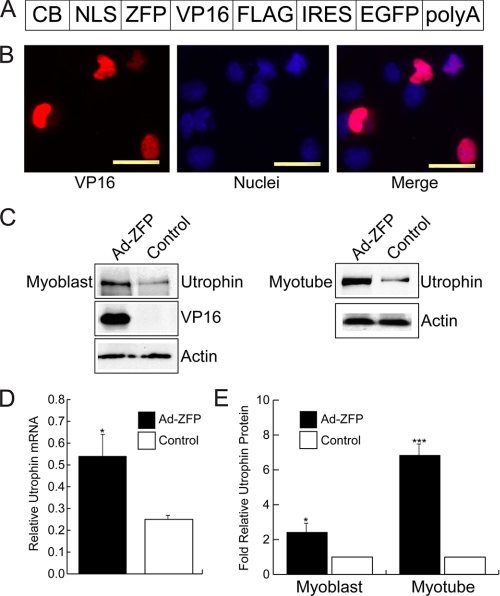
FIGURE 4.
In vitro characterization of Ad-ZFP. A, schematic representation of the ZFP construct inserted into the helper-dependent adenovirus vector (Ad-ZFP). The promoter was changed to the stronger chicken β-actin/CMV enhancer hybrid (CB), and EGFP was expressed via an IRES sequence. B, nuclear localization of ZFP/VP16 fusion protein in Ad-ZFP-infected C2C12 myoblasts. Scale bar = 10 μm. C and D, increased utrophin expression in Ad-ZFP-infected C2C12 myoblasts and myotubes as compared with control cells as determined by Western blot analysis (C and D, right panel showing quantitation for myoblasts) and by quantitative real-time RT-PCR (D, left panel). *, p < 0.05; ***, p < 0.001.
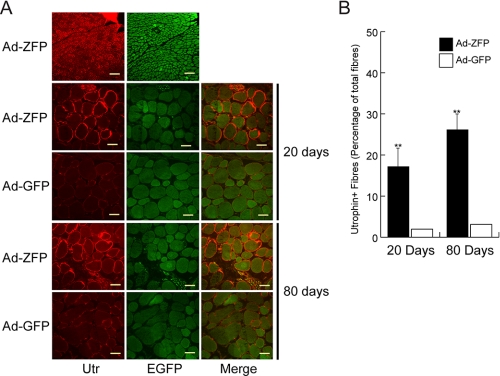
In Vivo Effects of ZFP51 in Skeletal Muscle—To determine whether utrophin up-regulation could be achieved in the dystrophic muscle of mdx mice, a single dose of Ad-ZFP was injected into the tibialis anterior (TA) muscles of mdx neonates. The contralateral control TA received Ad-EGFP. Visualization of EGFP post-administration of adenoviral vector provided an estimation of the extent of muscle transduction and showed equivalent vector distribution in both groups (data not shown). Injected muscle nuclei were immunopositive for VP16, indicating that the ZFP51 was properly localized to nuclei (Fig. 5A). In the TA, ZFP51 expression was associated with an increase of ∼3-fold in total utrophin protein levels as determined by Western blot analysis (Fig. 5, B and C). The 4- to 5-fold increase in utrophin transcripts was sustained throughout the course of the study (Fig. 5D). Immunocytochemistry showed utrophin to be present throughout the sarcolemma in Ad-ZFP-treated muscle (Fig. 6A). The Ad-ZFP-injected group had a significantly higher number of utrophin-positive fibers, ranging from 250 ± 58 fibers (∼15% of total number of fibers of the entire section) at 20 days to 438 ± 47 fibers (∼26% of total fibers) at 80 days, compared with control side TA, which had 28 ± 11 (∼2%) and 58 ± 10 (∼3%), respectively (Fig. 6B).
FIGURE 5.
Ad-ZFP induced utrophin expression in vivo. A, in Ad-ZFP-injected skeletal muscle, ZFP51 fusion protein localizes to myonuclei as shown by the merge (right panel) of VP16 immunoreactivity (red, left panel) and nuclear staining with Hoechst dye (blue, middle panel). Scale bar = 5 μm. B, Western blot analysis of utrophin expression in Ad-ZFP-injected skeletal muscle at 80 days as compared with that in Ad-GFP-injected contralateral muscle. VP16 expression was used as surrogate for ZFP51 and vinculin was used as loading control. C, quantitation of utrophin protein levels by Western blot analysis at 80 days post-Ad injection. Values were obtained from three independent muscles analyzed in triplicate. *, p < 0.05 versus control group. D, quantitation of utrophin transcripts by real-time RT-PCR of Ad-ZFP-injected skeletal muscle, normalized to glyceraldehyde-3-phosphate dehydrogenase, and expressed as relative to the utrophin transcript levels in Ad-GFP-injected contralateral muscle. At days 20 and 80, **, p < 0.01 versus control group, respectively. Values were obtained from three independent muscles analyzed in triplicate.
FIGURE 6.
Immunolocalization of utrophin in Ad-ZFP injected mdx muscle. A, immunostaining for utrophin and GFP expression in frozen muscle sections. Sections were photographed under the same exposure conditions. Note that utrophin is expressed in a continuous pattern around the sarcolemma only in the Ad-ZFP injected muscles. Scale bar = 140μm (top panel); 40 μm (day 20) and 80 μm (day 80). B, quantitation of the number of utrophin-positive fibers in mdx muscle sections injected with Ad-ZFP as compared with Ad-GFP. Relative value was obtained by normalizing to total fiber number per section. Day 20, **, p < 0.01, n = 6 and day 80, **, p < 0.01, n = 15. The difference in the number of utrophin-positive fibers observed at 20 and 80 days was significant, *, p < 0.05.
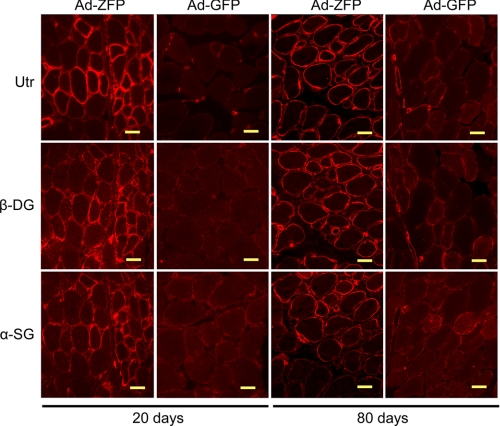
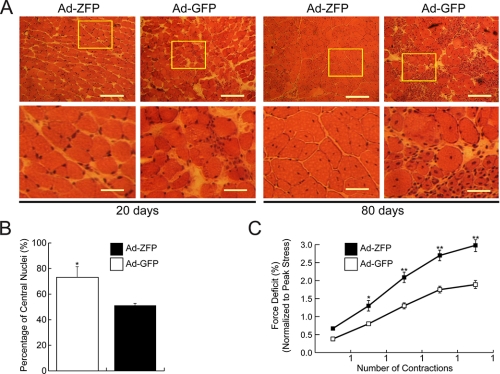
Functional Effects of ZFP51 Gene Transfer to Dystrophic Muscle—Lack of dystrophin in Duchenne muscular dystrophy patients and mdx mice also results in loss of sarcolemmal dystrophin-associated protein complex. Utrophin up-regulation through administration of Ad-ZFP restored the dystrophin-associated protein complex as demonstrated by the immunofluorescent localization of β-dystroglycan and α-sarcoglycan (Fig. 7). Moreover, sarcolemmal expression of dystrophin-associated protein complex was maintained at least for 80 days. Histological analysis of the adenovirus-treated muscles revealed that the TAs, which had a received a single injection of Ad-ZFP, had fewer areas of necrosis and infiltrating cells than the contralateral sides injected with Ad-GFP (Fig. 8A). The functional consequence of utrophin up-regulation was also reflected by the decrease in the number of muscle fibers, which had central myonuclei at 80 days post Ad-ZFP administration (Fig. 8B), implying that Ad-ZFP reduced the ongoing necrosis and regeneration in the mdx TA muscle. (At 20 days, no central nuclei are observed as the muscle fibers are in the pre-necrotic state (35)). A characteristic feature of dystrophin-deficient muscle is an abnormal susceptibility to injury caused by mechanical stress. To evaluate the overall physiological effect of Ad-ZFP in these muscles, we also assessed certain relevant contractile characteristics such as the magnitude of the force deficits induced by imposing a series of eccentric contractions on the TA. Importantly, at 80 days after Ad-ZFP administration, although overall force was not improved (data not shown), utrophin up-regulation protected against the characteristic force deficits that were observed after eccentric contraction (Fig. 8C).
FIGURE 7.
Restoration of the dystrophin protein complex in Ad-ZFP mdx-treated muscle. Serial sections were immunostained with antibodies to utrophin, β-dystroglycan, and α-sarcoglycan. Scale bar = 20 μm (day 20) and 40 μm (day 80).
FIGURE 8.
Functional consequences of Ad-ZFP injection of mdx muscles. A, appearance of Ad-ZFP- or Ad-GFP-injected mdx skeletal muscle after staining with hematoxylin and eosin. Enlarged views of the boxed areas are shown in the lower panels. Less necrosis is seen in the Ad-ZFP-injected muscle. B, the number of fibers containing central nuclei was counted in utrophin-positive and utrophin-negative fibers in Ad-ZFP- and Ad-GFP-injected muscle. Relative value was obtained by normalizing to the number of total muscle fibers per section, n = 15; p < 0.05. C, improvement of the resistance to contraction-induced injury in mdx TA muscle injected with Ad-ZFP as compared with Ad-GFP-injected contralateral muscle. Significant differences were detected after contraction number 2 (*, p < 0.05) and contractions 3, 4, and 5, **, p < 0.01, n = 15.
DISCUSSION
In this report we demonstrate the feasibility of regulating the utrophin gene in vivo using a custom-designed artificial transcription factor. We chose to target the utrophin A promoter for several reasons. Regulation of these promoter sequences have been studied extensively (36, 37). The utrophin A promoter is also the promoter that is reactivated during skeletal muscle regeneration, leading to transcription of the utrophin gene from extrasynaptic nuclei (38). The latter observation suggests that this genomic sequence may be in a “permissive” environment, a point that is supported by extrasynaptic expression of utrophin following administration of a small peptide from the heregulin ectodomain to mdx mice, in which expression was no longer restricted to the nuclei that are the usual targets of heregulin signaling (39). Moreover, it has previously been demonstrated that there is a clear difference between ZFP transcription factors targeted to accessible regions versus inaccessible regions in terms of increasing gene expression from endogenous loci (40).
Based on the results of DNase I protection assay, we targeted the sequences around –100 to –400 (numbering according to Perkins et al. (41)), and we designed several ZFPs to test activities in vitro and in vivo. We validated the ZFPs through a variety of assays: demonstrating specificity of binding to the chosen cognate DNA sequence, in vitro reporter assays to ensure transcriptional activity on the utrophin A-luciferase construct following transient transfection into HEK 293, NIH-3T3, COS-1, and C2C12 cells, assays for levels of utrophin transcript and protein following stable transfection into C2C12 myoblasts. These represent experiments of increasing rigor: a ZFP may bind its cognate sequence but may not be able to transactivate the utrophin A promoter carried on a plasmid backbone; furthermore, transactivation of the episomal reporter gene may not guarantee a similar activity on the endogenous utrophin A promoter, which was the ultimate goal of these studies. Although the ZFPs behaved differently in different cell lines, depending on the cell of origin, one ZFP (ZFP51) invariably performed the best in transient regulation of reporter gene linked to 1.3 kb of utrophin A upstream sequence. In our cell culture experiments (Fig. 2), ZFP51 was also superior to the previously described Jazz ZFP, which targets a 9-bp sequence (GCTGCTGCG) centered around –441 that is conserved in the mouse and human utrophin A upstream region (41). Similar to our results, variable transcriptional efficiency of artificial ZFP targeted to the γ-globin locus has been reported in which only one of three designed ZFP showed potent activity (42). Within the utrophin A promoter, ZFP51 bound close to the most proximal AP-2/Sp1 sequence that has been characterized upstream of the major initiation sites (41). It remains to be seen whether targeting multiple ZFPs to this region (+1 to –400) will result in additive or synergistic activation of the utrophin A promoter.
ZFP51 up-regulated transcript levels to a similar degree whether evaluated in vitro in pooled stable transfectants of myoblast cells (Figs. 3 and 4) or in vivo (Fig. 5) in injected skeletal muscle (4- to 5-fold). The increased transcript levels also led to increases in utrophin protein levels as determined by Western blot analysis (3- to 4-fold). Interestingly, when the ZFP Jazz was expressed in a muscle-specific fashion in transgenic mice, the animals with the highest transgene expression also showed a 3- to 4-fold increase in utrophin levels (43). It is unclear how much utrophin is required to compensate fully for the lack of dystrophin. Characterization of mdx utrophin transgenic mouse lines expressing varying levels of utrophin has shed some light on this: mice (the Fiona line), which express 3-fold (9, 44) to 10-fold (44) the level detected in mdx (regenerating fibers) have restored physiological function as opposed to partial correction in lines that express <2-fold (9). The levels we have achieved exceed the lower theoretical threshold value of 3-fold.
Importantly, this is the longest reported duration of ZFP function in vivo following gene transfer and leading to manifest physiological changes (45). (Because the ZFP Jazz transgenics were generated in wild-type background, the functional effects on dystrophic pathology could not be evaluated (43)). The present studies in mdx mice revealed a time-dependent increase in utrophin transcription and protein accumulation, resulting in the subsequent increase in the numbers of fibers expressing extrasynaptic utrophin of sufficient magnitude to restore the dystrophin-associated protein complex to the sarcolemma, prevents extensive necrosis and mechanical stress-induced injury. The results obtained are comparable to the effects observed following gene transfer of full-length utrophin cDNA using a helper-dependent adenovirus vector (32) in terms of mitigation of pathology, and protection from membrane damage following eccentric contractions. Of interest, these results are also similar to what has been reported with gene transfer of dystrophin constructs (46–48).
One intriguing observation is the continued increase in the utrophin transcript and protein levels between 20 and 80 days. We have previously observed a delay in gene expression following gene transfer using helper-dependent adenovirus vectors, as was the case in the present study (31). It is also conceivable that the targeting of ZFP51 to a GC-rich region of the utrophin A promoter, within an open chromatin configuration, induced additional mechanisms such as induction of demethylation (49) or even further recruitment of other DNA-binding proteins (50). Furthermore, as the utrophin promoter may also be activated in satellite cells prior to fusion, adenovirus-transduced satellite cells may provide a source of ZFP51 during muscle regeneration, leading to the appearance of regenerated fibers which express robust levels of utrophin.4
Our in vivo experiments were carried out for 80 days, and no overt toxicity was observed in any of the treated animals. In fact, as demonstrated, the injected muscles were improved according to several criteria. However, specificity of artificial ZFP transcription factors is an important consideration. As mentioned previously, some specificity is conferred upon by the availability of the cognate binding sequences. Not all recognition sequences are within transcriptionally active regions, able to interact with the basal transcriptional machinery. Even if they are properly located, not all of them will be accessible (DNase I hypersensitivity). Specificity can further be increased by increasing the length of the binding sequence (for example, to 18 bp, using a six-finger ZFP) so as to render the sequence statistically unique in the genome. This has been modeled in the context of the ZFP-mediated repression of the checkpoint kinase 2 gene (51). In two different cell lines examined by microarray analysis, besides robust repression of chk2 of over 10-fold, no other significant changes in gene expression were detected (<2-fold change in probes demonstrating 100% confidence call). Similar studies have also been undertaken with ZFP-mediated up-regulation of endogenous human vascular endothelial growth factor gene using a three-finger ZFP (52). In this case, ∼1% of the examined genes were altered in expression, and most of these were known to be regulated by vascular epidermal growth factor itself, involved in endothelial cell biology, angiogenesis, or hypoxia. Taken together, these analyses suggest that widespread dysregulation of gene expression will not occur through artificial ZFP function. Ultimately, ZFPs could be engineered as inducible, tissue-specific genes to further counter any unwanted effects (53).
Our results also compare favorably with the previous attempts at up-regulation of endogenous utrophin in dystrophic mdx mice by administration of the small peptide heregulin (39) or the substrate for nitric-oxide synthase, l-arginine (54). The ZFP expression cassette is small enough to be inserted into the adeno-associated virus to generate a recombinant adeno-associated virus for systemic delivery in vivo (47). These studies highlight the considerable therapeutic potential of artificial ZFPs for treating Duchenne muscular dystrophy in providing a convenient way of up-regulating the endogenous utrophin A promoter to compensate for dystrophin deficiency.
Acknowledgments
We thank Carol Allen and Stephen Prescott for technical help.
This work was supported in part by the United States Muscular Dystrophy Association and the Canadian Institutes of Health Research (Grant MOP-5366). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ZF, zinc finger; ZFP, zinc finger protein; CMV, cytomegalovirus; GFP, green fluorescent protein; EGFP, enhanced GFP; TA, tibialis anterior; RT, reverse transcription; PBS, phosphate-buffered saline; NLS, nuclear localization factor; Ad, adenovirus.
Y. Lu, G. Karpati, and J. Nalbantoglu, unpublished data.
References
- 1.Schofield, J., Houzelstein, D., Davies, K., Buckingham, M., and Edwards, Y. H. (1993) Dev. Dyn. 198 254–264 [DOI] [PubMed] [Google Scholar]
- 2.Karpati, G., Carpenter, S., Morris, G. E., Davies, K. E., Guerin, C., and Holland, P. (1993) J. Neuropathol. Exp. Neurol. 52 119–128 [DOI] [PubMed] [Google Scholar]
- 3.Ohlendieck, K., Ervasti, J. M., Matsumura, K., Kahl, S. D., Leveille, C. J., and Campbell, K. P. (1991) Neuron 7 499–508 [DOI] [PubMed] [Google Scholar]
- 4.Khurana, T. S., Watkins, S. C., Chafey, P., Chelly, J., Tome, F. M., Fardeau, M., Kaplan, J. C., and Kunkel, L. M. (1991) Neuromuscul. Disord. 1 185–194 [DOI] [PubMed] [Google Scholar]
- 5.Nguyen, T. M., Ellis, J. M., Love, D. R., Davies, K. E., Gatter, K. C., Dickson, G., and Morris, G. E. (1991) J. Cell Biol. 115 1695–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karpati, G. (1997) Nat. Med. 3 22–23 [DOI] [PubMed] [Google Scholar]
- 7.Clerk, A., Morris, G. E., Dubowitz, V., Davies, K. E., and Sewry, C. A. (1993) Histochem.J. 25 554–561 [PubMed] [Google Scholar]
- 8.Taylor, J., Muntoni, F., Dubowitz, V., and Sewry, C. A. (1997) Neuropathol. Appl. Neurobiol. 23 399–405 [PubMed] [Google Scholar]
- 9.Tinsley, J., Deconinck, N., Fisher, R., Kahn, D., Phelps, S., Gillis, J. M., and Davies, K. (1998) Nat. Med. 4 1441–1444 [DOI] [PubMed] [Google Scholar]
- 10.Gilbert, R., Nalbantoglu, J., Petrof, B. J., Ebihara, S., Guibinga, G. H., Tinsley, J. M., Kamen, A., Massie, B., Davies, K. E., and Karpati, G. (1999) Hum. Gene Ther. 10 1299–1310 [DOI] [PubMed] [Google Scholar]
- 11.Wakefield, P. M., Tinsley, J. M., Wood, M. J., Gilbert, R., Karpati, G., and Davies, K. E. (2000) Gene Ther. 7 201–204 [DOI] [PubMed] [Google Scholar]
- 12.Dennis, C. L., Tinsley, J. M., Deconinck, A. E., and Davies, K. E. (1996) Nucleic Acids Res. 24 1646–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burton, E. A., Tinsley, J. M., Holzfeind, P. J., Rodrigues, N. R., and Davies, K. E. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 14025–14030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gramolini, A. O., Angus, L. M., Schaeffer, L., Burton, E. A., Tinsley, J. M., Davies, K. E., Changeux, J. P., and Jasmin, B. J. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 3223–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Briguet, A., and Ruegg, M. A. (2000) J. Neurosci. 20 5989–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galvagni, F., Capo, S., and Oliviero, S. (2001) J. Mol. Biol. 306 985–996 [DOI] [PubMed] [Google Scholar]
- 17.Schaeffer, L., de Kerchove, d. A., and Changeux, J. P. (2001) Neuron 31 15–22 [DOI] [PubMed] [Google Scholar]
- 18.Gyrd-Hansen, M., Krag, T. O., Rosmarin, A. G., and Khurana, T. S. (2002) J. Neurol. Sci. 197 27–35 [DOI] [PubMed] [Google Scholar]
- 19.Khurana, T. S., Rosmarin, A. G., Shang, J., Krag, T. O., Das, S., and Gammeltoft, S. (1999) Mol. Biol. Cell 10 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angus, L. M., Chakkalakal, J. V., Mejat, A., Eibl, J. K., Belanger, G., Megeney, L. A., Chin, E. R., Schaeffer, L., Michel, R. N., and Jasmin, B. J. (2005) Am. J. Physiol. Cell Physiol. 289 C908–C917 [DOI] [PubMed] [Google Scholar]
- 21.Handschin, C., Kobayashi, Y. M., Chin, S., Seale, P., Campbell, K. P., and Spiegelman, B. M. (2007) Genes Dev. 21 770–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Briguet, A., Bleckmann, D., Bettan, M., Mermod, N., and Meier, T. (2003) Neuromuscul. Disord. 13 143–150 [DOI] [PubMed] [Google Scholar]
- 23.Weir, A. P., Burton, E. A., Harrod, G., and Davies, K. E. (2002) J. Biol. Chem. 277 45285–45290 [DOI] [PubMed] [Google Scholar]
- 24.Pabo, C. O., Peisach, E., and Grant, R. A. (2001) Annu. Rev. Biochem. 70 313–340 [DOI] [PubMed] [Google Scholar]
- 25.Jamieson, A. C., Miller, J. C., and Pabo, C. O. (2003) Nat. Rev. Drug Discov. 2 361–368 [DOI] [PubMed] [Google Scholar]
- 26.Ponder, K. P., Gupta, S., Leland, F., Darlington, G., Finegold, M., DeMayo, J., Ledley, F. D., Chowdhury, J. R., and Woo, S. L. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 1217–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, L., Spratt, S. K., Liu, Q., Johnstone, B., Qi, H., Raschke, E. E., Jamieson, A. C., Rebar, E. J., Wolffe, A. P., and Case, C. C. (2000) J. Biol. Chem. 275 33850–33860 [DOI] [PubMed] [Google Scholar]
- 28.Liu, Q., Xia, Z., Zhong, X., and Case, C. C. (2002) J. Biol. Chem. 277 3850–3856 [DOI] [PubMed] [Google Scholar]
- 29.Dreier, B., Beerli, R. R., Segal, D. J., Flippin, J. D., and Barbas, C. F., III (2001) J. Biol. Chem. 276 29466–29478 [DOI] [PubMed] [Google Scholar]
- 30.Corbi, N., Libri, V., Fanciulli, M., Tinsley, J. M., Davies, K. E., and Passananti, C. (2000) Gene Ther. 7 1076–1083 [DOI] [PubMed] [Google Scholar]
- 31.Gilbert, R., Liu, A.-B., Petrof, B. J., Nalbantoglu, J., and Karpati, G. (2002) Mol. Ther. 6 1–9 [DOI] [PubMed] [Google Scholar]
- 32.Deol, J. R., Danialou, G., Larochelle, N., Bourget, M., Moon, J. S., Liu, A. B., Gilbert, R., Petrof, B. J., Nalbantoglu, J., and Karpati, G. (2007) Mol. Ther. 15 1767–1774 [DOI] [PubMed] [Google Scholar]
- 33.Dudley, R. W., Lu, Y., Gilbert, R., Matecki, S., Nalbantoglu, J., Petrof, B. J., and Karpati, G. (2004) Hum. Gene Ther. 15 145–156 [DOI] [PubMed] [Google Scholar]
- 34.Gross, D. S., and Garrard, W. T. (1988) Annu. Rev. Biochem. 57 159–197 [DOI] [PubMed] [Google Scholar]
- 35.Disatnik, M. H., Dhawan, J., Yu, Y., Beal, M. F., Whirl, M. M., Franco, A. A., and Rando, T. A. (1998) J. Neurol. Sci. 161 77–84 [DOI] [PubMed] [Google Scholar]
- 36.Jasmin, B. J., Angus, L. M., Belanger, G., Chakkalakal, J. V., Gramolini, A. O., Lunde, J. A., Stocksley, M. A., and Thompson, J. (2002) J. Physiol. (Paris) 96 31–42 [DOI] [PubMed] [Google Scholar]
- 37.Khurana, T. S., and Davies, K. E. (2003) Nat. Rev. Drug Discov. 2 379–390 [DOI] [PubMed] [Google Scholar]
- 38.Galvagni, F., Cantini, M., and Oliviero, S. (2002) J. Biol. Chem. 277 19106–19113 [DOI] [PubMed] [Google Scholar]
- 39.Krag, T. O., Bogdanovich, S., Jensen, C. J., Fischer, M. D., Hansen-Schwartz, J., Javazon, E. H., Flake, A. W., Edvinsson, L., and Khurana, T. S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 13856–13860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, P. Q., Rebar, E. J., Zhang, L., Liu, Q., Jamieson, A. C., Liang, Y., Qi, H., Li, P. X., Chen, B., Mendel, M. C., Zhong, X., Lee, Y. L., Eisenberg, S. P., Spratt, S. K., Case, C. C., and Wolffe, A. P. (2001) J. Biol. Chem. 276 11323–11334 [DOI] [PubMed] [Google Scholar]
- 41.Perkins, K. J., Burton, E. A., and Davies, K. E. (2001) Nucleic Acids Res. 29 4843–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graslund, T., Li, X., Magnenat, L., Popkov, M., and Barbas, C. F., III (2005) J. Biol. Chem. 280 3707–3714 [DOI] [PubMed] [Google Scholar]
- 43.Mattei, E., Corbi, N., Di Certo, M. G., Strimpakos, G., Severini, C., Onori, A., Desantis, A., Libri, V., Buontempo, S., Floridi, A., Fanciulli, M., Baban, D., Davies, K. E., and Passananti, C. (2007) PLoS. ONE. 2 e774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rybakova, I. N., Patel, J. R., Davies, K. E., Yurchenco, P. D., and Ervasti, J. M. (2002) Mol. Biol. Cell 13 1512–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rebar, E. J., Huang, Y., Hickey, R., Nath, A. K., Meoli, D., Nath, S., Chen, B., Xu, L., Liang, Y., Jamieson, A. C., Zhang, L., Spratt, S. K., Case, C. C., Wolffe, A., and Giordano, F. J. (2002) Nat. Med. 8 1427–1432 [DOI] [PubMed] [Google Scholar]
- 46.DelloRusso, C., Scott, J. M., Hartigan-O'Connor, D., Salvatori, G., Barjot, C., Robinson, A. S., Crawford, R. W., Brooks, S. V., and Chamberlain, J. S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 12979–12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gregorevic, P., Blankinship, M. J., Allen, J. M., Crawford, R. W., Meuse, L., Miller, D. G., Russell, D. W., and Chamberlain, J. S. (2004) Nat. Med. 10 828–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilbert, R., Dudley, R. W. R., Liu, A.-B., Petrof, B. J., Nalbantoglu, J., and Karpati G. (2003) Hum. Mol. Genet. 12 1287–1299 [DOI] [PubMed] [Google Scholar]
- 49.Tang, D., Sivko, G. S., and Dewille, J. W. (2006) Breast Cancer Res. Treat. 95 161–170 [DOI] [PubMed] [Google Scholar]
- 50.Miura, P., Thompson, J., Chakkalakal, J. V., Holcik, M., and Jasmin, B. J. (2005) J. Biol. Chem. 280 32997–33005 [DOI] [PubMed] [Google Scholar]
- 51.Tan, S., Guschin, D., Davalos, A., Lee, Y. L., Snowden, A. W., Jouvenot, Y., Zhang, H. S., Howes, K., McNamara, A. R., Lai, A., Ullman, C., Reynolds, L., Moore, M., Isalan, M., Berg, L. P., Campos, B., Qi, H., Spratt, S. K., Case, C. C., Pabo, C. O., Campisi, J., and Gregory, P. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 11997–12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bae, K. H., Kwon, Y. D., Shin, H. C., Hwang, M. S., Ryu, E. H., Park, K. S., Yang, H. Y., Lee, D. K., Lee, Y., Park, J., Kwon, H. S., Kim, H. W., Yeh, B. I., Lee, H. W., Sohn, S. H., Yoon, J., Seol, W., and Kim, J. S. (2003) Nat. Biotechnol. 21 275–280 [DOI] [PubMed] [Google Scholar]
- 53.Beerli, R. R., Schopfer, U., Dreier, B., and Barbas, C. F., III (2000) J. Biol. Chem. 275 32617–32627 [DOI] [PubMed] [Google Scholar]
- 54.Voisin, V., Sebrie, C., Matecki, S., Yu, H., Gillet, B., Ramonatxo, M., Israel, M., and De La, P. S. (2005) Neurobiol. Dis. 20 123–130 [DOI] [PubMed] [Google Scholar]