Abstract
Lipin1 expression was induced at a late stage of differentiation of 3T3-L1 preadipocytes and maintained at high levels in mature adipocytes. Knockdown of expression of lipin1 by small interfering RNA in 3T3-L1 preadipocytes almost completely inhibited differentiation into adipocytes, whereas overexpression of lipin1 accelerated adipocyte differentiation, demonstrating that lipin1 is required for adipocyte differentiation. In mature adipocytes, transfection of lipin1-small interfering RNA decreased the expression of adipocyte functional genes, indicating the involvement of lipin1 in the maintenance of adipocyte function. Lipin1 increases the transcription-activating function of peroxisome proliferator-activated receptor γ2 (PPARγ2) via direct physical interaction, whereas lipin1 did not affect the function of other adipocyte-related transcription factors such as C/EBPα, liver X-activated receptor α, or sterol regulatory element binding protein 1c. In mature adipocytes, lipin1 was specifically recruited to the PPARγ-response elements of the phosphoenolpyruvate carboxykinase gene, an adipocyte-specific gene. C/EBPα up-regulates lipin1 transcription by directly binding to the lipin1 promoter. Based on the existence of a positive feedback loop between C/EBPα and PPARγ2, we propose that lipin1 functions as an amplifier of the network between these factors, resulting in the maintenance of high levels of the specific gene expression that are required for adipogenesis and mature adipocyte functions.
Adipose tissue plays an essential role in maintaining metabolic homeostasis (1). White adipose tissue takes up fatty acids derived from the diet or the liver as well as increases the uptake of glucose in response to insulin by recruiting glucose transporter 4 (GLUT4)2 to the plasma membrane. Then white adipose tissue stores the glucose or fatty acids as a form of triacylglyceride and releases free fatty acids during states of starvation. Recent studies have shown that adipose tissue secretes various humoral factors called adipocytokines which play numerous functions associated with food intake, insulin sensitivity, energy homeostasis, inflammatory responses, and atherogenesis (2).
In obese subjects adipocytes cannot function adequately, thereby causing various metabolic syndromes including insulin resistance, dyslipidemia, and coronary-vascular disease (3–6). Lipodystrophy leads to the same condition as obesity due to lack of adipocyte function (7–9). Thus, studying the molecular mechanisms that control adipose tissue development and function is important for understanding the pathophysiology of metabolic syndromes.
Adipogenesis is a process in which premature cells acquire adipocyte-specific functions. A complex network of transcription factors is developed during this process in response to extracellular adipogenic stimuli. In 3T3-L1 preadipocyte cells the CCAAT/enhancer-binding proteins β and δ (C/EBPβ and C/EBPδ) are induced immediately upon adipogenic hormonal stimuli, and they are expressed for approximately 2 days after which C/EBPα and peroxisome proliferator-activated receptor γ2 (PPARγ2) are induced (10, 11). Expression of C/EBPα and PPARγ2 is induced by C/EBPβ and C/EBPδ during the early stage of adipogenesis. During the late stages of adipogenesis when the expression of C/EBPβ and C/EBPδ has diminished, expression of C/EBPα and PPARγ2 is induced by each other, thereby keeping these proteins at high levels in mature adipocytes (12). C/EBPα and PPARγ2 activate the expression of genes important for adipocyte function, thereby maturing adipocytes and maintaining adipocyte-specific functions (13). Liver X-activated receptor α (LXRα) stimulates the expression of PPARγ2 by directly binding to the promoter region as well as up-regulating sterol regulatory element binding protein 1c (SREBP-1c), which induces PPARγ2 expression. Because PPARγ2 can up-regulate LXRα expression, the positive network between PPARγ2, LXRα, and SREBP-1c is also considered to be important during adipogenesis (14).
Lipin1 was discovered as a mutated gene in a fatty liver dystrophic (fld) mouse using a positional cloning approach (15). A null mutation in the lipin1 gene in these mice leads to lipodystrophy characterized by a severely reduced mass of adipose tissue and a deficiency in adipocyte differentiation (16, 17). Lack of adipose tissue in fld mice results in insulin resistance (16, 17) and circulating hyperlipidemia (18). In previous reports lipin1 was found to be expressed at a high level in adipose tissue (15), and the expression level of lipin1 was positively correlated with insulin sensitivity, which is associated with adipocyte function (19). These facts suggest that lipin1 plays an important role in adipogenesis and in the function of mature adipocytes. However, the molecular function and regulation of lipin1 remains unclear.
Recently, the yeast homolog of lipin1 was reported to have phosphatidic acid phosphohydrolase enzymatic activity (20, 21). Phosphatidic acid phosphohydrolase creates diacylglycerol from phosphatidic acid, which is the rate-limiting step in triacylglycerol synthesis. A defect in triacylglycerol synthesis is known to block adipocyte differentiation (22). Therefore, the phosphatidic acid phosphohydrolase activity of lipin1 relates to the molecular function of lipin1 in adipogenesis. However, there are several lines of evidence that suggest the possibility of a nuclear function for lipin1 as well. Two domains of lipin1 are highly conserved among species, the N-terminal conserved domain (NLIP) and the C-terminal conserved domain (CLIP). NLIP domain is known to interact with several nuclear proteins (23). Although lipin1 lacks a DNA binding domain, a yeast homolog of lipin1 was found in the promoter regions of genes involved in phospholipid biosynthesis (24), suggesting that lipin1 could be involved in the regulation of gene transcription by interacting with transcription factors. Furthermore, recent studies found that lipin1 acts as an amplifier of PPARα action by increasing the interaction between peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) and PPARα in liver (25).
In this study we investigated the expression of lipin1 and examined its potential role in adipogenesis. We also found that lipin1 increases the transcription-activating function of PPARγ2 via direct a physical interaction and is up-regulated at the transcriptional level by C/EBPα. These results provide compelling evidence that lipin1 reinforces the positive feedback loop between C/EBPα and PPARγ2, which is essential for adipogenesis and the maintenance of adipocyte functions.
MATERIALS AND METHODS
Cell Culture, in Vitro Differentiation—3T3-L1 and NIH3T3 cells were obtained from American Type Culture Collection. 3T3-L1 preadipocytes were propagated and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum. For in vitro differentiation, 2-days post-confluent (designated day 0) cells were treated with DMEM containing 10% fetal bovine serum, 520 μm 3-isobutyl-1-methylzanthine, 1 μm dexamethasone, and 167 nm insulin until day 2. The medium was then replaced and incubated with DMEM supplemented with 10% fetal bovine serum and 167 nm insulin until day 4. The medium was then replaced with DMEM supplemented with 10% fetal bovine serum, which was renewed daily until day 8. NIH3T3 cells were propagated and maintained in DMEM supplemented with 10% fetal bovine serum.
Western Blot Analysis—Cultured cells for Western blot analysis were washed once with cold phosphate-buffered saline and then harvested with whole cell lysis buffer (1% SDS, 60 mm Tris-HCl, pH 6.8), heated at 100 °C for 10 min, and centrifuged at 12,000 rpm for 5 min at room temperature. Supernatants were transferred to new tubes and used for Western blot analysis.
Plasmid Constructs, Transfections, and Luciferase Assays—To generate the lipin1β-V5 expression vector, full-length lipin1β (accession number BC018071) cDNA was amplified by PCR using the primers 5′-CACCATGAATTACGTGGGGCAGTTA-3′ and 5′-CGCTGAGGCAGAATGAATGTCCTG-3′ and cloned into the pcDNA3.1-V5/His-TOPO expression vector (Invitrogen) and pSG5-FLAG-tagged expression vector, resulting in pcDNA-lipin1β-V5 and pSG5-FLAG-lipin1β. The pSG5-FLAG-lipin1β (1–441) was generated by self-ligation after the removal of BamHI fragment from pSG5-FLAG-lipin1β. The pSG5-FLAG-lipin1-(425–926) were produced by cloning lipin1 cDNA fragment amplified using primers 5′-ACGAAGCCGACATCTTGGTGCT-3′ and 5′-CGCTGAGGCAGAATGAATGTCCTG-3′ into pSG5-FLAG-tagged expression vector.
The expression plasmids pCMV-rC/EBPα and pCMV-rC/EBPβ were gifts from Dr. J. R. Cardinaux (Centre de Neurosciences Psychiatriques, Lausanne, Switzerland). The expression plasmids pCMX-mPPARγ, pCMX-mRXRα, pCMX-mLXRα, and 3xLXRE-tk-LUC were gifts from Dr. D. Mangelsdorf (26). The pSG5-HA-PPARγ, pSG5-HA-PPARγ-(1–273), and pSG5-HA-PPARγ-(274–475) were generated from pCMX-mPPARγ by cloning respective mPPARγ cDNA into pSG5-HA-tagged expression vector. The expression plasmid pCMV7-hSREBP-1c was previously described (27). The 3xPPRE-tk-LUC plasmid, which contains three copies of the PPARγ-response element and a luciferase reporter gene, was received from Jae-Bum Kim (14). The human FAS promoter-luciferase reporter plasmid containing the sequences from -2.7 to +1.8 kilobases of the FANS gene, was previously described (28). The human G6Pase promoter-luciferase reporter plasmid, human glucose-6-phosphatase (-1227/+57), which contains a 1.2-kilobase-proximal promoter fragment extending from nt -1227 to +57, was a gift from Dr. D. Schmoll (29). The human lipin1 promoter-luciferase reporter plasmid, pGL3BA-hLipin1, was constructed by generating a lipin1 gene fragment extending from nt -1300 to +524 via PCR using the primers 5′-TCAGCAGTTTTGGGCAGCCGTGAT-3′ and 5′-AACTGCAGCGCTGGAATCCTAACT-3′ and then cloning the fragment into the pGL3-basic-acceptor luciferase vector. The pGL3-basic-acceptor luciferase was generated by inserting the splicing acceptor sequence consisting of 163 bp to the 3′ end of the first intron and 19 bp to the 5′ end of the 2nd exon of rat ACLY gene at HindIII site of pGL3-Basic. The pRL-SV40 construct, which was the expression vector of renilla luciferase, was obtained from Promega Corp. (Madison, WI).
For transfection of NIH3T3, cells were plated in 10-cm plates or 6-well plates 1 day before transfection. When the cells reached 60–80% confluency, they were transiently transfected with plasmids using Lipofectamine PLUS™ (Invitrogen) following the supplier's protocol. For transfection of 3T3-L1, cells were plated at 8 × 105 cells/60-mm dish or 3 × 105 cells per 30-mm dish 1 day before transfection. When the cells reached 90–95% confluency, they were transiently transfected with plasmids using FuGENE® HD Transfection Reagent (Roche Applied Science) following the supplier's protocol.
Luciferase activities in whole cell lysates were measured using a Dual-Luciferase® reporter assay system (Promega). The luciferase activity was normalized to the level of renilla luciferase activities of each cell lysate using a pRL-SV40 construct.
Small Interfering RNA (siRNA)—siRNA oligonucleotides specific for regions in the lipin1 mRNA were constructed according to the manufacturer's protocol for the Silencer® siRNA construction kit (Ambion, Austin, TX). Several siRNA oligonucleotides were screened and tested for their ability to silence lipin1 expression. The most active of these oligonucleotides for lipin1 (sense, 5′-GGAACUCUGUAGACAGAAUtt-3′; antisense, 5′-AUUCUGUCUACAGAGUUCCtc-3′) effectively blocked the expression of lipin1. Negative control #1 siRNA (Ambion) was used as a control. For transfection, 3T3-L1 preadipocytes were plated at 8 × 105 cells per 60-mm dish 1 day before transfection. Fifty nanomolar double-stranded 21-mer siRNA was transfected into 3T3-L1 cells using 15 μl of RNAiMAX (Invitrogen) according to the manufacturer's protocol. Confluency of the cells at the time of transfection was ∼90–95%. The cells were incubated in medium containing the siRNA-RNAiMAX complex for 6 h. The medium was then replaced with DMEM supplemented with 10% calf serum, and the cells were incubated for additional 12 h until they were just confluent. The medium was then replaced with that containing the conventional hormonal stimuli.
Reverse Transcription and Real-time PCR—Total RNA was purified from cultured cells with TRIzol® reagent according to the manufacturer's instructions (Invitrogen). First-strand cDNA synthesis was performed using oligo dT-primers with SuperScript™ II reverse transcriptase (Invitrogen) following the supplier's protocol. Quantitative real-time PCR analyses were performed on the 7300 Real Time PCR System (Applied Biosystems, Foster City, CA) using SYBR® Green PCR Master Mix (Applied Biosystems). Each assay was performed in triplicate and included four serial dilution points of a control cDNA, a no-template control, and each sample cDNA. The relative concentrations of the genes of interest, and the endogenous control (actin) were calculated by plotting the threshold cycle (Ct) versus the log of the serial dilution points. The relative expression of the gene of interest was determined after normalizing to the endogenous control. The sequences of primers used for real-time PCR can be found in Table 1.
TABLE 1.
Oligonucleotides used in PCR and ChIP assay
| Name | Sequence |
|---|---|
| Mouse lipin1 sense | 5′-CGA TGG ATG GAA GCA GAA CTC-3′ |
| Mouse lipin1 antisense α | 5′-AGC CTG AAG GAC TGG GGC TGG-3′ |
| Mouse lipin1 antisense β | 5′-AAG ACT GCA GAG GAC AAG AGC-3′ |
| Mouse lipin1 antisense Total | 5′-TGG GAT ACG ATG CTG ACT-3′ |
| Mouse C/EBPβ sense | 5′-CAA GCT GAG CGA CGA GTA CA-3′ |
| Mouse C/EBPβ antisense | 5′-CAG CTG CTC CAC CTT CTT CT-3′ |
| Mouse PPARγ2 sense | 5′-CCA GAG CAT GGT GCC TTC GCT-3′ |
| Mouse PPARγ2 antisense | 5′-CAG CAA CCA TTG GGT CAG CTC-3′ |
| Mouse C/EBPα sense | 5′-GAA CAG CAA CGA GTA CCG GGT A-3′ |
| Mouse C/EBPα antisense | 5′-GCC ATG GCC TTG ACC AAG GAG-3′ |
| Mouse aP2 sense | 5′-GAA CCT GGA AGC TTG TCT TCG-3′ |
| Mouse aP2 antisense | 5′-ACC AGC TTG TCA CCA TCT CG-3′ |
| Mouse PEPCK sense | 5′-TCA TCA TCA CCC AAG AGC AGA G-3′ |
| Mouse PEPCK antisense | 5′-CTT TCA TGC ACC CTG GGA AC-3′ |
| Mouse GLUT4 sense | 5′-TCG TCA TTG GCA TTC TGG TTG-3′ |
| Mouse GLUT4 antisense | 5′-AGC TCG TTC TAC TAA GAG CAC G-3′ |
| Mouse SREBP-1c sense | 5′-GGA GCC ATG GAT TGC ACA TT-3′ |
| Mouse SREBP-1c antisense | 5′-CTG AGT GTT TCC TGG AAG G-3′ |
| Mouse LXRα sense | 5′-GAG AAG CTG GTG GCT GCC CA-3′ |
| Mouse LXRα antisense | 5′-AGC TGT AGG AAG CCA GGG AG-3′ |
| Mouse β-actin sense | 5′-TTG TAA CCA ACT GGG ACG ATA TGG-3′ |
| Mouse β-actin antisense | 5′-CGA CCA GAG GCA TAC AGG GAC AAC-3′ |
| PEPCK (–1096/–665) sense | 5′-TCTGAACTCCGACAAGCAAGCTCT-3′ |
| PEPCK (–1096/–665) antisense | 5′-ACGTACATACTGACCCCTGCTCT-3′ |
| PEPCK (–665/–375) sense | 5′-AGAGCAGGGGTCAGTATGTACGTA-3′ |
| PEPCK (–665/–375) antisense | 5′-TGGACTTCATATGTTGCTGGCTGC-3′ |
| PEPCK (–2998/–2722) sense | 5′-GCCTCTCTCTCTATATATATAAAT–3′ |
| PEPCK (–2998/–2722) antisense | 5′-ATCTATCTATCTATCTACCCATCT-3′ |
| PPARγ2 (–537/–375) sense | 5′-TATCTGGTGTTTCATAACTTAGAGATTAAG-3′ |
| PPARγ2 (–537/–375) antisense | 5′-ATTACAAGGAAAACGTTGCTACATTGTCTC-3′ |
| SREBP–1c (–410/–170) antisense | 5′-GAACCAGCGGTGGGAACACAGAGC-3′ |
| SREBP–1c (–410/–170) asense | 5′-GACGGCGGCAGCTCGGGTTTCTC-3′ |
| FAS (–227/–50) sense | 5′-GACGGAAGCAGGCGGGGGCT-3′ |
Oil Red O Staining—Cultured cells for Oil Red O staining were washed once with phosphate-buffered saline then fixed with 3.7% formaldehyde for 2 min at room temperature. After 1 wash with water, cells were added to the Oil Red O working solution (0.5% Oil Red O in isopropanol:water, ratio 3:2) and incubated for 1 h at room temperature. The Oil Red O solution was then aspirated, and stained cells were washed with water and then observed.
Coimmunoprecipitation Assay—For coimmunoprecipitation assays, PPARγ and lipin1 were overexpressed in NIH3T3 cells as indicated in Fig. 6. Transfected cells were incubated for 2 days, then washed twice with phosphate-buffered saline, scraped off, and centrifuged at 5000 rpm for 2 min at 4 °C. Pellets were lysed in lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 0.2% Triton X-100, 0.3% Nonidet P-40) and centrifuged at 13,000 rpm for 10 min at 4 °C. Supernatants were transferred to a new tube after which 1-ml aliquots were rotated with 20 μl of protein A/G plus-agarose (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) for 10 min at 4 °C and centrifuged at 3000 rpm for 3 min at 4 °C. Supernatants were transferred to a new tube and incubated with rotation for 12 h at 4 °C with anti-V5 or anti-HA antibody (Santa Cruz Biotechnology). The lysates were then added to 20 μl of protein A/G plus-agarose (Santa Cruz Biotechnology) and rotated for 90 min at 4 °C. After centrifugation at 3000 rpm for 3 min at 4 °C, the pellets were resuspended in wash buffer (lysis buffer:phosphate-buffered saline, 1:2) then incubated with rotation for 10 min at 4 °C. This wash step was repeated three times, and then the immunoprecipitants were heated with SDS-containing sample buffer at 95 °C for 5 min after which Western blotting was performed using either an anti-PPARγ antibody or an anti-FLAG antibody (Sigma-Aldrich).
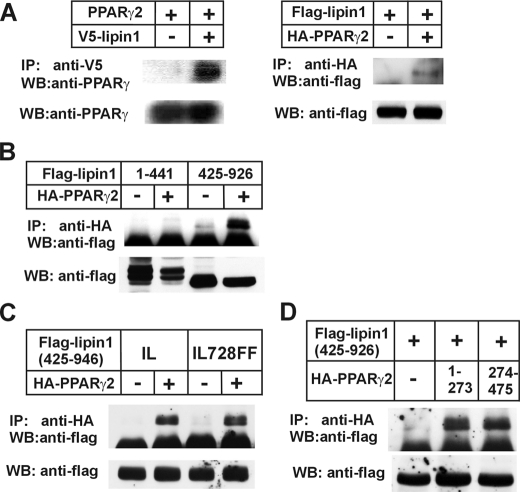
FIGURE 6.
Lipin1 physically interacts with PPARγ2. A, in the left panel NIH3T3 cells were transfected with PPARγ2 overexpression construct in company with pcDNA or pcDNA-lipin1β-V5. Two days later cells were lysed and subjected to immunoprecipitation. 2000 μg of the precleared cell lysate was incubated with 2 μg of anti-V5 antibody overnight at 4 °C. The next day 50 μl of protein A/G Plus-agarose were added and incubated for 2 h. The agarose was centrifuged, washed, and suspended in SDS-PAGE sample buffer. The immunoprecipitated (IP) protein was analyzed by immunoblot (WB) for the presence of PPARγ2. Whole cell lysates were also used for immunoblot analysis before immunoprecipitation using an anti-PPARγ2 antibody as input. In the right panel FLAG-tagged lipin1β was overexpressed with or without HA-tagged PPARγ2. After that the presence of FLAG-tagged lipin1β was analyzed in immunoprecipitates by an anti-HA antibody according to the above method. B, the pSG5-FLAG-lipin1-(1–441) or pSG5-FLAG-lipin1-(425–926) was transfected into NIH3T3 cells together with pSG5-HA-tagged PPARγ2 or empty control vector. C, NIH3T3 cells were transfected with pSG5-FLAG-lipin1-(425–926) or mutated construct containing IL692FF mutation together with HA-tagged PPARγ2 overexpression vector or empty control vector. D, pSG5-FLAG-lipin1-(425–926) was transfected together with pSG5 empty vector, pSG5-HA-tagged PPARγ2-(1–273), or pSG5-HA-tagged PPARγ2. The presence of FLAG-tagged lipin1 fragment were observed in immunoprecipitates by an anti-HA antibody from whole cell lysates. The expression of FLAG-lipin1 fragments in whole cell lysates were confirmed by Western blot using an anti-FLAG antibody.
Chromatin Immunoprecipitation (ChIP) Assay—To confirm lipin1 recruitment into the promoter region on which PPARγ or other factors bind, a ChIP assay was carried out using a rabbit polyclonal anti-lipin1 antibody (Millipore, Billerica, MA) and ChIP assay kit (Millipore) following the supplier's protocol with in vitro differentiated 3T3-L1 mature adipocytes (day 8) and 3T3-L1 preadipocytes (day 0). 3T3-L1 cells cultured in 6-cm dishes were treated with formaldehyde at the indicated time points. The cross-linked adducts were resuspended and sonicated, resulting in DNA fragments of 200 to 1000 bp. Then the fragmented chromatin samples were immunoprecipitated with anti-lipin1 antibody. Immunoprecipitated chromatin samples were reverse-cross-linked and purified and then analyzed by PCR. DNA prepared from chromatin samples served as a positive control (input). DNA prepared from chromatin samples immunoprecipitated without antibody served as a negative control. The sequences of the PCR primers can be found in Table 1.
Electrophoretic Mobility Shift Assay—To confirm the binding of C/EBPα to the putative C/EBPα-binding sequence in the lipin1 promoter, a gel retardation assay was performed. The oligonucleotide encoding the putative C/EBPα-binding site in the lipin1 promoter was used as probe. Wild-type and mutant oligonucleotides were used as competitors. The sequence of the wild-type oligonucleotide probe was (-647) 5′-ATAACGTCTGGTGGTGAAATGATTTCCATCAA-3′ (-678), and the sequence of the mutant oligonucleotide probe was (-647) 5′-ATAACGTCTGGTGGTAACCAGATTTCCATCAA-3′ (-678), where the italicized bases indicate the putative C/EBPα-binding site, and the underlined bases are mutated. The 32P-labeled probe was incubated with 5 μg of either cell extract from NIH3T3 cells that overexpressed C/EBPα or from cells that had been transfected with the empty vector. After loading the reaction mixtures onto a PAGE gel, the bands were examined and compared.
RESULTS
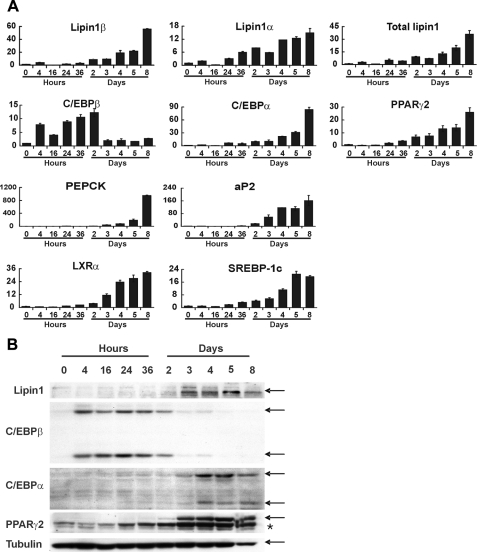
Lipin1 Is Induced in the Late Stages of Adipocyte Differentiation—To determine the function of lipin1 in white adipose tissue, lipin1 expression was examined during adipocyte differentiation of 3T3-L1 preadipocytes, which is a well characterized differentiation model after exposure of the cells to adipogenic hormonal stimuli (30). During adipocyte differentiation, expression of lipin1 mRNA began to increase 2 days after induction of differentiation and was continuously elevated further, thereby reaching a maximum level in mature adipocytes (Fig. 1A).
FIGURE 1.
Lipin1 is induced in the late stages of adipocyte differentiation. 3T3-L1 preadipocytes were differentiated to adipocytes using conventional hormonal stimuli. The expression levels of lipin1, adipogenic transcription factors, and adipocyte functional genes were then analyzed at different time points after the induction of differentiation. A, analysis of mRNA expression levels using real-time PCR with primers described in Table 1. The relative mRNA levels of each gene were normalized to the levels of actin and then further normalized with the respect to the level at 0 h. The data shown are representative of three independent experiments performed in triplicate. B, analysis of protein expression levels by Western blot. 3T3-L1 cells were lysed at each time points, and the total protein was prepared for immunoblot with the antibodies indicated. Tubulin was used as an internal loading control. Arrows indicate the positions of bands corresponding to each labeled protein; an asterisk indicates PPARγ1 protein bands.
Lipin1 mRNA undergoes alternative splicing creating two isoforms, lipin1α and lipin1β. Lipin1β is a longer form containing an insertion of 33 amino acids (31). During the maturation of adipocytes, the increase of lipin1α (the shorter isoform of lipin1) is less than that of lipin1β (Fig. 1A). As previously reported (10–13), expression of both C/EBPα and PPARγ2 showed a gradual increase during the maturation of 3T3-L1 cells (14, 32), similar to lipin1 (Fig. 1A). C/EBPα and PPARγ2 are known to induce adipocyte-specific genes such as phosphoenolpyruvate carboxykinase (PEPCK) and adipocyte/macrophage fatty acid-binding protein (aP2) (13, 33, 34). Additionally, the expression pattern of PEPCK and aP2 was similar to that of C/EBPα, PPARγ2, and lipin1 (Fig. 1A). Consistent with their involvement in adipocyte maturation (14), the expression of LXRα and SREBP-1c expression also persisted through the maturation of 3T3-L1 cells (Fig. 1A). The levels of lipin1 protein strictly coincide with those of its mRNA, increasing 2 days after the onset of hormonal stimulation and reaching high levels in mature adipocytes (Fig. 1B). The protein levels of C/EBPβ, C/EBPα, and PPARγ2 also match with those of their mRNA. Taken together, these findings indicate that lipin1 was induced at a late stage and maintained at a high level in mature adipocytes in a pattern that was similar to the late adipogenic factors C/EBPα and PPARγ2 rather than the early adipogenic factor, C/EBPβ.
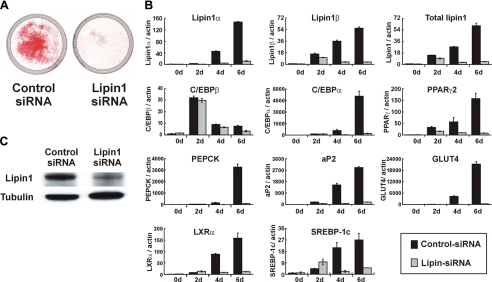
Lipin1 Is Necessary for 3T3-L1 Adipocyte Differentiation—The pattern of lipin1 expression in 3T3-L1 cells suggests that lipin1 is involved in adipocyte differentiation. To clarify the function of lipin1 more specifically, 3T3-L1 cells were transfected with lipin1-siRNA to block lipin1 function, and then the cells were hormonally stimulated to differentiate. Cells transfected with lipin1-siRNA showed significantly less expression of lipin1 than cells transfected with the control siRNA (Fig. 2B), confirming that the expression of lipin1 was blocked by the specific siRNA. The level of lipin1 protein was also reduced in cells transfected with lipin1-siRNA on day 5 (Fig. 2C). When lipid droplets were examined by Oil Red O staining on day 7 after induction, the accumulation of lipid droplets was almost completely blocked in cells transfected with lipin1-siRNA (Fig. 2A). Both C/EBPα and PPARγ2 as well as their target genes such as PEPCK, aP2, and GLUT4 (13, 33, 34) showed markedly reduced expression in lipin1-siRNA transfected cells (Fig. 2B), whereas expression of C/EBPβ was not affected (Fig. 2B). Expression of LXRα and SREBP-1c was also reduced in lipin1-siRNA-transfected cells (Fig. 2B). These findings suggest that lipin1 is indispensable to the adipocyte differentiation program in 3T3-L1 cells, and the involvement of lipin1 in adipocyte differentiation is through its effect on the expression of the late adipogenic factors C/EBPα and PPARγ2 and not of the early adipogenic factor C/EBPβ.
FIGURE 2.
Lipin1 is necessary for 3T3-L1 adipocyte differentiation. 3T3-L1 cells were transfected with lipin1-siRNA (50 nm) or control siRNA (50 nm) using RNAiMAX according to the manufacturer's instructions (Invitrogen). After that, cells were induced to differentiate under a suboptimal condition, which resulted in a lower level of induction as described under “Materials and Methods.” A, control siRNA transfected cells and lipin1-siRNA-transfected cells were stained with Oil Red O on day 7 after induction of differentiation. B, the expression levels of lipin1, adipogenic transcription factors, and adipocyte functional genes were analyzed by real-time PCR at the indicated times after induction of differentiation. Relative mRNA levels of each gene were normalized to the levels of actin and then further normalized to the level of siRNA-transfected cells at day 0. The data shown are representative of three independent experiments performed in triplicate. C, expression of lipin1 protein was analyzed by Western blot analysis. Protein was isolated from cells transfected with lipin1-siRNA or control siRNA at day 5 after induction of differentiation. Blots were probed with the indicated antibodies. Tubulin was used as an internal loading control.
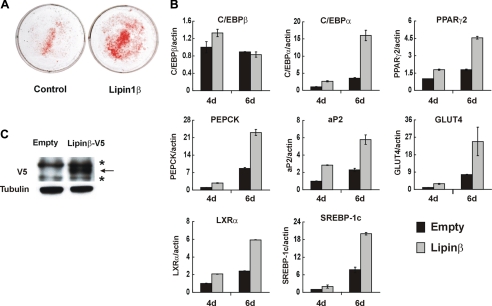
Lipin1 Accelerates 3T3-L1 Adipocyte Differentiation—To further analyze the importance of lipin1 in adipocyte differentiation, we investigated the effect of overexpression of lipin1 on adipocyte differentiation. When the expression levels of the two isoforms of lipin1 were compared during adipocyte differentiation, we found that the increase of lipin1 during terminal differentiation was mainly due to the increase in expression of lipin1β rather than lipin1α. Additionally, lipin1β was the major form in mature adipocytes, whereas lipin1α was almost undetectable in mature adipocytes (data not shown). These results suggest that the potential involvement of lipin1 in terminal differentiation seemed to be due to the action of lipin1β rather than of lipin1α. Therefore, we examined the effect of lipin1β overexpression in this study. 3T3-L1 cells were transfected with the plasmid overexpressing lipin1β-V5, and then the cells were hormonally stimulated to induce differentiation into adipocytes. The expression of lipin1β-V5 was confirmed by Western blot analysis using anti-V5 antibody (Fig. 3C). Overexpression of lipin1β-V5 resulted in accumulation of lipid droplets in a larger fraction of cells than was seen in the cells transfected with the empty control vector (Fig. 3A). Additionally, the expression of C/EBPα, PPARγ2, and their target genes such as PEPCK, aP2, and GLUT4 were much more increased in lipin1β-V5-overexpressing cells (Fig. 3B). The expression of LXRα and SREBP-1c was also increased in cells overexpressing lipin1β-V5, but expression of C/EBPβ was not significantly altered in these cells (Fig. 3B). These results suggest that during the process of differentiation into adipocytes, lipin1 affects the expression of terminal adipogenic factors such as C/EBPα and PPARγ2 but not that of the early adipogenic factor, C/EBPβ.
FIGURE 3.
Lipin1 accelerates 3T3-L1 adipocyte differentiation. 3T3-L1 cells were transfected with the pcDNA empty vector (control) or the pcDNA-lipin1β-V5 and induced to differentiate under a suboptimal condition without post-confluent incubation for 2 days. A, transfected cells were stained with Oil Red O on day 6 after induction of differentiation. B, the expression levels of lipin1, adipogenic transcription factors, and adipocyte functional genes were analyzed by real-time PCR from cells harvested at the indicated time points after induction of differentiation. Relative mRNA levels of each gene were normalized to the levels of actin and then further normalized to the level of pcDNA-transfected cells at day 4. The data shown are representative of three independent experiments performed in triplicate. C, 3T3-L1 cells were transfected with pcDNA or pcDNA-lipin1β-V5 and incubated for 3 days after transfection without hormonal stimuli. Proteins isolated from these cells were used for Western blot analysis, and blots were probed with indicated antibodies. Tubulin was used as an internal loading control. Arrowheads indicate the positions of bands corresponding to lipin1 protein; asterisks indicate nonspecific protein bands.
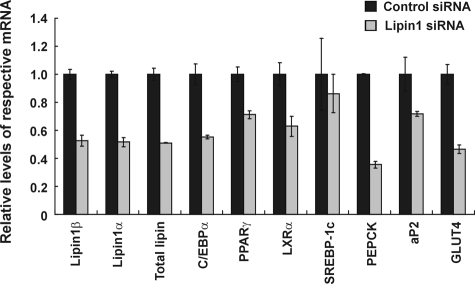
Lipin1 Maintains the Expression of Adipocyte Functional Genes in 3T3-L1 Mature Adipocytes—The persistent expression of lipin1 in mature adipocytes suggests that it is likely to be important not only in the process of adipocyte differentiation but in mature adipocytes as well. To investigate this further, lipin1-siRNA was transfected into mature adipocytes. Transfection of lipin1-siRNA reduced lipin1 mRNA levels (Fig. 4). In mature adipocytes transfected with lipin1-siRNA, the levels of C/EBPα and PPARγ2 were significantly decreased although not as much as during adipocyte differentiation (Fig. 4). Expression of PEPCK, aP2, and GLUT4 were markedly decreased in mature adipocytes that were transfected with lipin1-siRNA (Fig. 4). PEPCK, aP2, and GLUT4 are target genes of C/EBPα and PPARγ2 regulation (13), suggesting that lipin1 has a role in adipocyte function in mature adipocytes by influencing the expression of C/EBPα and PPARγ2.
FIGURE 4.
Lipin1 maintains the expression of adipocyte functional genes in 3T3-L1 mature adipocytes. 3T3-L1 cells were differentiated to adipocytes, and cells were transfected with lipin1-siRNA or control siRNA. After that cells were incubated for 2 days and harvested to examine the expression of the gene of interest. The expression levels of lipin1, adipogenic transcription factors, and adipocyte functional genes were analyzed by real-time PCR with primers described in Table 1. Relative mRNA levels of each gene were normalized to the levels of actin and then further normalized to the level of siRNA-transfected cells. The data shown are representative of three independent experiments performed in triplicate.
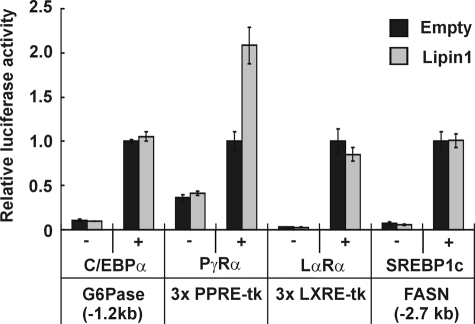
Lipin1 Functions by Interacting with and Activating PPARγ2—C/EBPα and PPARγ2 induce the expression of each other (12). LXRα and SREPB-1c are also able to induce expression of C/EBPα and PPARγ2 directly or indirectly during adipogenesis (12, 14). The expression of these transcription factors is increased during adipogenesis and maintained at a high level in mature adipocytes by a strong positive network between these factors. To examine whether lipin1 can enhance transcriptional activity of these factors, a transient transfection assay was performed in NIH3T3 fibroblasts using respective reporter constructs (Fig. 5). A promoter region of the glucose-6-phosphatase gene was stimulated by C/EBPα as previously reported (35, 36), but the promoter activity was not affected by addition of lipin1 regardless of the presence of C/EBPα, suggesting that lipin1 does not increase the transcription-activating function of C/EBPα.A 3xPPRE-tk construct containing three copies of the PPARγ response element (PPRE) responded well to PPARγ2 activation. Transfection of the plasmid expressing lipin1 alone did not stimulate luciferase activity; however, the activation by PPARγ2 was markedly enhanced by addition of lipin1. When the 3xLXRE-tk promoter and the FAS promoter were used as reporter constructs for LXRα and SREBP-1c, respectively, expression of lipin1 had no significant effect on the luciferase activity, which was induced by LXRα or SREBP-1c. In brief, lipin1 only increases the transcription activating function of PPARγ2, whereas it did not have any effect on the activity of C/EBPα, LXRα, and SREBP-1c. These results suggest that the critical role of lipin1 during adipogenesis and in mature adipocytes might be to enhance the action of PPARγ2, which is well known master regulator in maturing and maintaining adipocytes (10–13).
FIGURE 5.
Lipin1 activates the transactivation function of PPARγ2. NIH3T3 cells were cotransfected with each of the indicated luciferase constructs and a vector overexpressing the respective transcriptional factors in company with pcDNA or pcDNA-lipin1β-V5. Results are shown in relative luciferase activities. The data shown are representative of three independent experiments performed in triplicate. PγRα, PPARγ and retinoid X receptor α; LαRα, LXRα and retinoid X receptor α.
These findings led us to investigate whether lipin1 directly interacts with PPARγ2. When lipin1β-V5 and PPARγ2 were overexpressed in NIH3T3 cells, PPARγ2 was detected in the immunoprecipitants pulled down with anti-V5 antibody, whereas PPARγ2 was almost undetectable in precipitate from control cells (Fig. 6A, left panel), indicating PPARγ2 specifically bound to lipin1β-V5. And when FLAG-lipin1β and HA-PPARγ2 were overexpressed in NIH3T3 cells, lipin1 was detected in the immunoprecipitants pulled down with anti-HA antibody, whereas lipin1 was almost undetectable in the absence of overexpression of HA-PPARγ2 (Fig. 6A, right panel). This result is consistent with the result of previous IP (Fig. 6A, left panel), confirming that lipin1 interacts with PPARγ2 directly. These results suggest that lipin1 increases the transcription-activating function of PPARγ2 through a direct interaction with PPARγ2.
To define the region of lipin1 mediating the interaction with PPARγ2, two fragments of lipin1 containing either amino acids 1–441 or 425–926 were employed in the analysis for the interaction with HA-tagged PPARγ. Immunoprecipitates were obtained from whole cells lysates using an anti-HA antibody and subjected to immunoblot analysis for the presence of FLAG-lipin1-(1–441) or FLAG-lipin1-(425–926). Only FLAG-lipin1-(425–926) was coprecipitated with PPARγ2, but not FLAG-lipin1-(1–441), indicating the C terminus of lipin1 is involved in the interaction with PPARγ2 (Fig. 6B).
Lipin1 has a LXXIL motif (LGHIL; residues 725–729) in the C-terminal region, which plays a critical role in the phosphohydrolase activity and the interaction with PPARα (25). To identify whether PPARγ2 also binds on the LXXIL motif of lipin1, we introduced the mutation to change the LXXIL motif to LXXFF in FLAG-lipin1-(425–926). However, the mutation of IL728FF did not affect the interaction with PPARγ2 at all, indicating that PPARγ2 does not bind to the LXXIL motif not like PPARα (Fig. 6C). To define the region of PPARγ2 mediating the interaction with lipin1, we made two vectors expressing the N-terminal or C-terminal fragments of PPARγ2 containing the residues 1–273 or 274–475, respectively. HA-PPARγ-(1–273) contains AF1 and DNA binding domains, whereas HA-PPARγ2-(274–475) has ligand binding and AF2 domains. In immunoprecipitation analysis, FLAG-lipin1-(425–926) was coprecipitated with both N- and C-terminal fragments of PPARγ (Fig. 6D). This result suggests that the interaction of PPARγ with lipin1 was somehow different from the interaction of PPARα in which the major interaction region is reported to be the C-terminal region encompassing ligand binding and AF2 domains (25).
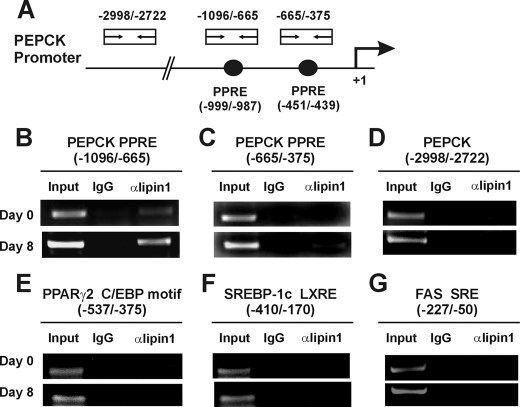
Next, the recruitment of lipin1 to a PPARγ2-controlled gene was examined in intact cells using ChIP assays. Chromatin samples were prepared from 3T3-L1 cells after induction of adipocyte differentiation and then immunoprecipitated with an anti-lipin1 antibody. We examined the recruitment of lipin1 to the PPRE in the promoter region of the PEPCK gene, which is one of the well known PPARγ2-controlled genes in adipocytes (34, 37). There are two functional PPREs in the PEPCK gene from nt -999 to -987 and from nt -451 to -439, of which the former is functional in adipocytes, whereas the latter is important in liver, thereby providing tissue-specific regulation of PEPCK (37). The binding of lipin1 to the adipocyte-specific PPRE (from nt -999 to -987) was almost undetectable on day 0 but was enhanced on day 8 (Fig. 7B). The binding of lipin1 to the liver-specific PPRE (from nt -451 to -439) was undetectable on day 0 and very weak on day 8 (Fig. 7C). There was no binding in the region between nt -2772 and -2998 of PEPCK gene, which does not contain a PPARγ binding motif and was used as a control (Fig. 7D). In light of the fact that lipin1 does not have a DNA binding domain (24, 31), these results demonstrate that lipin1 directly interacts with PPARγ2 and is recruited to the PPRE regions with PPARγ2, resulting in an increase of PPARγ2 transcriptional activity. Lipin1 was not recruited to the C/EBP-binding site of the PPARγ2 promoter, the LXR-binding site of SREBP-1c gene, or the SREBP-binding site of the FAS gene (Fig. 7, E–G). These results support the previous data which suggests that increasing PPARγ2 activity is a key mechanism of lipin1 action.
FIGURE 7.
Lipin1 is recruited around PPRE on the PPARγ2 target gene. Chromatin was immunoprecipitated from 3T3-L1 cells with an anti-lipin1 antibody at 0 and 8 days after induction of differentiation. PCR was performed using the immunoprecipitants as a template with the labeled primer pairs shown at the top of each figure. A, schematic representations of the positions of PPARγ binding elements and PCR primers used in ChIP assays on PEPCK promoter. B–D, PCR analyses were performed with primer pairs specific for the adipocyte-specific (B), liver-specific (C), or nonspecific (D) PPARγ binding elements within the PEPCK promoter. E–G, PCR analyses from immunoprecipitated chromatins were performed using primer pairs specific for the C/EBP binding motif within the PPARγ2 promoter (E), the LXR binding element (LXRE) within the SREBP-1c promoter (F), and the SREBP binding element (SRE) within the FAS promoter (G).
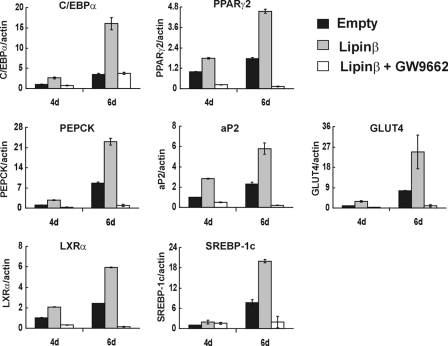
To further confirm this, the effect of lipin1 overexpression was examined after blocking PPARγ2 activity by GW9662, a synthetic antagonist of PPARγ. The ability of lipin1 to increase the expression of adipogenic factors or their target genes (Fig. 3B) was diminished when PPARγ2 activity was blocked by GW9662, demonstrating that lipin1 is an amplifier of PPARγ2 activity (Fig. 8). Taken together, these results show that lipin1 activates PPARγ2 via a direct interaction during adipogenesis and in mature adipocytes.
FIGURE 8.
Lipin1 action is attenuated by blocking PPARγ with a synthetic antagonist of PPARγ, GW9662, in 3T3-L1 cells. 3T3-L1 cells were transfected with pcDNA or pcDNA-lipin1β and induced to differentiate in the presence or absence of GW9662. The expression of adipogenic transcription factors and adipocyte functional genes were analyzed by real-time PCR with primers described in Table 1. The expression levels of each gene were normalized to actin and then further normalized with respect to the levels in pcDNA-transfected cells. The data shown are representative of three independent experiments performed in triplicate.
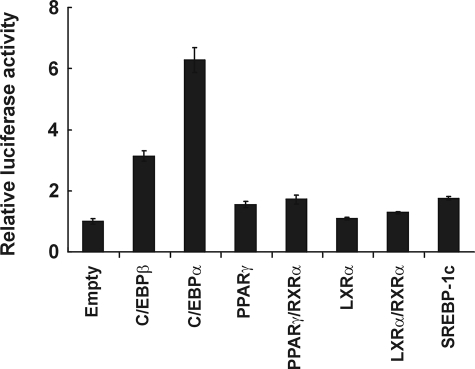
C/EBPα Directly Controls the Expression of Lipin1—A strong positive regulatory network between C/EBPα, PPARγ2, LXRα, and SREBP-1c is important to maintain a high level expression of these proteins in adipocytes. In view of the fact that the expression pattern of lipin1 during adipocyte differentiation is similar to that of these transcription factors and that lipin1 enhances PPARγ2 activity, we hypothesized that lipin1 might be a component in this network of transcription factors. Therefore, we attempted to show that the key regulator(s) of lipin1 expression is among these transcription factors. In this context, we examined which of these transcription factors could activate lipin1 promoter expression. In Fig. 9, we show that overexpression of C/EBPα activated the lipin1 promoter (from nt -1340 to +524) about 6-fold over the basal activity in a transient transfection assay, whereas overexpression of PPARγ2, LXRα, and SREBP-1c did not significantly increase lipin1 promoter activity. During the differentiation of adipocytes, lipin1 expression began to increase about 2 days after induction, when C/EBPβ was also actively expressed (Fig. 1A). Therefore, we also examined whether C/EBPβ could stimulate the lipin1 promoter. Overexpression of C/EBPβ also stimulated the lipin1 promoter about 3-fold over the basal activity (Fig. 9). These results indicate that C/EBPα may be the primary regulator of lipin1 expression during adipocyte maturation and in mature adipocytes, and in the early stages of differentiation C/EBPβ could induce lipin1 but to a lesser extent.
FIGURE 9.
C/EBPα activates the lipin1 promoter. A luciferase reporter construct, pGL3BA-hLipin1, containing a 1.3-kilobase lipin1 promoter fragment was cotransfected with empty vectors or vectors expressing each protein into NIH3T3 cells. Each sample was normalized to the level of renilla luciferase activities and expressed as -fold activation by each overexpressed transcription factor. The data shown are representative of three independent experiments performed in triplicate.
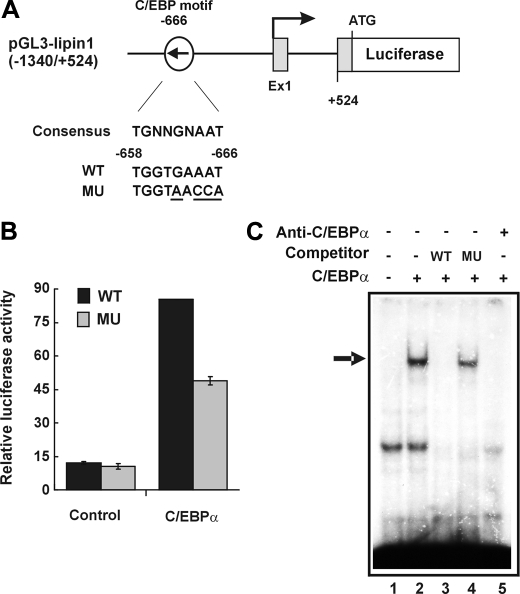
Sequence analysis of the lipin1 promoter region revealed a well conserved C/EBP binding motif (38) located at position -666 (Fig. 10A). Mutation of this C/EBP element attenuated the activation of the lipin1 promoter by C/EBPα (Fig. 10B). To confirm that C/EBPα can specifically bind to the conserved C/EBP binding motif at position -666, an electrophoretic mobility shift assay was performed. Indeed, a shifted band was formed when cellular extracts of C/EBPα-overexpressing cells were mixed with the probe containing the C/EBP binding motif (Fig. 10C, lane 2). The formation of shifted complexes was competitively inhibited by the presence of unlabeled probe but not by mutant oligonucleotides (Fig. 10C, lanes 3 and 4). The formation of DNA-protein complexes were blocked by anti-C/EBPα antibody (Fig. 10C, lane 5), indicating that the complexes were made by C/EBPα binding to the probe. Another well conserved C/EBP motif was found at position -1150, but it did not form a complex with C/EBPα in electrophoretic mobility shift assay (data not shown). These results indicate that the C/EBP element located at position -666 likely plays a critical role in C/EBPα-mediated activation of the lipin1 promoter.
FIGURE 10.
Identification of the C/EBPα-binding site in the lipin1 promoter. A, schematic representations showing lipin1 promoter-luciferase constructs containing a wild-type (WT) or mutant (MU) putative C/EBP-binding site. B, the lipin1 promoter luciferase constructs were cotransfected with the empty vector (Control) or pcDNA-C/EBPα in NIH3T3 cells. The results are expressed as a luciferase activity which was normalized to the level of renilla luciferase activities. The data shown are representative of three independent experiments performed in triplicate. C, whole cell lysates were harvested from transfected NIH3T3 cells with an empty vector (lane 1) or pCMV-C/EBPα (lanes 2–5) and were used in electrophoretic mobility shift assay of the putative C/EBP binding sequence on lipin1 promoter. Wild-type oligonucleotide (WT) (lane 3) and mutant oligonucleotide (MU) (lane 4) were used as competitors. For the supershift assay, 5 μg of anti-C/EBPα antibody was added into the reaction mixture in lane 5. The arrow indicates the band shifted by C/EBPα.
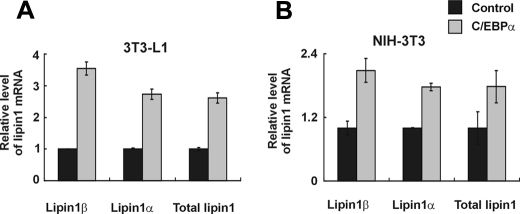
To assess the role of C/EBPα in the induction of endogenous lipin1 during adipocyte differentiation, we examined the effect of overexpressing C/EBPα. The levels of lipin1 mRNA were increased by the C/EBPα overexpression in differentiating 3T3-L1 cells (Fig. 11A). Additionally, C/EBPα increased the transcriptional levels of lipin1 in NIH3T3 fibroblasts (Fig. 11B). These results clearly demonstrate that C/EBPα controls the expression of lipin1 during adipogenesis.
FIGURE 11.
C/EBPα increases lipin1 expression in 3T3-L1 cells and NIH3T3 cells. A, 3T3-L1 cells were transfected with pcDNA-C/EBPα or the control vector and induced to differentiate using the conventional hormonal stimuli for 4 days. The expression level of lipin1 was analyzed by real-time PCR. The mRNA level was normalized to actin and then further normalized to the levels in control pcDNA-transfected cells. The data shown are representative of three independent experiments performed in triplicate. B, NIH3T3 cells were transfected with pcDNA-C/EBPα or the control vector and incubated for 3 days. Expression of lipin1 was analyzed by real-time PCR. The expression level of lipin1 mRNA was normalized to the level of actin mRNA and then further normalized to the levels in control pcDNA-transfected cells.
DISCUSSION
A complex regulatory network of transcription factors is important in keeping expression levels high for gene regulation during adipogenesis and in mature adipocytes (10, 11, 13, 14). In this network activation of one of these factors can induce all of them. C/EBPα, PPARγ2, LXRα, and SREBP-1c are representative regulators involved in this network. In addition to a positive feedback loop that exists between C/EBPα and PPARγ2 (12), PPARγ2, LXRα, and SREBP-1c are also in a positive network with each other (14). Previous studies established C/EBPα and PPARγ2 as key regulators for the differentiation of 3T3-L1 preadipocytes into adipocytes during the late stage and for the overall function of adipocytes (13, 33, 34). C/EBPα and PPARγ2 induce each other's expression in adipocytes, forming a positive feedback loop that is necessary for their high level expression (12). The results of our study identify lipin1 as another important component in the positive feedback loop between C/EBPα and PPARγ2.
One of the most typical phenotypes caused by null mutation of lipin1 in the fld mouse is lipodystrophy, characterized by a severely reduced mass and impaired development of white adipose tissue (16, 17). We report that in a siRNA-mediated knock-down of lipin1, adipocyte differentiation was completely blocked, and the induction of C/EBPα and PPARγ2 was severely attenuated, but the temporal increase of early stage C/EBPβ expression was not affected. In agreement with this, the exogenous overexpression of lipin1 markedly increased C/EBPα and PPARγ2 expression and accelerated adipocyte differentiation. These results indicate the importance of the precise regulation of lipin1 during adipocyte differentiation and suggest that lipin1 action is intertwined with the expression of C/EBPα and PPARγ2.
We subsequently found that lipin1 can physically interact with PPARγ2 and that lipin1 specifically activates PPARγ2 action in a reporter assay without any effect on C/EBPα, LXLα, and SREBP-1c action. Furthermore, lipin1 was recruited only to the adipocyte-specific PPARγ2 response element of the PEPCK gene in mature adipocytes but not to the binding elements of C/EBPα, LXRα, and SREBP-1c. In differentiating 3T3-L1 cells, lipin1 increased the expression of aP2 and GLUT4, which are also target genes of PPARγ2 regulation (13, 33, 34). These results provide strong evidence that lipin1 enhances PPARγ2 action.
We verified that the C terminus of lipin1 is involved in the interaction with PPARγ2, the same as that with PPARα (25). However, PPARγ2 does not bind to the LXXIL motif, which is crucial motif for the binding of PPARα. And also we found that FLAG-lipin1-(425–926) interacts with both N- and C-terminal fragments of PPARγ. With these results, we noticed that the binding of PPARγ2 on lipin1 is different from that of PPARα (25).
We found that exogenous expression of C/EBPα markedly up-regulated endogenous lipin1 expression in 3T3-L1 and NIH3T3 fibroblasts. In a reporter gene assay, the lipin1 promoter responded to C/EBPα as well as to C/EBPβ (to a lesser extent) but not to PPARγ2, LXR, or SREBP-1c. Therefore, C/EBPα regulates lipin1 expression, and by regulating the activity of PPARγ2, lipin1 affects the expression of C/EBPα, which forms a regulatory loop with PPARγ2. Therefore, lipin1 controls adipocyte function through inducing C/EBPα and PPARγ2.
The inhibition of adipogenesis in a C/EBPα knock-out experiment was compensated for by the addition of PPARγ2 (39, 40), but the inhibition of adipogenesis under PPARγ-deficient conditions was not compensated for by the addition of C/EBPα (40, 41). These facts suggest that the function of PPARγ2 is absolutely important for adipogenesis but C/EBPα is involved in adipogenesis by acting as a regulator of PPARγ2 rather than by direct action. In the present study we demonstrate that C/EBPα increases the activity of PPARγ2 by inducing lipin1 in addition to directly elevating PPARγ2 expression. In this scenario lipin1 seems to be involved in amplifying the activity of PPARγ2 and reinforcing C/EBPα as a regulator of PPARγ2 activity and expression.
An overload of lipid beyond the capacity of adipocytes or when the number of adipocytes is low induces the accumulation of lipid in extra-adipose tissues, such as muscle and liver, causing insulin resistance. Reduced levels of lipin1 in mature adipocytes decreased the expressions of genes which are responsible for adipocyte function, such as PEPCK, aP2, and GLUT4. This result provides the precise mechanism for the previous report that the expression level of lipin1 in adipocytes is positively correlated with insulin sensitivity (19).
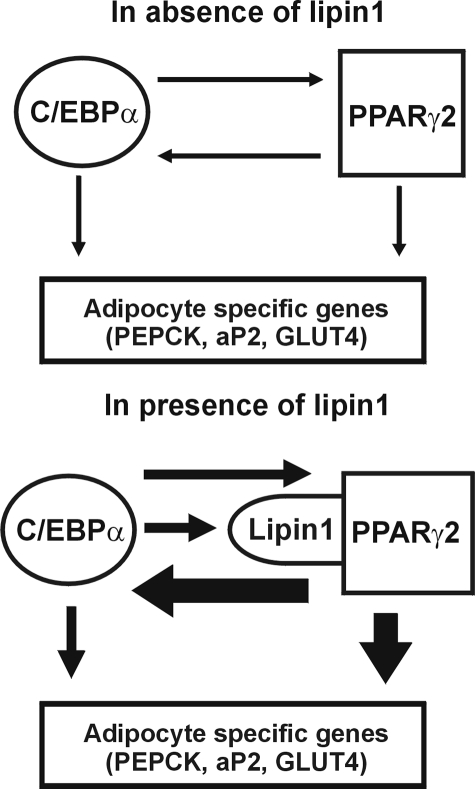
In summary, the present study provides compelling evidence of the pivotal role played by lipin1 in developing and mature adipocytes. Lipin1 expression is induced by C/EBPα and in turn controls the expression of adipocyte functional genes by increasing PPARγ2 activity, thus amplifying gene expression required for maturation and maintenance of adipocytes (Fig. 12). Future investigation of how the binding of lipin1 to PPARγ2 could increase the transcriptional activity of PPARγ2 should provide additional insight into adipocyte differentiation and its physiology.
FIGURE 12.
A schematic of the proposed functional roles of lipin1 in maturation and maintenance of adipocytes. Lipin1 reinforces the network between C/EBPα and PPARγ2, thereby increasing the induced level of adipocyte-specific genes.
This work was supported by Korea Science and Engineering Foundation Grant R13-2002-054-03001-0. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GLUT4, glucose transporter 4; DMEM, Dulbecco's modified Eagle's medium; HA, hemagglutinin; nt, nucleotide(s); PPAR, peroxisome proliferator-activated receptor; PPRE, PPARγ response element; C/EBP, CCAAT/enhancer-binding proteins β and δ; LXR, liver X-activated receptor α; SREBP-1c, sterol regulatory element binding protein 1c; siRNA, small interfering RNA; ChIP, chromatin immunoprecipitation; PEPCK, phosphoenolpyruvate carboxykinase; aP2, acid-binding protein.
References
- 1.Spiegelman, B. M., and Flier, J. S. (1996) Cell 87377 -389 [DOI] [PubMed] [Google Scholar]
- 2.Flier, J. S. (2004) Cell 116337 -350 [DOI] [PubMed] [Google Scholar]
- 3.Scott, C. L. (2003) Am. J. Cardiol. 9235 -42 [Google Scholar]
- 4.Must, A., Spadano, J., Coakley, E. H., Field, A. E., Colditz, G., and Dietz, W. H. (1999) J. Am. Med. Assoc. 2821523 -1529 [DOI] [PubMed] [Google Scholar]
- 5.Kress, A. M., Hartzel, M. C., and Peterson, M. R. (2005) Prev. Med. 41 63-69 [DOI] [PubMed] [Google Scholar]
- 6.Nowicki, E. M., Billington, C. J., Levine, A. S., Hoover, H., Must, A., and Naumova, E. (2003) Mil. Med. 168252 -256 [PubMed] [Google Scholar]
- 7.Schott, M., Scherbaum, W. A., and Bornstein, S. R. (2004) N. Engl. J. Med. 351103 -104 [PubMed] [Google Scholar]
- 8.Reitman, M. L., Arioglu, E., Gavrilova, O., and Taylor, S. I. (2000) Trends Endocrinol. Metab. 11 410-416 [DOI] [PubMed] [Google Scholar]
- 9.Hegele, R. A. (2003) Trends Endocrinol. Metab. 14371 -377 [DOI] [PubMed] [Google Scholar]
- 10.Christy, R. J., Kaestner, K. H., Geiman, D. E., and Lane, M. D. (1991) Proc. Natl. Acad. Sci. U. S. A 882593 -2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke, S. L., Robinson, C. E., and Gimble, J. M. (1997) Biochem. Biophys. Res. Commun. 240 99-103 [DOI] [PubMed] [Google Scholar]
- 12.Ramji, D. P., and Foka, P. (2002) Biochem. J. 365561 -575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farmer, S. R. (2006) Cell Metab. 4263 -273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seo, J. B., Moon, H. M., Kim, W. S., Lee, Y. S., Jeong, H. W., Yoo, E. J., Ham, J., Kang, H., Park, M. G., Steffensen, K. R., Stulnig, T. M., Gustafsson, J. A., Park, S. D., and Kim, J. B. (2004) Mol. Cell. Biol. 243430 -3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterfy, M., Phan, J., Xu, P., and Reue, K. (2001) Nat. Genet. 27121 -124 [DOI] [PubMed] [Google Scholar]
- 16.Reue, K., and Peterfy, M. (2000) Curr. Atheroscler. Rep. 2390 -396 [DOI] [PubMed] [Google Scholar]
- 17.Reue, K., Xu, P., Wang, X. P., and Slavin, B. G. (2000) J. Lipid Res. 411067 -1076 [PubMed] [Google Scholar]
- 18.Langner, C. A., Birkenmeier, E. H., Ben-Zeev, O., Schotz, M. C., Sweet, H. O., Davisson, M. T., and Gordon, J. I. (1989) J. Biol. Chem. 2647994 -8003 [PubMed] [Google Scholar]
- 19.van Harmelen, V., Ryden, M., Sjolin, E., and Hoffstedt, J. (2007) J. Lipid Res. 48 201-206 [DOI] [PubMed] [Google Scholar]
- 20.Han, G. S., Wu, W. I., and Carman, G. M. (2006) J. Biol. Chem. 2819210 -9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scavello, G. S., Jr., Paluru, P. C., Zhou, J., White, P. S., Rappaport, E. F., and Young, T. L. (2005) Mol. Vis. 1197 -110 [PubMed] [Google Scholar]
- 22.Gale, S. E., Frolov, A., Han, X., Bickel, P. E., Cao, L., Bowcock, A., Schaffer, J. E., and Ory, D. S. (2006) J. Biol. Chem. 28111082 -11089 [DOI] [PubMed] [Google Scholar]
- 23.Tange, Y., Hirata, A., and Niwa, O. (2002) J. Cell Sci. 1154375 -4385 [DOI] [PubMed] [Google Scholar]
- 24.Santos-Rosa, H., Leung, J., Grimsey, N., Peak-Chew, S., and Siniossoglou, S. (2005) EMBO J. 241931 -1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finck, B. N., Gropler, M. C., Chen, Z., Leone, T. C., Croce, M. A., Harris, T. E., Lawrence, J. C., Jr., and Kelly, D. P. (2006) Cell Metab. 4199 -210 [DOI] [PubMed] [Google Scholar]
- 26.Peet, D. J., Janowski, B. A., and Mangelsdorf, D. J. (1998) Curr. Opin. Genet. Dev. 8 571-575 [DOI] [PubMed] [Google Scholar]
- 27.Kim, H. I., Kim, J. W., Kim, S. H., Cha, J. Y., Kim, K. S., and Ahn, Y. H. (2000) Diabetes 491517 -1524 [DOI] [PubMed] [Google Scholar]
- 28.Lee, M. Y., Moon, J. S., Park, S. W., Koh, Y. K., Ahn, Y. H., and Kim, K. S. (2008) Biochem. J., in press
- 29.Schmoll, D., Walker, K. S., Alessi, D. R., Grempler, R., Burchell, A., Guo, S., Walther, R., and Unterman, T. G. (2000) J. Biol. Chem. 27536324 -36333 [DOI] [PubMed] [Google Scholar]
- 30.Ntambi, J. M., Buhrow, S. A., Kaestner, K. H., Christy, R. J., Sibley, E., Kelly, T. J., Jr., and Lane, M. D. (1988) J. Biol. Chem. 26317291 -17300 [PubMed] [Google Scholar]
- 31.Peterfy, M., Phan, J., and Reue, K. (2005) J. Biol. Chem. 28032883 -32889 [DOI] [PubMed] [Google Scholar]
- 32.Rosen, E. D. (2002) Ann. N. Y. Acad. Sci. 979143 -158 and 188-196 [DOI] [PubMed] [Google Scholar]
- 33.Bogacka, I., Xie, H., Bray, G. A., and Smith, S. R. (2004) Diabetes Care 271660 -1667 [DOI] [PubMed] [Google Scholar]
- 34.Duplus, E., Benelli, C., Reis, A. F., Fouque, F., Velho, G., and Forest, C. (2003) Biochimie (Paris) 851257 -1264 [DOI] [PubMed] [Google Scholar]
- 35.Yubero, P., Hondares, E., Carmona, M. C., Rossell, M., Gonzalez, F. J., Iglesias, R., Giralt, M., and Villarroya, F. (2004) Endocrinology 1454268 -4277 [DOI] [PubMed] [Google Scholar]
- 36.Pedersen, T. A., Bereshchenko, O., Garcia-Silva, S., Ermakova, O., Kurz, E., Mandrup, S., Porse, B. T., and Nerlov, C. (2007) EMBO J. 261081 -1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beale, E. G., Forest, C., and Hammer, R. E. (2003) Biochimie (Paris) 851207 -1211 [DOI] [PubMed] [Google Scholar]
- 38.Akira, S., Isshiki, H., Sugita, T., Tanabe, O., Kinoshita, S., Nishio, Y., Nakajima, T., Hirano, T., and Kishimoto, T. (1990) EMBO J. 91897 -1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, Z., Rosen, E. D., Brun, R., Hauser, S., Adelmant, G., Troy, A. E., McKeon, C., Darlington, G. J., and Spiegelman, B. M. (1999) Mol. Cell 3151 -158 [DOI] [PubMed] [Google Scholar]
- 40.Rosen, E. D. (2005) Prostaglandins Leukot. Essent. Fatty Acids 7331 -34 [DOI] [PubMed] [Google Scholar]
- 41.Rosen, E. D., Hsu, C. H., Wang, X., Sakai, S., Freeman, M. W., Gonzalez, F. J., and Spiegelman, B. M. (2002) Genes Dev. 1622 -26 [DOI] [PMC free article] [PubMed] [Google Scholar]