Abstract
Alix/AIP1 regulates cell death in a way involving interactions with the calcium-binding protein ALG-2 and with proteins of ESCRT (endosomal sorting complex required for transport). Using mass spectrometry we identified caspase-8 among proteins co-immunoprecipitating with Alix in dying neurons. We next demonstrated that Alix and ALG-2 interact with pro-caspase-8 and that Alix forms a complex with the TNFα receptor-1 (TNF-R1), depending on its capacity to bind ESCRT proteins. Thus, Alix and ALG-2 may allow the recruitment of pro-caspase-8 onto endosomes containing TNF-R1, a step thought to be necessary for activation of the apical caspase. In line with this, expression of Alix deleted of its ALG-2-binding site (AlixΔALG-2) significantly reduced TNF-R1-induced cell death, without affecting endocytosis of the receptor. In a more physiological setting, we found that programmed cell death of motoneurons, which can be inhibited by AlixΔALG-2, is regulated by TNF-R1. Taken together, these results highlight Alix and ALG-2 as new actors of the TNF-R1 pathway.
Endocytosis of cell surface receptors has long been described as an effective way of switching off extracellularly induced signals. Endocytosed activated receptors traffic through early endosomes and are sorted into intralumenal vesicles accumulating inside endosomes known as multivesicular bodies (MVBs).5 These MVBs fuse with lysosomes where the receptors meet their end by acid hydrolysis (1).
In some cases however, such as for neurotrophin-bound Trk receptors, activated receptors continue signaling inside endosomes (2). Also, in the case of death receptors, Schütze and co-workers (3) showed that endocytosis of TNF-R1, which occurs after binding to TNFα, is a necessary step for activation of caspases and consequently apoptosis. They found that the apical procaspase-8 is recruited and thereby activated on the surface of multivesicular endosomes containing activated TNF-R1.
Biogenesis of MVBs is under tight control by a set of proteins, making the so-called ESCRT-0 to -III (endosomal sorting complex required for transport), which sequentially associate on the cytosolic surface of endosomes (4). A partner of ESCRT proteins, which also regulates the making of MVBs, is the protein Alix/AIP1, first characterized as an interactor of the calcium-binding protein ALG-2 (apoptosis-linked gene-2) (5–7). Enveloped viruses, like human immunodeficiency virus, type 1, use Alix to recruit the ESCRT machinery to deform membranes and allow fission during viral budding (8, 9). Furthermore, two recent reports have claimed that Alix together with ESCRT proteins might also be involved in the abscission stage of cytokinesis (10, 11).
Besides Tsg101 and CHMP-4B of ESCRT-I and –III, respectively, Alix interacts with lysobisphosphatidic acid, a phospholipid involved in intralumenal vesiculation of endosomes (12), and with regulators of endocytosis (CIN85 and endophilins) (13, 14). However, the precise role of Alix on endosomes remains largely unclear because neither we nor other laboratories have found any striking effect of Alix on endocytosis and degradation of EGF or transferrin receptors (15, 16).
We and others have gathered evidence that Alix plays a role in cell death. In particular, expression of a mutant lacking the N-terminal part (Alix-CT) blocks death of HeLa cells induced by serum starvation (7) and of cerebellar neurons deprived of potassium (17). In this latter paradigm, Alix-CT, whose protecting activity was strictly correlated with its capacity to bind ALG-2, accumulated inside cytoplasmic aggregates together with ALG-2 and caspases. We also demonstrated, using electroporation in the chick embryo, that Alix mutants block programmed cell death of motoneurons during normal development, depending on binding to ALG-2 and ESCRT-I and -III. Our interpretation of these results is that some truncated forms of Alix behave as dominant negative mutants, blocking the formation of an ALG-2-Alix-ESCRT complex necessary for cell death (18). Therefore the Alix-ALG-2 complex could make a link between endosomes and a signaling or an execution step of neuronal death (19).
We have undertaken the present study to characterize this link and found that Alix and ALG-2 form a complex with endocytosed TNF-R1 and pro-caspase-8. The physiological relevance of these interactions was revealed by the demonstration that several Alix mutants inhibit TNF-R1-induced cell death both in vitro and in vivo.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies—Human recombinant TNFα-FLAG was from Alexis Biochemicals (Covalab). Mouse monoclonal anti-hemagglutinin (HA) antibody was from Cell Signaling (Ozyme); polyclonal antibodies against Myc, TNF-R1, and FADD were from Santa Cruz Biotechnology; polyclonal antibodies against LAMP1 and EEA1 were from AbCam; anti-FLAG monoclonal (M1 and M2) and polyclonal antibodies were from Sigma-Aldrich; GM130 and monoclonal anti-AIP1/Alix were from BD Transduction; anti-ALG-2 was from Swant; HSP70 mitochondria was from Affinity Bioreagents (Ozyme); anti-caspase 8 was from Biovision (Clinisciences); horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit Alexa594 antibody were from Jackson Laboratories; and biotinylated goat anti-rabbit antibody was from Vector Laboratories.
DNA Constructs—For expression in the chick embryo, human TNF-R1 (wt or mutants), a catalytically dead version of human pro-caspase-8 (mutation C360A), and baculovirus p35 were inserted into the pCAGGS expression vector (a gift of Tsuyoshi Momose, Nara Institute of Science and Technology). Mammalian expression vectors coding for DN-caspase-8-HA-tagged and p35 were a gift of P. Mehlen (INSERM, Lyon, France). Those coding for wt or mutated human TNF-R1 were kind gifts from W. Schneider-Brachert (University of Regensburg, Regensburg, Germany). Mutated ALG-2ΔEF1,3 was a gift of M. Maki (Nagoya University, Nagoya, Japan).
In Ovo Electroporation—Fertilized Isa Brown eggs (Société Française de Production Avicole, St. Marcellin, France) were electroporated at Hamburger-Hamilton (HH) stage 16. Plasmid DNA was electroporated as described in Ref. 18.
Histological Analysis—Chick embryos were collected 48 h after electroporation processed and cryo-sectioned as described previously in Ref. 18.
Immunohistochemistry and Immunofluorescence—Frozen sections were incubated with polyclonal anti-TNF-R1 or monoclonal anti-HA antibodies, diluted to 1/100 in Tris-buffered saline containing 1% GS, 0.02% saponine (TBSS) for 12–24 h at 4 °C. The sections were rinsed in TBSS and treated with a secondary anti-rabbit Alexa594 antibody or a biotinylated goat anti-rabbit secondary antibody, amplified using the ABC kit (Vector Laboratories) and revealed with 3,3′-diaminobenzidine and nickel intensification. The sections were rinsed in TBS, incubated 30 min at 37 °C in Hoechst 33342, 2 μg/ml (Sigma, France) before mounting in Mowiol (Calbiochem, France).
Terminal Deoxynucleotidyl Transferase-mediated dUTP-Biotin Nick End Labeling (TUNEL) Method—TUNEL analysis was performed using an in situ cell death detection kit (Roche Applied Science). Fluorescent positive cells were counted in every third section. Twelve sections/embryo were counted.
Reverse Transcription-PCR—RNA were extracted from whole chick embryo using TRIzol reagent (Invitrogen). cDNAs were synthesized with Moloney murine leukemia virus reverse transcriptase (Promega) and controlled with glyceraldehyde-3-phosphate dehydrogenase. TNF-R1 expression was further analyzed by amplification of a 695-nt fragment with the following oligonucleotides: 5′-GATACTGTGTGTGGCTGT-3′ and 5′-CGTAAATGTCGATGCTCC-3′ based on the chick TNF-R1 homologue (Gallus gallus accession number AJ720473).
Cell Culture and Transfection—HEK-293 cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (Invitrogen), 2 mm glutamine, 10 μg/ml streptamycin, and 10 units/ml penicillin. BHK-21 cells were maintained in Glasgow minimum essential medium (Invitrogen) containing 5% fetal bovine serum, 2.6 g/liter tryptose phosphate broth, 2 mm glutamine, 10 μg/ml streptamycin, and 10 units/ml penicillin. HEK 293 or BHK-21 cells were transfected using JetPEI (Ozyme).
Alix Knockdown in BHK Cells—The Alix small hairpin RNA was cloned downstream of the human H1 promoter in the vector pSuperGFP (Oligoengine). The sequences of the synthetic oligonucleotides (Invitrogen) used for Alix small hairpin RNA construct were the following: 5′-GATCCCCGCCGCTGGTGAAGTTCATCTTCAAGAGAGATGAACTTCACCAGCGGCTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGTTCATCCAGCAGACTTACTCTCTTGAAGTAAGTCTGCTGGATGAACGGG-3′. Annealed oligonucleotides were ligated into the BglII cleavage site within the pSuperGFP vector linearized with the same restriction enzymes.
BHK cells were transfected with pSuper/shAlix plasmid or pSuperGFP vector for control using the JetSi transfection reagent (Polyplus Transfection). Transfected cells were selected using 800 μg/ml of G418. After 15 days, the clones were isolated and selected for the best reduction in Alix expression. Cells (pSuper/shAlix and control) were grown in the presence of 800 μg/ml of G418.
Mass Spectrometry and Protein Identification—Cultures of mouse cerebellar granule neurons were prepared as described previously (17) and cultured in basal medium Eagle containing 25 mm potassium. The medium was changed for basal medium Eagle containing 5 mm potassium, and the cells were lysed 4 h later in RIPA buffer (150 mm NaCl, 50 mm Tris, pH 8.0, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS containing a protease inhibitor mixture (Roche Applied Science). Immunoprecipitations were performed using polyclonal anti-Alix antibody and protein G-coupled Sepharose beads. After separation by SDS-PAGE, discrete bands were excised from the Coomassie Blue-stained gel. In-gel digestion was performed as previously described (20). Gel pieces were then sequentially extracted with 5% (v/v) formic acid solution, 50% acetonitrile, 5% (v/v) formic acid, and acetonitrile. After drying, the tryptic peptides were resuspended in 0.5% aqueous trifluoroacetic acid. For MALDI-TOF mass spectrometry analyses, a 0.5-μl aliquot of peptide mixture was mixed with 0.5 μl of matrix solution (cyano-4-hydroxycinnamic acid at half-saturation in 60% acetonitrile, 0.1% trifluoroacetic acid (v/v)). The resulting solution was spotted on a MALDI-TOF target plate, dried, and rinsed with 2 μl of 0.1% trifluoroacetic acid. Peptide mixtures were then analyzed with a MALDI-TOF mass spectrometer (Autoflex, Bruker Daltonik, Germany) in reflector/delayed extraction mode over a mass range of 0–4200 Da. The spectra were annotated (XMass software), and the peptide mass fingerprints obtained were finally submitted to data base searches against the Swissprot Trembl data base with an intranet 1.9 version of MASCOT software.
Immunoprecipitation and Western Blotting—Twenty-four hours after transfection, HEK-293 or BHK-21 cells were lysed in RIPA buffer. The cell lysates were cleared by a 14,000 × g centrifugation for 15 min and incubated overnight at 4 °C with anti-FLAG monoclonal M2 antibody. Immune complexes were precipitated with protein G-Sepharose (Amersham Biosciences), and the beads were washed with RIPA buffer. Immunoprecipitated proteins were separated by 10% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Millipore). The membranes were blocked with 5% milk in TBS containing 0.1% Tween and incubated with the appropriate antibodies. The bands were revealed using the ECL detection reagent (Perbio).
In the case of immunoprecipitation between endogenous Alix and TNF-R1, HeLa cells were lysed in RIPA buffer. The cell lysates were cleared by a 14,000 × g centrifugation for 15 min followed by two incubations of 30 min with protein G-Sepharose beads. The cell lysates containing 5 mg of total proteins were incubated for 1 h at 4 °C with the polyclonal antibody against TNF-R1. Immune complexes were precipitated with protein G-Sepharose (Amersham Biosciences), and the beads were washed with RIPA buffer. Immunoprecipitated proteins were separated by 8% SDS-PAGE and treated as described above. Alix was detected using the monoclonal anti-AIP1/Alix from BD Transduction Laboratories.
Quantification of Cell Death Induced by TNF-R1—Twenty hours after transfection, HEK-293 were washed in PBS, pH 7.4, and fixed in 4% paraformaldehyde for 20 min at 4 °C. The cells were stained with polyclonal anti-TNF-R1 antibody (1/100) and anti-rabbit Alexa594 antibody (1/500). The cells were rinsed in TBS, incubated for 30 min at 37 °C in 2 μg/ml Hoechst 33342 (Sigma), and mounted in Mowiol. Cell viability was scored on the basis of nuclear morphology, with condensed or fragmented nuclei being counted as dead.
In some cases, tetrazolium salt 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma) was added to cells at a final concentration of 1 mg/ml and incubated for 30 min at 37 °C. Dimethyl sulfoxide was used to dissolve the formazan product, and absorbance was measured in each well at 540 nm, using a measure at 630 nm as reference, in a Biotek Elx-800 microplate reader (Mandel Scientific Inc.).
TNFα Internalization—BHK cells co-expressing TNF-R1 and the indicated proteins were incubated for 1 h at 4 °C with 1 ng of 125I-labeled human recombinant TNFα (specific activity, 2160 kBq/μg) (PerkinElmer Life Sciences). The cells were then incubated at 37 °C to allow endocytosis for the indicated times. The amount of intracellular 125I-TNF receptor complexes formed at 37 °C was estimated after washing cells for 5 min in cold acetic acid buffer (200 mm acetic acid, 500 mm NaCl, pH 2.5). After two washes in PBS, the cells were lysed in RIPA buffer. Total amount of cell-associated 125I-TNFα was determined on cells, washed only with PBS, instead of the acetic acid buffer. The amount of internalized (pH 2.5 resistant) 125I-TNF was calculated as a percentage of 125I-TNF bound at pH 7.4.
Magnetic Isolation of Endosomes Containing TNF-R1 Bound to TNFα—HEK 293 cells transfected with TNF-R1 and Alix-Myc were incubated in a total volume of 1 ml of cold D-PBS (0.9 mm CaCl2, 0.493 mm MgCl2, 2.67 mm KCl, 1.47 mm KH2PO4, 138 mm NaCl, 8 mm Na2HPO4) containing 3% bovine serum albumin, 100 ng/ml TNFα-FLAG, and 10 μg/ml anti-FLAG monoclonal M1 for 1 h at 4 °C. They were then washed twice in cold D-PBS and incubated for 1 h at 4 °C in 1 ml of cold D-PBS containing 50 μl of protein G microbeads (μMACS Protein G Microbeads, MACS Molecular, Miltenyi Biotec). The cells were then washed twice in cold D-PBS and incubated in Dulbecco's modified Eagle's medium containing 10% SVF and kept at 4 °C or incubated for 30 min at 37 °C. Removal of surface-bound M1 antibody was achieved by washing the cells three times for 5 min in cold PBS containing 1 mm EDTA. A post-nuclear supernatant was prepared in 8% sucrose supplemented in 3 mm imidazole, pH 7.4, and 2× protease inhibitor mixture. The magnetic immune complex was passed over a column placed in the magnetic field of a MACS Separator. The labeled TNFα receptosome was retained within the column, whereas unbound material was washed away with 8% sucrose, 3 mm imidazole, pH 7.4. The magnetic fractions were collected by removing the column from the magnetic field and analyzed by SDS-PAGE and Western blotting. Solubilization of endosomes was performed using a solution of 8% sucrose, 3 mm imidazole, pH 7.4, containing 0.5% Triton X-100.
RESULTS
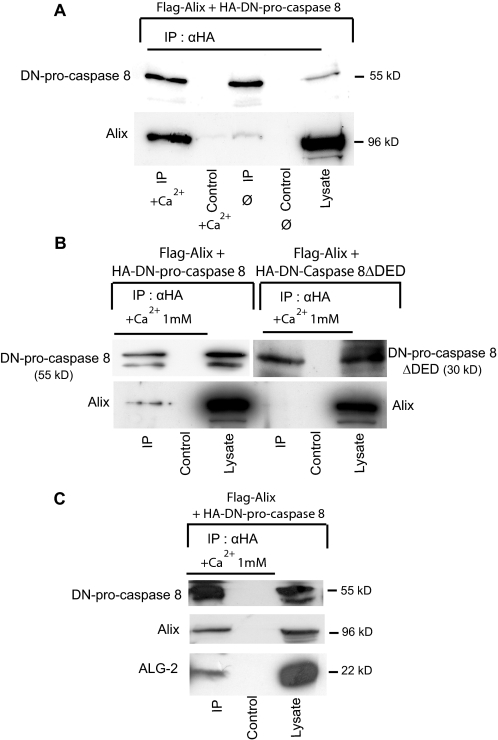
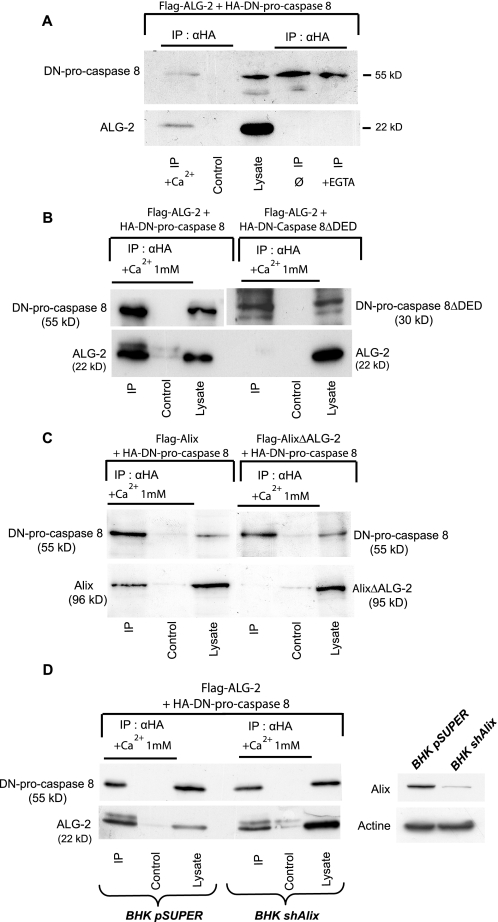
Alix Interacts with Pro-caspase-8 through ALG-2—To understand how Alix might control cell death, we first characterized some of the proteins interacting with it during apoptosis. For this, we used cerebellar granule cells, which survive in absence of serum when cultured in high extracellular potassium (25 mm) but undergo apoptosis soon after they are incubated in a medium containing normal extracellular potassium concentrations (5 mm). We previously observed that expression of Alix-CT blocks caspase activation in neurons deprived of potassium (17). We used a polyclonal anti-Alix antibody to immunoprecipitate the endogenous protein from cell lysates made from neurons incubated for 4 h in 5 mm potassium. Using peptide mass fingerprinting (see “Experimental Procedures”), we found caspase-8 among the proteins co-immunoprecipitated with Alix, suggesting that the protease may physically associate with Alix. We further showed that in BHK-21 cells, FLAG-tagged Alix (FLAG-Alix) and an HA-tagged, catalytically inactive version of pro-caspase-8 (HA-DN-pro-caspase-8) (Fig. 1) co-immunoprecipitated. This demonstrates that the two proteins exist in a complex and that activity of the caspase is not required for this interaction (Fig. 2A). Furthermore, deletion of the prodomain of pro-caspase-8 containing the two death effector domains (DEDs) abolished the capacity of the caspase to interact with Alix (Fig. 2B). The addition of 1 mm Ca2+ in the lysates strikingly enhanced the amount of Alix co-immunoprecipitating with pro-caspase-8 (Fig. 2A), and we detected endogenous ALG-2 in immunoprecipitates containing overexpressed Alix and pro-caspase-8 (Fig. 2C). This prompted us to test a potential interaction of pro-caspase-8 with ALG-2, whose binding to Alix depends on calcium. In the case where ALG-2 was co-expressed with DN-pro-caspase-8, ALG-2 co-immunoprecipitated with the caspase in presence of 1 mm CaCl2 (Fig. 3A). Here again, this interaction required the pro-domain of the caspase (Fig. 3B). FLAG-Alix deleted of the sequence 802PPYPTYPGYPGY813 necessary for the binding of ALG-2 (17) (AlixΔALG-2) (Fig. 1) did not immunoprecipitate with DN-pro-caspase-8 (Fig. 3C). On the other hand, ALG-2 could be co-immunoprecipitated with DN-pro-caspase-8 equally well from lysates of wt BHK cells or from cells in which Alix expression had been down-regulated using a pSuper vector coding for an small hairpin RNA against Alix, indicating that ALG-2 can interact with pro-caspase-8 independently of Alix (Fig. 3D).
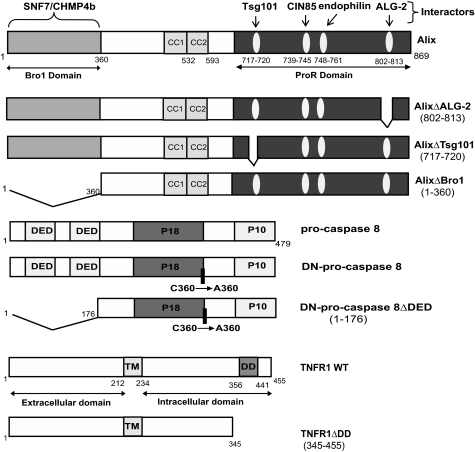
FIGURE 1.
Schematic representation of Alix pro-caspase-8 and TNF-R1 and their mutants used throughout this study.
FIGURE 2.
Alix co-immunoprecipitates with DN-pro-caspase-8. A, Alix/pro-caspase-8 co-IP requires calcium. BHK cells co-expressing FLAG-Alix and HA-DN-pro-caspase-8 were lysed and immunoprecipitated with anti-HA antibody in RIPA buffer supplemented or not with 1 mm CaCl2. Immunoprecipitates were blotted and analyzed with polyclonal antibodies against Alix (lower panel) and against HA (upper panel). B, co-IP of Alix with DN-pro-caspase-8 requires the pro-domain of the zymogen. BHK cells co-expressing FLAG-Alix together with HA-DN-pro-caspase-8 or HA-DN-caspase-8-ΔDED were lysed and immunoprecipitated with anti-HA in RIPA buffer containing 1 mm CaCl2 (CaRIPA) and analyzed as in A. C, endogenous ALG-2 is detected in pro-caspase-8/Alix co-immunoprecipitates. BHK cells co-expressing FLAG-Alix together with HA-DN-pro-caspase-8 were lysed and immunoprecipitated as in B; immunoprecipitates were analyzed by Western blot with polyclonal anti-HA, anti-ALG-2, and anti-FLAG antibodies (from top to bottom).
FIGURE 3.
ALG-2 co-immunoprecipitates with DN-pro-caspase-8. A, ALG-2/pro-caspase-8 co-IP requires calcium. BHK cells co-expressing FLAG-ALG-2 and HA-DN-pro-caspase-8 were lysed and immunoprecipitated with anti-HA in RIPA buffer (φ) or RIPA buffer containing either 3 mm EGTA or 1 mm CaCl2. Immunoprecipitates were analyzed using polyclonal antibodies against HA (upper panel) and FLAG (lower panel). B, co-IP of ALG-2 with DN-pro-caspase-8 requires the pro-domain of the zymogen. BHK cells co-expressing FLAG-ALG-2 together with HA-DN-pro-caspase-8 or HA-DN-caspase-8-ΔDED were lysed and immunoprecipitated with anti-HA in CaRIPA and analyzed as in A. C, co-IP of Alix with DN-pro-caspase-8 requires its ALG-2 binding domain. BHK cells were co-transfected with either FLAG-Alix WT or FLAG-AlixΔALG-2 and HA-DN-pro-caspase-8. IP with anti-HA antibody were performed in CaRIPA. Immunoprecipitates were analyzed by Western blot using polyclonal antibodies against HA (upper panel) and Alix (lower panel). D, ALG-2 co-immunoprecipitates with DN-pro-caspase-8 in cells depleted of Alix. BHK cells expressing shAlix to down-regulate expression of the protein were transfected with FLAG-ALG-2 and HA-DN-pro-caspase-8 lysed in CaRIPA and immunoprecipitated with an anti-HA antibody. Immunoprecipitates were analyzed by Western blot using anti-FLAG (lower panel) and HA polyclonal antibodies (upper panel). Right inset, Western blot analysis of lysates using a polyclonal antibody anti-Alix and a monoclonal anti-actin shows the decrease of endogenous Alix expression in BHK shAlix cells.
Alix Immunoprecipitates with TNF-R1 Independently of ALG-2—The demonstration that TNF-R1 needs endocytosis to recruit pro-caspase-8 prompted us to test whether Alix and ALG-2 act as adaptors bringing procaspase-8 into close vicinity of endosomes containing TNF-R1. First, we examined whether Alix and ALG-2 can form a complex with TNF-R1.
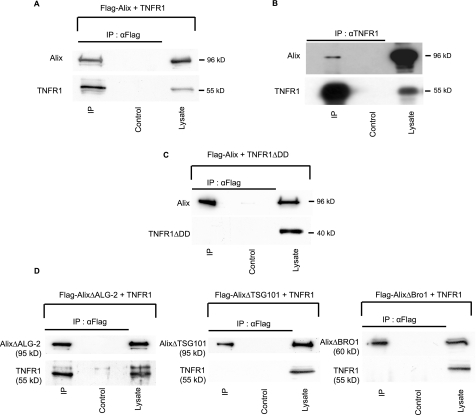
These experiments were performed in HEK 293 cells transiently transfected with expression vectors coding for TNF-R1 and Alix. As reported (21), TNF-R1 overexpression was sufficient to induce apoptosis, even in the absence of TNFα. Western blot analysis of Alix or TNF-R1 immunoprecipitates (Fig. 4A and not shown) revealed the presence of TNF-R1 and Alix, respectively, suggesting the existence of a complex containing both proteins. We further proved the existence of such a complex by showing that endogenously expressed Alix is pulled down with endogenous TNF-R1 immunoprecipitated from HeLa cells (Fig. 4B). In HEK 293 cells, TNF-R1 mutant deleted of the death domain (TNF-R1ΔDD) (Fig. 1) did not co-immunoprecipitate with Alix, indicating that the interaction depends on the integrity of this region (Fig. 4C). AlixΔALG-2, which does not interact with pro-caspase-8, was capable of co-immunoprecipitating with TNF-R1 (Fig. 4C). This underscores the fact that binding of Alix to a TNF-R1 complex is independent of ALG-2 and of pro-caspase-8. Interestingly, Alix mutants lacking the Bro1 domain, required for interaction with CHMP-4B, or four amino acids necessary for binding to Tsg101, were not capable of co-precipitating with TNF-R1 (Fig. 4D). Thus, co-immunoprecipitation of Alix with TNF-R1 requires its ability to bind ESCRT proteins.
FIGURE 4.
Alix co-immunoprecipitates with TNF-R1. A, Co-IP of overexpressed Alix and TNF-R1. RIPA lysates of HEK 293 cells co-expressing FLAG-Alix and TNF-R1 were immunoprecipitated with anti-FLAG antibodies and blots probed with anti-Alix (upper panel) and anti-TNF-R1 (lower panel). B, co-IP of endogenous Alix and TNF-R1. RIPA lysates of HeLa cells were immunoprecipitated using a polyclonal anti-TNF-R1. The IP was analyzed by Western blot using a monoclonal antibody against human Alix. C, co-IP of Alix with TNF-R1 requires the death domain of the receptor. RIPA lysates of HEK 293 cells co-expressing FLAG-Alix and TNF-R1 deleted from its death domain (TNF-R1ΔDD) were immunoprecipitated with anti-FLAG or with anti-TNF-R1, and blots were probed with the appropriate polyclonal antibodies as in A. D, co-immunoprecipitation of Alix with TNF-R1 requires its binding site to Tsg101 and Bro1 but not to ALG-2. Cells co-expressing TNF-R1 and either FLAG-AlixΔTsg101, FLAG-AlixΔBro1, or FLAG-AlixΔALG-2 were solubilized in RIPA buffer, and total lysates were immunoprecipitated with anti-FLAG; Western blots were probed with the appropriate polyclonal antibodies as in A and C.
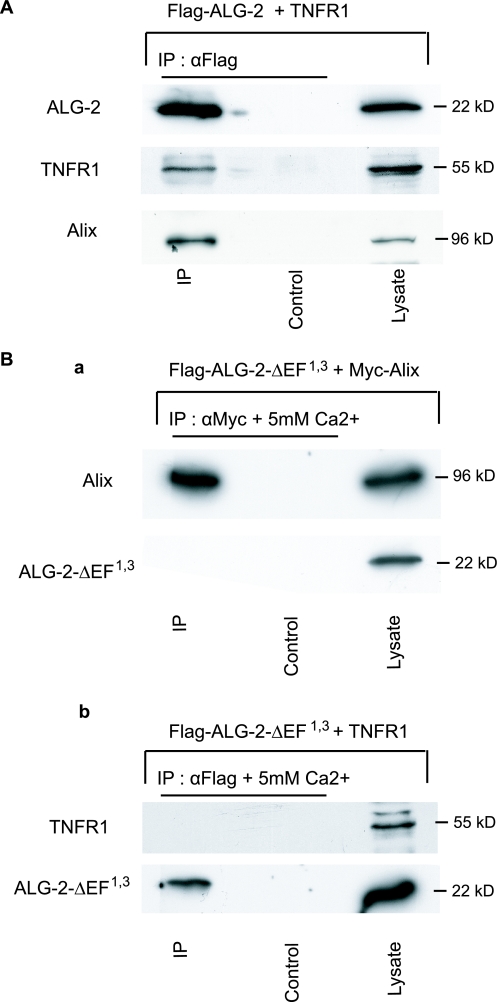
ALG-2 Immunoprecipitates with TNF-R1 in a Way That Tightly Depends on Its Capacity to Bind Calcium—We next asked whether ALG-2 can associate to TNF-R1. Lysates from HEK 293 cells co-transfected with FLAG-ALG-2 and wild type TNF-R1 expression vectors were immunoprecipitated with anti-FLAG (Fig. 5A) or with anti-TNF-R1 antibodies (not shown). In both cases, Western blots revealed co-immunoprecipitation of TNF-R1 with ALG-2. Furthermore, endogenous Alix was detected in the ALG-2 immunoprecipitates containing TNF-R1 (Fig. 5A).
FIGURE 5.
ALG-2 co-IP with TNF-R1 requires its capacity to bind calcium. A, ALG-2 co-IP with TNF-R1 and Alix. HEK293 cells were co-transfected with FLAG-ALG-2 and TNF-R1. RIPA cell lysates were immunoprecipitated with anti-FLAG or with anti-TNF-R1, and blots were probed with the appropriate antibodies. The bottom panel shows the presence of endogenous Alix within the immunoprecipitates as revealed with an anti-Alix antibody. B, calcium binding-deficient ALG-2 mutant does not co-IP with Alix (panel a) or with TNF-R1 (panel b). HEK293 cells co-expressing FLAG-Alix (panel a) or TNF-R1 (panel b) together with FLAG-ALG-2ΔEF1,3, a point mutant unable to bind Ca2+, were solubilized and immunoprecipitated in CaRIPA; Western blots were probed with the appropriate polyclonal antibodies.
We also used a mutated form of ALG-2, ALG-2ΔEF1,3, harboring point mutations in the first and third EF hands, which abolish its capacity to bind calcium. In accordance to published observations (22), this calcium binding-deficient ALG-2 did not co-immunoprecipitate with Alix (Fig. 5B, panel a). ALG-2ΔEF1,3 was also unable to interact with TNF-R1, stressing the fact that ALG-2 must be able to bind Ca2+ to co-immunoprecipitate with TNF-R1 (Fig. 5B, panel b). These results show that even though formation of a complex containing TNF-R1/Alix does not necessitate interaction with ALG-2, the latter bound to calcium can associate with it, possibly by binding to Alix.
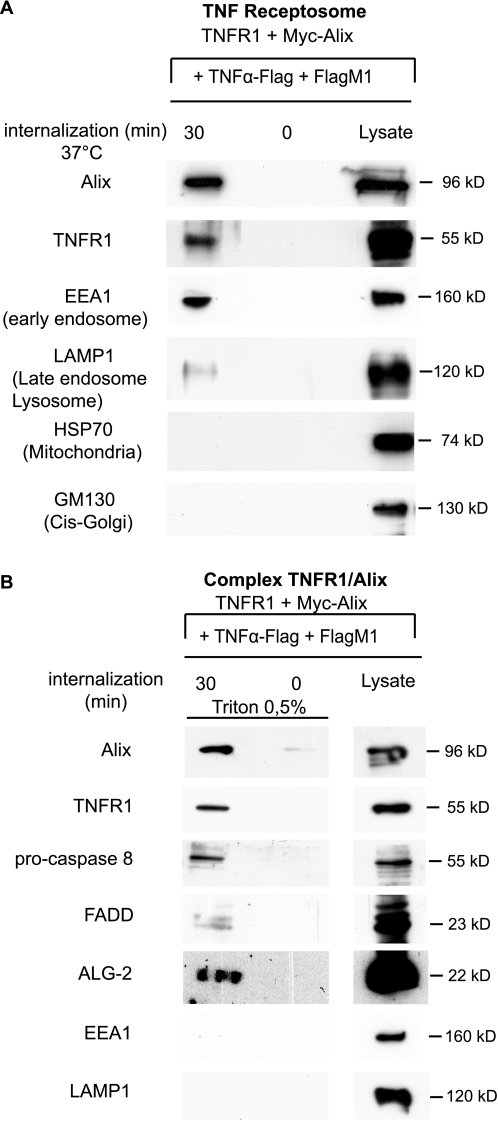
Alix Is Associated with TNF-R1 Receptosomes—Our observation that deletion of ESCRT-interacting domains from Alix impairs its capacity to bind to TNF-R1-containing complexes could suggest that interaction of the protein with the TNF-R1 complex occurs on endosomes. To examine this possibility, we used magnetic beads coupled to protein G to immunoisolate TNFα containing endosomes, a method modified from that published by Schütze and co-workers (3); FLAG-tagged TNFα was applied to HEK 293 cells overexpressing TNF-R1 at 4 °C in presence of M1, an anti-FLAG antibody binding only in presence of calcium. The cells were incubated at 37 °C for 30 min to allow endocytosis and then treated briefly with EDTA to remove surface-bound antibody. The resulting immunoisolated membrane fractions contained TNF-R1, two markers of endosomes, EEA1 and Lamp-1, but neither the mitochondrial marker HSP70, nor cis-Golgi marker GM130, stressing the endosomal origin of these fractions (Fig. 6A). None of the proteins tested could be detected in magnetic particles recovered from cells left at 4 °C and treated with EDTA (Fig. 6A, lanes 0), thus demonstrating the efficiency of the washing procedure for removing the M1 anti-FLAG antibody from TNFα bound to TNF-R1 remaining on the cell surface.
FIGURE 6.
Alix and ALG-2 are present with pro-caspase-8 and FADD on TNF receptosomes. A, Western blot analysis of FLAG-TNFα containing endosomes prepared from HEK 293, over-expressing TNF-R1 and Myc-Alix. Lane 0, cells maintained at 4 °C were washed with EDTA prior to endosome preparation. Lane 30, cells incubated for 30 min at 37 °C to induce endocytosis of TNFα bound to TNF-R1 before lysis. Magnet-isolated endosomes were run on SDS-PAGE and immunoblotted to reveal the presence of EEA1 (early endosomes), LAMP1 (late endosomes and lysosomes), GM130 (cis-Golgi), or HSP70 (mitochondria). B, solubilization of TNF-R1 receptosomes leaves intact a complex containing Alix, ALG-2, FADD, and pro-caspase-8. TNF-R1 receptosomes prepared as in A were solubilized on the affinity column using 0.5% Triton X-100 and analyzed by Western blotting using the appropriate antibodies. The lack of EEA1 and LAMP1 confirms membrane solubilization of TNF receptosomes.
Triton X-100 solubilization of the endosomal membranes, witnessed by the loss of EEA1 and Lamp-1 labeling, left intact TNF-R1, Alix, ALG-2, pro-caspase-8, and FADD immunoreactivity (Fig. 6B). This result indicates that these proteins interact within complexes containing endocytosed TNF-R1 bound to TNFα.
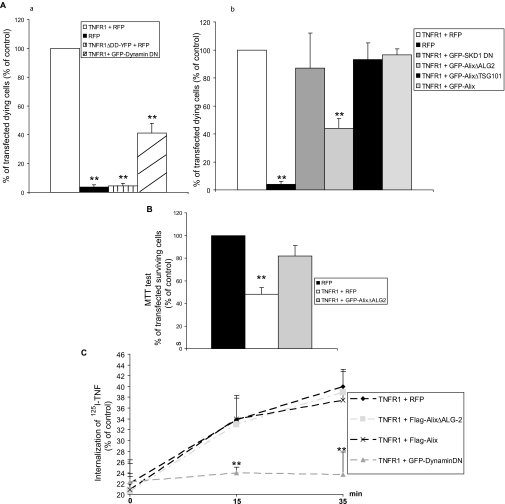
AlixΔALG-2 Protects HEK 293 Cells from TNF-R1-induced Cell Death without Affecting Internalization of the Receptor—HEK 293 cells die soon after transfection with an expression vector coding for TNF-R1 but not with one encoding TNF-R1ΔDD (Fig. 7A, panel a). Blocking clathrin-dependent endocytosis by a dominant negative mutant of dynamin 2a significantly reduced TNF-R1-induced cell death (Fig. 7A, panel a). AlixΔALG-2, which interacts with TNF-R1 but not with procaspase-8, inhibited cell death induced by TNF-R1, estimated by Hoechst nuclear staining (Fig. 7A, panel b) and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (Fig. 7B). Alix-wt or AlixΔTSG101, which cannot interact with TNF-R1, did not protect against TNF-R1-induced cell death (Fig. 7A, panel b).
FIGURE 7.
AlixΔALG-2 protects HEK293 from TNF-R1-induced cell death without impairing internalization of the receptor. A, TNF-R1 overexpression triggers apoptosis, which is inhibited by AlixΔALG-2 but not by AlixΔTsg101 or SKD1 DN. Panels a and b, HEK 293 cells transfected with expression vectors coding for RFP or TNF-R1 and the indicated proteins. Viability was scored on the basis of nuclear morphology; cells with condensed or fragmented nuclei were counted as dead. The values are the means ± S.D. of dying cells from three independent experiments. **, t test, p < 0.001. B, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide metabolic colorimetric test confirms the protective effect of AlixΔALG-2. Each value represents the mean ± S.D. of triplicate wells from three independent experiments. **, t test, p < 0.001. C, TNF-R1/TNFα internalization is impaired by a dominant negative form of dynamin but not by Alix or AlixΔALG-2. TNF-R1 internalization was analyzed using [125I]TNFα in TNF-R1 expressing cells. Each point represents the mean ± S.D. of triplicate wells from three independent experiments. **, t test, p < 0.001.
Using 125I-radiolabeled TNFα, we found no difference in TNFα internalization between TNF-R1 overexpressing cells co-transfected with control plasmid, Alix, or AlixΔALG-2, whereas internalization was abolished by co-transfection with a dominant negative mutant of dynamin 2a (Fig. 7C). These results indicate that the protective effect of AlixΔALG-2 is not due to blockade of receptor internalization.
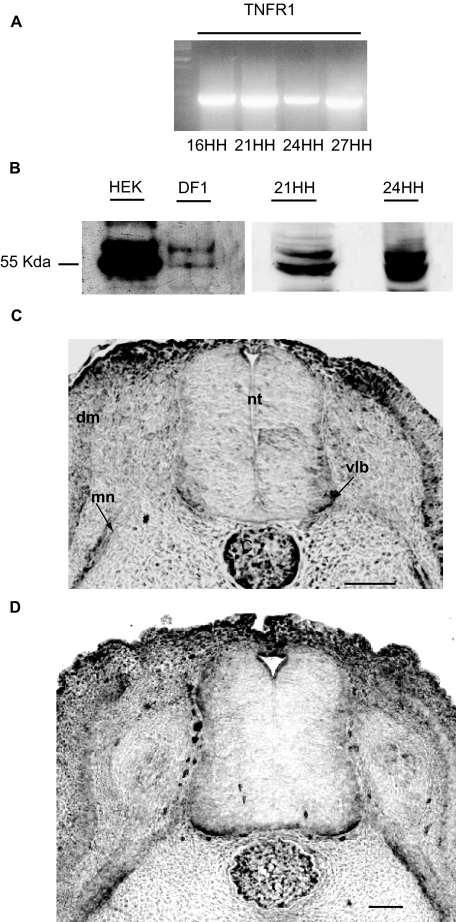
AlixΔALG-2 Blocks Cell Death of Cervical Motoneurons, Which Is Regulated by TNF-R1—We finally tested whether the participation of Alix and ALG-2 in the TNF-R1 death pathway demonstrated in vitro may also apply in vivo. For this we searched whether programmed cell death of cervical motoneurons (MTN) in the chick embryo, which is efficiently blocked by expression of AlixΔALG-2 (18), depends on TNF-R1. Reverse transcription-PCR analysis revealed expression of the TNF-R1 mRNA in whole chick embryo extracts from HH stage 16 to HH stage 27 (Fig. 8A). We next used a polyclonal antibody, raised against the extracellular part of human TNF-R1, which we verified to recognize the chick counterpart (40.5% identity on the 222 extracellular amino acids). Indeed, the antibody revealed bands at the expected 55 kDa in blots prepared from chick fibroblasts (DF1 cell line) and chick embryo extracts (Fig. 8B). Immunostaining revealed expression of TNF-R1 in the dermomyotome but not in the neural tube of HH stage 16 chick embryo. Labeling of the ventral-most part of the ventral horn and of MTN axons growing out of the tube was detected at HH stages 21 and 24 (Fig. 8, C and D) and decreased at HH stage 27 (not shown). Taken together, these results show that TNF-R1 is transiently expressed in MTN during a short period corresponding to that of programmed cell death. In comparison, Alix is expressed in the same areas of the neural tube but appearing earlier (HH stage 16) and for longer during development (23).
FIGURE 8.
TNF-R1 is expressed during chick neural tube development. A, reverse transcription-PCR analysis shows gene expression in whole extracts from chick embryos from HH stage 16 to HH stage 27. B, polyclonal anti-TNF-R1 recognizes a main band at 55 kDa in HEK cells and a doublet with the lower band corresponding to 55 kDa in DF1 chick fibroblasts. A similar pattern is observed in chick embryo extracts (21HH, 24HH). C and D, cross-sections of HH stage 21 (C) or 24 (D) chick embryo neural tubes immunostained with polyclonal anti-TNF-R1. Scale bar, 50 μm. C, cord; dm, dermomyotome; mn, motoneuron axons; nt, neural tube; vlb, ventro-lateral border.
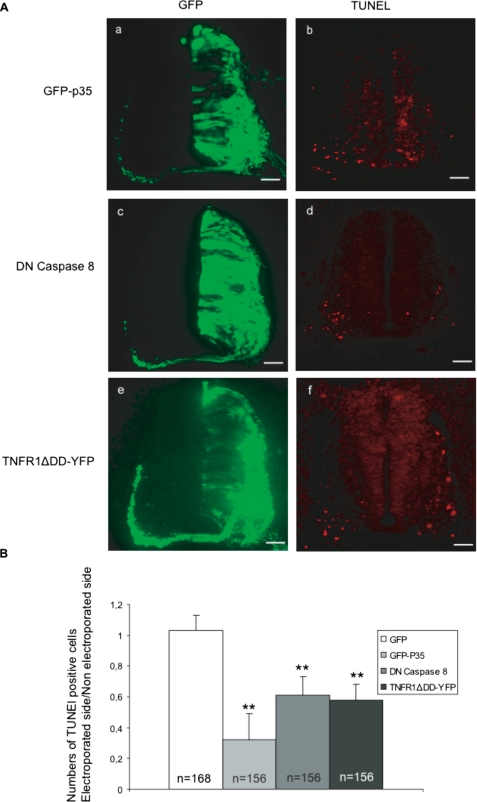
We first tested the involvement of caspase-8 during death of MTN, using electroporation of the chick neural tube. This technique allows expression of the protein of interest in one half of the embryo, the other half being left intact for comparison. p35, a baculovirus protein inhibiting most caspases (24), was electroporated into the cervical neural tube of HH stage 16 embryos, which were sacrificed 48 h later (Fig. 9A, panel a). Comparison between transfected and non transfected ventral horns of the same embryo revealed a reduction by half of the number of TUNEL positive neurons in the p35 expressing ventral horn (Fig. 9, A, panel b, and B). Using the same technique, we found that expression of DN-procaspase-8 (Fig. 9, A, panels c and d, and B) also reduced the number of dying neurons to a similar degree as AlixΔALG-2 (18). This indicates a role of Alix/ALG-2 and of the death receptor-regulated apical caspase-8 in MTN programmed cell death during development.
FIGURE 9.
TNF-R1 is involved in early cell death of cervical motoneurons. A, cross-sections of HH stage 24 chick embryo neural tubes electroporated with pCAGGS expression vectors coding for: GFP-p35 (panels a and b), DN-pro-caspase-8 (panels c and d), TNFR1ΔDD-YFP (panels e and f). Panels a, c, and e, GFP expression 48 h after electroporation. Panels b, d, and f, TUNEL labeling of adjacent sections. Scale bar, 50 μm. B, ratio of TUNEL-positive cells in the electroporated versus nonelectroporated side of the neural tube. The mean numbers ± S.D. of TUNEL-positive cells in the neural tube of electroporated embryos, n = number of sections analyzed; 12 sections/embryo were counted. **, t test, p < 0.001.
Overexpression of human wild type TNF-R1 is sufficient, like Alix overexpression, to induce massive apoptosis throughout the neural tube (data not shown). On the contrary, expression of TNF-R1ΔDD halved the number of TUNEL-positive nuclei in the ventral part of the tube (Fig. 9, A, panels e and f, and B). This, together with our demonstration that the endogenous receptor is expressed at the time of programmed cell death, indicates that activation of TNF-R1 regulates the death of cervical motoneurons. Taken together, these in vivo data strongly suggest that AlixΔALG-2 blocks programmed cell death by interfering with TNF-R1 death signaling.
DISCUSSION
We previously showed that overexpressed Alix is sufficient to activate caspases and apoptosis and that this activity requires binding to ALG-2. On the contrary, expression of the C-terminal half of Alix blocked apoptosis depending on its capacity to bind ALG-2, suggesting that it acts by titrating out ALG-2 (17, 18). Interestingly, this truncated form of Alix accumulates in cytoplasmic inclusions, which also contain ALG-2 and activated caspases (17). Rao et al. (25) have shown that ALG-2 is necessary for caspase activation following an abnormal rise in cytosolic calcium. Their observations suggested that ALG-2 belongs to a complex allowing caspase-9 activation following cytosolic calcium elevation because of a stress to the reticulum. Our present results demonstrate that ALG-2 can also form a complex with pro-caspase-8, independently of Alix, and that this requires the prodomain of the zymogen but not the catalytic activity of the protease.
Historically, caspase-8 was described as an initiator caspase activated by members of the TNF-R1 family of death receptors stimulating the extrinsic pathway of apoptosis (26). These latter recruit, through an 80-amino acid-long death domain (DD), other DD-containing cytoplasmic proteins that participate in the formation of the death-inducing signaling complex (DISC) recruiting and activating pro-caspase-8 or 10. We observed that the DD of TNF-R1 is mandatory for the interaction with Alix and therefore with the Alix/ALG-2 complex. The TNFα ligand is known to induce, via the intracellular domain of the receptor, the rapid formation of complex I made of the adaptor TRADD, the protein kinase RIP1, and the signal transducer TRAF2. This complex signals cell survival through c-Jun N-terminal kinase (JNK) and IκB kinase, the latter activating transcription factor NFκB, whose targets can mediate cell survival. Complex I can dissociate from the receptor and, together with the adaptor protein FADD, recruit and activate pro-caspase-8 (27). Schütze and co-workers (3) introduced a twist to this model when they published convincing data showing that aggregation of TRADD, FADD, and caspase-8 can occur on TNF-R1 but that this is critically dependent on receptor endocytosis. In line with this, we observed that blocking clathrin-dependent endocytosis significantly inhibited apoptosis induced by TNF-R1. In their model, Schneider-Brachert et al. (3) claim that TNF-R1-containing endosomes would establish the platform on which the DISC may form. Because Alix can bind TNF-R1 on one side and ALG-2, which can interact with caspase-8, on the other, it may act as an adaptor for recruiting pro-caspase-8 on the surface of TNF-R1-containing endosomes. In favor of this model, we found that binding of Alix to TNF-R1 is totally abolished upon deletion of its binding domains to CHMP-4B and Tsg-101 of ESCRT-III and ESCRT-I, respectively, suggesting that stabilization of Alix binding to the TNF-R1 complex occurs through interactions with these endosomal proteins. Furthermore we could identify TNF-R1, Alix, ALG-2, caspase-8, and FADD within a common, Triton X-100-resistant complex in endosomal fractions containing TNFα bound to its endocytosed receptor. Thus, AlixΔALG-2 may block death induced by TNF-R1 by impairing the recruitment of pro-caspase-8 onto surface of TNF-R1-containing endosomes. The protecting effect of AlixΔALG-2 is not simply due to blocking of endocytosis of the activated receptor, because the latter process is not influenced by the mutant. Nor is it due to an impairment of the degradation of the receptor inside lysosomes because the dead version of the ATPase SKD-1, which leads to gross abnormalities in multivesicular endosomes (28), had no protecting activity against TNF-R1.
We have tested the effect of Alix/ALG-2 down-regulation on HeLa cells treated with 100 ng/ml of TNFα and 1 mg/ml cycloheximide and on Jurkat cells in which the NFκB survival pathway is inhibited by knockdown of the IKK Nemo (a kind gift of O. Micheau, Dijon, France; data not shown).
Down-regulation of Alix or ALG-2 did not afford statistically significant protection against TNF-R1-induced cell death in either case. The apparent discrepancy between these small interfering RNA results and those using mutant expressions might have two explanations: 1) Alix down-regulation was never complete, being reduced by 90% at best, and ALG-2 extinction never attained more than 50%. Thus, the remaining Alix and ALG-2 might be sufficient for recruiting and activating caspase-8. 2) In the absence of Alix/ALG-2, other proteins might allow the recruitment of the caspase onto TNF-R1-containing endosomes. A similar argument could be offered to explain why down-regulating FADD has no protective effect on TNF-R1-induced cell death (29). The fact that FADD is still widely accepted as being essential for TNF-R1-induced cell death, even after the very solid demonstration of Jin and El-Deiry, begs caution in the use of small interfering RNAs in the field of caspase activation.
We strengthened the notion of a role of Alix/ALG-2 in regulating TNF-R1-induced cell death, by demonstrating that programmed cell death of chick cervical MTN, which we had previously found to be inhibited by AlixΔALG-2 (18), is driven by TNF-R1. Among death receptors of the TNF-R family, P75NGFR, Fas, and TNF-R1 (30) have been postulated to play an active role during MTN developmental death. Sedel et al. (31) have shown that death of an embryonic day 12–13 rat MTN is regulated by TNFα and that death of mouse MTN is significantly reduced in double knock-out for TNFα and TNF-R1. In the chick embryo, TNFα was detected by immunohistochemistry from HH stage 18 (embryonic day 3) with a peak at HH stage 24–25 (embryonic day 4.5–5) (32), and we found that TNF-R1 is transiently expressed during programmed cell death of cervical MTN with a pattern reminiscent of that of Alix. The TNF-R1 pathway seems to be instrumental in controlling death of the MTN during this developmental period; indeed expression of TNF-R1ΔDD or DN-procaspase-8, which both act as dominant negative mutants of the cognate proteins, reduced apoptosis of the MTN as much as the pan-caspase inhibitor p35. The fact that AlixΔALG-2 confers a similar protection of the same MTN (18) strongly suggests that this protection is due to a modification of the TNF-R1 pathway in ovo. It is here noteworthy that CIN85, another interactor of Alix, has been reported to be involved in TNF-R1 signaling (33), and we have observed that deletion of the CIN85-binding site impairs the death blocking activity of the C-terminal half of Alix in chick MTN.6
de Gassart et al. (34) have reported caspase activity in exosomes that correspond to intralumenal vesicles of MVBs and are enriched in Alix. Exosomes also bear TNF-R1, indicating that the receptor is a cargo of the intralumenal vesicles, but without its interactors TRADD, RIP, and TRAF2 (35). It may therefore be speculated that Alix and ALG-2, which accompany and get entrapped into vesicles budding inside MVBs, could pull TNF-R1 and caspase-8 inside endosomes. This would efficiently isolate the activated caspase from its cytosolic substrates and lead to its rapid degradation inside lysosomes fusing with MVBs. Thus, one reason for why activation of pro-caspase-8 in the vicinity of Alix and ALG-2 occurs on the surface of endosomes containing TNF-R1 is that it may allow a potent control to rapidly tune down caspase-8 activity.
Because ALG-2 binding to Alix is strictly calcium-dependent, our results also beg the question of the relationship between cytosolic calcium and TNF-R1. The present literature remains controversial about the role of cytosolic calcium in TNFα-induced cell death. Some investigators have failed to find evidence for a TNFα-induced calcium response in U937 monocytes (36) or in KYM-1 and HeLa cells overexpressing the TNF receptor (37). In contrast, Bellomo et al. (38) and Kong et al. (39) found an increase in intracellular calcium using BT-20 and L929 cells, respectively. More recently, Draper et al. (40) found a sustained rise in cytosolic calcium following cycloheximide/TNFα treatment of C3HA fibroblast cells. Fas, which also requires endocytosis for caspase-8 activation, has been shown to induce calcium release from internal stores. It is also noteworthy that endosomes are known to contain high concentrations of calcium and calcium leakage from endosomes regulates their maturation (41). Thus, a rise in calcium in the close vicinity of endosomes might allow ALG-2 to bind to Alix and recruit caspase-8 on the surface of endosomes. This is even more relevant considering that Fas-mediated apoptosis requires calcium release from the endoplasmic reticulum (42), and we will now test whether Fas-mediated cell death also involves Alix/ALG-2 during activation of caspase-8. We will also need to examine whether the interaction of ALG-2 and Alix to pro-caspase-8 and TNF-R1, respectively, is direct or through proteins of the DISC. Interactions within this complex are homophilic between DD or DED. We showed that the DD of TNF-R1 and the DED containing pro-domain of pro-caspase 8 are obligatory for binding to Alix and ALG-2, respectively, and that FADD is present in the Alix/ALG-2/pro-caspase-8 complex. Because apparently, neither Alix nor ALG-2 contain death adaptor domains, we now need to characterize the new module that allows their interaction to the DISC.
Acknowledgments
We thank Patrick Mehlen, Wulf Schneider-Brachert, and Masatoshi Maki for reagents; Yves Goldberg, Karin Sadoul, and Fiona Hemming for critical reading of the manuscript and numerous suggestions throughout this work; Gilles Bodon for help with the receptosome experiment; and Thomas Mellier for patience.
This work was supported in part by INSERM, the University Joseph Fourier, and grants from the Association Française contre les Myopathies, the Association pour la Recherche sur la Sclérose Latérale Amyotrophique, and the Association pour la Recherche contre le Cancer. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MVB, multivesicular body; DD, death domain; DED, death effector domain; HH, Hamburger-Hamilton; TNF, tumor necrosis factor; TNF-R1, TNF receptor 1; TUNEL, terminal transferase dUTP nick end labeling; HA, hemagglutinin; wt, wild type; RIPA, radioimmune precipitation assay; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; TBS, Tris-buffered saline; PBS, phosphate-buffered saline; MTN, motoneurons; DISC, death-inducing signaling complex; CaRIPA, RIPA buffer containing 1 mm CaCl2; IP, immunoprecipitation; FADD, Fas-associated protein with death domain.
A.-L. Mahul-Mellier, unpublished observation.
References
- 1.van der Goot, F. G., and Gruenberg, J. (2006) Trends Cell Biol. 16 514-521 [DOI] [PubMed] [Google Scholar]
- 2.Barker, P. A., Hussain, N. K., and McPherson, P. S. (2002) Trends Neurosci. 25 379-381 [DOI] [PubMed] [Google Scholar]
- 3.Schneider-Brachert, W., Tchikov, V., Neumeyer, J., Jakob, M., Winoto-Morbach, S., Held-Feindt, J., Heinrich, M., Merkel, O., Ehrenschwender, M., Adam, D., Mentlein, R., Kabelitz, D., and Schütze, S. (2004) Immunity 21 415-428 [DOI] [PubMed] [Google Scholar]
- 4.Williams, R. L., and Urbe, S. (2007) Nat. Rev. Mol. Cell. Biol. 8 355-368 [DOI] [PubMed] [Google Scholar]
- 5.Missotten, M., Nichols, A., Rieger, K., and Sadoul, R. (1999) Cell Death Differ. 6 124-129 [DOI] [PubMed] [Google Scholar]
- 6.Vito, P., Lacana, E., and D'Adamio, L. (1996) Science 271 521-525 [DOI] [PubMed] [Google Scholar]
- 7.Vito, P., Pellegrini, L., Guiet, C., and D'Adamio, L. (1999) J. Biol. Chem. 274 1533-1540 [DOI] [PubMed] [Google Scholar]
- 8.Strack, B., Calistri, A., Craig, S., Popova, E., and Gottlinger, H. G. (2003) Cell 114 689-699 [DOI] [PubMed] [Google Scholar]
- 9.von Schwedler, U. K., Stuchell, M., Muller, B., Ward, D. M., Chung, H. Y., Morita, E., Wang, H. E., Davis, T., He, G. P., Cimbora, D. M., Scott, A., Krausslich, H. G., Kaplan, J., Morham, S. G., and Sundquist, W. I. (2003) Cell 114 701-713 [DOI] [PubMed] [Google Scholar]
- 10.Carlton, J. G., and Martin-Serrano, J. (2007) Science 316 1908-1912 [DOI] [PubMed] [Google Scholar]
- 11.Morita, E., Sandrin, V., Chung, H. Y., Morham, S. G., Gygi, S. P., Rodesch, C. K., and Sundquist, W. I. (2007) EMBO J. 13 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuo, H., Chevallier, J., Mayran, N., Le Blanc, I., Ferguson, C., Faure, J., Blanc, N. S., Matile, S., Dubochet, J., Sadoul, R., Parton, R. G., Vilbois, F., and Gruenberg, J. (2004) Science 303 531-534 [DOI] [PubMed] [Google Scholar]
- 13.Chatellard-Causse, C., Blot, B., Cristina, N., Torch, S., Missotten, M., and Sadoul, R. (2002) J. Biol. Chem. 277 29108-29115 [DOI] [PubMed] [Google Scholar]
- 14.Chen, B., Borinstein, S. C., Gillis, J., Sykes, V. W., and Bogler, O. (2000) J. Biol. Chem. 275 19275-19281 [DOI] [PubMed] [Google Scholar]
- 15.Cabezas, A., Bache, K. G., Brech, A., and Stenmark, H. (2005) J. Cell Sci. 118 2625-2635 [DOI] [PubMed] [Google Scholar]
- 16.Schmidt, M. H., Hoeller, D., Yu, J., Furnari, F. B., Cavenee, W. K., Dikic, I., and Bogler, O. (2004) Mol. Cell. Biol. 24 8981-8993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trioulier, Y., Torch, S., Blot, B., Cristina, N., Chatellard-Causse, C., Verna, J. M., and Sadoul, R. (2004) J. Biol. Chem. 279 2046-2052 [DOI] [PubMed] [Google Scholar]
- 18.Mahul-Mellier, A.-L., Hemming, F. J., Blot, B., Fraboulet, S., and Sadoul, R. (2006) J. Neurosci. 26 542-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadoul, R. (2006) Biol. Cell 98 69-77 [DOI] [PubMed] [Google Scholar]
- 20.Ferro, M., Seigneurin-Berny, D., Rolland, N., Chapel, A., Salvi, D., Garin, J., and Joyard, J. (2000) Electrophoresis 21 3517-3526 [DOI] [PubMed] [Google Scholar]
- 21.Todd, I., Radford, P. M., Draper-Morgan, K. A., McIntosh, R., Bainbridge, S., Dickinson, P., Jamhawi, L., Sansaridis, M., Huggins, M. L., Tighe, P. J., and Powell, R. J. (2004) Immunology 113 65-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibata, H., Yamada, K., Mizuno, T., Yorikawa, C., Takahashi, H., Satoh, H., Kitaura, Y., and Maki, M. (2004) J. Biochem. (Tokyo) 135 117-128 [DOI] [PubMed] [Google Scholar]
- 23.Fraboulet, S., Hemming, F. J., Mahul, A. L., Cristina, N., and Sadoul, R. (2003) Gene Expr. Patterns 3 139-142 [DOI] [PubMed] [Google Scholar]
- 24.Bump, N. J., Hackett, M., Hugunin, M., Seshagiri, S., Brady, K., Chen, P., Ferenz, C., Franklin, S., Ghayur, T., Li, P., Licari, P., Mankovich, J., Shi, L., Greenberg, A. H., Miller, L. K., and Wong, W. W. (1995) Science 269 1885-1888 [DOI] [PubMed] [Google Scholar]
- 25.Rao, R. V., Poksay, K. S., Castro-Obregon, S., Schilling, B., Row, R. H., del Rio, G., Gibson, B. W., Ellerby, H. M., and Bredesen, D. E. (2004) J. Biol. Chem. 279 177-187 [DOI] [PubMed] [Google Scholar]
- 26.Muzio, M., Chinnaiyan, A. M., Kischkel, F. C., O'Rourke, K., Shevchenko, A., Ni, J., Scaffidi, C., Bretz, J. D., Zhang, M., Gentz, R., Mann, M., Krammer, P. H., Peter, M. E., and Dixit, V. M. (1996) Cell 85 817-827 [DOI] [PubMed] [Google Scholar]
- 27.Micheau, O., and Tschopp, J. (2003) Cell 114 181-190 [DOI] [PubMed] [Google Scholar]
- 28.Fujita, H., Yamanaka, M., Imamura, K., Tanaka, Y., Nara, A., Yoshimori, T., Yokota, S., and Himeno, M. (2003) J. Cell Sci. 116 401-414 [DOI] [PubMed] [Google Scholar]
- 29.Jin, Z., and El-Deiry, W. S. (2006) Mol. Cell. Biol. 26 8136-8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raoul, C., Pettmann, B., and Henderson, C. E. (2000) Curr. Opin. Neurobiol. 10 111-117 [DOI] [PubMed] [Google Scholar]
- 31.Sedel, F., Bechade, C., Vyas, S., and Triller, A. (2004) J. Neurosci. 24 2236-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wride, M. A., and Sanders, E. J. (1995) Anat. Embryol. (Berl.) 191 1-10 [DOI] [PubMed] [Google Scholar]
- 33.Narita, T., Nishimura, T., Yoshizaki, K., and Taniyama, T. (2005) Exp. Cell Res. 304 256-264 [DOI] [PubMed] [Google Scholar]
- 34.de Gassart, A., Geminard, C., Fevrier, B., Raposo, G., and Vidal, M. (2003) Blood 102 4336-4344 [DOI] [PubMed] [Google Scholar]
- 35.Hawari, F. I., Rouhani, F. N., Cui, X., Yu, Z. X., Buckley, C., Kaler, M., and Levine, S. J. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 1297-1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasegawa, Y., and Bonavida, B. (1989) J. Immunol. 142 2670-2676 [PubMed] [Google Scholar]
- 37.McFarlane, S. M., Anderson, H. M., Tucker, S. J., Jupp, O. J., and MacEwan, D. J. (2000) Mol. Cell Biochem. 211 19-26 [DOI] [PubMed] [Google Scholar]
- 38.Bellomo, G., Perotti, M., Taddei, F., Mirabelli, F., Finardi, G., Nicotera, P., and Orrenius, S. (1992) Cancer Res. 52 1342-1346 [PubMed] [Google Scholar]
- 39.Kong, S. K., Fung, K. P., Choy, Y. M., and Lee, C. Y. (1997) Oncology 54 55-62 [DOI] [PubMed] [Google Scholar]
- 40.Draper, D. W., Harris, V. G., Culver, C. A., and Laster, S. M. (2004) J. Immunol. 172 2416-2423 [DOI] [PubMed] [Google Scholar]
- 41.Saito, M., Hanson, P. I., and Schlesinger, P. (2007) J. Biol. Chem. 282 27327-27333 [DOI] [PubMed] [Google Scholar]
- 42.Wozniak, A. L., Wang, X., Stieren, E. S., Scarbrough, S. G., Elferink, C. J., and Boehning, D. (2006) J. Cell Biol. 175 709-714 [DOI] [PMC free article] [PubMed] [Google Scholar]