Abstract
We report an analysis of the reaction mechanism of ornithine 4,5-aminomutase, an adenosylcobalamin (AdoCbl)- and pyridoxal l-phosphate (PLP)-dependent enzyme that catalyzes the 1,2-rearrangement of the terminal amino group of d-ornithine to generate (2R,4S)-2,4-diaminopentanoic acid. We show by stopped-flow absorbance studies that binding of the substrate d-ornithine or the substrate analogue d-2,4-diaminobutryic acid (DAB) induces rapid homolysis of the AdoCbl Co–C bond (781 s–1, d-ornithine; 513 s–1, DAB). However, only DAB results in the stable formation of a cob(II)alamin species. EPR spectra of DAB and [2,4,4-2H3]DAB bound to holo-ornithine 4,5-aminomutase suggests strong electronic coupling between cob(II)alamin and a radical form of the substrate analog. Loading of substrate/analogue onto PLP (i.e. formation of an external aldimine) is also rapid (532 s–1, d-ornithine; 488 s–1, DAB). In AdoCbl-depleted enzyme, formation of the external aldimine occurs over long time scales (∼50 s) and occurs in three resolvable kinetic phases, identifying four distinct spectral intermediates (termed A–D). We infer that these represent the internal aldimine (λmax 416 nm; A), two different unliganded PLP states of the enzyme (λmax at 409 nm; B and C), and the external aldimine (λmax 426 nm; D). An imine linkage with d-ornithine and DAB generates both tautomeric forms of the external aldimine, but with d-ornithine the equilibrium is shifted toward the ketoimine state. The influence of this equilibrium distribution of prototropic isomers in driving homolysis and stabilizing radical intermediate states is discussed. Our work provides the first detailed analysis of radical-based catalysis in this Class III AdoCbl-dependent enzyme.
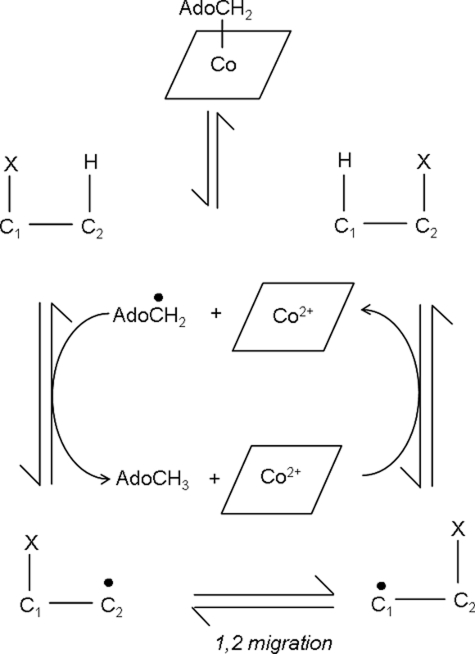
Adenosylcobalamin (AdoCbl3; coenzyme B12) serves as a radical repository for a group of enzymes that catalyze unusual isomerizations, whereby a hydrogen atom (H) is interchanged with an electron-withdrawing group (X) on a neighboring carbon atom (Scheme 1) (1–4). To date, 11 AdoCbl-dependent isomerases have been identified and grouped into three classes: (i) Class I (mutases) that catalyze carbon skeletal rearrangements; (ii) Class II (eliminases) that catalyze, with one exception, the migration and elimination of a heteroatom; and (iii) Class III (aminomutases) that catalyze intramolecular 1,2-amino shifts. Turnover for all AdoCbl-dependent isomerases begins with substrate-induced homolysis of the AdoCbl Co–C bond and formation of two paramagnetic centers: the 5′-deoxyadenosyl radical (Ado•) and cob(II)alamin. The highly reactive Ado• species propagates radical formation by abstracting a hydrogen atom from the substrate (or an amino acid side chain in the case of ribonucleotide reductase) (5), generating deoxyadenosine and a substrate radical. The latter carbon-centered radical isomerizes to a product radical intermediate, which then reabstracts a hydrogen atom to form Ado•. Geminate recombination between Ado• and cob(II)alamin regenerates AdoCbl and primes the enzyme for another catalytic cycle.
SCHEME 1.
Minimal reaction mechanism for AdoCbl-isomerase catalysis.
Studies on AdoCbl-dependent isomerases have shown that the first step in the catalytic cycle (homolytic rupture of the Co–C bond) is coupled kinetically to hydrogen abstraction by Ado• (6–9). Thus, the highly reactive Ado• species is short lived due to rapid neutralization by hydrogen abstraction, and this species has yet to be observed. The second paramagnetic center, cob(II)alamin, “lingers” in the active site, until product is formed, after which it recombines with Ado• to form the resting enzyme. In the presence of substrate, cob(II)alamin accumulates at a steady-state concentration, which can be observed by EPR and UV-visible spectroscopy. Class I and Class II isomerases have been extensively studied, and the cob(II)alamin spectroscopic signature has been valuable in studies of mechanism (6, 8, 10). Lysine 5,6-aminomutase (5,6-LAM) is the only Class III enzyme for which detailed studies of mechanism are reported. Cob(II)alamin does not accumulate in steady-state turnover as the enzyme undergoes rapid “suicide inactivation” involving removal of an electron from cob(II)alamin by a substrate and/or product radical intermediate (11). Irreversible formation of cob(III)alamin results, and cob(II)alamin is unable to recombine with Ado• to reform AdoCbl in the resting form of the enzyme.
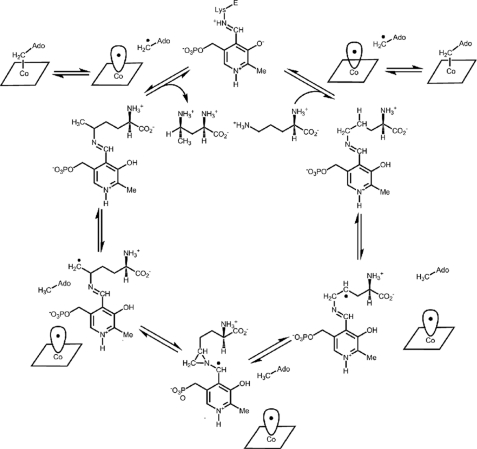
Ornithine 4,5-aminomutase (OAM; EC 5.4.3.5) is a Class III AdoCbl-dependent isomerase. In the Gram-positive anaerobe Clostridium sticklandii, it participates in the oxidative fermentation of l-ornithine (12) by interconverting d-ornithine to 2,4-diaminopentanoic acid (13). OAM is a α2β2 heterodimer comprising two strongly associating subunits, OraS (12.8 kDa) and OraE (82.9 kDa) (13). The enzyme contains AdoCbl and PLP (12), and both are required in the catalytic mechanism (Fig. 1). In the resting enzyme, a Schiff base (internal aldimine) is formed between an internal lysine residue and PLP (14, 15). Substrate displaces this lysine residue to form a new Schiff base (the external aldimine). Substrate binding results in breakage of the AdoCbl Co–C bond and radical propagation to the PLP-bound substrate. Rearrangement to the product-like radical intermediate is thought to proceed through a cyclic (azacyclopropylcarbinyl radical) intermediate (16–19). After “ring opening,” hydrogen is reabstracted from deoxyadenosine by the product-like radical intermediate, regenerating Ado•, which in turn recombines with cob(II)alamin. The transformed amino acid is released when PLP reforms the internal aldimine linkage. Although the precise role of PLP is not fully understood, computational studies suggest that the electron-withdrawing properties of the pyridine ring stabilize high energy radical intermediates formed in the catalytic cycle (20).
FIGURE 1.
Proposed mechanism and kinetic scheme for the reversible rearrangement of d-ornithine to 2,4-diaminopentanoic acid. In the mechanism, the PLP forms a Schiff base with the substrate, d-ornithine.
Important challenges are faced by B12-dependent isomerases. Precise timing of Co–C bond homolysis is crucial, as is appropriate “steering” of the reactivity of radical intermediates formed in the catalytic cycle. The “misfiring” of radicals within the active site of 5,6-LAM, leading to suicide inactivation, illustrates the danger of poor control of reactivity in B12-dependent enzymes (11). In this paper, we report an analysis of the reaction chemistry catalyzed by OAM to probe the stability, steering, and reactivity of radical species. We have used a combination of UV-visible and EPR spectroscopies in reactions with the natural substrate (d-ornithine) and a substrate analog/inhibitor DAB. We report the differential stabilization of radical species with d-ornithine and DAB, analysis of Co–C bond homolysis, and formation of the external aldimine with both compounds. We also report studies with AdoCbl-depleted OAM, which have allowed us to identify intermediates formed with PLP cofactor in the absence of spectral overlap from AdoCbl. Our work has enabled us to derive new and important mechanistic information for this Class III aminomutase, which we discuss in terms of a proposed mechanism for OAM (Fig. 1).
EXPERIMENTAL PROCEDURES
Materials—AdoCbl, PLP, d-ornithine, dl-2,4-diaminobutryic acid, l-2,4-diaminobutryic acid, and dl-2,3-diaminopropanioc acid were obtained from Sigma. [2,4,4-2H3]d-2,4-diaminobutryic acid was from QMX Laboratories. The Ni2+-nitrilotriacetic acid column and Q-Sepharose high performance resin were from GE Biosciences. Restriction endonucleases were from New England Biolabs, and Pfu Turbo DNA polymerase was purchased from Stratagene.
Cloning and Expression of OAM—C. sticklandii (ATCC 12662) was grown in reinforced clostridial media (OXNOID) in an anaerobic environment at 33 °C. The medium was used immediately after autoclaving to keep the oxygen content to a minimum. Cells were harvested, and genomic DNA was isolated using the Promega Wizard Genomic DNA purification kit. Adjacent genes encoding OAM (oraS and oraE) were amplified from the genomic DNA using Pfu Turbo DNA polymerase and the primers 5′-GGG GGG GCC ATG GAA AGA GCA GAC GAT-3′ (forward primer) and 5′-GGG GGG GAT CCT CAT TAT TTC CCT TCT-3′ (reverse primer), which contain NcoI and BamHI restriction sites (shown in boldface type), respectively. The PCR cycle parameters were as follows: 94 °C for 2 min followed by 30 cycles of 94 °C for 45 s, 50 °C for 1 min, and 72 °C for 6 min. The PCR product was digested with BamHI and NcoI and inserted into the pET23d vector, which had been digested with the same restriction enzymes. The ligation mixture was transformed into Escherichia coli strain XL1 Blue (Stratagene). A hexahistidine tag was inserted at the C terminus of OraE (by changing the stop codon to encode for a serine residue) by site-directed mutagenesis using the following primers: 5′-GAT GAG AGA AGG GAA ATC AGA GGA TCC GAA TTC-3′ (forward primer) and 5′-CGA ATT CGG ATC CTC TGA TTT CCC TTC TCT CAT-3′ (reverse primer). The plasmid containing oraE and oraS with a C-terminal hexahistidine tag was designated pETOAMH2. Sequencing of the plasmid by MWG Biotech AG (Covent Garden, United Kingdom) confirmed that no PCR-induced errors had occurred. The plasmid was transformed into E. coli strain Rosetta(DE3)pLysS (Novagen). A single colony was used to inoculate LB medium (5 ml) containing ampicillin (100 μg/ml), and the culture was grown for 8 h at 37 °C. This culture was used to inoculate LB medium (200 ml) with ampicillin (100 μg/ml), which was subsequently grown at 37 °C for 16 h. An initial culture (5 ml) was used to inoculate Terrific Broth (0.5 liters) containing ampicillin (100 μg/ml). The culture was grown at 25 °C, with shaking (220 rpm) until the culture reached an optical density at 600 nm of 1.0, at which time isopropyl β-d-1-thiogalactopyranoside (0.1 mm) was added. The temperature of the incubator was reduced to 20 °C, and the culture was allowed to grow for a further 16 h. Cells were harvested by centrifugation (4000 × g, 15 min, 4 °C), and the cell pellet was stored at –20 °C until purification.
Purification of OAM—All purification steps were performed at 4 °C. Cells (20 g, wet weight) were resuspended in 50 ml of 50 mm Tris-HCl, pH 7.5, 1 mm phenylmethylsulfonyl fluoride, and one EDTA-free protease inhibitor tablet (Roche Applied Science). Cells were disrupted by sonication (10-s pulses for 30 min with a 50-s interval, power setting 45%). The cell suspension was clarified by centrifugation at 25,000 × g for 50 min. Imidazole (20 mm) and NaCl (0.5 m) were added to the supernatant, which was then applied to a 5-ml Ni2+-nitrilotriacetic acid column equilibrated with 50 mm Tris-HCl, pH 7.5, 0.5 m NaCl, and 20 mm imidazole. The column was washed with 30 ml of 50 mm Tris-HCl, 0.5 m NaCl, pH 7.5, and 20 mm imidazole, and the protein was eluted with 200 mm imidazole. Fractions containing OAM were pooled and dialyzed against 50 mm Tris-HCl, pH 7.5, for 16 h at 4 °C. The dialysate was loaded onto a 58-ml Q-Sepharose HP column (2.6 × 11 cm) equilibrated in 50 mm Tris-HCl, pH 7.5. The column was washed with 120 ml of 50 mm Tris-HCl, pH 7.5, and the protein was eluted with a 0.9-liter linear gradient of 0–0.5 m NaCl at a flow rate of 4 ml/min. Fractions containing OAM, as judged by SDS-PAGE, were pooled, concentrated, and flash-frozen in liquid nitrogen in 20% glycerol and stored at –80 °C. Protein concentrations were determined by the Bradford method (21) using bovine serum albumin as a standard.
Anaerobic UV-visible Spectroscopy—Anaerobic UV-visible spectroscopic assays were performed in a Belle Technology glove box (O2 < 5 ppm) using a Hitachi U-1800 spectrophotometer at 25 °C. Buffer (100 mm NH4EPPS, pH 8.5) was purged for 2 h with nitrogen and then brought into the glove box and allowed to equilibrate for 18 h in an oxygen-free environment. Solid AdoCbl, PLP, d-ornithine, and the substrate analogues (dl-2,4-diaminobutryic acid, l-2,4-diaminobutryic acid, and dl-2,3-diaminopropanioc acid) were introduced to the glove box and dissolved in anaerobic buffer. A concentrated protein sample (2 ml of 12 mg/ml) was introduced into the glove box and gel-filtered using a 10-ml Econo-pack column (Bio-Rad) equilibrated with anaerobic buffer. A reference spectrum was recorded using buffer alone. For reactions involving the holoenzyme, a 1-ml reaction mixture contained 15 μm apo-OAM, 15 μm PLP, and 15 μm AdoCbl in 100 mm NH4EPPS, pH 8.5. The reaction was initiated by the addition of 2.5 mm d-ornithine or 2.5 mm substrate analogue, and spectra were recorded from 700 to 300 nm every 60 s for 20 min. For spectral studies involving only the PLP-bound form of the enzyme, a 1-ml reaction contained 15 μm apo-OAM and 15 μm PLP in 100 mm NH4EPPS, pH 8.5. The spectrum of the PLP-bound form of the enzyme was recorded from 700 to 300 nm, and then 2.5 mm d-ornithine, dl-2,4-diaminobutryic acid, l-2,4-diaminobutryic acid, or dl-2,3-diaminopropanoic acid was added to the 1-ml solution. Following incubation (5 min), the spectrum of the substrate/inhibitor-bound form of the enzyme was recorded.
Aerobic UV-visible Spectroscopy—Aerobic UV-visible spectral assays were performed on a Cary 50 UV-visible spectrophotometer (Varian) at 25 °C. The 1-ml reaction contained 15 μm apo-OAM, 15 μm PLP, and 15 μm AdoCbl in 100 mm NH4EPPS, pH 8.5. The reaction was initiated by the addition of 2.5 mm d-ornithine or 2.5 mm dl-2,4-diaminobutryic acid, and the spectra were recorded from 700 to 300 nm every minute for 20 min.
Pre-steady-state Kinetic Analysis—Stopped-flow studies were performed using an Applied Photophysics SX.17 MV stopped-flow spectrophotometer under anaerobic conditions. The temperature of the mixing chamber was maintained at 25 °C with a circulating water bath. The sample-handling unit of the stopped-flow instrument was contained within an anaerobic glove box (Belle Technology). Transient kinetic experiments were performed in 100 mm NH4EPPS, pH 8.5. Anaerobic solutions of buffer, enzyme, cofactors, substrate, and substrate analogues were prepared by the same method as described for UV-visible spectroscopy (see above). All solutions containing AdoCbl were kept in the dark to prevent photolysis of the cofactor. Immediately prior to single wavelength stopped-flow studies, the holoenzyme solution was generated by adding an equimolar mixture of AdoCbl, PLP, and the apoenzyme solution. A 50 μm solution of holoenzyme was then rapidly mixed with 5 mm substrate or substrate analogue, and the change in absorbance was monitored at 528 nm (AdoCbl homolysis), 470 nm (cob(II)alamin formation), and 416 nm (external aldimine formation). The substrate/analogue and enzyme were diluted 2-fold after mixing. An average of 7–9 traces were averaged at each wavelength and used to fit to a single exponential equation to extract the observed rate constants.
For photodiode array spectroscopic studies involving the PLP-bound form of the enzyme, 35 μm PLP was premixed with 35 μm apo-OAM. Following incubation (5 min), the PLP-bound enzyme was rapidly mixed with 5 mm d-ornithine or dl-2,4-diaminobutryic acid; both solutions were diluted 2-fold after mixing. The photodiode array detector was used to monitor spectral changes from 1.28 ms to 100 s postmixing. The time-dependent spectral changes were evaluated using singular value decomposition associated with Pro-K software (Applied Photophysics Ltd.). A set of output matrices comprising ordered data sets of basis spectra and time-dependent amplitudes was obtained along with corresponding weighting factors in the form of singular values. The number of singular values is a model-free indication of the number of the independent components present in the original data set. A kinetic model was built based on the number of independent spectra predicted by this method.
EPR Spectroscopy—Samples were prepared in an anaerobic glove box in 100 mm NH4EPPS, at 25 °C and under very dim light to minimize photolysis of AdoCbl. Anaerobic buffer and stock solutions of substrates and cofactors were prepared by the method described for anaerobic UV-visible spectroscopy. A 2-ml concentrated protein sample (19 mg/ml) was introduced into the glove box and gel-filtered using a 10-ml Econo-pack column (Bio-Rad) equilibrated with anaerobic buffer. The protein solution was then concentrated to 0.6 mm in the anaerobic glove box using a Vivaspin 500 column (Vivascience-Sartorius Group) and then mixed with an equimolar amount of PLP and AdoCbl. Following incubation (5 min), 5 mm d-ornithine or 5 mm dl-2,4-diaminobutryic acid was added to 0.56 mm of holo-OAM. The samples were loaded into EPR tubes and frozen in liquid nitrogen. EPR spectra were recorded using a Bruker ELEXSYS E500 spectrometer equipped with an ESR900 cryostat (Oxford Instruments). The sample temperature was 20 K. Spectra were recorded at nonsaturating microwave power using a 100-kHz modulation frequency and 0.5-mT modulation amplitude.
RESULTS
Expression and Purification of OAM—The gene products comprising OAM, OraS, and OraE were co-expressed in E. coli and co-purified via Ni2+-nitrilotriacetic acid affinity and ion exchange chromatography. A 15% SDS-polyacrylamide gel of purified OAM (data not shown) revealed two proteins of 83 and 13 kDa, corresponding to OraE and OraS, respectively.
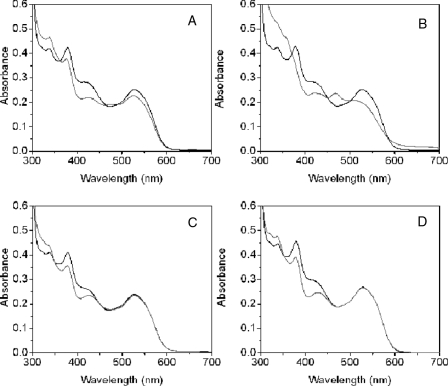
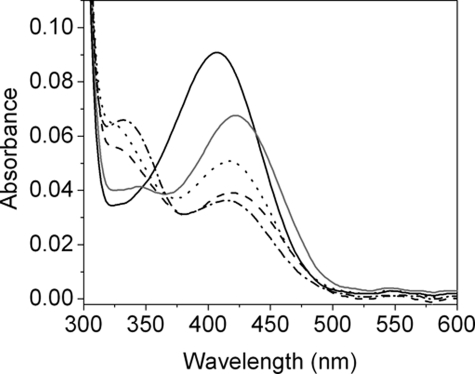
Steady-state Anaerobic UV-visible Spectral Analysis of Holo-OAM—The UV-visible spectrum of holo-OAM shows the overlapping absorbance of the AdoCbl and PLP cofactors. The absorbance peaks at 335, 378, and 528 nm originate from AdoCbl, and a broad absorbance shoulder at 416 nm corresponds to PLP in the internal aldimine state (Fig. 2). The addition of the natural substrate, d-ornithine, to the holoenzyme leads to transimination, as indicated by a decrease in the absorbance shoulder at 416 nm coupled with formation of an absorbance peak at 425 nm (Fig. 2A). The UV-visible spectrum also shows a small decrease in absorbance at 528 nm (reflecting homolysis of the AdoCbl C–Co bond), accompanied by an even smaller increase in absorbance at 470 and 311 nm (signifying cob(II)alamin formation). The difference spectra, in which the contribution from the PLP cofactor (both internal and external aldimine components) is subtracted from holo-OAM spectra (Fig. S1), allows closer examination of absorbance changes at 470 and 311 nm. In contrast, the addition of a mixture of the d- and l-stereoisomers of 2,4-diaminobutryic acid results in a substantial rise in absorbance between 310 and 360 nm along with the formation of a discrete peak at 470 nm (indicating the formation of cob(II)alamin). The substrate analogue induces a larger fraction of the holoenzyme to undergo AdoCbl homolysis, since it causes a more pronounced decrease in absorbance at 528 nm (Figs. 2B and S1B). On the basis of a Δε value of 4.8 mm–1 cm–1 (5), the change in absorbance at 528 nm indicates that ∼18 and ∼39% of the holoenzyme lacks AdoCbl at steady-state with d-ornithine and dl-2,4-diaminobutryic acid, respectively. As with the natural substrate, dl-2,4-diaminobutryic acid also forms an imine linkage with the PLP cofactor. Interestingly, only the d-isomer of 2,4-diaminobutryic acid causes homolysis, since the addition of l-2,4-diaminobutryic acid to the holoenzyme only induces spectral shifts in the UV-visible spectrum that correspond to formation of an external aldimine (Fig. 2C). The same spectral shift in the holoenzyme is also observed for dl-2,3-diaminopropanoic acid, indicating that although this compound can induce transimination, it is incapable of causing homolysis (Fig. 2D). The time trace from UV-visible spectra of the holoenzyme following prolonged anaerobic incubation with the substrate shows an increase in absorbance at 356 nm with a rate constant of ∼6 × 10–4 s–1 (data not shown), probably reflecting cob(III)alamin formation (Fig. 3A). Minimal absorbance change occurs at 356 nm with D,L-2,4-diaminobutyric acid, under similar conditions (Fig. 3B). The presence of oxygen greatly accelerates cob(III)alamin formation with either the substrates or inhibitor (Fig. 3, C and D), as demonstrated by a large absorbance change at 358 nm and the distinctive double “humps” appearing at 505 and 534 nm, the hallmark spectral feature of hydroxycob(III)alamin.
FIGURE 2.
Anaerobic spectral changes of holo-OAM induced by binding of substrate and substrate analogues. The holoenzyme solution contained 100 mm NH4-EPPS, pH 8.5, 30 μm OAM, 30 μm PLP, and 30 μm AdoCbl in a total volume of 1 ml. Spectral changes in holo-OAM were recorded at 25 °C, before (black lines) and 1 min after (gray lines) the addition of 5 mm d-ornithine (A), 5 mm dl-2,4-diaminobutryic acid (B), 5 mm l-2,4-diaminobutryic acid (C), and 5 mm dl-2,3-diaminopropanioic acid (D).
FIGURE 3.
Change in UV-visible spectra of holo-OAM induced by binding of substrate and a substrate analogue. The experimental procedures are the same as those described for Fig. 2, except that 15μm apo-OAM, 10μm PLP, and 10 μm AdoCbl were mixed in a total volume of 1 ml. Spectral changes for holo-OAM were recorded at 25 °C, at 0, at 10 s, and then at every 60 s up to 20 min following the addition of 2.5 mm d-ornithine (A) or 2.5 mm dl-2,4-diaminobutryic acid (B). C and D are the same as those described for A and B, respectively, except spectra were recorded under aerobic conditions. For clarity, only representative spectra are shown. The arrows indicate the direction of absorbance change over time.
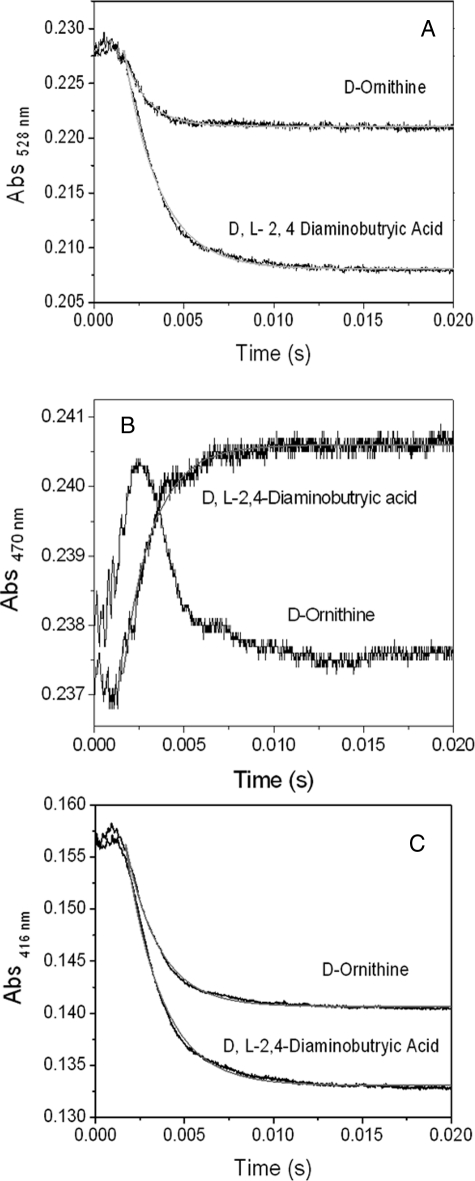
Analysis of AdoCbl Homolysis and External Aldimine Formation in Holo-OAM—The time course for AdoCbl homolysis in B12-dependent enzymes has been monitored by following a decrease in absorbance at 528 nm (disappearance of AdoCbl) and/or an increase in absorbance at 470 nm (formation of cob(II)alamin). We have adopted a similar strategy with OAM. Fig. 4 shows the stopped-flow absorbance trace at 528 nm (Fig. 4A) or 470 nm (Fig. 4B) following rapid mixing of 5 mm d-ornithine or 5 mm dl-2,4-diaminobutryic acid with 50 μm holo-OAM. Both the substrate and substrate analogue cause a rapid decrease in absorbance a 528 nm. Analysis of the data (from 1.6 to 20 ms) by fitting to a monoexponential change in absorption gave observed rate constants of 781 ± 12 s–1 and 513 ± 2 s–1 for d-ornithine and dl-2,4-diaminobutryic acid, respectively. The change in absorbance over this time domain was ∼2-fold greater for dl-2,4-diaminobutryic acid compared with d-ornithine. For the natural substrate (d-ornithine), the calculated amplitude change at 528 nm corresponds to 5.1 μm enzyme (20%) undergoing AdoCbl homolysis compared with 9.8 μm (39%) in the presence of the inhibitor. These values obtained for the percentage of enzyme undergoing Co–C bond homolysis are similar to those obtained under steady-state conditions. The time course for the absorbance changes at 470 nm (Fig. 4B) reveals very different traces for the inhibitor and substrate. For dl-2,4-diaminobutryic acid, there is a very rapid pre-steady-state (single exponential) appearance of cob(II)alamin, and a fit of the absorbance trace between 1.6 and 20 ms using a single exponential expression produces an observed rate constant of 488 ± 8 s–1. The stopped-flow trace following rapid mixing of d-ornithine with holo-OAM is characterized by a rapid “up” phase that is nearly complete within the dead time of the stopped-flow instrument (1.6 ms), followed by a slower “down” phase. Although, the up phase may be an artifact of the stopped-flow instrument, it may represent transient formation cob(II)alamin, and the down phase may represent its decay. When the reaction was repeated at 10 °C, the stopped-flow trace appears over a longer time domain (Fig. S2); however, because of the small amplitude change and fast time frame, we cannot unequivocally assign the absorbance change to a discrete kinetic step. Transimination with the natural substrate and inhibitor also occur on the same time scale as homolysis, since the absorbance change at 416 nm decreases rapidly upon mixing with the holoenzyme. Analysis of the data in Fig. 4C gave rate constants of 532 ± 10 s–1 and 488 ± 12 s–1 for d-ornithine and dl-2,4-diaminobutryic acid, respectively.
FIGURE 4.
Stopped-flow absorbance changes following mixing of holo-OAM (50 μm; before mixing) with d-ornithine (5 mm; before mixing; A) and dl-2,4-diaminobutryic acid (5 mm before mixing; A). A, time course absorbance change at 528 nm, which monitors AdoCbl Co-C bond homolysis. B, stopped-flow trace at 470 nm monitoring cob(II)alamin formation. C, change in absorbance at 416 nm monitoring the formation of the external aldimine. The kinetic traces were fitted to a single exponential equation.
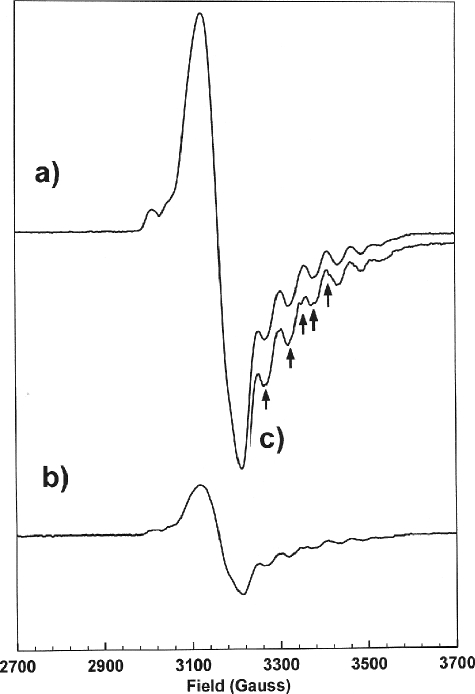
EPR Spectroscopic Characterization of Holo-OAM—EPR spectroscopy has proven to be a valuable method for providing information on the relative positions of the paramagnetic centers formed during the catalytic cycle. During steady-state turnover of AdoCbl-dependent enzymes, these generally correspond to the low spin Co2+ atom of cob(II)alamin and a substrate/product-like radical intermediate. Magnetic interactions between Co2+ and the organic radical create mutual perturbations in the EPR spectra of the component radicals, which provide information on the level of spin-spin coupling between the paramagnetic centers and hence their relative positions (13). The EPR spectrum of the holoenzyme mixed with d-ornithine did not show any evidence of a paramagnetic species; the EPR spectrum closely resembled the control sample, which contained only the holoenzyme and is essentially a base-line spectrum (data not shown). However, for dl-2,4-diaminobutryic acid, there is very clear evidence of cob(II)alamin species coupled to an organic radical (Fig. 5). The EPR spectrum is very intense and has a g⊥ value of 2.11 and A∥ value of 54 G (arising from the cobalt-59 splitting). The EPR spectrum is visually similar to that observed for Class I mutases, which comprises a strongly coupled spin system consisting of a low spin Co2+ ion and an organic radical in a hybrid triplet state (10, 22, 23). Hyperfine splittings of the gz line, resulting from coupling of the unpaired electron on the cobalt nucleus (I = 7/2), are evident in the high field region of the spectrum, where six of the eight splittings are observed.
FIGURE 5.
EPR spectra of holo-OAM samples mixed with dl-2,4-diaminobutyric acid (a) or [2,4,4-2H3]d-2,4-diaminobutyric acid (b). Spectra are shown on the same vertical scale. c, a section of the spectrum recorded with [2,4,4-2H3]d-2,4-diaminobutyric acid on an enlarged vertical scale to facilitate comparison with a; the arrows indicate the magnetic fields at which the deuteration of the inhibitor perturbs the spectrum. Sample preparation and experimental procedures are outlined under “Experimental Procedures.”
The EPR spectrum obtained upon mixing the holoenzyme with [2,4,4-2H3]d-2,4-diaminobutryic acid (Fig. 5), shows perturbation in the hyperfine structure and indicates that the organic radical is derived from the substrate analog. Partial deuteration of DAB also reduces the yield of this EPR-detectable species to 22% of that observed with the fully protonated molecule. The diminution of signal intensity may arise from isotopic substitution (i.e. breaking of a C–H versus a C–D bond) perturbing the equilibrium toward the diparamagnetic state. This type of kinetic isotope effect has been observed previously for AdoCbl-dependent isomerases (6, 7, 22).
Anaerobic Spectral Analysis of PLP-OAM—The absorbance maximum of the free PLP cofactor at pH 8.5 is at 397 nm (data not shown). The addition of equimolar OAM to the PLP solution causes the absorbance peak to shift to 408 nm over several minutes, reflecting the formation of an internal aldimine between a lysine residue and PLP. Transimination occurs with the addition of d-ornithine to PLP-OAM as the UV-visible absorbance maxima red-shifts from 408 to 418 nm (Fig. 6). There is also a small increase in absorbance between 320 and 340 nm, suggesting that the external aldimine exists in the enolimine and ketomine state, with the equilibrium strongly favoring the latter tautomer. The addition of dl-2,4-diaminobutryic acid, l-2,4-diaminobutryic acid, or dl-2,3-diaminopropanoic acid also results in transimination. However, the addition of either of these substrate analogues shifts the tautomeric equilibrium (to varying degrees) toward the enolimine state. Nonenzymatic condensation of PLP and dl-2,4-diaminobutryic acid at pH 7.0 or 8.5 generates both tautomers (data not shown), but at lower pH, the equilibrium shifts more toward the ketoimine tautomer.
FIGURE 6.
UV-visible spectral changes associated with formation of an external aldimine in PLP-OAM. An equimolar mixture of apo-OAM (15 μm) and PLP (15 μm) was mixed in a total volume of 1 ml in 100 mm NH4EPPS, pH 8.5, at 25 °C in an anaerobic environment. The UV-visible spectrum (black line) was recorded from 300 to 700 nm following a 5-min incubation of PLP with the apoenzyme. The UV-visible spectrum was also recorded after the addition of 2.5 mm d-ornithine (gray line), 2.5 mm dl-2,4-diaminobutryic acid (dashed line), 2.5 mm l-2,4-diaminobutryic acid (dotted line), and 2.5 mm dl-2,3-diaminopropanoic acid (dashed and dotted line). The experimental conditions are described under “Experimental Procedures.”
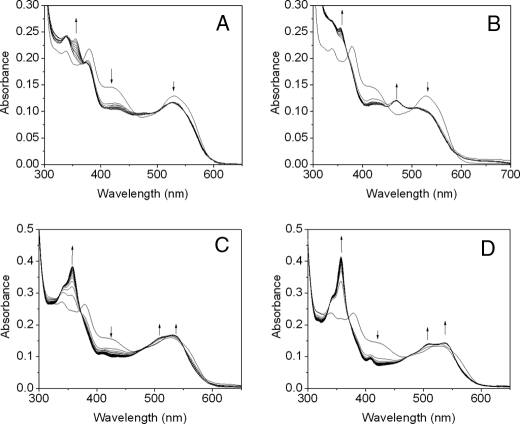
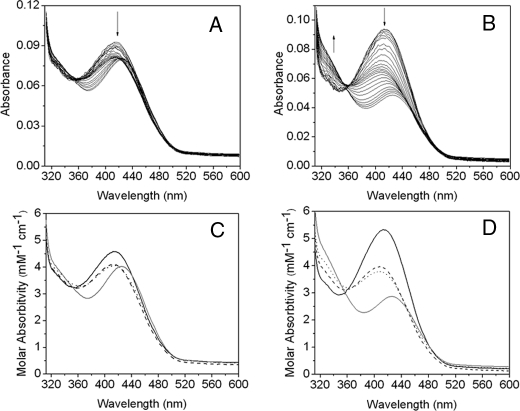
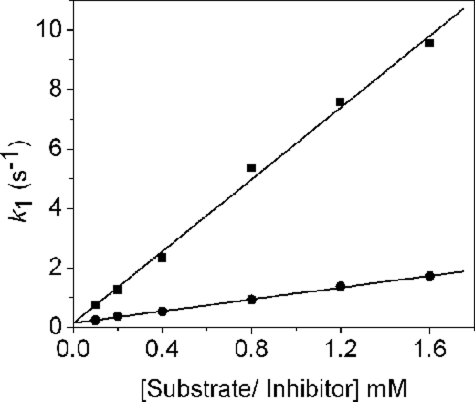
Stopped-flow Analysis of the Reaction of PLP-OAM with Substrate and Inhibitor—Absorbance changes resulting from rapid mixing of PLP-OAM (i.e. AdoCbl-depleted enzyme) with either d-ornithine or dl-2,4-diaminobutryic acid under pseudo-first order conditions are shown in Fig. 7, A and B, respectively. Global analysis of the singular value decomposition-transformed data for both set of spectra (Fig. 7, C and D) reveals the presence of four components. The spectral signature of the first component (λmax = 414 nm) is assigned to the internal aldimine. This species decays to components B and the C, which elicit similar spectral properties with λmax at 406 nm. These two spectral species may represent the PLP cofactor in a “free” state (i.e. unliganded to either the lysine residue or the terminal amine of the substrate/substrate analogue). The absence of a λmax at 320 nm, which would otherwise indicate formation of a gem-diamine, suggests that transimination involves a PLP-free state of the cofactor. The final spectral component has an absorbance maximum at 425 nm and corresponds to the external aldimine. The observed rate constants for interconversion of each of the spectral components with d-ornithine and dl-2,4-diaminobutryic acid are listed in Table 1. The rate of formation of species B (i.e. the PLP-free form of OAM) is linearly dependent on both the concentration of d-ornithine and dl-2,4-diaminobutryic acid and yielded a bimolecular (pseudo-first order) rate constant of 6.04 ± 0.21 and 0.99 ± 0.02 mm–1 s–1, respectively (Fig. 8). The rate constants for the formation of components C and D were independent of substrate/substrate analogue concentration.
FIGURE 7.
Spectral changes resulting from the addition of d-ornithine and dl-2,4-diaminobutryic acid to PLP-OAM. The apoenzyme (35 μm) was preincubated with an equimolar amount of PLP (35 μm) and then rapidly mixed with 5 mm d-ornithine (A) and dl-2,4-diaminobutryic acid (B) in 100 mm NH4EPPS, pH 8.5, at 25 °C in an anaerobic environment. The UV-visible absorption changes were recorded from 1.28 ms to 100 s after mixing. For clarity, only subsequent select spectra are shown. Time-dependent singular value decomposition analysis of the data shown in A and B revealed the presence of four spectral species. Data were fitted globally to a three-step reversible model (A ↔ B ↔ C ↔ D). The deconvoluted spectra of the intermediates from global analysis of A and B are shown in C and D, respectively. The four spectral intermediates, A, B, C, and D, shown in C and D are represented by the solid, dashed, dotted, and gray lines, respectively. The observed rate constants for interconversion of the spectral intermediates are shown in Table 1.
TABLE 1.
Rate constants obtained for the formation of the external aldimine in PLP-OAM determined by global analysis of the singular value decomposition-transformed PDA data shown in Fig. 7
Experimental conditions are described under “Experimental Procedures.”
| d-Ornithine | dl-2,4-Diaminobutryic acid | |
|---|---|---|
| k1 (s-1) A > B | 7.86 ± 0.03 | 3.93 ± 0.09 |
| k-1 (s-1) B > A | 0.03 ± 0.01 | 0.02 ± 0.01 |
| k2 (s-1) B > C | 0.45 ± 0.01 | 0.77 ± 0.01 |
| k-2 (s-1) C > B | 3.1 × 10-3 ± 2 × 10-4 | 9.6 × 10-5 ± 6 × 10-6 |
| k3 (s-1) C > D | 4.7 × 10-2 ± 2 × 10-4 | 6.2 × 10-2 ± 1 × 10-4 |
| k-3 (s-1) D > C | 1.7 × 10-4 ± 1.7 × 10-4 | 1.7 × 10-6 ± 3 × 10-7 |
FIGURE 8.
Dependence of the apparent rates for the formation of the PLP-free form (i.e. species B) on the concentration of d-ornithine (▪) and dl-2,4-diaminobutryic acid (•).
DISCUSSION
Cob(II)alamin Is Formed Only Transiently upon Reaction with the Substrate Ornithine—Our studies reveal that although d-ornithine is able to induce AdoCbl homolysis in holo-OAM, the reaction does not lead to a detectable steady-state level of cob(II)alamin. The paramagnetic center is short lived (<5 ms) following substrate-induced homolysis, since stopped-flow traces show a rapid appearance and then disappearance in absorbance at 470 nm, which may represent its transient formation. Therefore, in contrast with Class I and II isomerases, the metallo-paramagnetic center does not linger in the active site of OAM, during steady-state turnover. We have also demonstrated that following prolonged incubation (30 min) with substrate, hydroxycobalamin is formed (judging by the small increase in absorbance at 356 nm) from cob(II)alamin. We have also shown that the rate of cob(III)alamin formation with the natural substrate is 10-fold slower than that observed with 5,6-LAM (11). Of interest is the finding that formation of hydroxycobalamin in OAM is greatly accelerated in an aerobic environment, which apparently is not the case with 5,6-LAM (11). The oxygen lability of the catalytic reaction indicates that it proceeds through transient formation of cob(II)alamin and that although suicide inactivation (akin to that observed in 5,6-LAM) is a feature of OAM turnover, it is relatively minor. The lack of a detectable level of cob(II)alamin during steady-state turnover by OAM is probably attributable to the position of rate-limiting steps in the catalytic cycle. For example, if release of product from PLP is rate-limiting and occurs after recombination of Ado• and cob(II)alamin, then the latter paramagnetic species will not be a long lived intermediate during turnover, making detection difficult. Alternatively, the rate-limiting step may be hydrogen abstraction by Ado• from the PLP-bound substrate, as suggested for 5,6-LAM (24).
Despite the similarities in the sequences of OAM and 5,6-LAM (reflected also in their identical cofactor requirements and analogous reaction mechanisms), the two enzymes exhibit very different kinetic behavior. This might be attributable to differences in the allosteric/dynamic control of the reaction chemistry defined by the structures of the two enzymes. Purified from Clostridia species, 5,6-LAM comprises two components, the core enzyme (E1) and the activating enzyme (E2). Although the E1 component is functional, the enzyme loses activity in the absence of ATP (an allosteric activator) and E2 (11, 12).4 The function of E2 and ATP in maintaining 5,6-LAM activity is unknown, but it may be analogous to the ATP-driven chaperone-like system observed for Class II eliminases (27).
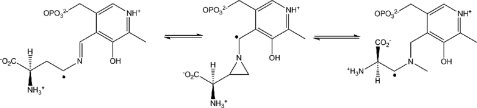
dl-2,4-Diaminobutryic Acid Forms a Stable Radical in OAM—Although many B12-isomerases are able to use alternative substrates (24, 28), OAM is unable to turn over the competitive inhibitor, dl-2,4-diaminobutryic acid (15). The binding of DAB to holo-OAM probably leads to mechanism-based inactivation of the enzyme that entails AdoCbl-mediated radical propagation, leading to the formation of a PLP-bound radical intermediate. Overstabilization of this intermediate may prevent it from reacting further and reabstracting a hydrogen from deoxyadenosine, to regenerate Ado• and eventually the resting state of the coenzyme. This type of mechanism-based inactivation has previously been observed with ethanolamine ammonia lyase following incubation with (i) hydroxyethylhydrazine (leading to formation of stable hydrazine radical) (29) and (ii) glycolaldehyde (leading to formation of a cis-ethanesemidione anion radical) (30). The stable radical is likely to be one of the three PLP-bound forms shown in Scheme 2. Although precise assignment will require detailed characterization using EPR spectroscopy, computational studies indicate that the cyclic intermediate is the most stable (31).
SCHEME 2.
Proposed PLP-bound radical intermediates following the reaction of d-2,4-diaminobutryic acid with holo-OAM.
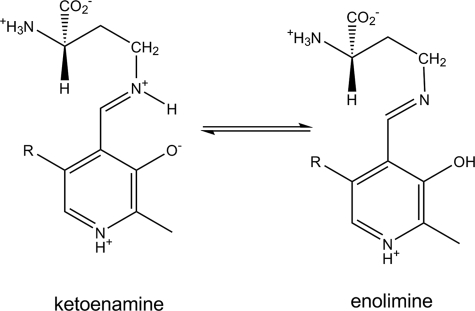
Overstabilization of the DAB-derived radical intermediate may explain why the inhibitor induces a larger fraction of holo-OAM to undergo homolysis, compared with the natural substrate. The extent of Co–C bond homolysis may be linked to the formation of a stable PLP-bound radical species, effectively pulling the reaction toward a diparamagnetic state. The larger degree of homolysis might also be influenced by the equilibrium distribution of the PLP-DAB prototropic isomers (Scheme 3). Formation of the external Schiff base with dl-2,4-diaminobutryic acid results in a nearly equal mixture of the ketoimine and enolimine tautomers, whereas the Schiff base formed with d-ornithine results mainly in the formation of the ketoimine state. At this stage, it is unclear why the equilibrium is shifted toward the enolimine state with the inhibitor, but this could be related to the protonation state of the terminal amine group. The enolimine tautomer may be more efficient at driving AdoCbl homolysis, because (i) hydrogen abstraction by Ado• is more favorable with a deprotonated imine group or (ii) the enolimine tautomer may ultimately lead to a more stable PLP radical. The latter possibility is contrary to most other PLP-dependent enzymes, since studies have shown the enzyme can enhance reactivity by promoting the prototropic tautomer in which the proton is formally bonded to the Schiff base (i.e. the ketoimine state) (32, 33). For the majority of B6-enzymes, catalytic steps post-transimination lead to the formation of carbanionic intermediates. Thus, the positively charged imine nitrogen enhances their stability by allowing more resonance stabilization.
SCHEME 3.
Prototropic isomers of the d-2,4-diaminobutyric acid-PLP complex.
Ab initio molecular modeling studies have shown that protonation of the migrating group facilitates many 1,2-shifts in B12-mediated catalysis (16, 17, 34). That said, the opposite is true for 1,2-amino shifts catalyzed by aminomutases. Molecular modeling studies have shown that stabilization of the radical intermediate, in particular the cyclic azacyclopropylcarbinyl radical, is obtained through a synergistic combination of π -electron withdrawal by the pyridine ring and electron donation to the radical center by the lone pairs on the imine nitrogen (19, 31). The latter component of this captodative stabilization would benefit from a deprotonated state of the imine nitrogen and the formal protonation of the phenolic hydroxyl. Thus, computational data support the view that the enolimine tautomer leads to a more stable radical intermediate.
The Paramagnetic Centers in OAM Are Strongly Coupled—EPR spectroscopy has revealed that the degree of electron spin-spin coupling between the paramagnetic centers (i.e. the low spin Co2+ and the substrate/product intermediate) formed during turnover of AdoCbl-dependent enzymes is a function of their separation and relative orientation (10). Analysis of the EPR signatures from Class I and II isomerases reveals that the former have strong exchange-coupled spin systems, indicating an interspin distance of <6 Å (22, 35, 36), whereas the latter exhibit a weakly coupled spin system, reflecting a larger distance (11–13 Å) between the paramagnetic centers (30, 37). The EPR spectrum of DAB-bound holo-OAM is visually similar to the Class I mutases, suggesting that the radical pair is strongly spin-coupled and separated by <6 Å. The narrow spacing in the hyperfine structure (A∥ value of 54 G) is consistent with a “base-off/His-on”5 mode of AdoCbl binding (36). This observation agrees with the finding that mutagenesis of a histidine localized to the (DXHXXG) “base-off/His-on” binding motif to a glycine residue disrupts AdoCbl binding in holo-OAM (13). The perturbation in the hyperfine structure induced by the binding of [2,4,4,2H3]DAB to holo-OAM indicates that the organic radical coupled metallo-paramagnetic center is derived from a DAB intermediate. Further isotopic perturbation experiments are required to determine the structure of the radical intermediate (Scheme 2) and the atom where the radical is centered.
Interestingly, the crystal structure of the closely related substrate-free form of 5,6-LAM6 shows a distance of 25 Å between the pyridine ring of PLP and cob(II)alamin (14). However, EPR studies with OAM suggest a distance of <6 Å between the substrate-derived radical and Co2+ during intermediate steps of the catalytic cycle (e.g. intramolecular isomerization). Should OAM be structurally similar to 5,6-LAM, this would require major protein conformational change in OAM to coincide with external aldimine formation and Co–C bond breakage. Such large scale conformational dynamics (previously not seen with B12-isomerases) would significantly complicate the challenges faced by the enzyme in controlling Co–C bond homolysis and the trajectories of high energy radical intermediates within the active site.
Transaldimination and Co–C Bond Homolysis Are either Concerted or Coupled—The first and common step with all PLP-dependent enzymes is transimination. This is a multistep process and is frequently rapid compared with subsequent slower catalytic steps. Formation of the external aldimine in the holoenzyme (with either the substrate or inhibitor) occurs on the same time scale as homolysis, suggesting that the two catalytic steps are either concerted or coupled. The presence of AdoCbl in OAM clearly affects the rate of transimination, since the reaction is considerably slower (2500-fold) when only PLP is bound (i.e. lacking the AdoCbl cofactor). Binding of AdoCbl may induce conformational change in OAM, improving substrate access to PLP and increasing rates of external aldimine formation. Analysis of photodiode array stopped-flow data indicates (at least for the PLP-bound form of OAM) that transimination occurs in three spectrally resolved steps. The first step is release of the ε-amino of Lys629 (15) from PLP. The observed rate constant for this initial step shows a linear dependence on substrate/inhibitor concentration. The second and third reaction components probably represent a state in which PLP is unliganded to protein as the spectra are blue-shifted. Individually, these may correspond to different ionization or conformational states of the protein. The final spectral component represents the external aldimine, since there is a significant red shift in its corresponding spectral component. It is unclear if this multistep transimination mechanism involving a PLP-free state exists for the holoenzyme.
A direct comparison of rate constants for Co–C bond breakage between OAM and other B12-dependent isomerases is difficult, since prior stopped-flow studies on glutamate mutase, methylmalonyl-CoA mutase, and ethanolamine ammonia lyase provide only estimates (>600 s–1 at 25 °C), since the reaction is essentially complete within the dead time of the stopped-flow instrument (6, 8, 38). However, we are able to track homolytic rupture in OAM using the stopped-flow method, suggesting that this step is slower compared with Class I and II isomerases. Should Co–C bond breakage be genuinely attenuated in OAM (and aminomutases in general), this might be related to the requirement for protein conformational change and/or transimination prior to bond homolysis. OAMs, like all B12-isomerases, enhance Co–C bond homolysis (∼1012-fold) over the comparable reaction in the absence of enzyme (39). The mechanism(s) by which this is achieved remains an integral and longstanding question in B12 enzymology.
Conclusions—According to the preposed mechanism for aminomutases, there are a total of at least 12 steps, six steps of the catalytic cycle shown in Fig. 1 plus binding and release of substrate/product, two transimination steps, homolysis, and recombination (24). Our spectral studies of holo-OAM have revealed that, when the holoenzyme reacts with its natural substrate, (i) homolysis and transimiation are probably concerted or coupled events; (ii) the rate of homolysis appears to be attenuated in OAM, compared with related isomerases, which may be due to conformational change and/or coupling Co–C breakage to external aldimine formation; and (iii) cob(II)alamin is a short lived catalytic intermediate that does not accumulate to detectable levels at steady-state. The latter suggests that either the release of product from PLP occurring subsequent to AdoCbl recombination or hydrogen abstraction by Ado• is the rate-limiting step in catalysis. Moreover, transimination may involve multiple steps involving a PLP-free state.
The absence of cob(II)alamin as a “spectator” during catalytic turnover with the physiological substrate is one of the main mechanistic differences between OAM and Class I and II isomerase. Cob(II)alamin is stabilized, however, with the analog dl-2,4-diaminobutryic and is strongly spin-coupled to an organic radical. This radical probably originates from the PLP-DAB intermediate, possibly the cyclic azacyclopropylcarbinyl radical in which the imine nitrogen is unprotonated. We have identified key differences in the mechanism between OAM, the related 5,6-LAM, and other classes of AdoCbl-dependent enzyme systems and identified a need for conformational coupling of the reaction chemistry. Our analysis provides deeper understanding of the reaction chemistry of OAM, which, in conjunction with future structural and computational evaluations of the enzyme chemistry, should advance our understanding of radical-based catalysis in B12 enzyme systems.
Supplementary Material
The work was funded by the United Kingdom Biotechnology and Biological Sciences Research Council (to N. S. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: AdoCbl, adenosylcobalamin; Ado•, 5′-deoxyadenosyl radical; 5,6-LAM, lysine 5,6-aminomutase; NH4EPPS, ammonium N-(2-hydroxyethyl)piperazine-N′-3-propanesulfonic acid; OAM, ornithine 4,5-aminomutase; PLP, pyridoxal l-phosphate; DAB, d-2,4-diaminobutryic acid.
The E1 component comprises 30- and 51-kDa subunits (encoded by KamD and KamE, respectively) and, according to Barker and Stadman (25), a 12.8-kDa protein not encoded in the Kam operon. Heterologous expression of the KamD and KamE gene products in E. coli produces an α2β2 heterotetrameric protein that, despite being able to turnover substrate, succumbs to suicide inactivation (11). Interestingly, sequence analysis of the 12.8-kDa protein co-purifying with the KamD and KamE gene products indicates that it is OraS (26). Tseng et al. (26) recently demonstrated that the addition of the S subunit of OAM to the recombinantly expressed 5,6-LAM restores the ATP-dependent allosteric activation of the enzyme. It is unclear, however, if OraS alone prevents deleterious inactivation of 5,6-LAM, but it may act in conjunction with E2 to reactivate 5,6-LAM.
In the “base-off/His-on” binding mode for AdoCbl, a histidine residue from the protein replaces the dimethylbenzimidazole base as the lower axial ligand.
OraE shows 28% identity and 39% similarity to the gene product of KamD and 35% identity and 47% similarity to protein encoded by KamE.
References
- 1.Toraya, T. (2003) Chem. Rev. 103 2095–2127 [DOI] [PubMed] [Google Scholar]
- 2.Marsh, E. N. G., and Drennan, C. L. (2001) Curr. Opin. Chem. Biol. 5 499–505 [DOI] [PubMed] [Google Scholar]
- 3.Banerjee, R. (2003) Chem. Rev. 103 2083–2094 [DOI] [PubMed] [Google Scholar]
- 4.Brown, K. L. (2005) Chem. Rev. 105 2075–2149 [DOI] [PubMed] [Google Scholar]
- 5.Licht, S., Gerfen, G. J., and Stubbe, J. (1996) Science 271 477–481 [DOI] [PubMed] [Google Scholar]
- 6.Padmakumar, R., Padmakumar, R., and Banerjee, R. (1997) Biochemistry 36 3713–3718 [DOI] [PubMed] [Google Scholar]
- 7.Cheng, M. C., and Marsh, E. N. (2007) Biochemistry 46 883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marsh, E. N., and Ballou, D. P. (1998) Biochemistry 37 11864–11872 [DOI] [PubMed] [Google Scholar]
- 9.Bandarian, V., and Reed, G. H. (2000) Biochemistry 39 12069–12075 [DOI] [PubMed] [Google Scholar]
- 10.Reed, G. H., and Mansoorabadi, S. O. (2003) Curr. Opin. Struct. Biol. 13 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang, K. H., Chang, C. H., and Frey, P. A. (2001) Biochemistry 40 5190–5199 [DOI] [PubMed] [Google Scholar]
- 12.Baker, J. J., van der Drift, C., and Stadtman, T. C. (1973) Biochemistry 12 1054–1063 [DOI] [PubMed] [Google Scholar]
- 13.Chen, H. P., Wu, S. H., Lin, Y. L., Chen, C. M., and Tsay, S. S. (2001) J. Biol. Chem. 276 44744–44750 [DOI] [PubMed] [Google Scholar]
- 14.Berkovitch, F., Behshad, E., Tang, K. H., Enns, E. A., Frey, P. A., and Drennan, C. L. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 15870–15875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, H. P., Hsui, F. C., Lin, L. Y., Ren, C. T., and Wu, S. H. (2004) Eur. J. Biochem. 271 4293–4297 [DOI] [PubMed] [Google Scholar]
- 16.Smith, D. M., Golding, B. T., and Radom, L. (1999) J. Am. Chem. Soc. 121 9388–9399 [Google Scholar]
- 17.Smith, D. M., Golding, B. T., and Radom, L. (1999) J. Am. Chem. Soc. 121 5700–5704 [Google Scholar]
- 18.Smith, D. M., Golding, B. T., and Radom, L. (1999) J. Am. Chem. Soc. 121 1037–1044 [Google Scholar]
- 19.Wetmore, S. D., Smith, D. M., and Radom, L. (2000) J. Am. Chem. Soc. 122 10208–10209 [Google Scholar]
- 20.Wetmore, S. D., Smith, D. M., Golding, B. T., and Radom, L. (2001) J. Am. Chem. Soc. 123 7963–7972 [DOI] [PubMed] [Google Scholar]
- 21.Bradford, M. M. (1976) Anal. Biochem. 72 248–254 [DOI] [PubMed] [Google Scholar]
- 22.Zelder, O., Beatrix, B., Leutbecher, U., and Buckel, W. (1994) Eur. J. Biochem. 226 577–585 [DOI] [PubMed] [Google Scholar]
- 23.Mansoorabadi, S. O., Padmakumar, R., Fazliddinova, N., Vlasie, M., Banerjee, R., and Reed, G. H. (2005) Biochemistry 44 3153–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang, K. H., Casarez, A. D., Wu, W., and Frey, P. A. (2003) Arch. Biochem. Biophys. 418 49–54 [DOI] [PubMed] [Google Scholar]
- 25.Barker, J. J., and Stadman, T. C. (1984) in B12, Vol. 2, pp. 203–231, John Wiley and Sons, Inc., New York [Google Scholar]
- 26.Tseng, C. H., Yang, C. H., Lin, H. J., Wu, C., and Chen, H. P. (2007) FEMS Microbiol. Lett. 274 148–153 [DOI] [PubMed] [Google Scholar]
- 27.Mori, K., and Toraya, T. (1999) Biochemistry 38 13170–13178 [DOI] [PubMed] [Google Scholar]
- 28.Roymoulik, I., Chen, H. P., and Marsh, E. N. (1999) J. Biol. Chem. 274 11619–11622 [DOI] [PubMed] [Google Scholar]
- 29.Bandarian, V., and Reed, G. H. (1999) Biochemistry 38 12394–12402 [DOI] [PubMed] [Google Scholar]
- 30.Abend, A., Bandarian, V., Reed, G. H., and Frey, P. A. (2000) Biochemistry 39 6250–6257 [DOI] [PubMed] [Google Scholar]
- 31.Wetmore, S. D., Smith, D. M., and Radom, L. (2001) J. Am. Chem. Soc. 123 8678–8689 [DOI] [PubMed] [Google Scholar]
- 32.Toney, M. D. (2001) Biochemistry 40 1378–1384 [DOI] [PubMed] [Google Scholar]
- 33.Bach, R. D., Canepa, C., and Glukhovtsev, M. N. (1999) J. Am. Chem. Soc. 121 6542–6555 [Google Scholar]
- 34.Smith, D. M., Golding, B. T., and Radom, L. (1999) J. Am. Chem. Soc. 121 1383–1384 [Google Scholar]
- 35.Michel, C., Albracht, S. P., and Buckel, W. (1992) Eur. J. Biochem. 205 767–773 [DOI] [PubMed] [Google Scholar]
- 36.Padmakumar, R., and Banerjee, R. (1995) J. Biol. Chem. 270 9295–9300 [DOI] [PubMed] [Google Scholar]
- 37.Bandarian, V., and Reed, G. H. (2002) Biochemistry 41 8580–8588 [DOI] [PubMed] [Google Scholar]
- 38.Jones, A. R., Hay, S., Woodward, J. R., and Scrutton, N. S. (2007) J. Am. Chem. Soc. 129 15718–15727 [DOI] [PubMed] [Google Scholar]
- 39.Finke, R. G., and Hay, B. P. (1984) Inorg. Chem. 23 3041–3043 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.