Abstract
The red light-sensing photoreceptor FphA from Aspergillus nidulans is involved in the regulation of developmental processes in response to light. Here we present extended biochemical and spectroscopic characterization of recombinant FphA using a synthetic gene with host-adapted codon usage. The recombinant photosensory domain FphAN753 was shown to display all features of a bona fide phytochrome. It covalently binds biliverdin as chromophore and undergoes red/far-red light-inducible photoconversion with both parent states being protonated. The large N-terminal variable extension of FphA exerts a stabilizing effect on the active Pfr state. Upon substitution of the highly conserved histidine 504, involved in the hydrogen-bonding network of the protein moiety and the chromophore, chromophore attachment and photoreversibility were completely impaired. FphA is a functional sensor histidine kinase with a strong red-light-dependent autophosphorylation activity. Furthermore, intermolecular trans-phosphorylation to the response regulator domain of a second monomer could be demonstrated. Interestingly, co-incubation of FphA and FphA variants led to enhanced autophosphorylation, including the “inactive” Pr form. The latter observed phenomenon might suggest that auto- and trans-phosphorylation activity is modulated by additional interaction partners leading to variable phosphorylation events that trigger a specific output response.
Fungi are capable of responding to radiance from UV-C to far-red light (1). The first photoreceptors described in fungi belong to the family of blue light receptors, which can be subdivided into phototropins and cryptochromes. The phototropin-like photoreceptor and DNA-transcription factor WHITE COLLAR 1 (WC-1)2 from Neurospora crassa is the best characterized blue light receptor in fungi so far. It has been shown that WC-1 in complex with WC-2 mediates almost all blue light responses such as photoadaptation processes as well as the entrainment of the circadian clock (2, 3).
Only recently, the discovery of phytochromes in the genomes of heterotrophic bacteria and several fungi (4–6) confirmed the existence of a red light-sensing system beyond photosynthetic organisms. Phytochromes belong to the family of red/far-red light sensing photoreceptors that are able to reversibly photoisomerize between two stable conformations, a red light-absorbing Pr form and a far-red light-absorbing Pfr form. This photointerconversion is mediated by a linear tetrapyrrole prosthetic group (bilin), which undergoes a Z-E isomerization around the C15–C16 double bond. The apo-protein autocatalytically assembles the bilin chromophore to form a covalent thioether linkage with a conserved cysteine residue. Interestingly, in bacteria and fungi the chromophore attachment site as well as the chromophore itself differ from cyanobacterial and plant phytochromes indicating the prokaryotic origin of fungal phytochromes (4). The bilins of plant and cyanobacterial phytochromes are the structurally related linear tetrapyrroles phytochromobilin and phycocyanobilin, respectively. Both are coupled via the A-ring ethylidene side chain (C31) to a conserved cysteine in the GAF domain of the phytochrome. On the other hand, all bacterial and fungal phytochromes thus far identified, assemble the less reduced linear tetrapyrrole biliverdin (BV) through their A-ring vinyl-group (C32) to a conserved cysteine within the PAS domain (7, 8).
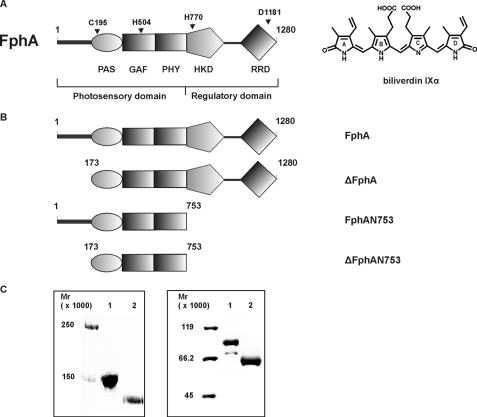
For the fungal phytochrome (Fph) FphA of Aspergillus nidulans cysteine 195 within the PAS domain was shown to be the site of attachment for BV (4) (Fig. 1A). All phytochromes share a common domain organization. They typically consist of an N-terminal photosensory domain, harboring the bilin chromophore and a C-terminal output module. The photosensory domain comprises a PAS (PER-ARNT-SIM), GAF (cGMP-specific phosphodiesterases; cyanobacterial adenylate cyclases; formate hydrogen lyase), and PHY (phytochrome) domain (9). In particular fungal phytochromes possess an N-terminal variable extension (NTE) preceding the PAS domain (Fig. 1) (4,6), which is not homologues to the serine-rich N-terminal P1 domain of phytochromes from plants (9). The C-terminal output modules are usually more diverged, but most bacterial and fungal phytochromes consist of light-regulated histidine kinase domains (4, 8, 10, 11). Fungal phytochromes are multifunctional proteins combining a histidine kinase domain and a response regulator domain (RRD) in one protein (Fig. 1A). Genes coding for corresponding response regulators in bacteria are most often encoded separately within the bacteriophytochrome-encoding (bph) operon (12). The presence of a histidine kinase domain in combination with a response regulator domain indicates that fungal phytochromes originate from an early bacterial hybrid kinase (13).
FIGURE 1.
A, domain organization of the fungal phytochrome FphA. Dark gray bar, variable N-terminal extension; PAS, PAS domain; GAF, GAF domain; and PHY, phytochrome domain (GAF-related domain) comprising the N-terminal photosensory domain. The C-terminal regulatory domain consists of histidine kinase domain (HKD) and response regulator domain (RRD). The following residues either essential for chromophore binding or coordination as well as for the signal output were assigned. C195, biliverdin IXα binding residue; H504, essential for chromophore coordination; H770, autophosphorylation site; and D1181, trans-phosphorylation site. The chemical structure of the covalently bound chromophore biliverdin IXα is shown on the right. B, domain structure of FphA variants. FphA, full-length protein; ΔFphA, protein without N-terminal variable extension; FphAN753, photosensory domain; ΔFphAN753, photosensory domain without N-terminal variable extension. C, SDS-PAGE analysis of recombinant, Streptactin-affinity purified holo-FphA variants. Left panel: 1, holo-FphA; 2, holo-ΔFphA_H770A; right panel: 1, holo-FphAN753; 2, holo-ΔFphAN753. The samples were either loaded onto a 7.5% SDS-gel (left panel) or onto a 10% SDS-gel (right panel), and the proteins were visualized by staining with Coomassie Brilliant Blue.
Red light-sensing has been extensively studied in the filamentous ascomycete A. nidulans. In addition, putative fungal phytochrome genes have been identified in the genomes of other ascomycetous fungi such as N. crassa, Gibberella zeae, Cochliobolus heterostrophus, and A. fumigatus, and also in the basidiomycetes Ustilago maydis and Cryptococcus neoformans (4, 14).
A. nidulans requires red light for asexual sporulation (15), but the involved red light photoreceptor was discovered and initially characterized only a few years ago (4). An A. nidulans fphA null mutant in comparison to the wild-type (wt) strain showed a clear developmental phenotype. Red light conditions led to enhanced sexual development in the ΔfphA mutant. Initial experiments revealed a cytoplasmic localization of the phytochrome (4), whereas further studies also identified a fraction of FphA in the nucleus (16). It was shown that FphA is part of a large blue- and red light-sensing protein complex, thereby interacting with the WC-2 homologue LreB and with the global regulator Velvet (VeA) (16). Recombinant FphA was characterized as a bona fide phytochrome with absorbance maxima at 707 and 754 nm for the Pr and Pfr forms, respectively. FphA most probably integrates the signal light via two-component signaling phosphorelays, because the protein contains both elements of a two-component regulatory system (Fig. 1A).
Although initial characterization on the putative FphA activity has been performed, proceeding experiments appeared extremely difficult because of unfavorable expression rates (data not shown). In this study we present an extended photobiochemical characterization of FphA using a synthetic fphA version with a host-adapted codon usage. This synthetic gene enabled us to produce adequate amounts of several FphA variants for stepping deeper into spectroscopic and functional analyses.
EXPERIMENTAL PROCEDURES
Bilin Preparations—BV IXα was purchased from Frontier Scientific (Carnforth, UK) and dissolved in DMSO prior to use.
Construction of Expression Vectors—A synthetic gene encoding the full-length FphA was adapted to the codon bias of Escherichia coli genes to provide a successful expression. The synthetic gene fphA of 3840 bp was assembled from synthetic oligonucleotides (GENEART GmbH, Regensburg, Germany). The synthetic DNA sequence has been deposited in GenBank™ under the accession number EU433998.
fphAN753s encoding the first 753 amino acids containing photosensory domain was amplified by PCR using primers 5′-CCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGAGCGAGCTGCCGAGC-3′ (fwd) and 5′-CCCTCGAGCGCTTCCTGCTGACGCCACAC-3′ (rev) and cloned after XbaI and XhoI restriction (restriction sites underlined in primer sequence) into pASK-IBA3 to get the E. coli expression vector pASK-fphAN753s. The site-directed mutants C195A and H504A were constructed using the QuikChange® site-directed mutagenesis kit (Stratagene) with the following primers (the underlined codon represents the introduced mutations): C195A, 5′-CCGTGGATAGCTTTAAAGCGGCCGAAGATGAACCGATTCATATTCC-3′ and H504A, 5′-CGTGCGATGAGCCCGATTGCTATCAAATATCTGGCCAACATGC-3′; the second primer was the complement. To yield an expression plasmid containing fphAN753s without the coding region for the N-terminal variable extension (173 amino acids), ΔfphAN753s, the PAS-GAF-PHY coding region was amplified via PCR and cloned into XbaI, XhoI restriction sites (in sequence underlined) in pASK-IBA3. Primers were the following: 5′-CCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGTGCGAAGATGAACCGATTCATATTCCG-3′ (fwd) and 5′-CCCTCGAGCGCTTCCTGCTGACGCCACAC-3′ (rev).
The full-length gene fphAs was cloned into pASK-IBA3 after XbaI and NcoI restriction to yield pASK-fphAs. The full-length gene without the coding region for the N-terminal variable extension, namely ΔfphAs, was amplified by using primers 5′-CCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGTGCGAAGATGAACCGATTCATATTCCG-3′ (fwd) and 5′-CGCCATGGTCGCTATGGGTATACGGGGTAAACC-3′ (rev) and cloned into pASK-IBA3 into XbaI and NcoI restriction sites to yield pASK-ΔfphAs.
The variant D1181A was constructed using the primer 5′-GGGATGTGGTGCTGATGGCTATTCAGATGCCGATTCTGG-3′, the second primer was the complement, and the underlined codons represent the exchange. With the purpose of hampering autophosphorylation the variant H770A of pASK-ΔfphAs was constructed with the primer 5′-GCTGGCCAACAGCGCGGCTGAAGTGCGTACCCCG-3′, the second primer was the complement, and the underlined codon represents the exchange. The integrity of the resultant plasmid constructs as well as the variants were confirmed by DNA sequencing (MWG, Ebersberg, Germany).
Protein Production and Purification—Recombinant A. nidulans FphA and all variants were produced using a tet promoter-driven C-terminal Strep tag expression system (IBA, Göttingen, Germany) in the E. coli strain BL21(λDE3). Cells were grown at 37 °C in Luria-Bertani medium containing ampicillin (100 μg·ml–1) to an A578 of 0.5. Gene expression was induced by the addition of 0.2 μg·ml–1 anhydrotetracycline, and cells were incubated for further 3 h at 30 °C. The bacterial pellet of 3 liters of culture was resuspended in elution and washing buffer (EW, 50 mm Tris/HCl, pH 7.8, 300 mm NaCl, 10% glycerol, 0.05% Tween 20; 1 mm dithiothreitol, and 0.25 mm 4-(2-aminoethyl) benzenesulfonyl fluoride were added freshly) (6 ml buffer per gram of cell weight) and disrupted by sonication. Cell debris was removed by centrifugation (90 min, 28,000 × g, 4 °C), and the supernatant was incubated with 40 μg·ml–1 avidin (final concentration) for at least 10 min on ice. Subsequently, the supernatant was loaded on to a Streptactin-Sepharose column (3 ml), which had previously been incubated with EW buffer. The purification was performed according to the instructions supplied by the manufacturer. Fractions containing the desired protein were combined and concentrated to a protein concentration of ∼100 μm using Vivaspin concentrator devices (MWCO 10,000, Sartorius Stedim Biotech, Göttingen, Germany). To obtain holo-FphA variants E. coli BL21(λDE3) cells were cotransformed with the plasmids pASK-fphAs, pASK-fphAs_D1181A, pASK-fphAN753s, or pASK-ΔfphAN753s and pAT-BV encoding hemeoxygenase 1 from Synechocystis sp. PCC 6803 under control of an isopropyl-β-thiogalactopyranoside-inducible Plac promoter as described before (4, 17). For holo-phytochrome synthesis the medium contained 50 μg·ml–1 ampicillin and 25 μg·ml–1 kanamycin. Bilin biosynthesis was induced using 250 μm isopropyl-β-thiogalactopyranoside 1 h before induction of the FphA synthesis to provide a sufficient amount of chromophore for the assembly. During production and purification of holo-FphA all steps were carried out under a green safelight or in the dark. Purified protein was subsequently concentrated to a final concentration of ∼100 μm (as described before) and either used directly or frozen in liquid nitrogen and stored at –20 °C.
Protein Determination—Protein concentration was determined by measuring A280 using the calculated ε280 nm for each individual protein: ε280 nm = 72,400 m–1·cm–1 for FphAN753, ε280 nm = 60,310 m–1·cm–1 for ΔFphAN753, ε280 nm = 113,650 m–1·cm–1 for FphA, and ε280 nm = 101,560 m–1·cm–1 for ΔFphA (18).
Chromophore-Protein Interactions—All experiments were performed under a green safelight. For photochemical analyses of FphAN753- or ΔFphAN753-BV in vivo reconstituted and purified holo-phytochrome was adjusted to ∼10 μm in buffer M (50 mm Tris/HCl, pH 8.0, 20 mm KCl) as judged by absorbance at 280 nm. Spectral analyses were performed with an 8453 UV/visible System (Agilent Technologies, Boeblingen, Germany) equipped with a temperature-controlled cuvette holder. For irradiation, light from a tungsten-halogen source (LOT-ORIEL, Darmstadt, Germany) was passed through an interference filter (LOT-ORIEL) of 636 nm (636FS10–50) and through a 750 nm filter (Wratten No. 87, Kodak, Stuttgart, Germany) with light intensities of 12 μmol·m–2·s–1 and 4 μmol·m–2·s–1, respectively. Absorbance spectra were obtained after 3-min illumination with red light at 636 nm (Pfr spectrum) and after 3-min incubation with far-red light at 750 nm (Pr spectrum) in a volume of 500 μl, and the difference was calculated. To characterize the different forms of holo-FphAN753 spectroscopically, absorbance spectra between 250 and 800 nm were measured at ∼18 °C during incubation in the dark or during irradiation with red and far-red light, respectively.
In vitro chromophore assembly of FphAN753 and the variants FphAN753_C195A and -H504A or ΔFphAN753 with BV was generally tested using 10 μm of recombinant apo-protein incubated with 10 μm chromophore in EW buffer for 30 min in the dark at ∼20 °C (final volume, 500 μl), and again absorbance spectra between 250 and 800 nm were taken during incubation in the dark or under irradiation with red and far-red light. Covalent chromophore attachment to the FphA variants was tested by visualizing covalently bound bilins via zinc-induced red fluorescence as described previously (19).
Resonance Raman Spectroscopy—Resonance Raman (RR) spectra were recorded with 1064 nm excitation (Nd-YAG cw laser, line width <1cm–1) using an RFS 100/S (Bruker Optics, Ettlinger, Germany) Fourier-transform Raman spectrometer (4 cm–1 spectral resolution). The near-infrared excitation line was sufficiently close to the first electronic transition to generate a strong pre-resonance enhancement of the chromophoric vibrational bands such that Raman bands of the protein matrix remained very weak in the spectra of the parent states. All spectra were measured at –140 °C using a liquid-nitrogen cooled cryostat (Linkam, Waterfield, Surrey, UK). The laser power at the sample was set at ∼700 milliwatts, which did not damage the chromoproteins as checked by comparing the spectra of the samples obtained before and after RR data acquisition. The total accumulation time was <2 h for each spectrum. For all RR spectra shown in this work, a polygonal baseline was subtracted. Photoproducts were accumulated by irradiating the samples for a few minutes at the specified temperatures. These raw RR spectra included substantial contributions from residual Pr, which was subtracted using the characteristic RR bands of Pr as a reference. Further RR experimental details have been described previously (20).
Protein Kinase Assay—Autophosphorylation was performed as described for Pseudomonas aeruginosa BphP (21). Before the addition of [32P]ATP[γP] 10 μg of FphA_D1181A-BV was either irradiated with red (636 nm) or far-red (750 nm) light for 30 min at 18 °C and incubated in darkness at room temperature for different periods of time. The reaction was stopped with SDS sample buffer without incubation at 95 °C, and the samples were subsequently loaded onto a 7.5% SDS-PAGE and blotted onto a polyvinylidene difluoride membrane (Roth, Karlsruhe, Germany). Radioisotope imaging was monitored by using a hyperscreen x-ray film (Fuji Photo Film Europe, Düsseldorf, Germany), thereafter the blot was stained with Ponceau S to confirm equal protein load.
In Vitro His-Asp Phosphotransfer—To demonstrate phosphotransfer from the conserved histidine residue of the histidine kinase domain to the conserved aspartate of the RRD, holo-FphA_D1181A photoconverted to the Pr and Pfr form was autophosphorylated for 1.5 h as described under “Protein Kinase Assays.” Afterwards the samples were mixed with 10 μg of holo-ΔFphA_H770A or apo-ΔFphA_H770A and incubated for 1.5 h in the dark at room temperature. Further procedures were the same as described under “Protein Kinase Assay.” As negative control one sample was treated the same without addition of holo-FphA_D1181A.
RESULTS
Low Production Rates of Recombinant FphA Are Due to an Unfavorable Codon Usage—Initial characterization on the putative phytochrome activity has been performed, but proceeding with distinct experiments was extremely difficult because of unfavorable expression rates. Minor expression rates were due to an unadapted codon usage toward E. coli. Application of the Codon Adaptation Tool (JCAT) of the PRODORIC data base (www.prodoric.de) indicated a Codon Adaptation Index-Value (CAI-Value) of ∼0.23 for fphA to E. coli K12 (42). Consequently, a synthetic gene of 3840 kb with an optimized CAI-Value of 0.98 was constructed (accession number EU433998) to overcome expression limitations. Based on this construct several variants were cloned and expressed in E. coli (Fig. 1B). All FphA variants were purified (Fig. 1C) and used for further photobiological characterization.
FphAN753 Behaves Like a True Phytochrome—A. nidulans FphAN753, comprising only the photosensory domain (Fig. 1, A and B) was either purified as holo-phytochrome assembled with BV in vivo or as apo-protein. For holo-phytochrome production in E. coli a dual plasmid system was employed as described for the biosynthesis of holo-BphP (21). A band migrating at ∼86,000 was obtained on SDS-PAGE, which correlates with the predicted molecular mass from the amino acid composition of the photosensory domain containing an additional Strep-tag (Fig. 1C, right panel, lane 1). Because in vivo reconstituted holo-FphAN753 revealed a greater stability than the in vitro assembled holo-protein, the purified in vivo complex was used for further photochemical analyses. The yield of purified holo-FphAN753 was typically 2.5 mg per liter of bacterial culture.
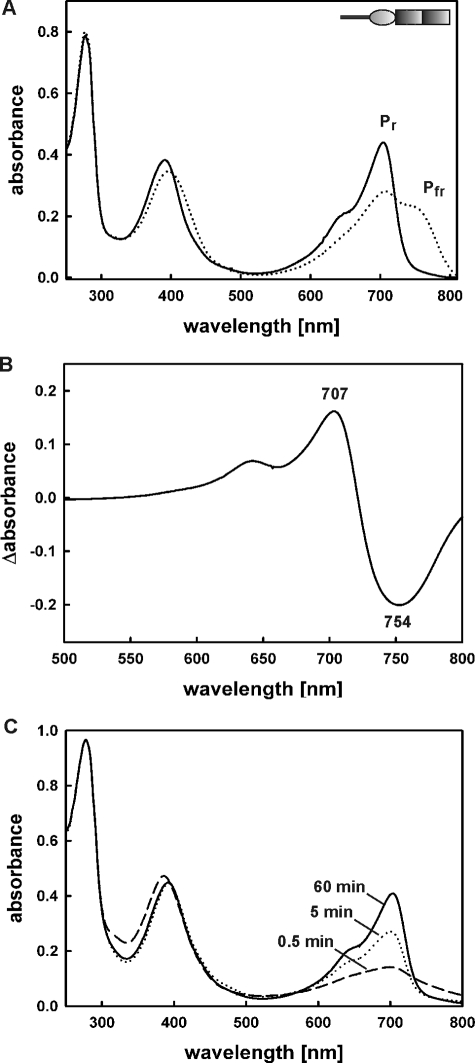
Recombinant holo-FphAN753 assembled with BV, most likely the natural chromophore, exhibits a characteristic red/far-red light reversible phytochrome signature. Illumination with saturated red light (636 nm) resulted in formation of the Pfr form (Amax 754 nm), which could be converted back into the Pr form (Amax 707 nm) through illumination with far-red light (750 nm) (Fig. 2A). The resultant calculated Pr-Pfr difference spectrum confirms the photoactivity of holo-FphAN753 (Fig. 2B). Both forms show absorption maxima at longer wavelengths characteristic of BV-binding bacterial and fungal phytochromes (8, 12, 21–24). The form initially synthesized in the dark is the Pr form (Fig. 2C). Neither the Pr- nor the Pfr form undergo a defined dark reversion, known to be a thermal relaxation process of non-photochemical Pfr-to-Pr dark reversion especially found by dicot plant phytochromes to regulate the signal output (25, 26) (Fig. 3A).
FIGURE 2.
Spectral properties of holo-FphAN753. A, absorbance spectra of recombinant FphA(N753)-BV obtained by holo-expression. Pr, Pr form established after illumination with far-red light (750 nm) (solid line); Pfr, Pfr-enriched form obtained after illumination with red light (636 nm) (long dashed line). The inset presents the domain organization of the photosensory domain (Fig. 1B). B, calculated Pr-Pfr difference spectrum of purified recombinant FphAN753-BV. Absorption maximum and minimum are indicated in nanometers. C, absorbance spectrum changes within 60 min in the dark during assembly of 10 μm apo-FphAN753 with 10 μm BV.
FIGURE 3.
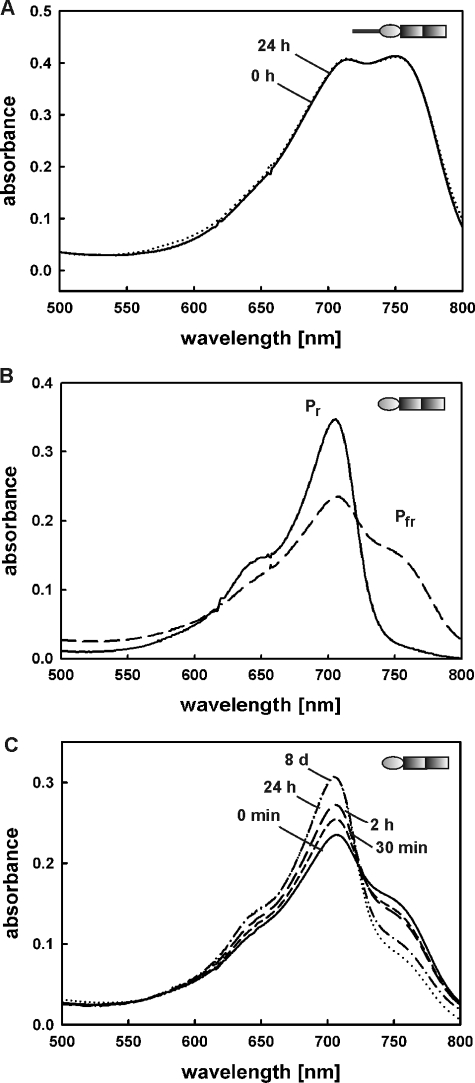
Spectral properties of BV-holoproteins FphAN753 and ΔFphAN753. A, dark reversion of recombinant FphAN753-BV after saturating illumination with red light. B, absorbance spectra of recombinant ΔFphAN753-BV obtained by holo-expression. Pr, Pr form established after illumination with far-red light (750 nm) (solid line); Pfr, Pfr-enriched form obtained after illumination with red light (636 nm) (long dashed line). C, dark reversion of ΔFphAN753-BV after saturating illumination with red light. Absorbance spectra were recorded at the indicated time points.
The N-terminal Variable Extension Stabilizes the Pfr Form—Fungal phytochromes identified so far also possess an N-terminal variable extension, which is not homologous to those of plant phytochromes. In case of A. nidulans phytochrome FphA the NTE is composed of ∼172 amino acids and comprises no conserved serine-residues. To investigate the role of this large subdomain (calculated Mr 18,300) on FphA photoreversibility or stability, NTE deletion mutants of the photosensory domain and the full-length protein (ΔFphAN753 and ΔFphA) were constructed (Fig. 1B). Holo-ΔFphAN753 also senses red and far-red light through photointerconversion between both photoreversible states, Pr and Pfr (Fig. 3B). Both forms showed slightly blue-shifted absorbance maxima in comparison to the complete photosensory domain and displayed a characteristic phytochrome difference spectrum (data not shown). Additionally, irradiation with red light resulted in a lower yield of the Pfr form, producing a mixture of ∼60% Pr and only 40% Pfr (Fig. 3B), indicating that the NTE might exert a stabilizing effect on the Pfr form and pointing to a lower thermal stability of the truncated protein. The involvement of the N-terminal variable extension in Pfr stability was further supported by the fact that inhibition of Pfr-to-Pr dark reversion was released (Fig. 3C). After photoconversion into the Pfr form followed by incubation at 15 °C in the dark for 24 h, a significant reduction of the Pfr absorption maximum at 750 nm was observed. Further incubation in the dark for additional 6 days at 4 °C did not alter the spectrum (Fig. 3C). Although the FphA N-terminal variable extension is not homologous to those of plant phytochromes, this result is consistent with investigations on the plant phytochrome phytochrome A where deletion of 100 N-terminal amino acids led to an enhanced dark reversion (25).
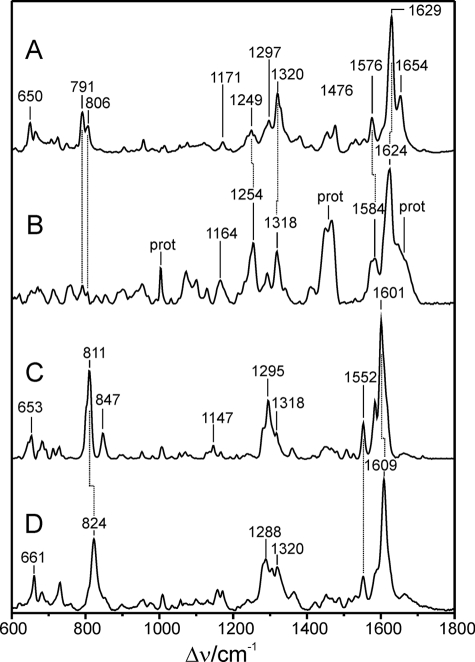
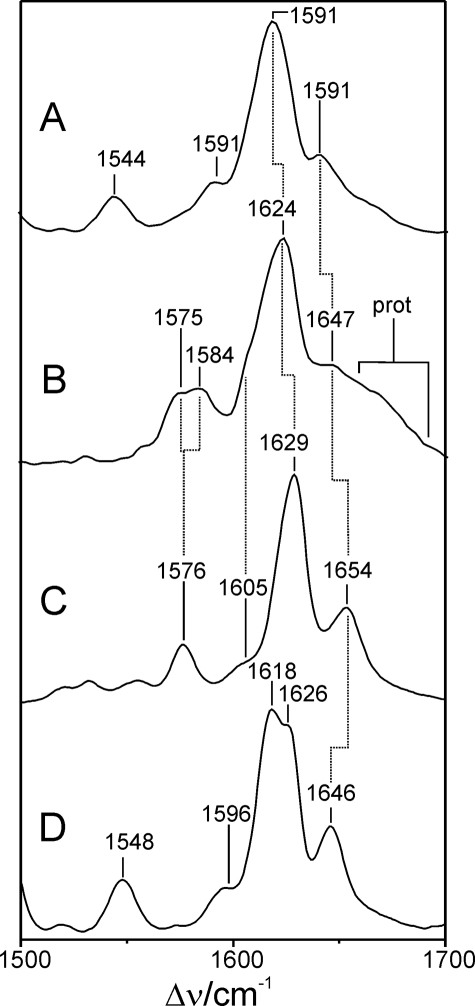
Resonance Raman Spectroscopy of FphAN753-BV Confirmed Protonation of the Chromophore during Photoconversion—The RR spectrum of the Pr state of FphAN753 is dominated by bands in the region between 1200 and 1700 cm–1, which is characteristic of all phytochromes (Fig. 4). However, there are considerable differences compared with the RR spectrum of the Pr state of bacterial phytochrome DrBphP, which is shown for comparison (20). These differences partly result from the higher spectral contribution of the protein matrix such as the broad hump at ∼1660 cm–1 and the intense doublet at ∼1460 cm–1, attributable to the amide I modes of the peptide backbone and methylene/methyl deformation modes of amino acid side chains, respectively. The substantial interference with Raman bands of the apo-protein is further reflected by the sharp band at ∼1005 cm–1 that originates from the ring-breathing modes of the Phe residues. Spectral differences solely attributable to the RR bands of the chromophore refer to the C–H out-of-plane modes at 791 and 806 cm–1, which exhibit much lower intensity in FphAN753, the modes including C–H and N–H in-plane bending and internal coordinates of the pyrrole substituents (1150 and 1350 cm–1), and the so-called marker band region between 1500 and 1700 cm–1. The bands in this latter region are known to be particularly sensitive for the configuration (Z/E) and conformation (syn/anti) of the tetrapyrrole geometry and for the protonation state of the chromophore. In the Pr state of FphAN753, these bands are distinctly broader than in DrBphP (Fig. 5). The strongest band at 1624 cm–1, attributable to the C=C stretching of the C–D methine bridge, exhibits a bandwidth of >20 cm–1 as compared with ∼15 cm–1 of the 1629-cm–1 band of DrBphP. Furthermore, the band is downshifted by 5 cm–1 in FphAN753. A comparable downshift is noted for the A–B stretching mode, which is observed at 1654 cm–1 in DrBphP and can be identified at 1647 cm–1 in FphAN753, partly obscured by the broad amide I band. Most remarkable, however, is the N–H in-plane bending of the pyrrole rings B and C, which gives rise to the band at 1576 cm–1 of DrBphP. Upon H/D exchange, this band disappears, and the corresponding N–D in-plane bending is observed at ∼1070 cm–1 (20). Thus, the 1576-cm–1 band is considered as a characteristic marker band of the protonated (cationic) form of tetrapyrroles. The counterpart of this band in the spectrum of FphAN753 is a doublet with band components at 1575 and 1584 cm–1, which both vanish in D2O (Fig. 5). This splitting can be attributed to the coexistence of two protonated chromophore species that differ in hydrogen bond interactions of the pyrrole rings B and C, and possibly also in details of the tetrapyrrole geometry, thereby accounting for the broadening of the bands in the methine bridge stretching region. However, in view of the overall similarities of the vibrational band pattern, specifically in the region of the marker bands, these structural differences are beyond the level of the methine bridge configuration and conformation, which for both Pr species of FphAN753 is thus likely to be the same as for the Pr chromophore in DrBphP.
FIGURE 4.
RR spectra of the Pr and Pfr states of FphAN753 and DrBphP from D. radiodurans, measured from samples dissolved in H2O. A, Pr and DrBphP; B, Pr and FphAN753; C, Pfr and FphAN753; and D, Pfr and DrBphP. Spectra of DrBphP were taken from a previous study (20).
FIGURE 5.
RR spectra of the Pr states of FphAN753 and DrBphP from D. radiodurans, measured from samples dissolved in H2O and D2O. The spectra display the marker band region in an expanded view. A, FphAN753 and D2O; B, FphAN753 and H2O; C, DrBphP and H2O; and D, DrBphP and D2O. Spectra of DrBphP were taken from a previous study (20).
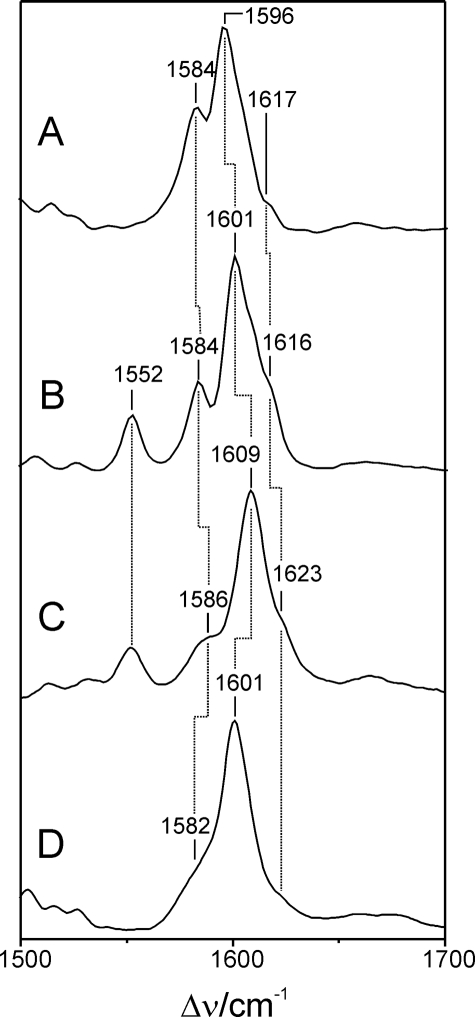
Upon light absorption of the Pr state, FphAN753 undergoes a photocycle similar to other phytochromes. In the intermediate Meta-Rc, cryogenically trapped at –30 °C, the chromophore is deprotonated as demonstrated by the lack of the N–H in-plane bending of the rings B and C in the RR spectrum, in analogy to the corresponding intermediate state of DrBphP (20) (supplemental Fig. S1). Also the RR spectra of the Pfr states are similar in FphAN753 and DrBphP (Fig. 6) and, unlike to the Pr state, there is no heterogeneity associated with the N–H in-plane bending of the rings B and C, which is observed in both cases at 1552 cm–1. However, we note striking differences in the same spectral regions as for Pr. In FphAN753, the prominent torsional mode at 653 cm–1 and the characteristic C–H out-of-plane (HOOP) mode of the C–D methine bridge at 811 cm–1 are downshifted by 8 and 13 cm–1, respectively. In addition, the band at 847 cm–1 that is assigned to the A–B HOOP mode has gained considerable intensity in FphAN753 pointing to a larger torsional dihedral of the A–B methine bridge as compared with DrBphP. Band intensities and frequencies between 1250 and 1350 cm–1 differ strongly between both proteins. In the marker band region the pattern of the methine bridge-stretching modes is preserved, that is a dominant band at 1601 cm–1 (C–D stretching) and a shoulder at 1616 cm–1 as well as band at 1584 cm–1, presumably attributable to the A–B and B–C stretching, respectively. However, the frequencies of these stretching modes are downshifted by 3–8 cm–1 in FphAN753.
FIGURE 6.
RR spectra of the Pfr states of FphAN753 and DrBphP from D. radiodurans, measured from samples dissolved in H2O and D2O. The spectra display the marker band region in an expanded view. A, FphAN753 and D2O; B, FphAN753 and H2O; C, DrBphP and H2O; and D, DrBphP and D2O. Spectra of DrBphP were taken from a previous study (20).
Because the crystal structure of Deinococcus radiodurans chromophore binding domain (DrCBD) (PDB code 1ZTU) in its Pr state gave more insights into protein-chromophore interactions, highly conserved amino acids flanking the phytochrome binding pocket were identified that are most probably essential for correct binding and coordination of the chromophore (27, 28). Among these amino acids the highly conserved aspartate Asp-207 (Asp-197 in Agp1 from Agrobacterium tumefaciens) was observed to play a critical role in proton release while the conserved histidine His-260 (His-250) is involved in proton uptake during photointerconversion from Pr to Pfr (20, 29). The corresponding mutation H504A in A. nidulans FphAN753 toward Agp1_H250A led to a complete loss of the chromophore-binding ability (supplemental Fig. S2), indicating that we eliminated an indispensable amino acid contributing to the hydrogen-bonding network in FphAN753.
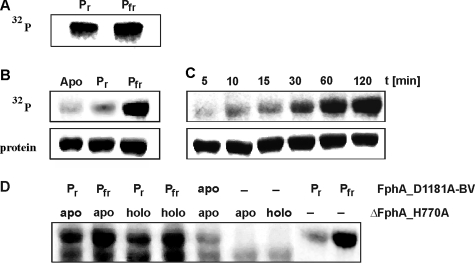
FphA Is a Functional Histidine Protein Kinase—The protein kinase activities of cyanobacterial and most bacterial phytochromes are light-regulated and dependent on the presence of the tetrapyrrole prosthetic group (8, 11, 12). Most probably protein phosphorylation plays an important role in integrating the light signal. FphA harbors a histidine-kinase module and additionally a response regulator domain, both elements of a functional two-component-system (Fig. 1A). Light-dependent autophosphorylation was examined using full-length FphA. Initial studies presented phosphorylation activity of holo-FphA in both light-inducible states (Fig. 7A). In view of the fact that FphA harbors a putative functional response regulator domain, the phosphorylation signal cannot clearly be assigned to either autophosphorylation or trans-phosphorylation activity or even to both. To hamper potential trans-phosphorylation activity, the conserved aspartate residue of the RRD was mutagenized. Both spectroscopic forms of purified holo-FphA_D1181A as well as apo-FphA_D1181A were incubated with [32P]ATP[γP] and autophosphorylation was visualized via autoradiography. The Pfr form phosphorylated very efficiently, whereas no phosphorylation signal could be detected in the Pr form or in the apo-protein (Fig. 7B). Autophosphorylation of the Pfr form occurred with slow kinetics and was almost saturated after 60 min (Fig. 7C).
FIGURE 7.
Autophosphorylation of holo-FphA variants and trans-phosphorylation of the response regulator within ΔFphA_H770A. A, autoradiogram after [32P]ATP[γP] labeling of holo-FphA, SDS-PAGE, and electroblotting. Radioactive labeling was only obtained in the dimeric form of holo-FphA. B, autoradiogram after [32P]ATP[γP] labeling of apo-FphA_D1181A and holo-FphA_D1181A in its Pr and Pfr form and (C) time course of holo-FphA_D1181A autophosphorylation in its Pfr form.10 μg of each protein was employed. D, autoradiogram of mixed holo-FphA_D1181A and ΔFphA_H770A samples after [32P]ATP[γP] labeling. For detection of phosphotransfer to the conserved aspartate of ΔFphA_H770A the autophosphorylated sample holo-FphA_D1181A, either in the Pr or the Pfr state, was mixed with 10 μg of apo- or holo-ΔFphA_H770A. Samples were further incubated for 1.5 h at room temperature. Autophosphorylation of holo-FphA_D1181A again was confirmed via [32P]ATP[γP] labeling of both forms. Apo- and holo-ΔFphA_H770A were also exclusively incubated with [32P]ATP[γP].
FphA Is Capable of His-Asp Phosphotransfer—Hybrid kinases are found in some prokaryotic and in many eukaryotic organisms. They usually contain multiple phosphodonor and -acceptor sites, which allow them to promote multistep phosphorelay schemes. To examine whether phosphotransfer to the RRD occurs, we intended to express and purify the RRD by itself, but purification failed due to highly insoluble protein (data not shown). Therefore, trans-phosphorylation experiments were performed with two FphA variants. FphA_D1181A served as autophosphorylating histidine kinase and ΔFphA_H770A, missing the conserved histidine responsible for autophosphorylation, was substituted for the RRD. Because these two variants differ in their relative molecular weight (MrFphA_D1181A ≈ 143,000; MrΔFphA_H770A ≈ 126,000), both proteins could be separated on a 7.5% SDS-PAGE. As control each protein was solely exposed to radioactive labeled [32P]ATP[γP]. When incubated with [32P]ATP[γP], again holo-FphA_D1181A extensively autophosphorylated only in its Pfr state (Fig. 7D, lane 9). Radiolabeled ΔFphA_H770A, as expected, showed only background levels of phosphorylation (Fig. 7D, lanes 6 and 7). To detect trans-phosphorylation, ΔFphA_H770A as holo- or apo-protein was added to phosphorylated holo-FphA_D1181A in its Pfr and Pr forms. Interestingly, the Pr form of holo-FphA_D1181A, which usually showed rather any phosphorylation signal, displayed a strong phosphorylation signal after incubation with holo- or apo-ΔFphA_H770A (Fig. 7D, lanes 1 and 3). In this case, the interaction of FphA_D1181A with a protein partner containing an “active” RRD stimulated the autophosphorylation of the Pr form. Trans-phosphorylation from FphA_D1181A-BV to ΔFphA_H770A occurred independent of bilin in the acceptor protein (Fig. 7D, lanes 1–4). Please note that holo-ΔFphA_H770A does not display a defined Pr or Pfr form due to the lack of the stabilizing NTE. Therefore, it was only designated “holo” here.
DISCUSSION
The Photosensory Domain of A. nidulans FphA Does Behave Like Bacterial Phytochromes—FphA is a hybrid kinase, a multifunctional protein combining the phytochrome region and histidine kinase domain in a single protein with a C-terminal response regulator domain. It is among the first fungal phytochromes to be discovered, and contrary to the N. crassa phytochromes PHY-1/PHY-2 which are still of unknown function, it is known to be involved in regulation of the asexual and sexual reproduction cycle of A. nidulans. In 1990 it had already been shown that by exposure to red light conidiation in A. nidulans is elicited while sexual development is repressed, but the corresponding photoreceptor had not been identified yet (15). A few years later, genome analyses of several fungi revealed putative phytochrome sequences in several genomes, and the deletion of the presumed phytochrome encoding gene fphA from A. nidulans indeed led to an enhanced de-repression of sexual development under red light irradiation pointing to a photoperceptive function of FphA (4). Initial characterization on the supposed phytochrome revealed that the protein acts as a photoactive red/far-red light sensor after assembly with BV. However, weak expression rates hampered a detailed biochemical and photobiological characterization. Employing a synthetic version of fphA significantly increased expression rates and yielded a sufficient amount of stable protein for further characterization. In agreement with phytochromes from other organisms we demonstrate that the photosensory domain by itself (FphAN753) assembled with BV acts as a bona fide phytochrome with all characteristics of a red/far-red light-reversible photoreceptor. Both phytochromic states showed red-shifted absorbance maxima in accordance with the BV-binding bacterial phytochromes compared with cyanobacterial and plant phytochromes (10, 11, 21). Because assembly of FphAN753 with BV in the dark always resulted in formation of a Pr state (Fig. 2C), the physiological ground state is established by the Pr form. This is consistent with the hypothesis of the Pfr form to be the light-activated state repressing sexual development (4). One of the interesting features of all phytochromes that has been extensively studied photobiochemically but remains to be exactly determined on the physiological level is the process of thermodynamic dark reversion of the active state to the ground state. Known from several bacterial phytochromes dark reversion occurs in time ranges from minutes to hours, indicative for controlling the signal output (8, 21, 30). Remarkably, neither the Pr nor the putative active Pfr form of FphA undergoes a defined dark reversion, although the Fph family appears to be most related to bacterial phytochromes. Instead the behavior of FphAN753 in the dark is reminiscent of monocot phytochrome A, in which proteolytic removal of 50–100 amino acids of the serine-rich N terminus led to enhanced dark reversion (25, 31). In agreement with most other phytochromes FphAN753 exists as an extended obligate dimer as demonstrated by gel permeation chromatography (data not shown).
BV Appears to Be the Natural Chromophore of FphA—Although several fungi contain heme oxygenases (32, 33) and no genes encoding bilin reductases it is still unclear from the genome sequence of A. nidulans which of the one or more enzymes provide the tetrapyrrole chromophore. Therefore, we can only speculate that BV is the natural chromophore of FphA. FphAN753 also binds phycocyanobilin but in a non-covalent manner (data not shown and Ref. 4). In this case, photoconversion toward the Pfr form displayed an extremely weak absorbance change similar to PHY domain-truncated phytochromes. Therefore, phycocyanobilin appears not to be the natural chromophore of FphA. Moreover, different bacterial phytochromes such as PsBphP from Pseudomonas syringae or DrBphP attached phycocyanobilin, but developed uncharacteristic phytochrome spectra (12). A recent study about blue- and red light-sensing in A. nidulans demonstrated that asexual development is connected to mycotoxin formation, which appeared to be more effective at light of 700 nm (16). This is in good agreement with the absorption maximum of the Pr state of FphA-BV. But to ascertain the exact nature of the chromophore, FphA needs to be purified directly from A. nidulans.
Resonance Raman Spectroscopy Confirmed Protonation of the Tetrapyrrole Chromophore during Photoconversion—RR spectroscopy confirmed that FphAN753-BV undergoes a full photocycle with a transient deprotonation of the chromophore, whereas the chromophore in the parent states Pr and Pfr is protonated as already shown for plant phytochromes and bacterial phytochromes DrBphP from D. radiodurans and Agp1 from A. tumefaciens (34, 35). The gross chromophore structures in the parent states are similar to DrBphP and other BV-binding proteins, which can be concluded from the similarities of the marker band region between 1500 and 1700 cm–1. The fact that all marker bands display lower frequencies seems to indicate that the differences do not exclusively originate from a localized structural change in the chromophore but suggests a perturbation of the electron delocalization, because the absorption maxima of the Pr and Pfr states of FphAN753 are slightly red-shifted (4–9 nm) in comparison to DrBphP (20). Moreover, FphAN753 did not appear to be fully saturated with BV, as reflected by the relative strong contributions of the protein Raman bands. However, the remarkable finding of a protonation marker doublet in the Pr state of FphAN753 indicates two species that differ with respect to hydrogen bonding interactions of the protein with rings B and C. Especially in this respect we have noted striking similarities with Agp2 from A. tumefaciens (36), which belongs to the bathy Bphs working in reverse, whereas for all other BV-binding phytochromes we have studied so far the Pr spectra are more similar to that of DrBphP. This heterogeneity associated with the N–H in-plane bending of the rings B and C detected in the Pr state was not observed for the Pfr form of FphAN753. In the Pfr state, the RR spectroscopic differences can be related to geometric differences between FphAN753 and DrBphP such as a higher dihedral angle of the C–D methine bridge, which is reflected by the large downshift of the respective C–H out-of-plane mode.
The Conserved Histidine 504 Is a Key Amino Acid for the Hydrogen Bonding Network and Photoreversibility of FphA—One crucial amino acid contributing to the hydrogen-bonding network that coordinates the chromophore in the phytochrome binding pocket and which probably serves as potential proton donor for the BV chromophore is His-260 in DrBphP (27). His-260 forms a hydrogen bond to an oxygen of the propionic acid side chain of ring C and also to the pyrrole water (Wat-12) that mediates the H-bonding network to the pyrrole rings A, B, and C (20). The phytochrome variants H250A (Agp1) and H260A (DrBphP) were still able to incorporate BV covalently and to create a Wt-like Pr form (20, 29). RR spectroscopic analyses confirmed that the chromophore was still protonated. However, the photocycle was blocked in a meta-Rc-like state, the formation of which is coupled to the release of a proton to the medium. Substitution of this conserved histidine by alanine prevents reprotonation of the chromophore (29). Upon substitution of the corresponding histidine 504 to alanine in FphAN753, the phytochrome variant fails to bind BV covalently and even non-covalently, and to generate a Pr-like state. A similar effect has been described for phytochrome A but seems to be dependent on the type of substitution. Although a H to G exchange has a drastic effect like that seen for the His-504 mutation in FphA, exchange of H with Q has no effect on phytochrome A photochromism, because the H-bonding groups are retained (37). Additionally, not only FphA_H504A but also FphA_C195A impairs BV binding (data not shown and Ref. 4) to form cofactor-protein adducts with absorption spectra characteristic of Pr and Pfr. This is in contrast to analogous mutants such as Agp1_C20A, which was even able to undergo photointerconversion (38).
Holo-FphA Is a Functional Sensor Histidine Kinase and Transphosphorylates the RRD of Another FphA Molecule—Most of the phytochromes identified so far are sensor histidine kinase proteins autophosphorylating at a conserved histidine residue in the regulatory domain. Some of them have even been demonstrated to trans-phosphorylate response regulators most often located in the bph operon (10, 12, 39). To exclusively analyze the light dependence of autophosphorylation the conserved aspartate residue of the RRD was exchanged to an alanine to hinder trans-phosphorylation. Accordingly, strong red light-dependent autophosphorylation was observed that so far have also been described for the P. syringae BphP and Agp2 from A. tumefaciens (10, 12) (Fig. 7, B and C). Most other bacterial or cyanobacterial phytochromes such as CphA and CphB from Calothrix sp. PCC7601 as well as Agp1 and BphP show autophosphorylation signals in the apo and in both holo forms, thereby displaying a more intense signal in the Pr form (8, 21, 22, 39). Considering the existence of the RRD in FphA, it is most likely that it takes part in light-dependent signal transduction. However, trans-phosphorylation experiments revealed somewhat unexpected results. Autophosphorylation of the Pr form was induced through interaction with ΔFphA_H770A. Thus the enhanced autophosphorylation activity of the Pr form is dependent on a functional RRD in the interacting protein, which could be also concluded from the protein kinase assays by the use of wt FphA (Fig. 7A). In addition trans-phosphorylation was independent of a chromophore in the receiver protein. These results point to the involvement of other interacting proteins that might modulate phosphorylation activity, thereby enabling a fine-tuning of specific output signals.
In this respect, the histidine-containing phosphotransfer protein (HPt) YpdA, which also serves in mitogen-activated protein kinase cascades, was shown to retrograde phosphorylate apo-FphA at aspartate Asp-1181 (40). Furthermore, the interaction of FphA with other proteins like the WC-2 homolog LreB and the regulator velvet (VeA) has been demonstrated (16, 41). It was found that the blue light-sensing complex LreA-LreB works as an antagonist of FphA by stimulating sexual development. Thereby, direct protein-protein interactions involving LreA and LreB, holo-FphA and LreB, and remarkably LreB and apo-FphA were observed. Through physical interactions with other proteins of the large signaling protein complex, FphA could be specifically modulated in its auto- and trans-phosphorylation behavior. Therefore, the tested physical interactions have to be analyzed in phosphorylation assays. However, because red light-dependent autophosphorylation exclusively occurred when Asp-1181 was mutagenized, it can be speculated that the RRD of holo-FphA is blocked under red light-sensing physiological conditions. Taken also into account the critical time slot during which A. nidulans must be exposed to light to elicit conidiation after becoming competent and being transferred from liquid to surface culture (15), FphA may not necessarily be sensitive toward light and specifically to red light all the time. Consequently, the red light sensitivity has urgently to be controlled as well. The results recently presented by Purschwitz and coworkers (16) combined with the designation of FphA to be a multitasking phytochrome protein depicts a tight regulatory interplay between blue and red light sensing in A. nidulans for exact tuning of asexual and sexual reproduction in response to light.
Supplementary Material
Acknowledgments
We are indebted to J. C. Lagarias for the gift of pAT-BV.
This work was supported by a grant from the Volkswagen Foundation (to N. F. D.) and funds from the Deutsche Forschungsgemeinschaft (Grant SFB 498 to P. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact. The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) EU433998.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: WC-1, -2, WHITE COLLAR-1 and -2; BV, biliverdin; DrCBD, D. radiodurans chromophore binding domain; Fph, fungal phytochrome; GAF, cGMP-specific phosphodiesterases, cyanobacterial adenylate cyclases, formate hydrogen lyase; NTE, N-terminal extension; PAS, PER-ARNT-SIM; RR, resonance Raman; RRD, response regulator domain; wt, wild type.
References
- 1.Purschwitz, J., Muller, S., Kastner, C., and Fischer, R. (2006) Curr. Opin. Microbiol. 9 566–571 [DOI] [PubMed] [Google Scholar]
- 2.He, Q., Cheng, P., Yang, Y., Wang, L., Gardner, K. H., and Liu, Y. (2002) Science 297 840–843 [DOI] [PubMed] [Google Scholar]
- 3.He, Q., and Liu, Y. (2005) Genes Dev. 19 2888–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumenstein, A., Vienken, K., Tasler, R., Purschwitz, J., Veith, D., Frankenberg-Dinkel, N., and Fischer, R. (2005) Curr. Biol. 15 1833–1838 [DOI] [PubMed] [Google Scholar]
- 5.Davis, S. J., Vener, A. V., and Vierstra, R. D. (1999) Science 286 2517–2520 [DOI] [PubMed] [Google Scholar]
- 6.Froehlich, A. C., Noh, B., Vierstra, R. D., Loros, J., and Dunlap, J. C. (2005) Eukaryot. Cell 4 2140–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamparter, T., Carrascal, M., Michael, N., Martinez, E., Rottwinkel, G., and Abian, J. (2004) Biochemistry 43 3659–3669 [DOI] [PubMed] [Google Scholar]
- 8.Lamparter, T., Michael, N., Mittmann, F., and Esteban, B. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 11628–11633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montgomery, B. L., and Lagarias, J. C. (2002) Trends Plant Sci. 7 357–366 [DOI] [PubMed] [Google Scholar]
- 10.Karniol, B., and Vierstra, R. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 2807–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh, K. C., Wu, S. H., Murphy, J. T., and Lagarias, J. C. (1997) Science 277 1505–1508 [DOI] [PubMed] [Google Scholar]
- 12.Bhoo, S. H., Davis, S. J., Walker, J., Karniol, B., and Vierstra, R. D. (2001) Nature 414 776–779 [DOI] [PubMed] [Google Scholar]
- 13.Pao, G. M., and Saier, M. H., Jr. (1997) J. Mol. Evol. 44 605–613 [DOI] [PubMed] [Google Scholar]
- 14.Karniol, B., Wagner, J. R., Walker, J. M., and Vierstra, R. D. (2005) Biochem. J. 392 103–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mooney, J. L., and Yager, L. N. (1990) Genes Dev. 4 1473–1482 [DOI] [PubMed] [Google Scholar]
- 16.Purschwitz, J., Muller, S., Kastner, C., Schoser, M., Haas, H., Espeso, E. A., Atoui, A., Calvo, A. M., and Fischer, R. (2008) Curr. Biol. 18 255–259 [DOI] [PubMed] [Google Scholar]
- 17.Gambetta, G. A., and Lagarias, J. C. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 10566–10571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill, S. C., and von Hippel, P. H. (1989) Anal. Biochem. 182 319–326 [DOI] [PubMed] [Google Scholar]
- 19.Berkelman, T. R., and Lagarias, J. C. (1986) Anal. Biochem. 156 194–201 [DOI] [PubMed] [Google Scholar]
- 20.Wagner, J. R., Zhang, J., von Stetten, D., Günther, M., Murgida, D. H., Mroginski, M. A., Walker, J. M., Forest, K. T., Hildebrandt, P., and Vierstra, R. D. (2008) J. Biol. Chem. 283 12212–12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tasler, R., Moises, T., and Frankenberg-Dinkel, N. (2005) FEBS J. 272 1927–1936 [DOI] [PubMed] [Google Scholar]
- 22.Jorissen, H. J., Quest, B., Remberg, A., Coursin, T., Braslavsky, S. E., Schaffner, K., de Marsac, N. T., and Gartner, W. (2002) Eur. J. Biochem. 269 2662–2671 [DOI] [PubMed] [Google Scholar]
- 23.Lamparter, T., Mittmann, F., Gartner, W., Borner, T., Hartmann, E., and Hughes, J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 11792–11797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, L., and Lagarias, J. C. (1992) J. Biol. Chem. 267 19204–19210 [PubMed] [Google Scholar]
- 25.Vierstra, R. D., and Quail, P. H. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 5272–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eichenberg, K., Baurle, I., Paulo, N., Sharrock, R. A., Rüdiger, W., and Schäfer, E. (2000) FEBS Lett. 470 107–112 [DOI] [PubMed] [Google Scholar]
- 27.Wagner, J. R., Brunzelle, J. S., Forest, K. T., and Vierstra, R. D. (2005) Nature 438 325–331 [DOI] [PubMed] [Google Scholar]
- 28.Wagner, J. R., Zhang, J., Brunzelle, J. S., Vierstra, R. D., and Forest, K. T. (2007) J. Biol. Chem. 282 12298–12309 [DOI] [PubMed] [Google Scholar]
- 29.von Stetten, D., Seibeck, S., Michael, N., Scheerer, P., Mroginski, M. A., Murgida, D. H., Krauss, N., Heyn, M. P., Hildebrandt, P., Borucki, B., and Lamparter, T. (2007) J. Biol. Chem. 282 2116–2123 [DOI] [PubMed] [Google Scholar]
- 30.Giraud, E., Zappa, S., Vuillet, L., Adriano, J. M., Hannibal, L., Fardoux, J., Berthomieu, C., Bouyer, P., Pignol, D., and Vermeglio, A. (2005) J. Biol. Chem. 280 32389–32397 [DOI] [PubMed] [Google Scholar]
- 31.Cherry, J. R., Hondred, D., Walker, J. M., and Vierstra, R. D. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 5039–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, D., Yukl, E. T., Moenne-Loccoz, P., and Montellano, P. R. (2006) Biochemistry 45 14772–14780 [DOI] [PubMed] [Google Scholar]
- 33.Pendrak, M. L., Chao, M. P., Yan, S. S., and Roberts, D. D. (2004) J. Biol. Chem. 279 3426–3433 [DOI] [PubMed] [Google Scholar]
- 34.Borucki, B., von Stetten, D., Seibeck, S., Lamparter, T., Michael, N., Mroginski, M. A., Otto, H., Murgida, D. H., Heyn, M. P., and Hildebrandt, P. (2005) J. Biol. Chem. 280 34358–34364 [DOI] [PubMed] [Google Scholar]
- 35.Kneip, C., Hildebrandt, P., Schlamann, W., Braslavsky, S. E., Mark, F., and Schaffner, K. (1999) Biochemistry 38 15185–15192 [DOI] [PubMed] [Google Scholar]
- 36.Von Stetten, D. (2008) Investigation of the chromophore structure in plant and bacterial phytochromes by comparison of experimental and calculated Raman spectra. Doctoral dissertation, Technische Universität, Berlin
- 37.Bhoo, S. H., Hirano, T., Jeong, H. Y., Lee, J. G., Furuya, M., and Song, P. S. (1997) J. Am. Chem. Soc. 119 11717–11718 [Google Scholar]
- 38.Lamparter, T., and Michael, N. (2005) Biochemistry 44 8461–8469 [DOI] [PubMed] [Google Scholar]
- 39.Hübschmann, T., Börner, T., Hartmann, E., and Lamparter, T. (2001) Eur. J. Biochem. 268 2055–2063 [DOI] [PubMed] [Google Scholar]
- 40.Azuma, N., Kanamaru, K., Matsushika, A., Yamashino, T., Mizuno, T., Kato, M., and Kobayashi, T. (2007) Biosci. Biotechnol. Biochem. 71 2493–2502 [DOI] [PubMed] [Google Scholar]
- 41.Purschwitz, J., and Fischer, R. (2008) Mol. Gen. Genom., in press
- 42.Grote, A., Hiller, K., Scheer, M., Muench, R., Noertemann, B., Hempel, D. C., and Jahn, D. (2005) Nucleic Acids Res. 33 526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.