Abstract
mRNA trafficking and local protein translation are associated with protrusive cellular domains, such as neuronal growth cones, and deregulated control of protein translation is associated with tumor malignancy. We show here that activated RhoA, but not Rac1, is enriched in pseudopodia of MSV-MDCK-INV tumor cells and that Rho, Rho kinase (ROCK), and myosin II regulate the microtubule-independent targeting of RNA to these tumor cell domains. ROCK inhibition does not affect pseudopodial actin turnover but significantly reduces the dynamics of pseudopodial RNA turnover. Gene array analysis shows that 7.3% of the total genes analyzed exhibited a greater than 1.6-fold difference between the pseudopod and cell body fractions. Of these, only 13.2% (261 genes) are enriched in pseudopodia, suggesting that only a limited number of total cellular mRNAs are enriched in tumor cell protrusions. Comparison of the tumor pseudopod mRNA cohort and a cohort of mRNAs enriched in neuronal processes identified tumor pseudopod-specific signaling networks that were defined by expression of M-Ras and the Shp2 protein phosphatase. Pseudopod expression of M-Ras and Shp2 mRNA were diminished by ROCK inhibition linking pseudopodial Rho/ROCK activation to the localized expression of specific mRNAs. Pseudopodial enrichment for mRNAs involved in protein translation and signaling suggests that local mRNA translation regulates pseudopodial expression of less stable signaling molecules as well as the cellular machinery to translate these mRNAs. Pseudopodial Rho/ROCK activation may impact on tumor cell migration and metastasis by stimulating the pseudopodial translocation of mRNAs and thereby regulating the expression of local signaling cascades.
Regulation of mRNA translation through PI3K/Akt/mTOR signaling and downstream eIF4E-binding protein and S6 kinases play key roles in tumorigenesis (1, 2). In particular, the initiation factor eIF4E induces tumorigenicity and has recently been effectively targeted to inhibit tumor growth (3, 4). In various nonmammalian and mammalian cell systems, mRNA localization is a driving force for cell polarization and a key mechanism in post-transcriptional gene regulation (5). Local translation in dendrites plays a critical role in synaptic plasticity and long term potentiation via mechanisms that involve expression of the initiation factor eIF2, the elongation factor eEF2 or the Fragile X mental retardation RNA-binding protein (6–8). RNA and RNA-binding proteins are associated with nascent adhesions during cell spreading, the RNA-binding IMPs have been shown to regulate invadopodia formation, and the elongation factor eEF1A2 induces filopodia in breast cancer cells (9–11). However, although mRNAs localized to dendrites and fibroblast protrusions have been identified (12–15), mRNAs localized to tumor cell protrusions have yet to be characterized.
Formation of a stable complex between F-actin, β-actin mRNA, and elongation factor 1α anchors β-actin mRNA in cellular protrusions linking protein translation to the actin cytoskeleton (16). β-Actin mRNA is targeted to the leading edge of fibroblasts by the chicken orthologue of the IMP RNA-binding proteins, zip code-binding protein 1 (ZBP1),2 via an actin-dependent process mediated by Rho/ROCK signaling and myosin motor proteins (17). However, RNA granule interaction with the adenomatous polyposis coli tumor suppressor and Fragile X mental retardation protein regulates the microtubule-dependent targeting of RNA in fibroblasts (15). IMP stabilization of CD44 mRNA has been shown to regulate invadopodia formation (11); however, comparison of metastatic MTLn3 and nonmetastatic MTC rat mammary adenocarcinoma cells showed reduced expression of ZBP1 and the inability to target β-actin mRNA to cellular protrusions (18). The demonstration that Src phosphorylation of ZBP1 results in the translational derepression of β-actin mRNA (19) is indicative of the complex interplay between mRNA trafficking, protein translation, and signaling events that spatially regulate protein synthesis and expression in cellular domains.
Proteomic analysis has shown that the protein translation machinery, RNA-binding proteins, and signaling proteins, including ROCK and Src, are major components of tumor cell protrusions of MSV-MDCK-INV tumor cells (20, 21). In this tumor cell line, pseudopodial protrusion is dependent on autocrine HGF activation and regulated by Rho/ROCK signaling (20, 22). Activated Rho has been localized to the protrusions of motile cells and shown to regulate pseudopodial RNA expression (23–25), and Rho/ROCK signaling has been associated with nonproteolytic, blebbed tumor cell migration (26). More recently, pseudopod-localized tyrosine-phosphorylated caveolin-1 (pCav1) has been shown to be a critical regulator of Rho activation state, focal adhesion turnover, and ROCK- and Src-dependent tumor cell migration and invasion (21).
RhoA, and not Rac1, is shown here to be activated in the pseudopodia of MSV-MDCK-INV tumor cells. Dominant active Rac1 inhibits pseudopodial RhoA and ROCK inhibition activates pseudopodial Rac1. Like β-actin mRNA in fibroblasts (17), RNA delivery to tumor cell protrusions is microtubule-independent, ROCK-dependent and myosin 2 (MLC2)-dependent. Pseudopodial actin turnover is not affected by ROCK inhibition that dramatically restricts dynamic RNA turnover in these domains. Competitive signaling between RhoA and Rac1 therefore regulates the dynamic delivery of RNA to protrusive cellular domains of tumor cells. Gene array analysis identifies mRNAs for signaling proteins and the translation machinery as predominant pseudopod-enriched mRNAs. Comparative analysis with neuronal process-enriched mRNA (14) identified M-Ras and Shp-2 as tumor cell-specific components of pseudopodial signaling networks. RhoA/ROCK activation may thereby impact on tumor cell migration by stimulating pseudopodial targeting of select mRNAs and altering the expression of signaling cascades in tumor cell protrusions.
EXPERIMENTAL PROCEDURES
Antibodies, Plasmids, and Reagents—Anti-β-actin monoclonal antibody, Hoechst, nocodazole, and blebbistatin were purchased from Sigma-Aldrich. Anti-syntrophin monoclonal antibody was from Affinity Bioreagents, anti-HSP70 was from Upstate Technologies, and anti-fibronectin monoclonal antibody was from BD Sciences. Anti-M-Ras was a generous gift from Dr. J. Schroeder (University of British Columbia). Alexa 488-, 568-, and 647-conjugated secondary antibodies and the RNA-binding fluorescent dyes Syto RNASelect and Syto14 were purchased from Molecular Probes. Y27632 was from Calbiochem. Raichu FRET probes for activated RhoA and Rac1 were generously provided by Michiuki Matsuda (Osaka University) and dominant active V14RhoA plasmid by Nathalie Lamarche (McGill University).
Cell Culture, Immunofluorescent Labeling, and Pseudopod Purification—MDCK, MSV-MDCK (DoCl1), and MSV-MDCK-INV cells were cultured as previously described (27). Transfections were carried out using Effectene transfection reagent (Qiagen). Immunofluorescent labeling of methanol/acetone fixed cells was essentially as previously described (20) using 400 nm SytoRNASelect, Hoeschst, and anti-β-actin and Alexa 647-conjugated secondary antibodies. The cells were imaged on a FV1000 Olympus confocal microscope using 60× planapochromat objectives. Actin-rich pseudopodia were scored visually for SytoRNA labeling from images of three cells or more (>30 cells and three experiments/condition).
Pseudopod Purification, Western Blots, and Activated Rho Pulldown—Pseudopodia purification and Western blots of pseudopodia, cell body, and total cell lysates were performed as previously described (20, 28). Briefly, 107 MSV-MDCK-INV cells were plated on 100-mm 1-micron pore filters mounted between two 3–3.5-inch custom-made washers (Boulons Plus, Anjou, Canada) in a 100-mm Falcon Petri dish, and the exterior was sealed with agarose to prevent cell leakage to the bottom of the filter. After 24 h of culture, the filter was washed four times with cold phosphate-buffered saline containing 0.1 mm Ca2+ and 1 mm Mg2+, and the top (cell bodies) and bottom (pseudopodia) of the filter were scraped with a glass coverslip in ice-cold lysis buffer (20 mm Tris-HCl, pH 7.6, 0.5% Nonidet P-40, 250 mm NaCl, 3 mm EDTA, 3 mm EGTA containing freshly added 2 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride, 1 mm sodium vanadate, 2.5 mm sodium fluoride, and 1 μm leupeptin). GTP-RhoA pulldown assay was performed as described previously (21). Briefly, 46 μl (∼30 μg) of Rho assay reagent (Rhotekin RBD, agarose; Upstate, CA) slurry was used to pull down active Rho from cell lysates prepared according to the manufacturer's protocol. Both active and total RhoA samples were subjected to SDS-PAGE and Western blotting with anti-RhoA monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Affymetrix Gene Array—Biotinylated complimentary RNA was prepared from 100 ng of total RNA as per the Affymetrix GeneChip Technical Analysis Manual (Affymetrix, Santa Clara, CA) using the two-cycle target labeling assay. Double-stranded cDNA was synthesized using SuperScriptII (Invitrogen) and oligo(dT)24 primers. Biotin-labeled complimentary RNA was prepared by cDNA in vitro transcription using the BioArray high yield RNA transcript labeling kit (Enzo Biochem, New York, NY) incorporating biotinylated UTP and CTP. GeneChips were processed at the London Regional Genomics Centre (Robarts Research Institute, London, Canada). Six Affymetrix canine arrays (three pseudopodia, three cell body) were scanned with the Affymetrix GeneChip Scanner 3000 and signal intensities for genes generated with GCOS1.2 (Affymetrix) using the default values for statistical expression algorithm parameters, a target signal of 150 for all probe sets and a normalization value of 1. To determine the significant differences in gene expression levels between pseudopodia and cell body samples, one-way analysis of variance was performed using Partek Pro (St. Louis, MO). Significantly modulated genes were defined as those with absolute log2 fold change greater than 0.7. The genes were summarized together using hierarchical clustering by Z score-normalized data crossing all six samples. The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE13253. Gene lists of mRNAs up-regulated in tumor protrusions and neuronal processes (supplemental Table S1) (14) were analyzed and compared using Ingenuity Pathways Analysis (Ingenuity Systems, Redwood City, CA).
Fluorescence Resonance Energy Transfer—FRET experiments used single molecule RhoA (Raichu1298) and Rac1 (Raichu1026) activity sensing FRET plasmids (generically YFP-Rho GTPase-Rho GTPase effector-binding domain-CFP). GTP binding promotes interaction of the activated GTPase and the effector-binding domain bringing the N-terminal YFP and C-terminal CFP into proximity enabling FRET (29, 30). The cells were transfected with Raichu1298, Raichu1026, or Raichu1298 and dominant active V12Rac1 plasmid in 4-well Lab-Tek II chambered coverglasses (Nalge Nunc International) for 24 h and treated with 20 μm Y27632 for 1 h prior to imaging in phenol red-free Dulbecco's modified Eagle's medium. CFP and YFP emission of transfected cells excited with a 405-nm laser was first acquired on an Olympus FV1000 confocal microscope, cell regions were bleached using a 515-nm laser, and the CFP and YFP channels were reimaged. The average pixel intensity of the selected region was determined before and after bleaching, and FRET efficiency was calculated in the CFP channel taking into account background pixel intensity. The percentage of bleach value was obtained by subtracting the average pixel intensity following bleaching from the average pixel intensity prior to bleaching.
Fluorescence Recovery after Photobleaching—MSV-MDCK-INV cells stained with 400 nm Syto RNASelect for 1 h, transiently transfected with EGFP or MSV-MDCK-INV cells stably expressing EGFP-β-actin (20) were plated in chambered cover-glasses for 24 h and incubated with 20 μm Y27632 for 1 h in phenol red-free Dulbecco's modified Eagle's medium prior to imaging for 90s following bleaching of select actin-rich pseudopodia using a 488-nm laser at 100%.
In Situ Hybridization—Approximately 20,000 cells were plated per well in 8-well Lab-Tek chamber slides (Nalge Nunc International) for 24 h and, where indicated, treated with 20 μm Y27632 for 90 min. The cells were fixed, prehybridized, and hybridized with end tail DIG-labeled control or target specific oligonucleotides and immunodetected following the standard protocol supplied by the manufacturer (Roche Applied Science). Subsequently, the cells were washed with 95% ethanol and mounted with gelvatol. To quantify the distribution of specific mRNAs, pseudopodia were defined as that portion of cellular protrusions beyond the point of narrowing, and the number of DIG immunoreactive profiles (spots) in pseudopodia and cell body was determined.
RESULTS
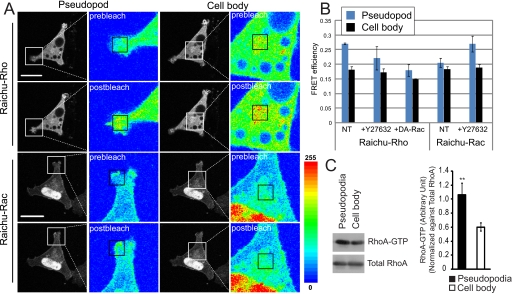
RhoA/ROCK Signaling Regulates Pseudopodial RNA Delivery and Turnover—Raichu FRET reporter constructs for RhoA and Rac1 have been extensively used to localize the active forms of these GTPases (29, 30). MSV-MDCK-INV cells were transfected with Raichu-Rho and Raichu-Rac and Rho and Rac activation, respectively, determined in the pseudopodia and cell bodies of the cells by photobleaching FRET (Fig. 1). RhoA activation is increased in pseudopodial regions compared with cell body regions, but no difference in Rac1 activation between these two regions of MSV-MDCK-INV cells was detected (Fig. 1, A and B). Interestingly, in cells treated for 1 h with the ROCK inhibitor Y27632, we observed selective activation of Rac1 in pseudopodial regions. Conversely, expression of dominant active Rac1 inhibited pseudopodial RhoA activation (Fig. 1B). To verify Rho activation in pseudopodial domains, we purified the pseudopodia of MSV-MDCK-INV cells and assessed the presence of activated Rho using a rhotekinin pulldown assay (21). As seen in Fig. 1C, activated Rho-GTP is enriched in the pseudopodial fraction of MSV-MDCK-INV cells purified on 1-micron pore filters (20) relative to the cell body. These results demonstrate a predominant role for RhoA signaling in MSV-MDCK-INV pseudopodia and are indicative of cross-talk between RhoA and Rac1 in this cellular domain.
FIGURE 1.
RhoA is selectively activated in the pseudopodia of MSV-MDCK-INV cells. A, cells were transfected with Raichu-Rho or Raichu-Rac FRET probes, as indicated. Fluorescence images of the CFP channel are taken before (prebleach) and after (postbleach) photobleaching the YFP acceptor with a 515-nm laser in either pseudopod or cell body regions, indicated by a square in the pseudocolor images. Raichu-Rho or Raichu-Rac activation was determined by the increase in average pixel density of CFP emission in the bleached region, visualized in the pseudo-colored images where regions of high intensity are colored in red and low intensity in blue. B, quantification of FRET efficiency in the pseudopodia (blue) and the cell body (black) is presented in graph form for Raichu-Rho or Raichu-Rac transfected cells untreated or treated with 20 μm Y27632 for 1 h prior to imaging or in cells cotransfected with Raichu-Rho and a dominant active Rac1 plasmid (DA-Rac) (means ± S.E.; n = 3; *, p < 0.01). C, MSV-MDCK-INV cells were plated on 1-micron pore filters, and Rho activation in purified pseudopodial and cell body fractions was analyzed by rotekinin pulldown. Densitometry of activated Rho was determined relative to total Rho and presented in graph form (n = 3; ± S.E.; **, p < 0.001). Scale bar, 20 μm.
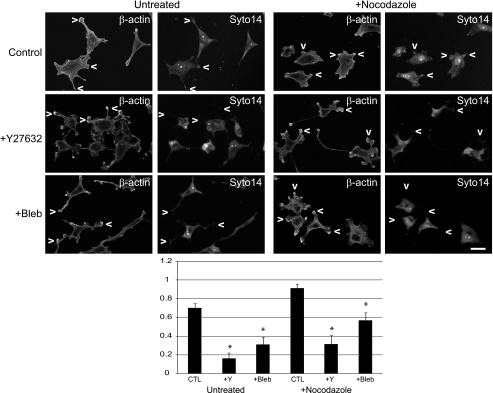
Generally, actin filaments and microtubules facilitate short and long distance mRNA transport, respectively (5). Disruption of cytoplasmic microtubules by nocodazole treatment of MSV-MDCK-INV cells results in cell rounding and blebbing followed by cell spreading and reformation of pseudopodia (31). In these cells, Syto14-labeled RNA extends to peripheral cellular protrusions (Fig. 2A). Scoring of actin-rich pseudopodia for Syto14 labeling showed that microtubule depolymerization increased the number of RNA-containing pseudopodia (Fig. 2B). RNA labeling of protrusions was prevented by treatment with Y27632 or the MLC2 inhibitor blebbistatin in both the absence and the presence of nocodazole (Fig. 2). Global RNA delivery to MSV-MDCK-INV protrusions therefore occurs via a microtubule-independent, Rho/ROCK/MLC2-dependent mechanism.
FIGURE 2.
Pseudopodial RNA translocation is microtubule-independent and ROCK and MLC2-dependent in MSV-MDCK-INV cell pseudopodia. A, MSV-MDCK-INV cells were either untreated (control, CTL); treated with the ROCK inhibitor Y27632 (20 μm), the myosin II inhibitor blebbistatin (7 μm), or the microtubule depolymerizing agent nocodazole (2 μm); or treated with both Y27632 and nocodazole or blebbistatin and nocodazole for 90 min. RNA was labeled with Syto14 and β-actin with specific antibodies, as indicated. The arrowheads point to actin-rich pseudopodia. B, pseudopodia were scored for the presence of RNA staining with Syto14 for all conditions (n > 30; ± S.E.; *, p < 0.001). Scale bar, 20 μm.
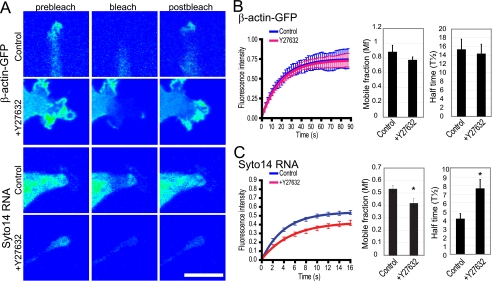
To further validate the effect of Rho/ROCK signaling on dynamic mRNA delivery to pseudopodial domains, we assessed turnover of pseudopodial actin and RNA using fluorescence recovery after photobleaching. ROCK inhibition did not impact on turnover of pseudopodial β-actin-GFP turnover (Fig. 3, A and B). However, ROCK inhibition not only reduces pseudopodial RNA content (Fig. 2A) (20) but also severely restricts the dynamics of the residual RNA present in these domains (Fig. 3, A and C). In the presence of the ROCK inhibitor, Y27632, the Syto14-labeled RNA showed a slower rate of recovery and a reduced mobile fraction (Fig. 3, A and C).
FIGURE 3.
ROCK-dependence of RNA but not actin turnover in MSV-MDCK-INV cell pseudopodia. A, MSV-MDCK-INV cells stably expressing pEGFP-actin (β-actin-GFP) and untransfected MSV-MDCK-INV cells labeled with the cell-permeant dye Syto14 (Syto14 RNA) were either left untreated (control) or treated with Y27632 for 60 min (+Y27632). Cells were imaged prior to photobleaching (prebleach), immediately after photobleaching of the pseudopodial region with the 488-nm laser (bleach), and 90 s after photobleaching (postbleach). A representative graph of three experiments shows the fluorescence intensity in the bleached region over time (t = 0 corresponds to the time immediately after photobleaching) for β-actin-GFP (B) or Syto14 labeled RNA (C). Untreated cells (control) are shown in blue, and Y27632-treated cells are in red. The values have been normalized to the prebleach intensity. The mobile fraction, representing the percent of fluorescence recovered in the bleached region, and half-time of recovery of control and Y27632 treated cells are shown in graph form for β-actin-GFP (B) and Syto14 RNA (C) (means ± S.E.; n = 3; *, p < 0.01). Scale bar, 20 μm.
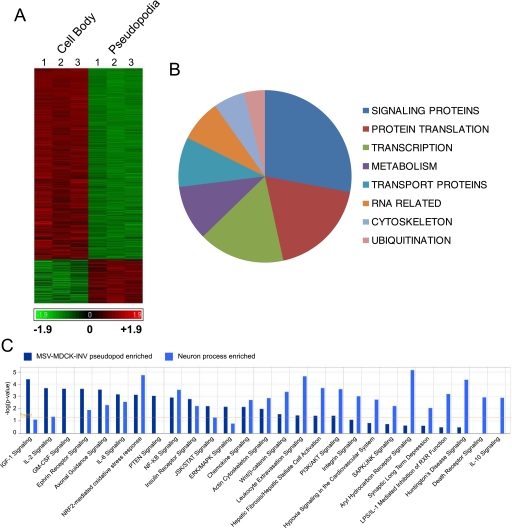
Tumor Cell Pseudopodia Contain a Distinct Spectrum of mRNA—To characterize the transcriptome of MSV-MDCK-INV pseudopodia, we performed gene array analysis of pseudopodia isolated on 1-micron pore polycarbonate filters (20, 28). Pseudopodial and cell body mRNA fractions were prepared and analyzed by Affymetrix microarray analysis. Of the 23,913 genes analyzed, only 1979 genes (7.3%) presented an absolute log2 fold change greater than 0.7 (∼1.6-fold) of mRNA expression in either the pseudopodial or cell body fraction with a minority (261; 13.2%) of mRNAs exhibiting increased expression in the pseudopodia relative to the cell body (Fig. 4A and supplemental Table S1). This is consistent with the idea that a select population of mRNAs is translocated to tumor cell pseudopodia.
FIGURE 4.
Identification of pseudopod-localized mRNAs. A, Affymetrix gene array analysis (23,913 genes) of mRNA from three preparations of cell bodies (left) and pseudopodia (right) of MSV-MDCK-INV cells presents up-regulated genes in red and down-regulated genes in green. See supplemental Table S1 for lists of log2 fold 0.7 up-regulated genes in pseudopodia and of log2 fold 2.0 up-regulated genes in the cell body. B, based on the classification of mRNAs up-regulated by >1.6-fold (log2 fold 0.7) in MSV-MDCK-INV pseudopodia (Table 1), the number of mRNAs per class is presented as a pie chart. C, gene lists of mRNAs up-regulated in MSV-MDCK-INV pseudopodia (supplemental Table S1; 261 pseudopod-enriched genes) and in neuronal processes (14) 159 process up-regulated genes) were analyzed and compared using Ingenuity Pathways Analysis. The yellow line indicates network expression above a threshold of p < 0.05. Networks significantly expressed in either tumor protrusions or neuronal processes are presented, and network components are listed in supplemental Table S2.
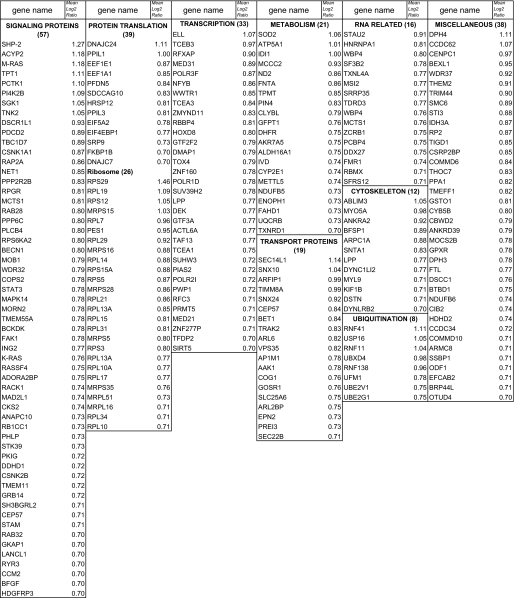
A cohort of 242 identified genes enriched 1.6-fold in the pseudopodial fraction is listed in Table 1 and functionally grouped. The number of genes/group is presented as a pie chart (Fig. 4B). Of particular interest is the abundance of proteins involved in signaling and protein translation. Indeed, a large number of pseudopod-enriched mRNAs encode ribosomal proteins (Table 1). Similarly, components of the translational machinery were among the most abundant mRNAs identified in a recent analysis of process-localized mRNAs in hippocampal neurons (14). Ingenuity network comparison of mRNAs enriched in neuronal protrusions compared with MSV-MDCK-INV pseudopodia revealed multiple common signaling pathways to be present in protrusions from normal neurons and cancer cells (Fig. 4C). However, a number of networks were identified to be selectively expressed in either tumor protrusions or neuronal processes indicative of the distinct mRNA composition of these two protrusive structures (Fig. 4C). Of particular interest, tumor protrusion networks were defined consistently by expression of mRNAs coding for M-Ras and Shp2 (PTPN11) that were not present in the neuronal process-enriched RNA cohort (supplemental Table S2).
TABLE 1.
mRNAs enriched 1.6-fold in the pseudopodial fraction relative to cell body
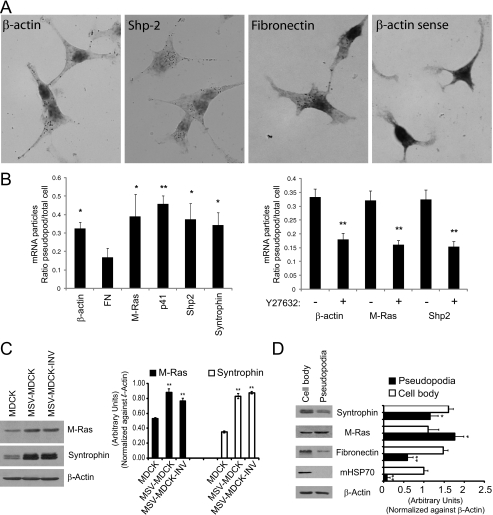
The Affymetrix data were validated by in situ hybridization of a select population of mRNAs including β-actin (1.4-fold up, log2 fold 0.49), Shp2 (Src homology 2 phosphotyrosyl phosphatase, PTPN11; 2.4-fold up, log2 fold 1.27), M-Ras (Ras small GTPase; 2.27-fold up, log2 fold 1.18), the p41 subunit of the Arp2/3 complex (ARPC1A, 1.84-fold up, log2 fold 0.88), and α-syntrophin (SNTA1, 1.78-fold up, log2 fold 0.83). β-Actin and Arp2/3 p41 subunit mRNA have been previously demonstrated to localize at or near membrane protrusions (17, 32). M-Ras and Shp2 define the networks selectively present in MSV-MDCK-INV pseudopodia (Fig. 4C). Syntrophin is part of the dystroglycan complex that is down-regulated in aggressive breast cancer cells (33). Fibronectin mRNA (10.8-fold down, log2 fold -3.44233), exhibits over 10-fold reduced expression in the pseudopodia relative to cell body by gene array and was included as a control.
At the mRNA level, in situ hybridization showed that for all of the probes tested, except fibronectin, a significant number of the DIG-immunoreactive profiles or spots were localized to pseudopodial protrusions (Fig. 5A). No labeling was detected for a control sense β-actin oligonucleotide probe. β-Actin mRNA showed 1.4-fold enrichment by gene array, corresponding to its pseudopodial distribution by in situ (Fig. 5B). For M-Ras, Arp2/3 p41 subunit, Shp2, and α-syntrophin, the in situ hybridization signal was enriched in the pseudopod fraction and significantly enhanced relative to fibronectin mRNA, which was enriched in the cell body (Fig. 5B). ROCK inhibition by treatment with the Y27632 compound resulted in the reduction of in situ labeling of pseudopodia for β-actin, M-Ras, and Shp2 to approximately the same levels as observed for fibronectin mRNA (Fig. 5B).
FIGURE 5.
Validation and expression of pseudopodial enriched mRNAs and gene products. A, MSV-MDCK-INV cells were in situ hybridized with oligonucleotides to the antisense strands of β-actin, Shp2, and fibronectin and the sense strand of β-actin (β-actin sense). In the bright field images, dark spots indicate the presence of DIG tailed oligonucleotide bound to its complementary mRNA molecule and were absent in cells hybridized with β-actin sense oligonucleotides. B, MSV-MDCK-INV cells were in situ hybridized with oligonucleotides to β-actin, fibronectin, M-Ras, p41 protein of the Arp2/3 complex, Shp2, and α-syntrophin (left panel). Alternatively, MSV-MDCK-INV cells, untreated or treated with 20 μm Y27632 for 90 min were in situ hybridized with oligonucleotides to β-actin, M-Ras, and Shp-2 (right panel). The total number of DIG-labeled mRNA profiles or spots in pseudopodia and cell body were counted, and the average ratio of DIG-labeled mRNA profiles for pseudopodia relative to cell body is presented in graphic form for each of the indicated probes (means ± S.E.; n = 3; *, p < 0.05; **, p < 0.005 relative to FN (left) or untreated cells (right)). C, total cell lysates of MDCK, MSV-MDCK, and MSV-MDCK-INV cells were probed by Western blot for M-Ras, syntrophin, and β-actin. D, lysates from cell bodies and pseudopodia of MSV-MDCK-INV cells were probed for fibronectin, M-Ras, syntrophin, mitochondrial HSP70 (mHSP70), and β-actin. The bands were quantified by densitometry relative to β-actin. (n = 3; ±S.E.; *, p < 0.01; **, p < 0.001).
Expression of both M-Ras and syntrophin is up-regulated in MSV-MDCK and MSV-MDCK-INV cells relative to MDCK cells (Fig. 5C). Pseudopodial and cell body fractions were prepared from MSV-MDCK-INV cells plated on 1-micron pore filters (20). The pseudopodia are depleted of mitochondrial HSP70, confirming the purity of the pseudopodial fraction (Fig. 5D). Interestingly, M-Ras is strongly expressed in the pseudopodial fraction relative to the cell body at levels equivalent to or greater than β-actin (Fig. 5D). However, like fibronectin, syntrophin shows reduced expression in the pseudopodial fraction (Fig. 5D). The reduced expression of α-syntrophin in the pseudopodial fraction relative to the cell body indicates that mRNA enrichment in pseudopodia is not necessarily associated with increased expression of its gene product.
DISCUSSION
Rho/ROCK-dependent RNA Translocation to Tumor Cell Pseudopodia—The repeated, directional extension of pseudopodia and their stabilization by substrate adhesive contacts constitutes the basic mechanism by which cells move over a substrate (34). Cell motility is therefore associated with the polarized formation of a distinct plasma membrane domain, the pseudopod, whose stabilization determines the directionality of cell movement (35). Receptor stimulation of Rac1/Cdc42 and phosphatidylinositol biphosphate activates WASp/Scar proteins at the leading edge recruiting Arp2/3 and actin monomers to induce actin filament branching and membrane protrusion (36). A recent proteomic analysis of neurites and soma localized a Cdc42/Rac signaling network to these protrusive domains of neurons (37). The demonstration here that Rho is the dominant GTPase activated in MSV-MDCK-INV pseudopodia is consistent with previous reports that Rho/ROCK signaling regulates pseudopodia protrusion in this cell line (20, 31) as well as with previous reports of Rho activation in protrusions of other motile cells (23–25).
The pseudopodia of MSV-MDCK-INV cells are highly blebbed, elongated protrusions and are morphologically distinct from lamellipodia (28). Inhibition of ROCK activates pseudopodial Rac1 (Fig. 1) and transforms the protrusive pseudopodia of MSV-MDCK-INV cells into lamellipodia without inhibiting cell motility (20) or, as shown here, pseudopodial actin turnover (Fig. 3, A and B). The ROCK-dependent delivery of RNA to pseudopodia in MSV-MDCK-INV cells is microtubule-independent and similar to the actin-mediated, Rho/ROCK- and MLC2-dependent pathway previously described for β-actin mRNA (17). ROCK inhibition prevents RNA accumulation in pseudopodia (Fig. 2), the dynamic turnover of pseudopodial RNA (Fig. 3) as well as the pseudopodial enrichment of distinct mRNA species (Fig. 5). Local Rho signaling therefore promotes the pseudopodial delivery of multiple RNA species via the Rho/ROCK/MLC2 actin-dependent pathway. Microtubule-dependent RNA granule translocation in neurons and fibroblasts (15, 38) suggests that different modes of RNA trafficking may be active in different cell types. The microtubule-independent pseudopodial delivery of RNA may therefore be associated with local activation of Rho/ROCK versus Rac signaling and be responsible for targeting of distinct cohorts of mRNAs to cellular protrusions of different cell types. This suggests that pseudopodial Rho versus Rac activation, although not necessarily impacting on pseudopodial actin dynamics or overall cell migration, may rather, by altering pseudopodial protein content through mRNA delivery and local translation, affect pseudopodial morphology and the migratory mode of the cells.
The Tumor Pseudopod Transcriptome—The cohort of mRNAs identified by gene array analysis to be enriched in MSV-MDCK-INV cell pseudopodia is consistent with the abundance of proteins involved in protein translation, RNA binding, and signaling that were identified in the proteomic analysis of the pseudopodia of these cells (20). Comparison of the proteome of fibroblast protrusions with a cohort of proteins enriched in metastatic prostate cancer cells supported an association between the pseudopodial localization of the translational machinery and tumor cell migration and metastasis (39). Interestingly, the major classes of proteins identified in the pseudopod proteome, cytoskeleton-associated, glycolytic enzymes, and chaperones (20) were not among the major classes of pseudopodial mRNAs identified. Comparison of the pseudopod proteome and transcriptome of MSV-MDCK-INV cells therefore suggests that local mRNA translation may regulate pseudopodial expression of less stable signaling molecules as well as the cellular machinery to locally translate these mRNAs.
It is, however, important to note that proteomic analysis detects the most abundant pseudopodial proteins. In addition, expression of the gene products of pseudopod localized mRNAs will depend both on the extent of translation of select mRNAs and on local degradation of the protein. For instance, reduction of pseudopodial RhoA activity by Smurf1-dependent ubiquitination promotes Cdc42/Rac1 activation, lamellipodia formation, and polarized cell movement; Smurf inhibition stimulates Rho-dependent activation of downstream ROCK and MLC2 and enhanced, random, nonproteolytic cell invasion (40, 41). The proteome of MSV-MDCK-INV cells (20) is enriched for proteasome-associated components consistent with the pseudopod as a site of active protein degradation. However, RhoA activation in the pseudopodia of MSV-MDCK-INV cells (Fig. 1) suggests that local ubiquitin-mediated degradation is associated with E3 ubiquitin ligases other than Smurf (41).
The fact that syntrophin mRNA, but not protein, is enriched in tumor pseudopodia demonstrates that pseudopodial mRNA enrichment does not necessarily translate into protein expression. Syntrophins bind to the actin-associated dystroglycan complex whose expression is down-regulated in breast cancer (33) but the functional significance of the pseudopodial enrichment of syntrophin mRNA is not clear. Similarly, the large number of mRNAs encoding proteins associated with transcription that were found to be enriched in the pseudopodial fraction (Table 1) may reflect alternate non-nuclear functions for these proteins, such as in RNA delivery to nascent focal adhesions (10). Alternatively, in muscle, disruption of intracellular junctions induced the Rho/ROCK/MLC2-dependent translocation of smooth muscle actin-inducing transcription factors (42). Sequestration of transcription factor mRNAs to actin-rich pseudopodial domains may function to regulate the activity of their gene products.
The Pseudopod Transcriptome and Signaling—The MSV-MDCK-INV transcriptome is not dissimilar to the previously reported transcriptome of neuronal growth cones (14). This is consistent with both domains being protrusive, motile cellular domains. Signaling network comparison of the transcriptome of MSV-MDCK-INV protrusions (Table 1) versus neuronal processes (14) showed that the majority of detected signaling networks were common to these protrusions derived from cancer and normal cells, respectively. These include actin regulatory and axonal guidance signaling networks that have also been mapped to chemotactic induced protrusions in fibroblasts (43). Interestingly, M-Ras and Shp2 were common components of the four networks significantly expressed in cancer cell pseudopodia but not in neuronal protrusions (supplemental Table S2). Pseudopodial expression of mRNAs for these two proteins was validated by in situ hybridization, and M-Ras protein was found to be enriched in MSV-MDCK-INV pseudopodia (Fig. 5). This potentially identifies a cancer cell pseudopod-specific signaling profile that may be related to the autocrine HGF and Rho/ROCK signaling that drives pseudopod protrusion in these cells (22, 31).
Rho/ROCK Signaling and Tumor Cell Motility—Both M-Ras and Shp2 have been linked to HGF-induced signaling (44, 45) that regulates pseudopodial protrusion in MSV-MDCK-INV cells (22). M-Ras induces actin-rich microspikes and multiple actin-rich foci upon injection into fibroblasts (46). Other studies have shown that overexpression of M-Ras is sufficient to induce epithelial-mesenchymal transition in epithelial cells and that this transformation is dependent on autocrine HGF signaling (44, 47). R-Ras regulates membrane protrusions through spatial regulation of Rho and Rac activity (48), and M-Ras could potentially play a similar role in response to autocrine HGF activation. Shp2 phosphatase activity is implicated in Ras/ERK and Src signaling that is associated with integrin signaling, adhesion turnover, and cell migration (49). Integrin dimerization has recently been shown to result in the binding and activation of both Shp2 and Src (50). Interestingly, both Shp2 and pCav1, a Src kinase substrate that also binds integrin (51, 52), promote stabilization of focal adhesion kinase within focal adhesions and focal adhesion disassembly (21, 53, 54). They have also both been reported to inactivate Src and/or p190RhoGAP, thereby promoting RhoA activation (55, 56). However, Shp2 also dephosphorylates and activates Src, whose phosphorylation of p190RhoGAP and RhoGDI regulates RhoGTPase activity (57, 58), suggesting that Src plays a central if complex role in integrin signaling. The delivery of RNA-binding proteins to nascent focal adhesions (10) and the derepression of β-actin mRNA translation by Src phosphorylation of ZBP1 (19) suggests that interplay between local signaling and mRNA translation may impact on adhesion turnover in protrusive cellular domains.
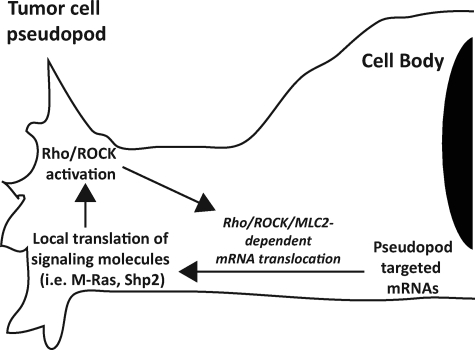
pCav1, pSrc (418) (21), pERK, RhoGDI (20), and activated RhoA (Fig. 1) are all enriched in MSV-MDCK-INV pseudopodia. pCav1 is a critical regulator of tumor cell Rho activation and of ROCK- and Src-dependent focal adhesion turnover and tumor cell migration and invasion (21). Together these data suggest that pseudopodial Rho/ROCK signaling may function to regulate pseudopodial signaling, morphology, and focal adhesion dynamics and thereby impact on tumor cell migration and invasion, potentially via promotion of a random mode of cell migration (31). Indeed, ROCK inhibition of pseudopodial expression and turnover of RNA is shown here to be associated with pseudopodial Rac1 activation and the formation of extended lamellipodia. Pseudopodial mRNA translocation and local translation of signaling molecules, including M-Ras and Shp2, may thereby constitute part of an autofeedback loop that potentiates the pseudopodial Rho/ROCK signaling required for mRNA delivery (Fig. 6).
FIGURE 6.
Pseudopodial Rho/ROCK signaling, mRNA targeting, and local signaling. Pseudopodial Rho/ROCK activation will enhance the pseudopodial delivery of mRNAs coding for signaling molecules (such as M-Ras and Shp2) as well as the protein translation machinery defining a feedback loop that may promote pseudopodial Rho/ROCK signaling through local mRNA delivery and translation.
RhoA/ROCK activation may thereby impact on tumor cell migration by regulating pseudopodial signaling through local RNA recruitment and protein translation in cellular protrusions. The data presented here suggest that a critical role for pseudopodial Rho activation might be the local enrichment of mRNAs coding for signaling molecules and regulators of protein translation. Gene array analysis of mRNAs enriched in this tumor cell pseudopodial domain identifies candidate mRNAs whose pseudopodial localization might be implicated in tumor cell migration.
Supplementary Material
Acknowledgments
We thank Justin Shih for assistance.
This work was supported by a Strategic Research Grant on Genomics and Proteomics of Metastatic Cancer from the Cancer Research Society Inc. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
Footnotes
The abbreviations used are: ZBP, zip code-binding protein; ROCK, Rho kinase; MDCK, Madin-Darby canine kidney; MSV, Moloney sarcoma virus; FRET, fluorescence resonance energy transfer; MLC2, myosin light chain 2; EGFP, enhanced green fluorescent protein; YFP, yellow fluorescent protein; CFP, cyan fluorescent protein; HGF, hepatocyte growth factor; DIG, digoxigenin.
References
- 1.Averous, J., and Proud, C. G. (2006) Oncogene 25 6423-6435 [DOI] [PubMed] [Google Scholar]
- 2.Mamane, Y., Petroulakis, E., LeBacquer, O., and Sonenberg, N. (2006) Oncogene 25 6416-6422 [DOI] [PubMed] [Google Scholar]
- 3.Lazaris-Karatzas, A., Montine, K. S., and Sonenberg, N. (1990) Nature 345 544-547 [DOI] [PubMed] [Google Scholar]
- 4.Graff, J. R., Konicek, B. W., Vincent, T. M., Lynch, R. L., Monteith, D., Weir, S. N., Schwier, P., Capen, A., Goode, R. L., Dowless, M. S., Chen, Y., Zhang, H., Sissons, S., Cox, K., McNulty, A. M., Parsons, S. H., Wang, T., Sams, L., Geeganage, S., Douglass, L. E., Neubauer, B. L., Dean, N. M., Blanchard, K., Shou, J., Stancato, L. F., Carter, J. H., and Marcusson, E. G. (2007) J. Clin. Investig. 117 2638-2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.St Johnston, D. (2005) Nat. Rev. Mol. Cell Biol. 6 363-375 [DOI] [PubMed] [Google Scholar]
- 6.Sutton, M. A., Taylor, A. M., Ito, H. T., Pham, A., and Schuman, E. M. (2007) Neuron 55 648-661 [DOI] [PubMed] [Google Scholar]
- 7.Pfeiffer, B. E., and Huber, K. M. (2007) J. Neurosci. 27 3120-3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa-Mattioli, M., Gobert, D., Stern, E., Gamache, K., Colina, R., Cuello, C., Sossin, W., Kaufman, R., Pelletier, J., Rosenblum, K., Krnjevic, K., Lacaille, J.-C., Nader, K., and Sonenberg, N. (2007) Cell 129 195-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amiri, A., Noei, F., Jeganathan, S., Kulkarni, G., Pinke, D. E., and Lee, J. M. (2006) Oncogene 26 3027-3040 [DOI] [PubMed] [Google Scholar]
- 10.de Hoog, C. L., Foster, L. J., and Mann, M. (2004) Cell 117 649-662 [DOI] [PubMed] [Google Scholar]
- 11.Vikesaa, J., Hansen, T. V., Jonson, L., Borup, R., Wewer, U. M., Christiansen, J., and Nielsen, F. C. (2006) EMBO J. 25 1456-1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberwine, J., Miyashiro, K., Kacharmina, J. E., and Job, C. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 7080-7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong, J., Zhang, T., and Bloch, L. M. (2006) BMC Neurosci. 7 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poon, M. M., Choi, S. H., Jamieson, C. A., Geschwind, D. H., and Martin, K. C. (2006) J. Neurosci. 26 13390-13399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mili, S., Moissoglu, K., and Macara, I. G. (2008) Nature 453 115-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, G., Grant, W. M., Persky, D., Latham, V. M., Jr., Singer, R. H., and Condeelis, J. (2002) Mol. Biol. Cell 13 579-592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latham, J., Vaughan M., Yu, E. H. S., Tullio, A. N., Adelstein, R. S., and Singer, R. H. (2001) Curr. Biol. 11 1010-1016 [DOI] [PubMed] [Google Scholar]
- 18.Shestakova, E. A., Wyckoff, J., Jones, J., Singer, R. H., and Condeelis, J. (1999) Cancer Res. 59 1202-1205 [PubMed] [Google Scholar]
- 19.Huttelmaier, S., Zenklusen, D., Lederer, M., Dictenberg, J., Lorenz, M., Meng, X., Bassell, G. J., Condeelis, J., and Singer, R. H. (2005) Nature 438 512-515 [DOI] [PubMed] [Google Scholar]
- 20.Jia, Z., Barbier, L., Stuart, H., Amraei, M., Pelech, S., Dennis, J. W., Metalnikov, P., O'Donnell, P., and Nabi, I. R. (2005) J. Biol. Chem. 280 30564-30573 [DOI] [PubMed] [Google Scholar]
- 21.Joshi, B., Strugnell, S. S., Goetz, J. G., Kojic, L. D., Cox, M. E., Gritffith, O. L., Chan, S. K., Jones, S. J., Leung, S. P., Masoudi, H., Leung, S., Wiseman, S. M., and Nabi, I. R. (2008) Cancer Res. 68 8210-8220 [DOI] [PubMed] [Google Scholar]
- 22.Vadnais, J., Nault, G., Daher, Z., Amraei, M., Dodier, Y., Nabi, I. R., and Noel, J. (2002) J. Biol. Chem. 277 48342-48350 [DOI] [PubMed] [Google Scholar]
- 23.Cardone, R. A., Bagorda, A., Bellizzi, A., Busco, G., Guerra, L., Paradiso, A., Casavola, V., Zaccolo, M., and Reshkin, S. J. (2005) Mol. Biol. Cell 16 3117-3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurokawa, K., and Matsuda, M. (2005) Mol. Biol. Cell 16 4294-4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pertz, O., Hodgson, L., Klemke, R. L., and Hahn, K. M. (2006) Nature 440 1069-1072 [DOI] [PubMed] [Google Scholar]
- 26.Sahai, E., and Marshall, C. J. (2003) Nat. Cell Biol. 5 711-719 [DOI] [PubMed] [Google Scholar]
- 27.Le, P. U., Nguyen, T. N., Drolet-Savoie, P., Leclerc, N., and Nabi, I. R. (1998) Cancer Res. 58 1631-1635 [PubMed] [Google Scholar]
- 28.Nguyen, T. N., Wang, H. J., Zalzal, S., Nanci, A., and Nabi, I. R. (2000) Exp. Cell Res. 258 171-183 [DOI] [PubMed] [Google Scholar]
- 29.Yoshizaki, H., Ohba, Y., Kurokawa, K., Itoh, R. E., Nakamura, T., Mochizuki, N., Nagashima, K., and Matsuda, M. (2003) J. Cell Biol. 162 223-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itoh, R. E., Kurokawa, K., Ohba, Y., Yoshizaki, H., Mochizuki, N., and Matsuda, M. (2002) Mol. Cell Biol. 22 6582-6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia, Z., Vadnais, J., Lu, M., Noel, J., and Nabi, I. R. (2006) Biol. Cell 98 337-351 [DOI] [PubMed] [Google Scholar]
- 32.Mingle, L. A., Okuhama, N. N., Shi, J., Singer, R. H., Condeelis, J., and Liu, G. (2005) J. Cell Sci. 118 2425-2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muschler, J., Levy, D., Boudreau, R., Henry, M., Campbell, K., and Bissell, M. J. (2002) Cancer Res. 62 7102-7109 [PubMed] [Google Scholar]
- 34.Lauffenberger, D. A., and Horwitz, A. F. (1996) Cell 84 359-369 [DOI] [PubMed] [Google Scholar]
- 35.Nabi, I. R. (1999) J. Cell Sci. 112 1803-1811 [DOI] [PubMed] [Google Scholar]
- 36.Pollard, T. D., and Borisy, G. G. (2003) Cell 112 453-465 [DOI] [PubMed] [Google Scholar]
- 37.Pertz, O. C., Wang, Y., Yang, F., Wang, W., Gay, L. J., Gristenko, M. A., Clauss, T. R., Anderson, D. J., Liu, T., Auberry, K. J., Camp, D. G., II, Smith, R. D., and Klemke, R. L. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 1931-1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bassel, G. J., Singer, R. H., and Kosik, K. S. (1994) Neuron 12 571-582 [DOI] [PubMed] [Google Scholar]
- 39.Wang, Y. C., Hanley, R., and Klemke, R. L. (2006) J. Proteome Res. 5 907-915 [DOI] [PubMed] [Google Scholar]
- 40.Sahai, E., Garcia-Medina, R., Pouyssegur, J., and Vial, E. (2007) J. Cell Biol. 176 35-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, H.-R., Zhang, Y., Ozdamar, B., Ogunjimi, A. A., Alexandrova, E., Thomsen, G. H., and Wrana, J. L. (2003) Science 302 1775-1779 [DOI] [PubMed] [Google Scholar]
- 42.Fan, L., Sebe, A., Peterfi, Z., Masszi, A., Thirone, A. C. P., Rotstein, O. D., Nakano, H., McCulloch, C. A., Szaszi, K., Mucsi, I., and Kapus, A. (2007) Mol. Biol. Cell 18 1083-1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, Y., Ding, S.-J., Wang, W., Jacobs, J. M., Qian, W.-J., Moore, R. J., Yang, F., Camp, D. G., II, Smith, R. D., and Klemke, R. L. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 8328-8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, K. X., Ward, K. R., and Schrader, J. W. (2004) Mol. Cancer Res. 2 242-255 [PubMed] [Google Scholar]
- 45.Duan, H. F., Qu, C. K., Zhang, Q. W., Yu, W. M., Wang, H., Wu, C. T., and Wang, L. S. (2006) Cell Signal. 18 2049-2055 [DOI] [PubMed] [Google Scholar]
- 46.Matsumoto, K., Asano, T., and Endo, T. (1997) Oncogene 15 2409-2417 [DOI] [PubMed] [Google Scholar]
- 47.Ward, K. R., Zhang, K. X., Somasiri, A. M., Roskelley, C. D., and Schrader, J. W. (2004) Oncogene 23 1187-1196 [DOI] [PubMed] [Google Scholar]
- 48.Wozniak, M. A., Kwong, L., Chodniewicz, D., Klemke, R. L., and Keely, P. J. (2005) Mol. Biol. Cell 16 84-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, S. Q., Yang, W., Kontaridis, M. I., Bivona, T. G., Wen, G., Araki, T., Luo, J., Thompson, J. A., Schraven, B. L., Philips, M. R., and Neel, B. G. (2004) Mol. Cell 13 341-355 [DOI] [PubMed] [Google Scholar]
- 50.Merdek, K. D., Yang, X., Taglienti, C. A., Shaw, L. M., and Mercurio, A. M. (2007) J. Biol. Chem. 282 30322-30330 [DOI] [PubMed] [Google Scholar]
- 51.Wary, K. K., Mariotti, A., Zurzolo, C., and Giancotti, F. G. (1998) Cell 94 625-634 [DOI] [PubMed] [Google Scholar]
- 52.Wei, Y., Yang, X., Liu, Q., Wilkins, J. A., and Chapman, H. A. (1999) J. Cell Biol. 144 1285-1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Wichert, G., Haimovich, B., Feng, G. S., and Sheetz, M. P. (2003) EMBO J. 22 5023-5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goetz, J., Joshi, B., Lajoie, P., Strugnell, S., Scudamore, T., Kojic, L., and Nabi, I. (2008) J. Cell Biol. 180 1261-1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sordella, R., Jiang, W., Chen, G. C., Curto, M., and Settleman, J. (2003) Cell 113 147-158 [DOI] [PubMed] [Google Scholar]
- 56.Grande-Garcia, A., Echarri, A., de Rooij, J., Alderson, N. B., Waterman-Storer, C. M., Valdivielso, J. M., and del Pozo, M. A. (2007) J. Cell Biol. 177 683-694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DerMardirossian, C., Rocklin, G., Seo, J.-Y., and Bokoch, G. M. (2006) Mol. Biol. Cell 17 4760-4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arthur, W. T., Petch, L. A., and Burridge, K. (2000) Curr. Biol. 10 719-722 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.