Abstract
Membrane depolarization controls long lasting adaptive neuronal changes in brain physiology and pathology. Such responses are believed to be gene expression-dependent. Notably, however, only a couple of gene repressors active in nondepolarized neurons have been described. In this study, we show that in the unstimulated rat hippocampus in vivo, as well as in the nondepolarized brain neurons in primary culture, the transcriptional regulator Yin Yang 1 (YY1) is bound to the proximal Mmp-9 promoter and strongly represses Mmp-9 transcription. Furthermore, we demonstrate that monoubiquitinated and CtBP1 (C-terminal binding protein 1)-bound YY1 regulates Mmp-9 mRNA synthesis in rat brain neurons controlling its transcription apparently via HDAC3-dependent histone deacetylation. In conclusion, our data suggest that YY1 exerts, via epigenetic mechanisms, a control over neuronal expression of MMP-9. Because MMP-9 has recently been shown to play a pivotal role in physiological and pathological neuronal plasticity, YY1 may be implicated in these phenomena as well.
Neuronal depolarization is important not only for a transmission of information throughout the nervous system but also for an initiation of adaptive neuronal responses to incoming stimuli. Examples of the adaptive changes are long term potentiation and kindling-evoked epileptogenesis believed to underlie physiological (such as learning and memory) and pathological neuronal plasticity, respectively. These long lasting adaptive changes have been linked to an activation of gene expression. Indeed, a number of depolarization-driven gene responses were described over the last 20 years, and in almost all cases inducible transcription factors, like cAMP-response element-binding protein, Elk-1, AP-1, Egrs, etc. (for review see Ref. 1), were found to be responsible for the increased gene expression. However, it is also conceivable to consider a repression of transcription, in addition to its activation, as a means to drive depolarization-evoked gene expression. So far only a very limited number of such repressive molecules has been discovered (2-4).
In this study we set out to search for these transcriptional repressors. We focused on a regulation of Mmp-9 that codes for an extracellular matrix protease involved in physiological and pathological extracellular matrix remodeling. Aberrant, and usually excessive, Mmp-9 expression has been linked to numerous disorders of the central nervous system (5-7) as well as other devastating diseases such as tumors (8, 9). Hence, detailed knowledge of its transcriptional repression is of great importance for an understanding of those pathologies and for a potential development of novel therapeutic approaches.
Our previous reports have shown that in the nondepolarized rat brain MMP-93 is predominantly expressed in neurons (10-12). However, its expression levels in those cells are very low, which points toward a presence of an efficient mechanism(s) repressing its transcription in unstimulated neurons (10).
Molecular mechanisms directly controlling MMP-9 transcription in physiology and pathology of neurons remain unknown. Data from other cell types clearly indicate that MMP-9 expression is regulated primarily at the level of transcription (6). Numerous stimulating transcription factors implicated in Mmp-9 gene activation have been identified and well characterized (6). On the other hand, their repressive counterparts remain poorly defined (13-15).
It has been reported previously that the -522/+19-bp Mmp-9 promoter region is sufficient to drive an appropriate basal and inducible reporter gene expression in transgenic mice (16). Using the DNase I footprinting, a method for an identification of novel DNA regulatory proteins (17, 18), we have analyzed herein the rat Mmp-9 promoter fragments overlapping the -557/+18-bp region of the gene in unstimulated neurons to look for repressive transcription factors.
As a result, we have identified YY1, a transcription factor belonging to a Polycomb group of proteins (PcGs), as a potential Mmp-9 gene repressor. We have confirmed this finding with a collection of different approaches, employing ex vivo brain mRNA, protein and chromatin extracts, as well as in vitro neuronal cultures.
MATERIALS AND METHODS
Animals—The experiments on animals were performed on adult male Wistar rats (weighing 210-300 g), according to the rules established by the Ethical Committee on Animal Research of the Nencki Institute, based on national laws that are in a full agreement with the European Union directive on animal experimentation.
Pentylenetetrazole (PTZ) Stimulation—The animals were handled and injected with physiological saline daily for 3-4 days before the experimental treatment. Then pentylenetetrazole (50 mg/kg) was administered intraperitoneally. Animals were sacrificed 2 and 4 h after an onset of seizures. Only the animals displaying clear seizures were used for the experiments.
Plasmids—Rat Mmp-9 gene promoter fragments -290/+18 bp and -557/-282 bp were cloned separately into the BamHI site of pUC 18 and used for DNase I footprinting. Rat Mmp-9 gene promoter fragment -1369/+35 bp was cloned into MluI/BglII sites of pGL3(R2.1) (Promega) for reporter assays. Point mutation in the core of the footprinted YY1-binding site of the Mmp-9 gene promoter was generated with the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Rat YY1 full-length coding sequence was amplified from cDNA library obtained from the unstimulated rat hippocampus and cloned into AscI/EcoRI site of GW1 vector (the vector was a generous gift from Dr. J. Jaworski, International Institute of Molecular and Cell Biology, Warsaw, Poland). Construct expressing siRNA for YY1 was cloned by inserting at BglII/HindIII sites of pSUPER (19) the following double-stranded oligonucleotide directed for 994-1012-bp region of the rat YY1 mRNA: 994-1012 bp (sense), GATCCCCGTTGAGAGCTCAAAGCTAATTCAAGAGATTAGCTTTGAGCTCTCAACTTTTTGGAAA, and 994-1012 bp (antisense), AGCTTTTCCAAAAAGTTGAGAGCTCAAAGCTAATCTCTTGAATTAGCTTTGAGCTCTCAACGGG.
For FISH probe production we used the pCRII plasmid containing the rat MMP-9 full-length coding sequence amplified from a rat visual cortex cDNA library (12). Construct expressing cytomegalovirus promoter-driven GFP was a generous gift from Dr. K. Duniec (Nencki Institute, Warsaw, Poland). Oligonucleotides were synthesized by Sigma. All constructs were verified by DNA sequencing (ABI377, PerkinElmer Life Sciences).
Antibodies—The following antibodies and suppliers were used: anti-YY1 (H-10 and C-20) (Santa Cruz Biotechnology); anti-CtBP1 (Pharmingen); anti-HDAC1 (H-51) (Santa Cruz Biotechnology); anti-HDAC2 (H-54) (Santa Cruz Biotechnology); anti-HDAC2 (3F3) (Abcam); anti-HDAC3 (H-99) (Santa Cruz Biotechnology); glyceraldehyde-3-phosphate dehydrogenase (MAB374) (Chemicon); histone H3 (07-690) (Upstate/Millipore); anti-ubiquitin (P4D1) (Cell Signaling); anti-acetylhistone H3 (06-599; Upstate Biotechnology, Inc.); anti-acetylhistone H4 (06-866; Upstate Biotechnology, Inc.); anti-RNA polymerase II CTD repeat YSPTSPS (phospho-Ser-5) antibody (ab5131; Abcam); and anti-RNA polymerase II CTD repeat YSPTSPS (phospho-Ser-2) antibody (ab5095; Abcam).
RNA Extraction—Total RNA was extracted using TRI Reagent® (Sigma) according to the manufacturer's procedure.
RT-PCR—1 μg of RNA samples was subjected to RT reaction using SuperScript first-strand synthesis system for RT-PCR (Invitrogen) according to enclosed procedure. PCR was performed with TaqPCR core kit (Qiagen). Primers and PCR conditions for MMP-9 were the same as described previously (10) For YY1 cDNA amplification we used the following primers: ACCAAGAGGTGATTCTGGTG and CTGCTCTTCAACCACTGTTTC, which were annealed to the templates at 59 °C.
Nuclear Protein Lysates—Rat hippocampal nuclear protein lysates were prepared using CelLytic™ NuCLEAR™ extraction kit (Sigma) according to the manufacturer's instructions. Lysis buffer additionally contained 1 mm PMSF, 5 mm sodium fluoride, 1 mm sodium orthovanadate, 5 mm sodium butyrate, and 100× diluted protease inhibitor mixture (Sigma). Protein concentration was calculated using Bradford method.
DNase I Footprinting—The rat Mmp-9 gene promoter fragments were cut out of pUC 18. DNase I footprinting was conducted using Core Footprinting System (Promega) according to manufacturer's procedure. Products of the reaction were extracted with phenol/chloroform/isoamyl alcohol (25:24:1), ethanol-precipitated, and dissolved in gel loading solution containing formamide and urea, and then resolved by electrophoresis in a DNA sequencing gel. Dried gels were exposed to BioMax system (Eastman Kodak Co.).
MatInspector Analyses—We searched MatInspector's library (28) of weight matrices for transcription factor-binding sites. We analyzed matrix group of vertebrates. A matrix similarity higher than 0.85 was used as a cutoff for consideration of potential query sequence matches with known transcription factor recognition sequences.
Electromobility Shift Assay (EMSA)—For EMSA probe we used the following double-stranded oligonucleotide containing the footprinted YY1 site from rat Mmp-9 gene promoter. Sequences of probes were as follows: for the wild-type region (YY1-binding site is underlined) GACCTAGGACTAGATGGCCCCTCCACCA, and for the mutated region (mutated nucleotides are underlined) GACCTAGGACTAGATAATCCCTCCACCA. We prepared double-stranded oligonucleotide probes by annealing equimolar amounts (10 μm) of complementary single-stranded oligonucleotides in an solution containing 0.1 m Tris-HCl (pH 7.5), 0.5 m NaCl, 0.05 m EDTA. Oligonucleotides were placed at 65 °C for 10 min, slowly cooled down to room temperature, and then kept at 4 °C for overnight. 100 ng of double-stranded oligonucleotides were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (Promega) and purified with NucTrap probe purification columns (Stratagene). Binding reactions were performed in a 20-μl volume. They contained 20 μg of rat hippocampal nuclear protein lysate diluted in buffer containing 5% glycerol, 10 mm Tris-HCl (pH 7.6), 10 mm KCl, 1 mm EDTA, 5 mm MgCl2, 1 mm dithiothreitol, 10 μg/ml poly(dI·dC), 1 mm PMSF, 5 mm sodium fluoride, 1 mm sodium orthovanadate, 5 mm sodium butyrate, and 100× diluted protease inhibitor mixture (Sigma). Rat nuclear hippocampal protein lysates were extracted from six different control or PTZ-stimulated rats and then they were mixed together. Samples were incubated for 15 min on ice, and then 30,000 cpm of probe was added followed by final incubation for 30 min on ice. Immediately before loading of samples on the gel, we added ice-cold 5× gel loading buffer (2× Tris-glycine buffer, 50% glycerol, 0.2% bromphenol blue). Protein-DNA complexes were resolved by electrophoresis on nondenaturing 8% polyacrylamide gels in 1× Tris-glycine buffer and were visualized by autoradiography.
EMSA Supershift—EMSA binding reaction was performed as described above, but 2 μg of antibody anti-YY1 (H-10; Santa Cruz Biotechnology) was additionally included. Samples were incubated for 90 min on ice, and then 30,000 cpm of probe was added followed by a final incubation for 30 min on ice. Immediately before a loading of samples on the gel, we added ice-cold 5× gel loading buffer (2× Tris-glycine buffer, 50% glycerol, 0.2% bromphenol blue). Protein-DNA complexes were resolved by electrophoresis on nondenaturing 8% polyacrylamide gels in 1× Tris-glycine buffer and were visualized by autoradiography.
Immunoprecipitation—Immunoprecipitations were performed as described previously (20). Two micrograms of appropriate antibody was added into the samples.
Western Blot Analysis—The samples were separated by 10% SDS-PAGE in Tris-glycine running buffer (25 mm Tris, 250 mm glycine (pH 8.3), 0.1% SDS) and transferred to a polyvinylidene difluoride membrane (Millipore) in transfer buffer (48 mm Tris, 39 mm glycine, 0.037% SDS, 20% methanol). Membranes were blocked in 5% nonfat milk/TBST (25 mm Tris-HCl (pH 8.0), 125 mm NaCl, 0.1% Tween 20) for 2 h at room temperature. The membranes were then incubated with an appropriate primary antibody (YY1α (H10) 1:400; CtBP1α 1:5000; ubiquitin α 1:1000; HDAC1α 1:1000; HDAC2α 1:2000; HDAC3α 1:1000; glyceraldehyde-3-phosphate dehydrogenase-α 1:5000; histone H3α 1:1000) at 4 °C for overnight, and horseradish peroxidase-conjugated secondary antibody IgG (Vector Laboratories) was added at a dilution of 1:5000. Results were developed using ECL Plus™ reagent (Amersham Biosciences) on an x-ray film (Kodak).
Rat Primary Neuronal Culture—Rat primary cortical neurons were prepared from newborn (P0) Wistar rats as described (21). Briefly, dissociated cortical neurons were plated at a density of 106 cells/ml in the basal medium Eagle's supplemented with 10% heat-inactivated bovine calf serum, 35 mm glucose, 1 mm l-glutamine, 100 units/ml penicillin, and 0.1 mg/ml streptomycin and maintained in a humidified incubator with 6.5% CO2 at 37 °C. Plates were coated with poly-d-lysine. Cytosine-β-d-arabinofuranoside (2.5 mm; Sigma) was added to cultures on the 2nd day after seeding to inhibit the proliferation of non-neuronal cells. Previous studies demonstrated that >90% of the cells in this culture preparation are neurons (22). The cultures were lipofected at either 48 or 72 h after cytosine-β-d-arabinofuranoside introduction. Two days later, they were stimulated with 30 mm KCl.
Lipofection—DNA constructs were introduced into rat primary cultures by a lipofection with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Lipofection efficiency ranged from 10 to 15% depending on an experiment.
Luciferase Reporter Assay—Luciferase activity was evaluated using luciferase assay system with Reporter Lysis Buffer (Promega) according to enclosed protocol. Results were recorded using luminometer model 2030-000 (Turner Biosystems) and presented in relative light units.
Immunocytochemistry on Cultured Cells—The cultures were fixed in 4% paraformaldehyde, incubated for antigen retrieval in 1× SSC for 30 min at 85 °C, and then permeabilized in 1× PBS containing 0.1% Triton X-100 and 0.5% Nonidet P-40. The cultures were stained with anti-YY1 antibody (H-10; diluted 1:200) purchased from Santa Cruz Biotechnology. The staining was detected using goat anti-mouse Alexa Fluor 555-conjugated antibody (Invitrogen; diluted 1:200). Finally, cell nuclei were stained with DAPI (Vector Laboratories). The samples were examined by the fluorescent microscopy by an Olympus IX70.
Brain Immunohistochemistry—Animals were deeply anesthetized and perfused intracardially with cold 4% paraformaldehyde in 0.1 m phosphate-buffered saline (PBS) (pH 7.4). Brains were dissected from the skulls, fixed overnight in 4% paraformaldehyde at 4 °C, and transferred to 30% sucrose for 3 days. Frozen brains were cut in 40-μm-thick sections on a cryostat (Cryocut 1800, Leica) and kept in 1 m PBS containing 0.1% sodium azide. The stainings were performed on free-floating sections. The sections were washed in PBS (pH 7.4) containing 0.1% Triton X-100 for 10 min, incubated to retrieved antigens in 2× SSC buffer (sodium chloride/sodium citrate buffer, Sigma) for 30 min at 80 °C, and washed once in PBS/Triton X-100. Samples were blocked with 10% normal donkey serum for 1 h at room temperature, washed three times for 10 min with PBS/Triton X-100, and incubated overnight at 4 °C with primary anti-YY1 (H-10) or anti-CtBP1 antibody (both diluted 1:200). The sections were then washed three times in PBS/Triton X-100 for 10 min and incubated for 2 h with donkey anti-mouse Alexa Fluor 488-conjugated IgG (1:200, Invitrogen). After the incubation samples were washed in PBS/Triton X-100 for 10 min, incubated with TO-PRO-3 (1:3000, Molecular Probes) for 5 min at room temperature to stain cell nuclei, and washed once in PBS/Triton X-100. The sections were mounted on slides, air-dried, and covered with Vectashield (Vector Laboratories). Images were captured using a confocal laser scanning microscope (Leica).
Fluorescent in Situ Hybridization (FISH) for MMP-9 mRNA Combined with Immunocytochemical Staining for GFP—For MMP-9 mRNA detection, fluorescein-labeled cRNA probes were used. Probes were prepared using the in vitro transcription method. Probe labeling reactions were performed with fluorescein RNA labeling mix (Roche Applied Science) and catalyzed by T7 or SP6 RNA polymerase (both from Roche Applied Science) according to the manufacturer's instructions. pCRII plasmid containing the full-length coding sequence of rat MMP-9 was linearized and used as a template to generate antisense (complementary to mRNA) and sense (identical to mRNA) probes. Sense probe was used as a negative control. In situ hybridization reaction was performed in 4% paraformaldehyde-fixed cultures that were permeabilized with 1× SSC containing 1% Triton X-100 and subjected to a prehybridization. The samples were incubated for 1 h at room temperature in prehybridization solution (Sigma) diluted 1:1 with formamide and then hybridized overnight at 60 °C in hybridization solution (Sigma) containing one of the denatured probes diluted 1:100. After hybridization the specimens were incubated for 5 min at 37 °C in RNase A solution (20 μg/ml) prepared in 2× SSC. The post-hybridization washes (each for 15 min at 55 °C) were performed in the following solutions: 2× SSC, 2× SSC containing 50% formamide, 0.5× SSC, 50% formamide, 0.1× SSC, 50% formamide, and 0.1× SSC. Prior to probe detection the specimens were incubated with 3% H2O2 for 30 min at room temperature and blocked with blocking solution from cyanine 3-coupled tyramide signal amplification system (PerkinElmer Life Sciences) for 1 h at room temperature. Then peroxidase-coupled anti-fluorescein antibody (Roche Applied Science) diluted 1:200 in blocking solution (PerkinElmer Life Sciences) was applied to the samples for overnight at 4 °C. The hybridization signal was developed using the cyanine 3-coupled tyramide signal amplification system (PerkinElmer Life Sciences) according to manufacturer's instructions. The FISH procedure quenched GFP capacity to fluoresce; thus, one had to detect GFP expression using immunocytochemical staining. To this end, the samples after FISH were incubated for 2 h at room temperature with anti-GFP antibody (sc-9996; diluted 1:100) purchased from Santa Cruz Biotechnology and then detected with Alexa Fluor 488-conjugated secondary antibody (Invitrogen; diluted 1:200). Finally, cell nuclei were stained with DAPI (Vector Laboratories), and the resulting samples were examined by the fluorescent microscopy by an Olympus IX70.
Co-immunostaining for YY1 and CtBP1—The brain sections were prepared as described above. The sections were stained for YY1 (1:200; anti-YY, H-10) overnight at 4 °C and then the signal was detected by 2 h of incubation with Alexa Fluor 488 donkey anti-mouse IgG (1:200, Invitrogen). Next the samples were washed three times in PBS/Triton X-100 for 10 min at room temperature, blocked with 10% normal horse serum for 1 h, and incubated with CtBP1 (1:400) for overnight at 4 °C. After three washes in PBS/Triton X-100 for 10 min, the samples were incubated with a secondary antibody, Alexa Fluor 555 goat anti-mouse IgG (1:200, Invitrogen) in PBS/Triton X-100 for 2 h at room temperature. The sections were washed three times in PBS/Triton X-100 for 10 min, incubated with incubated with TO-PRO-3 (1:3000, Molecular Probes) in PBS/Triton X-100 for 5 min, and again washed once in PBS/Triton X-100 for 10 min. The sections were mounted on slides, air-dried, and covered with Vectashield (Vector Laboratories). Images were captured using a confocal laser scanning microscope (Leica).
Co-immunostaining for YY1 and NeuN—The brain sections used for the stainings were prepared as described above. Samples were incubated with anti-YY1 H-10 antibody (1:200; Santa Cruz Biotechnology) for overnight at 4 °C. After triple washes in PBS/Triton X-100 for 10 min, the sections were incubated with Alexa Fluor 555 goat anti-mouse IgG (1:200, Invitrogen) for 2 h at room temperature, washed three times in PBS/Triton X-100 for 10 min, and fixed in 4% paraformaldehyde for 10 min at room temperature. After further washes, the sections were blocked in 10% normal mouse serum and incubated with mouse anti-NeuN biotin-conjugated antibody (1:100, Chemicon) for 2 h at room temperature. Then the samples were washed three times in PBS/Triton X-100 for 10 min, incubated with streptavidin-Alexa Fluor 647 (1:200, Invitrogen) for 1 h at room temperature, and washed again analogically. The sections were mounted on slides, air-dried, and covered with Vectashield containing DAPI (Vector Laboratories). Images were captured using a confocal laser scanning microscope (Leica).
Chromatin Immunoprecipitation (ChIP)—For ChIP both rat hippocampi were immediately isolated, fragmented with a blue pipette tip in 1% formaldehyde, and incubated at 37 °C with agitation. Total time for a contact of the tissue with the fixative was 15 min., samples were washed six times in ice-cold 1× PBS containing 1 mm PMSF, 5 mm sodium fluoride, 1 mm sodium orthovanadate, 5 mm sodium butyrate, and 100× diluted protease inhibitor mixture (Sigma). Pellets were resuspended in SDS Lysis Buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl (pH 8.1) containing 1 mm PMSF, 5 mm sodium fluoride, 1 mm sodium orthovanadate, 5 mm sodium butyrate, and 100× diluted protease inhibitor mixture), homogenized, incubated on ice for 10 min with frequent vortexing, and subjected to sonication to get ∼1-kb chromatin fragments. Sonicated tissue was centrifuged (13,000 rpm, 10 min, 4 °C), and supernatants from six different rats of the same group (neuronally unstimulated rats versus PTZ-induced ones) were pooled together and diluted 10× in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl (pH 8.1), 167 mm NaCl containing 1 mm PMSF, 5 mm sodium fluoride, 1 mm sodium orthovanadate, 5 mm sodium butyrate, and 100 times diluted protease inhibitor mixture). The polled samples were then separated equally into four study samples. For input sample, an additional equivalent of 1% of the pooled sample volume was saved. ChIP was carried out with chromatin immunoprecipitation assay kit (Upstate Biotechnology, Inc.). Samples were precleared with agarose A beads. Next, 2 μg of anti-YY1α H-10 or C-20, 3 μg of CtBP1α and HDAC3α, 4 μg of RNA polymerase II antibodies were added into two of the study samples, and an equivalent amount of normal isotype control antibody to the others. Then, the procedure followed manufacturer's protocol. Each experiment was repeated in triplicate.
Real Time PCR—Samples containing purified ChIPed DNA were subjected to the real time PCR analysis using 7500 real time PCR system (Applied Biosystems). PCR was performed with TaqPCR core kit (Qiagen) according to the manufacturer's procedure. PCR additionally contained 40 ppm of SYBR Green I (Sigma). Primer pairs amplifying two different rat Mmp-9 gene proximal promoter regions were as follows: for -157/-7-bp region M10 (CTTTGGGCTGCCCAACAC) as well as M20 (AGCAGAATTTGCGGAGGTTTT), and for -187/-13-bp region C1 (GCATAAAGGAGTGGGTAGTG) and C2 (GTGAAGCAGAATTTGCGGAG). Annealing temperatures were 57 °C for M10/M20 primer pair and 55 °C for C1/C2. For amplification of the distal Mmp-9 promoter fragment, we used primers U1 (ACTCACCCAGGGAACATTTG) and U2 (CCCCAAACGCTGAAGTATGT) and annealing temperature 60 °C. These primers amplified the -3631/-3516 region of the Mmp-9 promoter. For amplification of the region downstream to Mmp-9 gene promoter we used primers D1 (GGTGCTGGAGAGGTAGGTGA) and D2 (AGTGAAAATGGACCCCACAG) and annealing temperature of 60 °C. These primers amplified a region situated in the last Mmp-9 intron (∼7 kb downstream to the Mmp-9 promoter). Each PCR was repeated at least six times. The Ct value for Mmp-9 gene promoter content in the input sample constituted a reference value to which were compared the Mmp-9 gene promoter Ct values evaluated for anti-YY1 and control antibody samples (20).
RESULTS
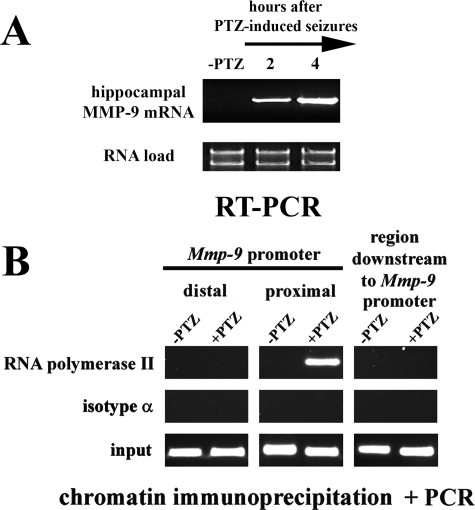
MMP-9 Gene Is Transcriptionally Activated after PTZ-evoked Neuronal Excitation in the Rat Hippocampus—We have previously shown that MMP-9 expression is strongly enhanced following treatment with the pronconvulsive and excitotoxic compound, kainate, in neurons of the rat hippocampal dentate gyrus (10). To test whether a similar phenomenon can be observed after a neuronal depolarization triggered by an agent that does not induce a neurodegeneration, we analyzed MMP-9 mRNA levels after seizures evoked by an antagonist of γ-aminobutyric acid, type A receptors, PTZ. PTZ induces a neuronal depolarization (23-25), and consequently, after its intraperitoneal application into rats, a strong generalized seizure attack is triggered (26). PTZ-induced seizures led to a substantial up-regulation of MMP-9 mRNA in the rat hippocampus (Fig. 1A). A marked mRNA up-regulation was observed at 2 h after an onset of the seizures (Fig. 1A), and reached even higher levels at the 4-h time point (Fig. 1A).
FIGURE 1.
Mmp-9 gene is transcriptionally activated after PTZ-evoked neuronal excitation in the rat hippocampus. A, MMP-9 mRNA is accumulated after PTZ-induced seizures in the rat hippocampus. RT-PCR analysis of RNA isolated from unstimulated (-PTZ) and PTZ-stimulated rat hippocampi. For each time point RNA samples obtained from six different rats were combined. For each analysis equal amounts of RNA samples were used. RT-PCR was repeated in triplicate. B, PCR-based evaluation of DNA samples obtained after immunoprecipitation of chromatin regions containing RNA polymerase II (form phosphorylated on serine 5 of C-terminal domain YSPTSPS repeats). Contents of the proximal and distal promoter fragments of the Mmp-9 gene as well as the region downstream to the Mmp-9 gene promoter were evaluated in the unstimulated (-PTZ) and in PTZ-stimulated (2 h after the onset of seizures; +PTZ) rat hippocampi. Control ChIP reactions were performed using an isotype antibody.
Next, we investigated whether induction of MMP-9 mRNA expression after PTZ administration may involve activation of transcriptional mechanisms. The C-terminal domain (CTD) of RNA polymerase II is composed of 52 tandem repeats of heptapeptide YSPTSPS that is phosphorylated at Ser-2 during transcriptional elongation (27). We immunoprecipitated chromatin bound to the C-terminal domain of RNA polymerase II phosphorylated on Ser-2 from the unstimulated and PTZ-treated rat hippocampi (2 h after seizure onset) and then analyzed the samples for DNA of Mmp-9 gene. In the immunoprecipitated samples, we have found a strong enrichment of the -187/-13-bp proximal Mmp-9 promoter fragments, whereas the distal Mmp-9 promoter DNA and the downstream one were not present in amounts higher than those found in controls (samples immunoprecipitated with an isotype antibody) (Fig. 1B). These data suggest that indeed the up-regulation of MMP-9 mRNA involves a transcriptional activation in the rat hippocampus.
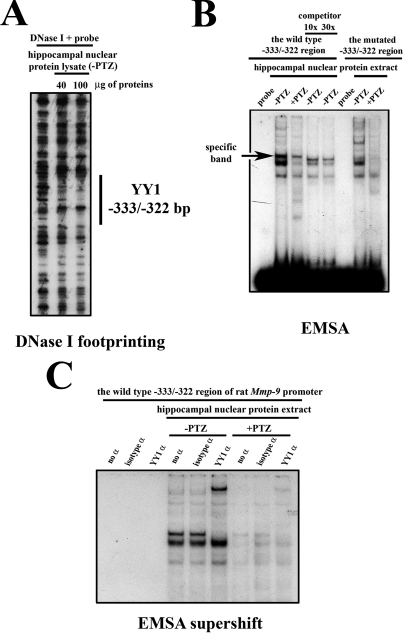
YY1 Occupies the Proximal Mmp-9 Promoter in Hippocampal Neurons—To screen for potential repressive DNA regulatory regions within the proximal -557/+18-bp fragment of the Mmp-9 promoter, we have performed DNase I footprinting analysis in the unstimulated rat hippocampus (Fig. 2A) and we have found a footprinted region at -333/-322-bp site (sequence ACTAGATGGCCC).
FIGURE 2.
YY1 occupies, both in vitro and in vivo, the -333/-322-bp site of the rat Mmp-9 promoter in the unstimulated rat hippocampus and is released as a result of neuronal depolarization. A, hippocampal nuclear proteins extracted from the unstimulated (-PTZ) rat hippocampi bind to the -333/-322-bp site of Mmp-9 promoter in vitro. B, in vitro binding of rat hippocampal nuclear proteins to the footprinted -333/-322-bp site of the rat Mmp-9 promoter is dependent on neuronal activity status being strongly reduced by neuronal depolarization. The specifically shifted band is indicated with the arrow indicated as “specific band; probe, free (nonincubated with proteins) probe; -PTZ, control; +PTZ, 2 h after PTZ-evoked neuronal excitation. C, EMSA supershift analysis revealed a strong binding of rat hippocampal YY1 to the -333/-322-bp region of Mmp-9 promoter in the unstimulated hippocampus (-PTZ) and its reduction at 2 h after PTZ (+PTZ). YY1 α, supershift with anti-YY1 antibody; no α, EMSA reaction with no antibody included; isotype α, control reaction conducted with an isotype antibody.
To confirm the capacity of the hippocampal proteins to bind to the footprinted sequence, we have employed EMSA using the DNA probe encompassing the footprinted region of the rat Mmp-9 promoter together with nuclear proteins extracted from the hippocampi before and at 2 h after neuronal activation evoked by PTZ. We identified three bands, with the strong uppermost protein-DNA complex disappearing in the competition experiment, thus proving itself as the specific DNA/protein interaction. Importantly, a similar conclusion was reached from the experiment with a mutated probe (Fig. 2B). Furthermore, we observed this complex to be much more abundant before than 2 h after the PTZ (Fig. 2B, compare lanes -PTZ and +PTZ).
The MatInspector (28) analysis revealed YY1 as a potential transcription factor that can bind to the DNA sequence identified in the footprinting experiment. To check whether indeed YY1 protein can bind to the identified promoter sequence, we incubated hippocampal nuclear protein lysates obtained from control as well as PTZ-treated animals with a YY1-specific antibody and then subjected to the EMSA reaction. ntibody super-shifted the specific band completely (Fig. 2C). No supershift was observed when a control antibody was used or no antibody was included or when hippocampal protein extract was omitted (Fig. 2C).
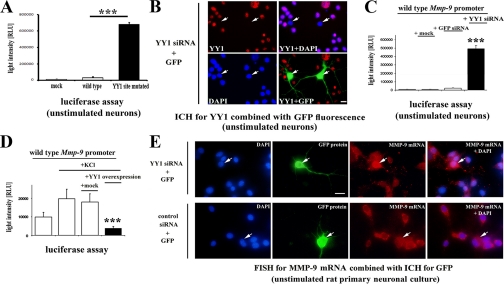
Footprinted -333/-322-bp Site of the Rat Mmp-9 Gene Promoter Exerts Strong Repressive Influence on Mmp-9 Gene Promoter Activity in Rat Neurons—Altogether, the results described so far suggested that YY1 may indeed be a repressor of Mmp-9 transcriptional activity in the brain neurons. To evaluate the impact exerted by the YY1 site onto Mmp-9 transcription, we introduced the construct containing a luciferase gene under the control of either the rat -1369/+35-bp Mmp-9 promoter fragment mutated at the YY1 site or its wild-type (WT) equivalent in rat primary neuronal cultures. The mutation robustly (over 30-fold) increased the promoter activity in the unstimulated cultures (Fig. 3A).
FIGURE 3.
YY1 acting via -333/-322-bp site of Mmp-9 promoter is a potent and essential transcriptional repressor of MMP-9 in neurons. A, footprinted -333/-322-bp site of the rat Mmp-9 promoter exerts a repressive influence on Mmp-9 promoter activity in rat neurons. Luciferase activities recorded after lipofection of the unstimulated rat primary neuronal cultures with constructs containing either WT or -333/-322-bp site mutated (YY1 site mutated) rat Mmp-9 promoters controlling luciferase gene. Mock indicates results obtained after a lipofection of a backbone of Mmp-9 promoter constructs. Error bars represent the mean ± S.D. (n = 5; ***, p < 0.001). B, YY1 immunocytochemical staining (in red) of the unstimulated rat primary neuronal culture co-lipofected with YY1 and GFP siRNA expression constructs. Cell nuclei are stained with DAPI (in blue); GFP is green. Scale, 20 μm. White arrows indicate the lipofected cells. C, depletion of YY1 leads to a robust induction of the rat Mmp-9 promoter activity in the unstimulated rat neurons. Luciferase activity recorded after co-lipofection of luciferase controlled by the WT rat Mmp-9 promoter with either empty vector (mock), or control siRNA (GFP siRNA) expression construct, or YY1 siRNA expression construct is presented. Error bars represent the mean ± S.D. (n = 5; ***, p < 0.001). D, YY1 overexpression results in a repression of the rat Mmp-9 promoter activity in the depolarized neurons. Luciferase activity recorded after co-lipofection of luciferase under control of the WT rat Mmp-9 promoter with one of the following constructs: GW1 empty vector (mock), YY1 expression construct (YY1 overexpression). Error bars represent the mean ± S.D. (n = 5; ***, p < 0.001). +KCl indicates the cultures at 2 h after KCl-evoked excitation. E, YY1 critically represses endogenous MMP-9 transcription in the unstimulated rat neurons. Fluorescent in situ hybridization for MMP-9 mRNA is shown along with immunocytochemistry for GFP in the unstimulated cultures co-lipofected with the GFP and with either YY1 or control (cortactin) siRNA expression constructs. MMP-9 mRNA is shown in red, and cell nuclei are presented in blue (DAPI) and GFP immunofluorescence in green. White arrows indicate the lipofected cells. Scale bar, 20 μm.
YY1 Is a Potent and Essential Transcriptional Repressor of MMP-9 in Neurons—In the next experiment, YY1 was depleted in the unstimulated neuronal cultures with siRNA directed against 994-1012-bp fragment of the rat YY1 mRNA. To visualize the transfected neurons, we have also co-transfected a GFP-expressing construct (see Fig. 3B). We observed YY1 expression that was localized exclusively to the cell nuclei in virtually all nontransfected neurons (Fig. 3B). In contrast, ∼85% of the transfected (GFP-positive) neurons were YY1-negative, in the next ∼10% of them YY1 was expressed at the lower level than in surrounding GFP-negative neurons, and in remaining ∼5% of GFP-positive cells the level of YY1 remained unchanged compared with the untransfected neurons.
To evaluate the effects of YY1 depletion on MMP-9 transcription, we co-transfected the YY1 siRNA-expressing construct with a plasmid harboring the WT Mmp-9 promoter, controlling luciferase expression. The YY1 depletion led to a very strong (∼50-fold) up-regulation of Mmp-9 promoter activity in the unstimulated neurons (Fig. 3C). Importantly, Mmp-9 promoter activity was not affected in neurons co-transfected with the rat WT Mmp-9 promoter-reporter and either GFP siRNA or the backbone for YY1 siRNA-expressing constructs (Fig. 3C).
To further confirm the repressive action of YY1 on MMP-9 transcription, we overexpressed the full-length rat YY1 protein along with a luciferase gene controlled by the WT Mmp-9 promoter. We found that YY1 led to a transcriptional repression of the promoter in neurons after its depolarization with KCl (Fig. 3D). Significantly, control co-transfection conducted with the rat WT Mmp-9 promoter reporter construct and the backbone for YY1-expressing construct did not affect Mmp-9 promoter activity (Fig. 3D).
Finally, we co-transfected the YY1 siRNA-expressing construct with the GFP-expressing construct in unstimulated cultures and evaluated the endogenous MMP-9 mRNA levels by fluorescent in situ hybridization (Fig. 3E). Around 27% of GFP-positive cells had markedly increased MMP-9 mRNA levels as compared with the nontransfected (GFP-negative) surrounding neurons. In 73% of neurons, MMP-9 mRNA levels were not appreciably up-regulated. For control, either empty pSuper vector or rat cortactin siRNA-expressing construct (29) was used, with no effect on MMP-9 mRNA levels. The construct expressing siRNA for GFP was not used as a control in the experiment, because it would quench GFP expression.
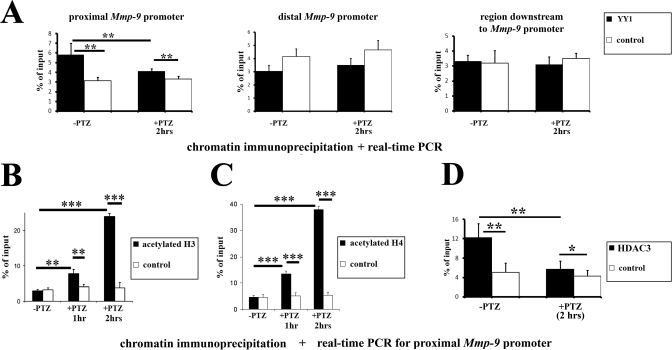
YY1 Is Contained within the Proximal Mmp-9 Gene Promoter Chromatin in Vivo, and Mmp-9 Repression Involves Histone Deacetylation—To address the question whether YY1 occupies proximal Mmp-9 promoter in vivo in the unstimulated rat hippocampus and to check if it is released after the PTZ-evoked neuronal activation, we carried out ChIP experiments using the hippocampi from unstimulated rats and animals at 2 h after an onset of the seizures (Fig. 4A). Real time PCR was used to quantify, in DNA purified from chromatin samples immunoprecipitated with either the YY1 antibody or its isotype control, the content of the proximal and distal Mmp-9 promoter fragments as well as a genome region situated downstream to the rat Mmp-9 gene promoter. We have found enrichment, over their isotype controls, of the proximal Mmp-9 promoter fragments (but not the other two) in YY1 immunoprecipitated samples, both in unstimulated as well as in stimulated hippocampi (Fig. 4A). However, the proximal Mmp-9 promoter DNA was enriched 2-fold in unstimulated rat hippocampal samples immunoprecipitated with anti-YY1 antibody, whereas in similarly prepared samples obtained from rats 2 h after PTZ-evoked seizures Mmp-9 promoter enrichment was very limited. Congruent results were obtained with two different anti-YY1 antibodies and two different sets of primers that amplify either -187/-13-bp or -157/-7-bp regions of the rat Mmp-9 promoter. Altogether, these data indicate that YY1 occupies the Mmp-9 promoter in vivo in the unstimulated rat hippocampus and is released after neuronal activation.
FIGURE 4.
In the rat hippocampus in vivo, neuronal depolarization triggers YY1 and HDAC3 dissociation from chromatin of the proximal Mmp-9 promoter as well as a hyperacetylation of histones H3 and H4 in this genomic fragment. A, neuronal depolarization triggers YY1 dissociation of the proximal Mmp-9 promoter in vivo. Evaluation was by real time PCR of the proximal and the distal Mmp-9 promoter contents as well as levels of the genomic fragment located downstream to the Mmp-9 gene promoter in DNA samples obtained because of immunoprecipitation of chromatin-bound YY1 from the unstimulated (-PTZ) as well as from the PTZ-stimulated (2 h, +PTZ) rat hippocampi. Control ChIP reactions were performed with an isotype antibody. Error bars represent the mean ± S.D. (n = 10; **, p < 0.01). B, neuronal depolarization induces hyperacetylation of histone H3 in the proximal Mmp-9 promoter of the rat hippocampal chromatin in vivo. Evaluation of Mmp-9 proximal promoter content by real time PCR in DNA samples obtained because of immunoprecipitation of chromatin-bound hyperacetylated histone H3 from the unstimulated (-PTZ) as well as from the PTZ-stimulated (2 h; +PTZ) rat hippocampi. Error bars represent the mean ± S.D. (n = 3; **, p < 0.01, ***, p < 0.001). C, neuronal depolarization induces hyperacetylation of histone H4 in the proximal Mmp-9 promoter of the rat hippocampal chromatin in vivo. Evaluation of Mmp-9 proximal promoter content by real time PCR in DNA samples obtained because of immunoprecipitation of chromatin-bound hyperacetylated histone H4 from the unstimulated (-PTZ) as well as from the PTZ-stimulated (2 h; +PTZ) rat hippocampi. Error bars represent the mean ± S.D. (n = 3; ***, p < 0.001). D, HDAC3 occupies the proximal Mmp-9 promoter in unstimulated hippocampus and is released after PTZ-stimulated neuronal depolarization. Evaluation by real time PCR of the proximal Mmp-9 promoter content in DNA samples obtained because of immunoprecipitation of chromatin bound to HDAC3 from the unstimulated (-PTZ) as well as from the PTZ-stimulated (2 h, +PTZ) rat hippocampi. Control ChIP reactions were performed with isotype antibody. Error bars represent the mean ± S.D. (n = 3; *, p < 0.05, **, p < 0.01).
YY1 can regulate transcription by affecting histone acetylation and chromatin condensation (30, 31). It has been shown that numerous HDACs (32-35) and acetylases (35-37) can bind to YY1. Therefore, we evaluated the levels of histone H3 and H4 hyperacetylation in the Mmp-9 proximal promoter chromatin by ChIP. We have not observed any differences in the amounts of the proximal Mmp-9 promoter fragments present in hyperacetyl-H3 and -H4 immunoprecipitated samples over their respective controls in the unstimulated hippocampi (Fig. 4, B and C). In contrast, at 1 h after PTZ, the differences begin to be noticeable, and at 2 h after the treatment, we have found a strong enrichment of the proximal Mmp-9 promoter fragments in hyperacetylated H3 (Fig. 4B) as well as H4 (Fig. 4C) immunoprecipitated samples over their isotype controls. Congruent results were obtained with two different sets of primers that amplify either -187/-13- or -157/-7-bp regions of the rat Mmp-9 promoter.
Then we have searched by ChIP for HDACs that operate onto the proximal Mmp-9 promoter in the rat hippocampus in vivo. We have analyzed occupancy of the promoter by HDAC1 to HDAC7 in the unstimulated rat hippocampus. As a result, we have discovered that HDAC3 was the only histone deacetylase present in the chromatin encompassing the MMP-9 genome region in the unstimulated hippocampus in vivo (Fig. 4D). Additionally, we have found that PTZ-dependent neuronal depolarization leads, 2 h after the seizures, to HDAC3 dissociation from the proximal Mmp-9 promoter in the rat hippocampus in vivo (Fig. 4D). These data suggest that YY1 could attract HDAC3 to the proximal Mmp-9 promoter in the unstimulated hippocampus in vivo and suggest that the neuronal excitation leads to a dissociation of the YY1/HDAC3 complex from the promoter, which may lead to the local histone hyperacetylation and thus loosen up the chromatin structure to favor transcription by RNA polymerase II.
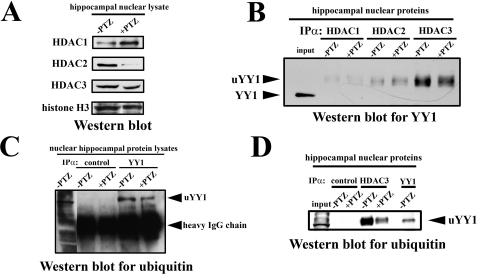
Neuronal Depolarization Reduces HDAC3/YY1 Interaction in the Cell Nuclei of the Rat Hippocampus—We evaluated by Western blot analysis the expression levels of HDACs before and 2 h after PTZ-evoked seizures in the cell nuclei of the rat hippocampal cells. We found that all analyzed HDACs (HDAC1, HDAC2, and HDAC3) were expressed in the unstimulated rat hippocampus; however, their expression was differently modulated by PTZ treatment (Fig. 5A). HDAC1 was up-regulated 2 h after PTZ-induced seizures, whereas HDAC2 and HDAC3 were down-regulated (Fig. 5A).
FIGURE 5.
Ubiquitinated YY1 physically interacts with HDAC3 in the nuclei of rat hippocampal cells; however, the interaction is markedly reduced after neuronal depolarization. A, HDAC1, HDAC2, and HDAC3 are expressed in cell nuclei of the rat hippocampal cells, and their expression is modified by PTZ treatment. Equal amounts (20 μg) of nuclear cell lysates obtained from the unstimulated (-PTZ) as well as from the PTZ-stimulated (2 h; +PTZ) rat hippocampi were analyzed by Western blot with antibodies against anti-HDAC1, -2, and -3. As a loading control, results of Western blot analysis for histone H3 are shown. B, YY1 physically interacts with HDACs in the nuclei of the rat hippocampal cells. Equal amounts (300 μg) of nuclear cell lysates obtained from the unstimulated (-PTZ) as well as from the PTZ-stimulated (2 h; +PTZ) rat hippocampi were immunoprecipitated (IPα) with anti-HDAC1, anti-HDAC2, or anti-HDAC3 antibody and analyzed for YY1 expression by Western blot. As a positive control, protein nuclear cell lysate obtained from the unstimulated hippocampi has been used (resolved in the input lane). The unmodified YY1 and ubiquitinated YY1 (uYY1) are indicated. C, after PTZ-evoked neuronal activation, an expression of the ubiquitinated form of YY1 is diminished in cell nuclei of the rat hippocampus. Equal amounts of nuclear cell lysates obtained from the unstimulated (-PTZ) as well as from the PTZ-stimulated (2 h; +PTZ) rat hippocampi were immunoprecipitated with anti-YY1 or control (isotype) antibody and analyzed for a ubiquitin expression by Western blot. As a positive control, protein nuclear cell lysates obtained from the unstimulated hippocampi have been used. Please note that the intensity of IgG bands reflects similar loading of immunoprecipitated samples. The ubiquitinated YY1 (uYY1) and IgG heavy chain bands are indicated. D, interaction of HDAC3 with ubiquitinated YY1 is neuronal activity-dependent in cell nuclei of the rat hippocampus. Equal amounts (300 μg) of nuclear cell lysates obtained from the unstimulated (-PTZ) as well as from the PTZ-stimulated (2 h; +PTZ) rat hippocampi were immunoprecipitated with anti-HDAC3 or control (isotype) antibody and analyzed by Western blot for ubiquitin. As a positive control, 20% of protein nuclear cell lysate immunoprecipitated with anti-YY1 antibody from the unstimulated hippocampi has been used. The ubiquitinated YY1 (uYY1) is indicated. In the input lane, 30 μg of hippocampal nuclear proteins has been resolved.
Then we investigated whether HDACs could associate with YY1 in the cell nuclei of rat hippocampal neurons in vitro. We immunoprecipitated HDAC3 as well as two other histone deacetylases HDAC1 and HDAC2 (which did not bind to the proximal Mmp-9 promoter, as observed by ChIP, see above) from rat hippocampal nuclear lysates, and we analyzed the resulting samples by Western blot for YY1 both in the unstimulated and PZT-treated (2 h after the seizures) rat hippocampi. In unstimulated hippocampal neurons, we found a very strong interaction between YY1 and HDAC3 and a much weaker interaction between YY1 and either HDAC1 or HDAC2 (Fig. 5B). Furthermore, decreased YY1 association with HDAC3, but not with HDAC1 and HDAC2, was most clearly dependent on neuronal depolarization as a substantial reduction of YY1/HDAC3 interaction occurred 2 h after PTZ-evoked seizures (Fig. 5B). Intriguingly, during analysis of the YY1 Western blot data, we observed that YY1 immunoprecipitated with HDACs migrates slower than expected (compare band migration in lanes containing HDAC immunoprecipitates to the band in the input lane in Fig. 5B). This unexpected finding indicated that HDACs can bind only a specific variant of YY1. The shift in migration indicated that both proteins differ by ∼10 kDa in their molecular mass, which could be a consequence of YY1 monoubiquitination.
To examine whether YY1 becomes ubiquinated, we immunoprecipitated hippocampal YY1 and probed for a ubiquitin using Western blot. We found a band of ubiquitinated protein migrating exactly at the same level as the retarded YY1 band found in the HDAC immunoprecipitates (Fig. 5C). We have also noticed that nuclear YY1 is apparently ubiquitinated to a greater degree before than at 2 h after the onset of seizures (Fig. 5C).
To confirm unequivocally that HDAC3 interacts with the ubiquitinated form of YY1 in the rat hippocampus, we immunoprecipitated HDAC3 from the hippocampal lysates and analyzed the samples by Western blot with an antibody specific for ubiquitin. We found that the bands observed in HDAC3 immunoprecipitates were of exactly the same size as ubiquitinated YY1 (compare HDAC3 IP lanes with YY1 IP lane at Fig. 5D). The results of these experiments indicate that there is a strong interaction between HDAC3 and ubiquitinated YY1 in the cell nuclei of the unstimulated hippocampus, which is substantially reduced at 2 h after the seizures (Fig. 5D).
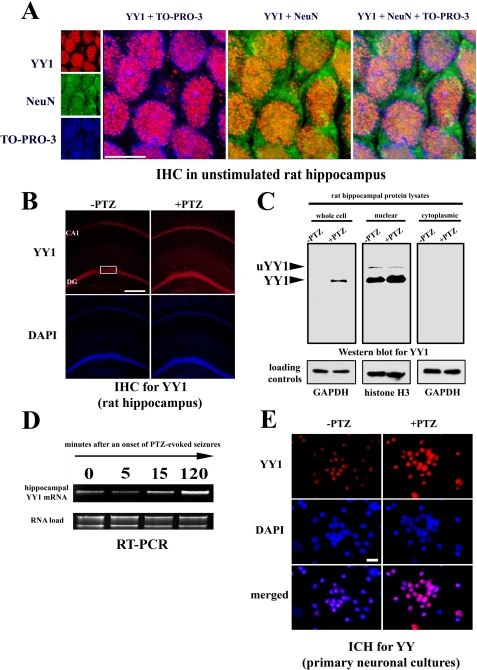
YY1 Protein and mRNA Are Localized to Neurons, and Their Expression Is Up-regulated after Neuronal Depolarization—Our previous studies showed that in the unstimulated hippocampus MMP-9 is predominantly expressed by neurons (10-12). Moreover, the data presented so far have consistently indicated that YY1 is a repressor of MMP-9 transcription. Thus, it could be expected that YY1 is expressed in the neuronal cell nuclei and is down-regulated by neuronal depolarization. To test this notion, we first co-immunostained sections of the unstimulated rat hippocampus for YY1 and a marker of neurons, NeuN, and we found extensive co-localization of the signals in cell nuclei (Fig. 6A). Next, we evaluated the influence of neuronal excitation onto YY1 expression. We unexpectedly found that YY1 expression is induced at 2 h after the PTZ seizures (Fig. 6B).
FIGURE 6.
YY1 is expressed in the cell nuclei of hippocampal neurons and is up-regulated after neuronal depolarization. A, YY1 co-localizes with the neuronal marker NeuN in the cell nuclei of the unstimulated rat hippocampus. The panel presents confocal photomicrographs of immunohistochemical staining for YY1 (in red) and neuronal marker NeuN (in green) in a fragment of hippocampal region marked with a white rectangle on B. Staining for cell nuclei with TO-PRO-3 is demonstrated in blue. Merged view of the staining is also included. Scale bar, 5 μm. B, confocal photomicrographs of YY1 immunohistochemical staining (in red) of the hippocampal sections obtained from control (unstimulated; -PTZ) rats and at 2 h after PTZ stimulation (+PTZ) are presented. CA1, CA1 field; DG, dentate gyrus. Cell nuclei are stained with DAPI (in blue). Scale bar, 200 μm. C, representative Western blot showing YY1 expression levels and YY1 intracellular distribution in the control (-PTZ) rat hippocampus and at 2 h after PTZ-induced neuronal depolarization (+PTZ). The unmodified YY1 and its ubiquitinated variant (uYY1) are indicated. Western blot data for loading controls were obtained by resolving (on an another gel) equal amounts of the same protein lysates as were analyzed on the YY1 Western blot. D, changes in YY1 mRNA expression after PTZ-induced seizures assessed by RT-PCR in the rat hippocampus. The intensity of rRNA bands was used as a loading control. E, immunocytological staining for YY1 (in red) of the unstimulated primary neuronal culture and at 2 h after an onset of a depolarization evoked by 30 mm KCl is demonstrated. Cell nuclei are stained with DAPI (in blue). Scale bar, 20 μm. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Furthermore, we have also demonstrated the same phenomenon of up-regulated YY1 expression at 2 h after the depolarization by means of Western blot approach (Fig. 6C). Moreover, we have observed that YY1 is expressed exclusively in the cell nuclear extract (Fig. 6C). Interestingly, in the nuclear cell lysates an additional, ∼10-kDa heavier form of YY1 was observed. Its expression was diminished at 2 h after the depolarization (Fig. 6C). It probably represents the monoubiquitinated variant of YY1. In addition, we have also shown that PTZ treatment provokes YY1 mRNA accumulation observed as quickly as 15 min after seizure onset (Fig. 6D). Finally, we have found that KCl-evoked neuronal depolarization in vitro also results in enhanced nuclear YY1 expression at 2 h after the treatment (Fig. 6E). Furthermore, it is also interesting to note that YY1 is expressed by almost all neurons in culture (Fig. 6E).
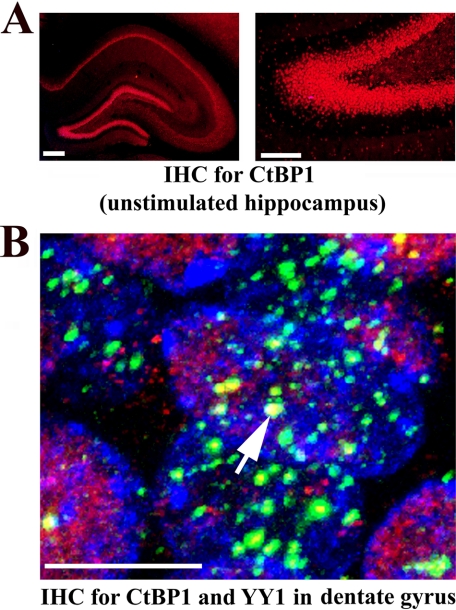
CtBP1 Is Strongly Expressed in the Cell Nuclei of the Rat Hippocampus and Co-localizes with YY1—To posit a mechanism responsible for the dissociation of YY1 from the proximal Mmp-9 promoter despite an up-regulation of YY1 expression in the neuronal nuclei of the PTZ-depolarized rat hippocampus, we have decided to follow the previous reports that YY1 critically requires the transcriptional co-regulator CtBP to repress genes in vivo (38). Moreover, it has also been shown that CtBP regulates YY1 binding to DNA (39). Thus, we tested in our experimental setting whether CtBP could regulate YY1 binding to the proximal Mmp-9 promoter. First, we immunostained sections of the unstimulated rat hippocampus with anti-CtBP1 antibody, and we found its strong expression in the cell nuclei of the neuronal body layers of all fields of the hippocampus proper and the dentate gyrus (Fig. 7A). Then we tested whether CtBP1 co-localized with YY1 in nuclei of the hippocampal cells on the immunostained (with anti-YY1 and anti-CtBP1) sections of the unstimulated rat hippocampus. In the case of CtBP1, we observed mostly a diffuse nuclear staining pattern, whereas YY1 nuclear distribution was clearly granular (Fig. 7B). We have noticed that CtBP1 is co-localized to the centers of some YY1 expression granules in cell nuclei of the hippocampus (Fig. 7B).
FIGURE 7.
CtBP1 is strongly expressed in the cell nuclei of the rat hippocampus and co-localizes with YY1. A, CtBP1 (in red) is expressed in the nuclei located in neuronal cell body layers of the unstimulated hippocampus. Fluorescent microscope photomicrograph on the left demonstrates CtBP1 immunolocalization throughout all areas of the rat hippocampus. Higher magnification of the same micrograph showing CtBP1 expression in a fragment of the dentate gyrus is shown on the right. Scale bars, for left photomicrograph 200 μm and for right photomicrograph 100 μm. B, CtBP1 (in red) is co-localized with YY1 expression bodies (in green) in the nuclei of the dentate gyral cells. High power confocal image of the cell nuclei from the granular layer of the unstimulated dentate gyrus is provided. One of the biggest co-localization centers is indicated with the white arrow. Cell nuclei are stained with TO-PRO-3 (in blue). Scale bar, 5 μm.
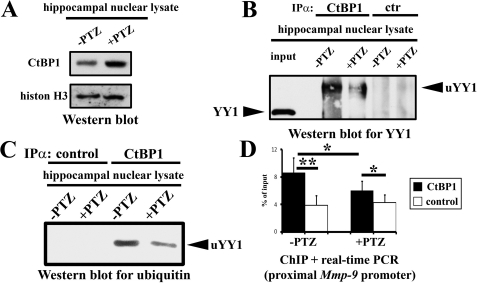
Neuronal Depolarization Leads to a Reduction of CtBP1/YY1 Interaction in Vitro and Triggers CtBP1 Dissociation from the Proximal Mmp-9 Promoter in Vivo in the Cell Nuclei of the Rat Hippocampus—We examined by Western blot analysis expression levels of CtBP1 before and 2 h after PTZ-evoked seizures in the cell nuclei of the rat hippocampal cells. We found that although CtBP1 was expressed in the unstimulated rat hippocampus, however, its expression was significantly up-regulated after PTZ-evoked seizures (Fig. 8A).
FIGURE 8.
Neuronal depolarization leads to a reduction of CtBP1/YY1 interaction in vitro and triggers CtBP1 dissociation from the proximal Mmp-9 promoter in vivo in the cell nuclei of the rat hippocampus. A, CtBP1 is expressed in cell nuclei of the rat hippocampal cells and up-regulated after PTZ-induced seizures. Equal amounts (20 μg) of nuclear cell lysates obtained from the unstimulated (-PTZ) as well as from the PTZ-stimulated (2 h; +PTZ) rat hippocampi were analyzed by Western blot with anti-CtBP1 antibody. As a loading control, results of Western blot analysis for histone H3 are shown. B, CtBP1 physically interacts with YY1 in a depolarization-dependent manner in cell nuclei of the rat hippocampus. Equal amounts (300 μg) of nuclear cell lysates obtained from the unstimulated (-PTZ) as well as from the PTZ-stimulated (2 h; +PTZ) rat hippocampi were immunoprecipitated (IPα) with anti-CtBP1 or control (isotype) antibody and analyzed for YY1 expression by Western blot. As a positive control, protein nuclear cell lysate obtained from the unstimulated hippocampi has been used (resolved in the input lane). Unmodified YY1 (YY1) and its ubiquitinated form (uYY1) are indicated. C, interaction of CtBP1 with ubiquitinated YY1 is neuronal activity-dependent in cell nuclei of the rat hippocampus. Equal amounts (300 μg) of nuclear cell lysates obtained from the unstimulated (-PTZ) as well as from the PTZ-stimulated (2 h; +PTZ) rat hippocampi were immunoprecipitated with anti-CtBP1 or control (isotype) antibody and analyzed for ubiquitin expression by Western blot. The ubiquitinated YY1 (uYY1) is indicated. D, CtBP1 occupies the proximal Mmp-9 promoter in the unstimulated hippocampus and is released after PTZ-evoked neuronal depolarization. Evaluation by real time PCR of the proximal Mmp-9 promoter content in DNA samples obtained because of immunoprecipitation of chromatin bound to CtBP1 from the unstimulated (-PTZ) as well as from the PTZ-stimulated (2 h; +PTZ) rat hippocampi. Control ChIP reactions were performed with isotype antibody. Error bars represent the mean ± S.D. (n = 3; *, p < 0.05; **, p < 0.01).
Next, we immunoprecipitated CtBP1 from the rat hippocampal nuclear lysates and then analyzed the resulting samples for YY1 expression by Western blot and found YY1 expression in CtBP1 immunoprecipitates but not in the control samples (Fig. 8B). This finding indicates the existence of a physical interaction between YY1 and CtBP1 in the cell nuclei of the rat hippocampus. This association was dependent on the depolarization. It was strong in unstimulated nuclei, while at 2 h after PTZ-evoked seizures CtBP1/YY1 complexes were markedly reduced (Fig. 8B). Moreover, YY1, which was immunoprecipitated with CtBP1, exhibited retarded migration compared with the native YY1 (Fig. 8B), having simultaneously the same molecular weight as the ubiquitinated form of YY1. Hence, it appears that CtBP1 binds only to the ubiquitinated YY1.
To confirm this hypothesis, we immunoprecipitated CtBP1 from the rat hippocampal nuclear lysates, and we analyzed resulting samples for ubiquitin expression by Western blot in the unstimulated and PZT-treated (2 h after the seizures) animals. We found that CtBP1 immunoprecipitates, but not the controls, contained ubiquitinated protein having the same size as the slower migrating variant of YY1 (Fig. 8C). Therefore, the data demonstrate that, similarly as in the case of HDAC3/YY1 association, YY1/CtBP1 complexes contain the monoubiquitinated form of YY1. Furthermore, we noticed a reduction in uYY1/CtBP1 interaction at 2 h after PTZ-induced seizures (Fig. 8C).
Finally, we analyzed by ChIP whether CtBP1 occupies the proximal Mmp-9 promoter in the unstimulated hippocampus. We immunoprecipitated hippocampal chromatin from unstimulated rats and animals 2 h after the onset of seizures using anti-CtBP1 antibody and a control isotype antibody. Then we quantified the amount of the proximal Mmp-9 promoter fragments in the samples, and we found over 2-fold enrichment of the proximal Mmp-9 promoter fragments in CtBP1 immunoprecipitates compared with their isotype controls in the unstimulated hippocampi (Fig. 8D). In samples obtained from the stimulated hippocampi, we observed only a very limited CtBP1 enrichment (Fig. 8D). These data suggest that CtBP1 is present in the proximal Mmp-9 promoter in unstimulated hippocampus in vivo, whereas 2 h after PTZ-evoked depolarization occupancy of the promoter by CtBP1 is markedly reduced. This finding suggests that enhanced neuronal activity triggers CtBP1 dissociation from this genomic region.
DISCUSSION
In this study, we show that in the unstimulated rat hippocampus in vivo, as well as in the nondepolarized brain neurons in primary culture, the transcriptional regulator YY1 is bound to the -333/-322-bp site of the Mmp-9 promoter and strongly represses Mmp-9 transcription. Furthermore, we demonstrate that YY1 critically regulates Mmp-9 mRNA synthesis in rat brain neurons by exerting control over the transcription, apparently via epigenetic mechanisms involving histone deacetylation.
With the aid of DNase I footprinting, we have discovered that in the unstimulated hippocampus there is a protein-occupied Mmp-9 promoter region that has been identified in silico to be capable of binding YY1. We have then confirmed YY1 binding to the identified Mmp-9 gene sequence by EMSA and EMSA supershift approaches and subsequently demonstrated, by ChIP, that YY1 associates with the proximal Mmp-9 promoter. Notably, we have shown that this interaction is much less pronounced following neuronal excitation. Decreased YY1 binding to the Mmp-9 promoter was concomitant with MMP-9 transcriptional activation as documented by RNA polymerase II association with the promoter and MMP-9 mRNA accumulation detected by RT-PCR. We have also shown by co-immunostaining that YY1 is expressed mainly in the cell nuclei of hippocampal neurons. Furthermore, using protein and chromatin immunoprecipitation approaches, we have shown that in the nonstimulated tissue YY1 interacts with histone deacetylase, HDAC3, and that this interaction and the promoter occupation by HDAC3 are diminished following seizures. On the other hand, after neuronal depolarization, the Mmp-9 promoter becomes gradually hyperacetylated at histone H3 and H4 lysine residues, as demonstrated by ChIP. We have also discovered by means of protein immunoprecipitation that a portion of YY1 in the cell nuclei is ubiquitinated and that only this modified variant of YY1 interacts with HDAC3. Moreover, the amount of ubiquitinated YY1 is decreased following neuronal excitation by PTZ, despite the fact of overall increased levels of unmodified YY1 after the occurrence of seizures. This apparent discrepancy can be resolved by our finding that ubiquitinated YY1 interacts in a depolarization-dependent manner with a transcriptional co-regulator CtBP1, which occupies the proximal Mmp-9 promoter in vivo and is expressed in the cell nuclei of neurons in the hippocampus. Importantly, it has been shown before that CtBP controls the capacity of YY1 to bind DNA (39).
The aforementioned data, indicating that YY1 exerts transcriptional repressive activity on the Mmp-9 promoter in vivo, are further strengthened by our in vitro studies on cultured primary neurons, in which we show that the MMP-9 gene promoter construct is not repressed in nondepolarized cells if the YY1-binding site is mutated. Furthermore, we demonstrate that in unstimulated neurons YY1 depletion by specific siRNA activates the wild-type Mmp-9 promoter construct as well as the endogenous MMP-9 gene. Accordingly, in depolarized neurons YY1 overexpression leads to a repression of stimulated wild-type Mmp-9 promoter activity.
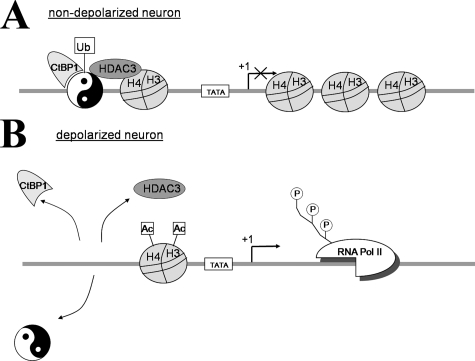
Altogether, our results allow us to propose the following model (Fig. 9) of the mechanisms governing the YY1 role in Mmp-9 gene silencing. In the unstimulated neurons in vivo, monoubiquitinated YY1 is bound to the Mmp-9 proximal promoter (presumably because of its interaction with CtBP1) and docks HDAC3 into the Mmp-9 genomic region, which keeps histones H3 and H4 of its chromatin in the hypoacetylated state. This in turn leads to the condensation of chromatin and as a consequence to active repression of Mmp-9 transcription. In contrast, in response to neuronal depolarization, YY1 becomes de-ubiquitinated, and CtBP1 dissociates from YY1 bound to the proximal Mmp-9 promoter, inducing as a consequence both YY1 and HDAC3 dissociation, hyperacetylation of histones H3 and H4, chromatin relaxation, attraction of RNA polymerase II, and initiation of Mmp-9 transcription.
FIGURE 9.
Model of Mmp-9 gene expression regulation by YY1 in neurons. A, in unstimulated neurons in vivo, YY1 is bound to the proximal Mmp-9 promoter because of YY1 ubiquitination and an interaction with CtBP1. uYY1/CtBP1 complex recruits HDAC3, which deacetylates local histones and compacts the chromatin structure, thereby inhibiting the binding of RNA polymerase II (Pol II) and repressing Mmp-9 transcription. B, after neuronal depolarization, YY1 ubiquitination is reduced, and CtBP1 is liberated from YY1 bound to the proximal Mmp-9 promoter, inducing as a consequence YY1 and HDAC3 dissociation, hyperacetylation of histones H3 and H4 leading to relaxation of chromatin around the Mmp-9 transcriptional start site and to RNA polymerase II binding and initiation of Mmp-9 transcription.
This model is in agreement with the findings that neuronal depolarization induces changes in the histone acetylation status of promoter chromatin of many different genes and, in this way, can regulate their transcription (40-42). Notably, we extend those studies by showing specific association of HDAC3 and not the other histone deacetylases with YY1 occupying the Mmp-9 promoter. Our model is also in a good agreement with YY1 functions established for other cell types. It has repeatedly been shown that YY1 can control transcription not only directly, but also indirectly, epigenetically, through a variety of mechanisms (34, 43-45). Interestingly, it has recently been reported that YY1 critically controls oligodendrocyte progenitor differentiation in the brain repressing genes coding for inhibitors of the maturation because of deacetylation of histones present in their promoters (46).
It should be mentioned that Mmp-9 gene regulation varies greatly, depending on species, tissue, cell type, stage of development, and differentiation as well as experimental setting (6, 47). Moreover, it should also be kept in mind that Mmp-9 transcriptional regulation is a very complex phenomenon and involves multiple elements of cell transcriptional machinery (6, 48). Thus, despite a strong inhibition of Mmp-9 transcription by YY1 in unstimulated hippocampal and cortical neurons, we cannot exclude the possibility that there are neuronal types in which Mmp-9 gene expression is not YY1-dependent. In our study we have focused on YY1-dependent regulation of Mmp-9 gene expression in neurons. Further studies are warranted to determine the YY1-dependent regulation of MMP-9 expression in other cell types.
The proposed molecular mechanism of MMP-9 gene regulation does not preclude participation of other transcriptional mechanisms in the control of MMP-9 gene expression. Noticeably, we observed that after a reduction of YY1 expression in neurons by YY1 siRNA, only more than a quarter of the cells displayed significant MMP-9 mRNA up-regulation, as evaluated by in situ hybridization, although siRNA depleted YY1 levels below or at the border of the anti-YY1 antibody detection limit in the majority of neurons. Consequently, we suggest that the up-regulation of MMP-9 expression levels is observed only in the cells with the most pronounced reduction in YY1 expression. Clearly, although most of the cells have YY1 expression down-regulated below the anti-YY1 antibody detection limit, they may still contain sufficient YY1 to exert a repressive influence on neuronal Mmp-9 promoter activity. Moreover, it is also conceivable that our in situ hybridization procedure detects only the most marked increases in mRNA levels, leaving less pronounced changes undetected.
It has been previously shown that YY1 can regulate protein ubiquitination directly (49, 50). Here we report on our finding that YY1 can itself be ubiquitinated and regulated by ubiquitination. Our data show that in the rat hippocampus, YY1 is apparently monoubiquitinated in a neuronal activity-dependent manner (see Fig. 5C and 6C), which is related to a change in its activity. To the best of our knowledge, this is the first demonstration of YY1 ubiquitination and points to a new mechanism for a regulation of YY1 transcriptional activity. In the model presented on the Fig. 9 we suggest that the de-ubiquitination may play a role in detaching YY1 from the gene promoter. Moreover, other groups have shown that the native YY1 can physically interact with HDAC3 (34, 35). In this aspect, it is an interesting observation that in the rat hippocampal neurons HDAC3 interacts only with ubiquitinated form of YY1.
YY1 is strongly expressed in the adult central nervous system being predominantly localized to neurons (51). Nevertheless, very little is known about YY1 actions in the adult brain cells, and no neuronal YY1 target genes were clearly defined therein in the adult central nervous system. Hence, MMP-9 is the first YY1-dependent gene identified in the adult neurons. It codes for an enzyme engaged in the regulation of neuronal plasticity (52-54) and can also contribute to neuronal cell death in pathological conditions (55). Thus, our data suggest that YY1 can also be implicated in neuronal plasticity as well as in neurodegeneration. Intriguingly, YY1 functions as a PcG in mammals (38). PcGs have already been implicated in the formation and maintenance of long term cellular memory as potent epigenetic regulators exerting long term transcriptional repression (56). PcGs work in multimeric protein complexes displaying diverse enzymatic activities transforming chromatin into compacted, regulatory inaccessible structures (56). YY1 is the only mammalian PcG able to bind DNA (30, 56) and to dock other PcGs on the genome (57). Therefore, it is tempting to speculate that also other PcGs could be involved in the regulation of synaptic plasticity, long term memory and neurodegeneration.
This work was supported by the Polish Ministry of Science and Higher Education Grant 2 P04A 081 29 and 7 FP European Union Grant MemStick. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MMP-9, matrix metalloproteinase-9; ChIP, chromatin immunoprecipitation; CtBP1, C-terminal binding protein 1; CTD, C-terminal domain; EMSA, electrophoretic mobility shift assay; GFP, green fluorescent protein; HDAC, histone deacetylase; PcG, Polycomb group proteins; PTZ, pentylenetetrazole; YY1, Yin Yang 1; FISH, fluorescent in situ hybridization; PMSF, phenylmethylsulfonyl fluoride; PBS, phosphate-buffered saline; DAPI, 4′,6-diamidino-2-phenylindole; RT, reverse transcription; siRNA, short interfering RNA.
References
- 1.Kaczmarek, L., and Robertson, H. (eds) (2002) Immediate Early Genes and Inducible Transcription Factors in Mapping of the Central Nervous System Function and Dysfunction, Elsevier Science Publishing Co., Inc., New York
- 2.Carrion, A. M., Link, W. A., Ledo, F., Mellstrom, B., and Naranjo, J. R. (1999) Nature 398 80-84 [DOI] [PubMed] [Google Scholar]
- 3.Chen, W. G., Chang, Q., Lin, Y., Meissner, A., West, A. E., Griffith, E. C., Jaenisch, R., and Greenberg, M. E. (2003) Science 302 885-889 [DOI] [PubMed] [Google Scholar]
- 4.Martinowich, K., Hattori, D., Wu, H., Fouse, S., He, F., Hu, Y., Fan, G., and Sun, Y. E. (2003) Science 302 890-893 [DOI] [PubMed] [Google Scholar]
- 5.Dzwonek, J., Rylski, M., and Kaczmarek, L. (2004) FEBS Lett. 567 129-135 [DOI] [PubMed] [Google Scholar]
- 6.van den Steen, P. E., Dubois, B., Nelissen, I., Rudd, P. M., Dwek, R. A., and Opdenakker, G. (2002) Crit. Rev. Biochem. Mol. Biol. 37 375-536 [DOI] [PubMed] [Google Scholar]
- 7.Yong, V. W. (2005) Nat. Rev. Neurosci. 6 931-944 [DOI] [PubMed] [Google Scholar]
- 8.Masson, V., de la Ballina, L. R., Munaut, C., Wielockx, B., Jost, M., Maillard, C., Blacher, S., Bajou, K., Itoh, T., Itohara, S., Werb, Z., Libert, C., Foidart, J. M., and Noel, A. (2005) FASEB J. 19 234-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao, T., Xia, W. H., Zheng, M. Q., Lu, C. Q., Han, X., and Sun, Y. J. (2008) Exp. Oncol. 30 60-64 [PubMed] [Google Scholar]
- 10.Szklarczyk, A., Lapinska, J., Rylski, M., McKay, R. D., and Kaczmarek, L. (2002) J. Neurosci. 22 920-930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilczynski, G. M., Konopacki, F. A., Wilczek, E., Lasiecka, Z., Gorlewicz, A., Michaluk, P., Wawrzyniak, M., Malinowska, M., Okulski, P., Kolodziej, L. R., Konopka, W., Duniec, K., Mioduszewska, B., Nikolaev, E., Walczak, A., Owczarek, D., Gorecki, D. C., Zuschratter, W., Ottersen, O. P., and Kaczmarek, L. (2008) J. Cell Biol. 180 1021-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konopacki, F. A., Rylski, M., Wilczek, E., Amborska, R., Detka, D., Kaczmarek, L., and Wilczynski, G. M. (2007) Neuroscience 150 31-39 [DOI] [PubMed] [Google Scholar]
- 13.Das, A., Fernandez-Zapico, M. E., Cao, S., Yao, J., Fiorucci, S., Hebbel, R. P., Urrutia, R., and Shah, V. H. (2006) J. Biol. Chem. 281 39105-39113 [DOI] [PubMed] [Google Scholar]
- 14.Sanceau, J., Boyd, D. D., Seiki, M., and Bauvois, B. (2002) J. Biol. Chem. 277 35766-35775 [DOI] [PubMed] [Google Scholar]
- 15.Yan, C., Wang, H., Toh, Y., and Boyd, D. D. (2003) J. Biol. Chem. 278 2309-2316 [DOI] [PubMed] [Google Scholar]
- 16.Mohan, R., Rinehart, W. B., Bargagna-Mohan, P., and Fini, M. E. (1998) J. Biol. Chem. 273 25903-25914 [DOI] [PubMed] [Google Scholar]
- 17.Antoniv, T. T., Tanaka, S., Sudan, B., De Val, S., Liu, K., Wang, L., Wells, D. J., Bou-Gharios, G., and Ramirez, F. (2005) J. Biol. Chem. 280 35417-35423 [DOI] [PubMed] [Google Scholar]
- 18.Barath, P., Poliakova, D., Luciakova, K., and Nelson, B. D. (2004) Eur. J. Biochem. 271 1781-1788 [DOI] [PubMed] [Google Scholar]
- 19.Brummelkamp, T. R., Bernards, R., and Agami, R. (2002) Science 296 550-553 [DOI] [PubMed] [Google Scholar]
- 20.Rylski, M., Welch, J. J., Chen, Y. Y., Letting, D. L., Diehl, J. A., Chodosh, L. A., Blobel, G. A., and Weiss, M. J. (2003) Mol. Cell. Biol. 23 5031-5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaworski, J., Mioduszewska, B., Sanchez-Capelo, A., Figiel, I., Habas, A., Gozdz, A., Proszynski, T., Hetman, M., Mallet, J., and Kaczmarek, L. (2003) J. Neurosci. 23 4519-4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hetman, M., Kanning, K., Cavanaugh, J. E., and Xia, Z. (1999) J. Biol. Chem. 274 22569-22580 [DOI] [PubMed] [Google Scholar]
- 23.Dubberke, R., Vasilets, L. A., and Schwarz, W. (1998) Pfluegers Arch. 437 79-85 [DOI] [PubMed] [Google Scholar]
- 24.Schormair, C., Bingmann, D., Wittkowski, W., and Speckmann, E. J. (1993) Brain Res. Bull. 32 329-338 [DOI] [PubMed] [Google Scholar]
- 25.Walden, J., Speckmann, E. J., and Witte, O. W. (1988) Brain Res. 473 294-305 [DOI] [PubMed] [Google Scholar]
- 26.Meldrum, B. (2002) Epilepsy Res. 50 33-40 [DOI] [PubMed] [Google Scholar]
- 27.Cheng, C., and Sharp, P. A. (2003) Mol. Cell. Biol. 23 1961-1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quandt, K., Frech, K., Karas, H., Wingender, E., and Werner, T. (1995) Nucleic Acids Res. 23 4878-4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hering, H., and Sheng, M. (2003) J. Neurosci. 23 11759-11769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luke, M. P., Sui, G., Liu, H., and Shi, Y. (2006) J. Biol. Chem. 281 33226-33232 [DOI] [PubMed] [Google Scholar]
- 31.Mokrani, H., Sharaf el Dein, O., Mansuroglu, Z., and Bonnefoy, E. (2006) Mol. Cell. Biol. 26 8551-8561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coull, J. J., Romerio, F., Sun, J. M., Volker, J. L., Galvin, K. M., Davie, J. R., Shi, Y., Hansen, U., and Margolis, D. M. (2000) J. Virol. 74 6790-6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sucharov, C. C., Langer, S., Bristow, M., and Leinwand, L. (2006) Am. J. Physiol. 291 C1029-C1037 [DOI] [PubMed] [Google Scholar]
- 34.Yang, W. M., Yao, Y. L., Sun, J. M., Davie, J. R., and Seto, E. (1997) J. Biol. Chem. 272 28001-28007 [DOI] [PubMed] [Google Scholar]
- 35.Yao, Y. L., Yang, W. M., and Seto, E. (2001) Mol. Cell. Biol. 21 5979-5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Austen, M., Luscher, B., and Luscher-Firzlaff, J. M. (1997) J. Biol. Chem. 272 1709-1717 [DOI] [PubMed] [Google Scholar]
- 37.Lee, T. C., Shi, Y., and Schwartz, R. J. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 9814-9818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atchison, L., Ghias, A., Wilkinson, F., Bonini, N., and Atchison, M. L. (2003) EMBO J. 22 1347-1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srinivasan, L., and Atchison, M. L. (2004) Genes Dev. 18 2596-2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taniura, H., Sng, J. C., and Yoneda, Y. (2006) Histol. Histopathol. 21 785-791 [DOI] [PubMed] [Google Scholar]
- 41.Huang, Y., Doherty, J. J., and Dingledine, R. (2002) J. Neurosci. 22 8422-8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia, Y. H., Zhu, X., Li, S. Y., Ni, J. H., and Jia, H. T. (2006) Neurosci. Lett. 403 103-108 [DOI] [PubMed] [Google Scholar]
- 43.Guo, B., Odgren, P. R., van Wijnen, A. J., Last, T. J., Nickerson, J., Penman, S., Lian, J. B., Stein, J. L., and Stein, G. S. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 10526-10530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inouye, C. J., and Seto, E. (1994) J. Biol. Chem. 269 6506-6510 [PubMed] [Google Scholar]
- 45.Kobrossy, L., Rastegar, M., and Featherstone, M. (2006) J. Biol. Chem. 281 25926-25939 [DOI] [PubMed] [Google Scholar]
- 46.He, Y., Dupree, J., Wang, J., Sandoval, J., Li, J., Liu, H., Shi, Y., Nave, K. A., and Casaccia-Bonnefil, P. (2007) Neuron 55 217-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bozdagi, O., Nagy, V., Kwei, K. T., and Huntley, G. W. (2007) J. Neurophysiol. 98 334-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma, Z., Shah, R. C., Chang, M. J., and Benveniste, E. N. (2004) Mol. Cell. Biol. 24 5496-5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sui, G., Affar el, B., Shi, Y., Brignone, C., Wall, N. R., Yin, P., Donohoe, M., Luke, M. P., Calvo, D., Grossman, S. R., and Shi, Y. (2004) Cell 117 859-872 [DOI] [PubMed] [Google Scholar]
- 50.Gronroos, E., Terentiev, A. A., Punga, T., and Ericsson, J. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 12165-12170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nowak, K., Lange-Dohna, C., Zeitschel, U., Gunther, A., Luscher, B., Robitzki, A., Perez-Polo, R., and Rossner, S. (2006) J. Neurochem. 96 1696-1707 [DOI] [PubMed] [Google Scholar]
- 52.Meighan, S. E., Meighan, P. C., Choudhury, P., Davis, C. J., Olson, M. L., Zornes, P. A., Wright, J. W., and Harding, J. W. (2006) J. Neurochem. 96 1227-1241 [DOI] [PubMed] [Google Scholar]
- 53.Nagy, V., Bozdagi, O., Matynia, A., Balcerzyk, M., Okulski, P., Dzwonek, J., Costa, R. M., Silva, A. J., Kaczmarek, L., and Huntley, G. W. (2006) J. Neurosci. 26 1923-1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wright, J. W., Masino, A. J., Reichert, J. R., Turner, G. D., Meighan, S. E., Meighan, P. C., and Harding, J. W. (2003) Brain Res. 963 252-261 [DOI] [PubMed] [Google Scholar]
- 55.Michaluk, P., and Kaczmarek, L. (2007) Cell Death Differ. 14 1255-1258 [DOI] [PubMed] [Google Scholar]
- 56.Orlando, V. (2003) Cell 112 599-606 [DOI] [PubMed] [Google Scholar]
- 57.Wilkinson, F. H., Park, K., and Atchison, M. L. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 19296-19301 [DOI] [PMC free article] [PubMed] [Google Scholar]