Abstract
Ipecac alkaloids produced in the medicinal plant Psychotria ipecacuanha such as emetine and cephaeline possess a monoterpenoid-tetrahydroisoquinoline skeleton, which is formed by condensation of dopamine and secologanin. Deglucosylation of one of the condensed products N-deacetylisoipecoside (1α(S)-epimer) is considered to be a part of the reactions for emetine biosynthesis, whereas its 1β(R)-epimer N-deacetylipecoside is converted to ipecoside in P. ipecacuanha. Here, we isolated a cDNA clone Ipeglu1 encoding Ipecac alkaloid β-d-glucosidase from P. ipecacuanha. The deduced protein showed 54 and 48% identities to raucaffricine β-glucosidase and strictosidine β-glucosidase, respectively. Recombinant IpeGlu1 enzyme preferentially hydrolyzed glucosidic Ipecac alkaloids except for their lactams, but showed poor or no activity toward other substrates, including terpenoid-indole alkaloid glucosides. Liquid chromatography-tandem mass spectrometry analysis of deglucosylated products of N-deacetylisoipecoside revealed spontaneous transitions of the highly reactive aglycons, one of which was supposed to be the intermediate for emetine biosynthesis. IpeGlu1 activity was extremely poor toward 7-O-methyl and 6,7-O,O-dimethyl derivatives. However, 6-O-methyl derivatives were hydrolyzed as efficiently as non-methylated substrates, suggesting the possibility of 6-O-methylation prior to deglucosylation by IpeGlu1. In contrast to the strictosidine β-glucosidase that stereospecifically hydrolyzes 3α(S)-epimer in terpenoid-indole alkaloid biosynthesis, IpeGlu1 lacked stereospecificity for its substrates where 1β(R)-epimers were preferred to 1α(S)-epimers, although ipecoside (1β(R)) is a major alkaloidal glucoside in P. ipecacuanha, suggesting the compartmentalization of IpeGlu1 from ipecoside. These facts have significant implications for distinct physiological roles of 1α(S)- and 1β(R)-epimers and for the involvement of IpeGlu1 in the metabolic fate of both of them.
Psychotria ipecacuanha Stokes (Rubiaceae) is a medicinal plant that is native to South and Central America. Its roots and rhizome have long been used as an emetic and an anti-amebic (1). These medicinal effects derive from its principal alkaloid emetine. In the plant, emetine occurs together with a number of related alkaloids, including cephaeline and ipecoside (2) (Fig. 1). They are collectively called Ipecac alkaloids and possess a monoterpenoid-tetrahydroisoquinoline skeleton. Some of the Ipecac alkaloids, including emetine and cephaeline, have also been found in Alangium lamarckii Thwaites (Alangiaceae), a medicinal plant distributed throughout India and Southeast Asia (2).
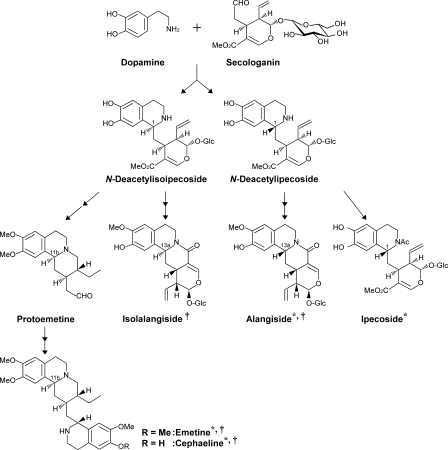
FIGURE 1.
Schematic representation of the biosynthetic pathway of emetine, cephaeline, and related alkaloidal glucosides found in P. ipecacuanha (*) and A. lamarckii (†).
Tetrahydroisoquinoline and fragmented monoterpenoid moieties of emetine and cephaeline originate from dopamine and secologanin, respectively (3–6). They are condensed in a manner of a Pictet-Spengler reaction to form two epimers, N-deacetylisoipecoside (1α(S)-epimer) and N-deacetylipecoside (1β(R)-epimer) (Fig. 1). Previously, it was revealed that the condensation reaction is catalyzed by two enzymes, N-deacetylisoipecoside synthase and N-deacetylipecoside synthase, each of which stereoselectively produces each epimer in Alangium (7, 8). The enzyme producing the 1β(R)-epimer was purified but not that producing the 1α(S)-epimer due to its lability (8). Tracer experiments using pure epimer clearly demonstrated that emetine and cephaeline are synthesized from the 1α(S)-epimer. On the other hand, the 1β(R)-epimer N-deacetylipecoside is converted to alkaloidal glucosides, ipecoside in Psychotria and alangiside in Alangium, with retention of configuration (5, 6) (Fig. 1). They are accumulated as the main alkaloidal glucosides.
The condensation reaction of dopamine and secologanin is similar to monoterpenoid-indole alkaloid biosynthesis where the reaction is launched from tryptamine and secologanin to form strictosidine but is different in terms of stereochemistry of the products. Strictosidine synthase that was characterized so far in many plants such as Rauvolfia serpentina (9, 10), Catharanthus roseus (11–13), Cinchona robusta (14), and Ophiorrhiza pumila (15) produced only strictosidine (3α(S)-epimer) but not vincoside (3β(R)-epimer) (see supplemental Fig. S1 for chemical structure). The enzyme activity catalyzing the formation of vincoside has not been detected. In accordance with these facts, exogenously administrated vincoside was not converted to any alkaloids, showing that vincoside is not the intermediate for terpenoid-indole alkaloid biosynthesis (16). Meanwhile strictosidine was demonstrated to serve as precursor for terpenoid-indole alkaloids not only with α-configuration, but also with β-configuration by inversion of the configuration at the corresponding chiral center in the biosynthetic process (17). Such inversion does not occur in Ipecac alkaloid biosynthesis (5, 6).
In terpenoid-indole alkaloid biosynthesis, strictosidine is deglucosylated by strictosidine β-glucosidase to form a highly reactive aglycon, which could be the precursor of a diverse array of indole alkaloids (18). Genes encoding strictosidine β-glucosidase have been cloned from C. roseus (19) and R. serpentina (20). Strictosidine β-glucosidase stereoselectively catalyzes the deglucosylation of strictosidine, but not its 3β(R)-epimer vincoside, which is consistent with the occurrence of only strictosidine in those plants. Some structures of the intermediates derived from strictosidine aglycon have been elucidated (20). In analogy with terpenoid-indole alkaloid biosynthesis, it has been predicted that N-deacetylisoipecoside is deglucosylated to form an intermediate of emetine biosynthesis.
Despite the occurrence of alkaloidal glucosides with both 1α(S)- and 1β(R)-configurations in Ipecac alkaloid biosynthesis, only the 1α(S)-epimer serves as precursor for emetine biosynthesis. Moreover non-glucosidic alkaloids with the β-configuration have not been found in P. ipecacuanha. These facts allowed us to hypothesize that the deglucosylation step plays important roles not only for the production of intermediate of emetine biosynthesis, but also for the metabolic channeling of the 1α(S)- and 1β(R)-epimers. In this report, we have isolated cDNAs encoding Ipecac alkaloid β-glucosidase from P. ipecacuanha. On the basis of enzymatic characterization and identification of the deglucosylated products, we propose a detailed biosynthetic process and discuss physiological significance of formation of the alkaloidal glucosides with distinct configurations in P. ipecacuanha.
EXPERIMENTAL PROCEDURES
Shoot and Root in Vitro Cultures—P. ipecacuanha in vitro plants were obtained from the Tropical Plant Biotechnology Laboratory, the Technological Institute of Costa Rica in San Carlos. Shoots were cultivated in LS medium (1650 mg/liter NH4NO3, 1900 mg/liter KNO3, 370 mg/liter MgSO4 · 7H2O, 170 mg/liter KH2PO4, 440 mg/liter CaCl2 · 2H2O, 41.3 mg/liter Na2-EDTA · 2H2O, 27.8 mg/liter FeSO4 7H2O, 6.2 mg/liter H3BO3, 16.9 mg/liter MnSO4 · H2O, 8.6 mg/liter ZnSO4 · 7H2O, 0.83 mg/liter KI, 0.25 mg/liter Na2MoO4 · 2H2O, 0.025 mg/liter CuSO4 · 5H2O, 0.025 mg/liter CoCl2 · 6H2O) (21), pH 5.75, containing 3% sucrose, 0.1 mg/liter thiamine hydrochloride, 0.5 mg/liter nicotinic acid, 0.5 mg/liter pyroxidine hydrochloride, 0.1 g/liter myo-inositol, 10 μg/liter 1-naphthaleneacetic acid, 3 mg/liter 6-benzylaminopurine, and 0.6% agar. Roots were cultivated in the same medium except for 0.5 mg/liter 1-naphthaleneacetic acid without 6-benzylaminopurine and agar. Shoot cultures were grown at 24 °C under a photoperiod of 16-h light (50 μmol/m2/s)/8-h dark, and maintained every 6–8 weeks. Root cultures were grown in darkness at 25–28 °C on an orbital shaker at 100 rpm. The root-culture medium was replenished every 3–4 weeks, and the root tissues were divided every 2 months.
Cloning of β-d-Glucosidase cDNAs—Total RNA was isolated from shoot and root cultures by the acid-guanidium-phenol-chloroform method (22) or by using an RNeasy Plant Mini Kit (Qiagen), and poly(A)+ RNA was purified from the total RNA using an Oligotex mRNA mini kit (Qiagen). A λ-ZAP cDNA library was constructed from root mRNA using a ZAP Express Kit (Stratagene). Inserts were amplified by PCR with universal primers and sequenced. Out of 1050 single-pass sequenced expressed sequence tags, a clone ph0212-60 showed high similarity to strictosidine β-glucosidase and raucaffricine β-glucosidase. The missing 5′-upper sequence was obtained by 5′-RACE using a SMART RACE3 cDNA amplification kit (Clontech) with the following gene-specific reverse primers: 5′-GCAGAGCTCCGCGAAGTCACGGAAGTCG-3′ (1st PCR) and 5′-CCAGGCAATATCCGTGGCCATGATATGGAG-3′ (2nd PCR). Then, PCR amplification of the cDNA, including the entire coding region was done using shoot cDNA as template by Pfu DNA polymerase with two sets of primers: 5′-GTGCAATCTCTCAACAACAACAACAATG-3′ (forward) and 5′-CCAATCTTCAATTGTCAAAAGGGACGC-3′ (reverse) for the 1st PCR, and 5′-CACCACAACAACAATGTCTAGTG-3′ (forward, underline is adaptor for TOPO cloning) and 5′-GTCAAAAGGGACGCTTTC-3′ (reverse) for the 2nd PCR. The products were inserted into pET100/D-TOPO (Invitrogen) and sequenced to obtain Ipeglu1 cDNA.
Because the Ipeglu1 coding sequence possessed 12 single nucleotide polymorphisms from the original partial glucosidase clone (ph0212-60) in the overlapped region, the presence of multiple copies of the glucosidase was predicted. To isolate additional glucosidase cDNAs, reverse transcription-PCR was performed using shoot and root cDNAs as template by Phusion HS-HF polymerase (Finnzymes) with the primers designed from the edge of 5′- and 3′-untranslated regions: 5′-GTCAACCAAAAGTTCCATTGCAAC-3′ (forward) and 5′-GAAAATAAAGAAGGAATTTATTCAACCGTTC-3′ (reverse). The products were inserted into pCR-Blunt II-TOPO (Invitrogen) and sequenced to obtain Ipeglu2-Ipeglu9.
Expression and Purification of β-d-Glucosidase in E. coli—The entire coding region of the glucosidase cDNA was ligated into pET100/D-TOPO (Invitrogen). The vector was introduced into the Escherichia coli strain PlusS (Expression Technologies) for gene expression. The recombinant E. coli was grown in 1 liter of LB medium containing 200 μg/ml ampicillin and 35 μg/ml chloramphenicol at 37 °C until the A600 reached 0.7. After cooling down the culture, isopropyl 1-thio-β-d-galactopyranoside was added (1 mm) to induce gene expression, followed by the overnight culture (∼18 h) at 18 °C. The cells were harvested (6,000 × g, 10 min) and resuspended in 40 ml of 50 mm NaPi buffer (pH 7.5) containing 10% glycerol and 0.5% protease inhibitor mixture (Sigma). After sonication (20 s at 50 watts, 6 times) and centrifugation (20,000 × g, 20 min), the supernatant containing soluble protein was collected for purification of N-terminal His-tagged IpeGlu protein.
The recombinant protein was purified with TALON His-Tag Purification Resin (Clontech). All operations were done at 4 °C. The resins (1 ml of bed volume per 20 ml of the supernatant) equilibrated in 50 mm NaPi buffer (pH 7.5) containing 0.5 m NaCl and 10% glycerol were mixed with the supernatant for 1 h. After the resin was washed with equilibration buffer, the recombinant IpeGlu protein was eluted in a stepwise manner by increasing the imidazole concentration (5, 50, and 200 mm) in 50 mm NaPi buffer (pH 7.0) containing 0.5 m NaCl and 10% glycerol. The 200 mm imidazole fraction was concentrated by ultrafiltration to 2.5 ml, and imidazole was removed using a PD10 column (GE Healthcare) equilibrated with the same buffer without imidazole. Purified protein was stored at –80 °C. The protein profile was analyzed by 10% SDS-PAGE. Protein concentration was measured by the method of Bradford (23) using bovine serum albumin as a standard.
General Conditions for Chemical Analysis—HPLC analysis was performed with a Hitachi D7000 system using an ODS column (Nova-Pak C18, 3.9 × 300 mm, 4 μm, Waters) at a flow rate of 0.8 ml/min. Elution was achieved with 0.1% trifluoroacetic acid (solvent A) and acetonitrile (solvent B). Preparative HPLC was performed with the same HPLC system using an ODS column (LiChrosorb RP18, 25 × 250 mm, 7 μm, Merck) at a flow rate of 8.0 ml/min. Unless otherwise noted, 0.1% trifluoroacetic acid (solvent A) and acetonitrile (solvent B) were used as the mobile phase. Fractions from preparative HPLC were concentrated in vacuo and freeze-dried.
For LC-MS/MS analysis, HPLC separation was conducted with a Shimadzu LC20A system. Mass spectra were obtained using a 4000 Q-TRAP triple stage quadruple mass spectrometer equipped with a TurboIonSpray ionization source (Applied Biosystems) in a positive ion mode.
Substrates for Enzyme Assay—Of the 30 compounds listed in Table 1 and supplemental Fig. S1, ipecoside, secologanin, loganin, galloyl-Glc, and esculin were from the chemical stock of Dr. Meinhart H. Zenk (Donald Danforth Plant Science Center). Raucaffricine and arbutin were gifts from Dr. Joachim Stöckigt (Johannes Gutenberg-Universität, Germany). Apigenin-7-O-Glc, naringenin-7-O-Glc, luteolin-7-O-Glc, luteolin-4′-O-Glc, quercetin-4′-O-Glc, and quercetin-3-O-Glc were gifts from Dr. Thomas Vogt (Leibniz Institute of Plant Biochemistry, Germany). Salicin was purchased from Sigma, and p-nitrophenyl (pNP)-α-Glc and pNP-β-Glc were from Acros Organics. Secologanin was repurified prior to use by preparative HPLC (eluent, A = 0.1% acetic acid, B = acetonitrile, gradient = 10–40% B/(A + B) in 36 min; detection, 254 nm). The other 14 compounds were prepared by chemical synthesis.
TABLE 1.
Substrate specificity of the recombinant IpeGlu1
Relative efficiency toward each substrate is the percent ratio of the kcat/Km values to that for N-deacetylipecoside.
| Substrate | Km | Vmax | kcat | kcat/Km | Relative efficiency |
|---|---|---|---|---|---|
| mm | nmol/min/mg | s-1 | s-1/mm | % | |
| N-Deacetylipecoside | 0.32 | 25,577 | 28.2 | 88.1 | 100 |
| N-Deacetylisoipecoside | 5.8 | 78,190 | 86.3 | 14.9 | 16.9 |
| 6-O-Methyl-N-deacetylipecoside | 0.39 | 28,157 | 31.1 | 79.7 | 90.5 |
| 6-O-Methyl-N-deacetylisoipecoside | 1.3 | 16,906 | 18.7 | 14.4 | 16.3 |
| 7-O-Methyl-N-deacetylipecoside | 12.7 | 45,580 | 50.3 | 4.0 | 4.5 |
| 7-O-Methyl-N-deacetylisoipecoside | 25.5 | 22,727 | 25.1 | 0.98 | 1.1 |
| 6,7-O,O-Dimethyl-N-deacetylipecoside | 13.5 | 35,732 | 39.4 | 2.9 | 3.3 |
| 6,7-O,O-Dimethyl-N-deacetylisoipecoside | 35.0 | 21,989 | 24.3 | 0.69 | 0.8 |
| Ipecoside | 0.10 | 46,729 | 51.6 | 516 | 586 |
| Demethylalangiside | NDa | ND | ND | ND | 0 |
| Demethylisoalangiside | ND | ND | ND | ND | 0 |
| Strictosidine | 0.19 | 1,945 | 2.1 | 11.1 | 12.6 |
| Vincoside | 0.29 | 5,786 | 6.4 | 22.1 | 25.1 |
| Strictosidine lactam | ND | ND | ND | ND | 0 |
| Vincoside lactam | ND | ND | ND | ND | 0 |
| Secologanin | 26.5 | 30,303 | 33.4 | 1.3 | 1.5 |
| Loganin | 33.2 | 19,881 | 21.9 | 0.66 | 0.7 |
| Raucaffricine | ND | ND | ND | ND | 0 |
| Galloyl-Glc | 65.1 | 6,024 | 6.6 | 0.10 | 0.1 |
| Apigenin-7-O-Glc | -b | 39.2c | - | - | - |
| Naringenin-7-O-Glc | - | 4.1c | - | - | - |
| Luteolin-7-O-Glc | - | 22.3c | - | - | - |
| Luteolin-4′-O-Glc | - | 28.4c | - | - | - |
| Quercetin-4′-O-Glc | - | 11.4c | - | - | - |
| Quercetin-3-O-Glc | ND | ND | ND | ND | 0 |
| Arbutin | ND | ND | ND | ND | 0 |
| Esculin | ND | ND | ND | ND | 0 |
| Salicin | ND | ND | ND | ND | 0 |
| pNP-β-Glc | 21.5 | 3,546 | 3.9 | 0.18 | 0.2 |
| pNP-α-Glc | - | 15.2d | - | - | - |
ND, not detected.
-, not determined.
Activity at 0.2 mm.
Activity at 20 mm.
N-Deacetylipecoside and N-deacetylisoipecoside were synthesized as described previously (3, 6) with modifications. Secologanin (60 mg) and dopamine hydrochloride (30 mg) were dissolved in 0.9 ml of 0.1 m citrate/0.2 m phosphate buffer (pH 5.0) and reacted for 3 days in darkness at room temperature under N2. The reaction mixture was directly subjected to preparative HPLC (10–25% B/(A + B) in 48 min; 230 nm) (obtained as trifluoroacetate). Their authenticity was confirmed as described in Ref. 6 and by MS/MS analysis.
Demethylalangiside and demethylisoalangiside were synthesized from N-deacetylipecoside and N-deacetylisoipecoside, respectively, according to Ref. 24 with modifications. Following the alkaline treatment of N-deacetylipecoside and N-deacetylisoipecoside, the reaction mixture was directly subjected to preparative HPLC (eluent, A = 0.1% formic acid, B = acetonitrile, gradient = 15–40% B/(A + B) in 45 min; 254 nm) (obtained as formate). Their authenticity was confirmed by comparing 1H NMR spectra with those reported by Ref. 24, as well as by MS/MS analysis. Configuration of C-13a (see Ref. 24) was confirmed by measuring nuclear Overhauser effect spectroscopy spectra detecting the correlation between H-13a and H-12a.
Preparation of 7-O-methyl-N-deacetylipecoside and 7-O-methyl-N-deacetylisoipecoside was performed by the condensation reaction of secologanin with 3-hydroxy-4-methoxyphenethylamine hydrochloride (25) with the same procedure as described for the reaction with dopamine, except for the HPLC condition (10–32% B/(A + B) in 50 min; 230 nm). Because condensation reactions of secologanin with 4-hydroxy-3-methoxyphenethylamine hydrochloride and with 3,4-dimethoxyphenethylamine were not successful, 6-O-methyl-N-deacetylipecoside, 6,7-O,O-dimethyl-N-deacetylipecoside, and their 1α(S)-epimers were synthesized as follows.
To a solution of N-deacetylipecoside-trifluoroacetate (50 mg, 78.5 μmol) and di-tert-butyldicarbonate (19 mg, 87 μmol) in 0.8 ml of methanol was added 20 μl (140 μmol) of triethylamine, and the mixture was reacted at room temperature until N-deacetylipecoside disappeared (2.5 h; monitored by HPLC). After the solvent was removed under N2 flow, the residue was treated with 0.8 ml of dilute HCl (7 mm) and extracted six times with 0.8 ml of ethyl acetate. The combined organic layer was washed with saturated NaCl and dried under N2 flow, followed by stirring in ether to precipitate the product as a white powder (N-tert-butoxycarbonyl deacetylipecoside, 42.2 mg, 67.7 μmol), containing small amount of impurities. It was dissolved in methanol (6.8 ml), and 1.7 ml (3.4 mmol) of (trimethylsilyl)diazomethane (in ether) was added. The reaction mixture in a sealed flask was stirred for 30 min at room temperature. Excess (trimethylsilyl)diazomethane was decomposed by adding 0.2 ml of acetic acid. After the solvent was evaporated under N2 flow, the residue was dissolved in methanol (1 ml) and subjected to preparative HPLC (eluent, A = water, B = acetonitrile, gradient = 20–48% B/(A + B) in 80 min; 230 nm). Four major peaks (material, and 6-O-methylated-, 7-O-methylated-, and 6,7-O,O-dimethylated products) were collected, and the material fraction was evaporated to dryness. Using the recovered material, reaction with (trimethylsilyl)diazomethane and purification were repeated twice. Combined HPLC fractions of the 6-O-methylated product were evaporated to dryness, and the residue was treated with trifluoroacetic acid (1 ml) for 5 min at room temperature to remove tert-butoxycarbonyl group. After removing trifluoroacetic acid under N2 flow, the residue was dissolved in 1 ml of 50% methanol/water and subjected to preparative HPLC (10–26% B/(A + B) in 90 min; 230 nm) to give 6-O-methyl-N-deacetylipecoside-trifluoroacetate (8.3 mg). Its 1α(S)-epimer 6-O-methyl-N-deacetylisoipecoside-trifluoroacetate (11.3 mg) was obtained by the same procedure from 50 mg of N-deacetylisoipecoside-trifluoroacetate.
Although 6,7-O,O-dimethyl-N-deacetylipecoside and its 1α(S)-epimer could be obtained from the HPLC fractions mentioned above, they were more efficiently prepared from 7-O-methyl-N-deacetylipecoside and 7-O-methyl-N-deacetylisoipecoside, respectively. Following the N-protection of 7-O-methyl-N-deacetylipecoside-trifluoroacetate (40 mg, 61.4 μmol), the O-methylation reaction was conducted using 100 eq of (trimethylsilyl)diazomethane for 1 h at room temperature. After deprotection with trifluoroacetic acid, the residue was purified by preparative HPLC (10–42% B/(A + B) in 72 min; 230 nm) to give 6,7-O,O-dimethyl-N-deacetylipecoside-trifluoroacetate (32 mg). Its 1α(S)-epimer 6,7-O,O-dimethyl-N-deacetylisoipecoside-trifluoroacetate (27.8 mg) was obtained by the same procedure from 40 mg of 7-O-methyl-N-deacetylisoipecoside-trifluoroacetate.
Strictosidine and vincoside were prepared according to Ref. 26 with modifications. Secologanin (20 mg) and tryptamine hydrochloride (10 mg) were dissolved in 0.15 ml of 0.1 m citrate/0.2 m phosphate buffer (pH 4.5) and reacted for 2.5 days in darkness at 37 °C under N2. The reaction mixture was directly subjected to preparative HPLC (eluent, A = 1% formic acid, B = acetonitrile, gradient = 20–30% B/(A + B) in 60 min; 254 nm) (obtained as formate).
Strictosidine lactam and vincoside lactam were prepared by the alkaline treatment of strictosidine (0.4 mg) and vincoside (0.4 mg) at 75 °C for 2 h (16). Because the reaction mixture hardly contained impurities, they were directly used for the enzyme assay after centrifugation.
Enzyme Assay—The enzyme assay of the recombinant IpeGlu1 for the screening of substrates (Table 1 and supplemental Fig. S1) was performed in 0.1 m citrate/0.2 m phosphate buffer (pH 5.0) containing 1 μg of enzyme and 1 mm substrate (0.2 mm for flavonoids and raucaffricine; 0.5 mm for demethylalangiside and demethylisoalangiside) in a total volume of 100 μl for 1 h at 30 °C. The reaction was terminated by the addition of 20 μl of 1 n HCl. After centrifugation (16,000 × g, 2 min), the supernatant was subjected to HPLC analysis. Conditions for UV detection and solvent gradient were changed appropriately with substrates. Decrease in substrate and/or generation of enzymatic products were monitored. When pNP-Glcs were used as substrate, the reaction was terminated by adding an equal volume of 0.2 m Na2CO3, and the product was monitored spectrophotometrically at 400 nm.
The reaction mixture (100 μl) for the determination of pH optimum contained 1 mm substrate (N-deacetylipecoside or N-deacetylisoipecoside) and enzyme (0.1 μg for N-deacetylipecoside and 0.5 μg for N-deacetylisoipecoside) in the following buffers; 0.1 m citrate/0.2 m phosphate (pH 2.5–7.0), 0.1 m KPi (pH 6.0–8.0), and 0.1 m Tris-HCl (pH 7.0–9.0). After incubation (30 °C, 10 min), the decrease in substrate was analyzed by HPLC (12–80% B/(A + B) in 30 min; 230 nm). The temperature optimum was determined using N-deacetylisoipecoside as substrate by the reaction in 0.1 m citrate/0.2 m phosphate buffer (pH 5.0) containing 1 mm substrate and 0.5 μg of enzyme in a total volume of 100 μl. After an incubation (25–70 °C, 10 min), decrease in substrate was analyzed by HPLC.
Effects of metal cations, EDTA and 2-mercaptoethanol, on the IpeGlu1 activity were examined by the reaction in 0.1 m sodium acetate buffer (pH 5.0) (instead of citrate/phosphate buffer to avoid formation of insoluble salt between metal cations and the phosphate ion) containing 1 mm N-deacetylipecoside, 0.1 μg of enzyme, and the additives listed in Table 2 in a total volume of 100 μl. After incubation (30 °C, 10 min), decrease in substrate was analyzed by HPLC.
TABLE 2.
Effects of EDTA, 2-mercaptoethanol, and metal cations on the enzyme activity of recombinant IpeGlu1
Enzyme activity was measured using 1 mm N-deacetylipecoside as substrate. Activity is shown as the percent ratio of the specific activity to that without additive (25.9 μmol/min/mg of protein).
| Additive (concentration) | Relative activity |
|---|---|
| % | |
| Control | 100 |
| EDTA (1 mm) | 101 |
| 2-Mercaptoethanol (4 mm) | 102 |
| Ag+ (1 mm) | 77 |
| Ca2+ (1 mm) | 95 |
| Co2+ (1 mm) | 86 |
| Cu2+ (1 mm) | 50 |
| Fe2+ (1 mm) | 46 |
| Mg2+ (1 mm) | 80 |
| Mn2+ (1 mm) | 86 |
| Ni2+ (1 mm) | 81 |
| Zn2+ (1 mm) | 64 |
The enzyme assay for the determination of kinetic parameters of IpeGlu1 was performed in 0.1 m citrate/0.2 m phosphate buffer (pH 5.0) at 30 °C. The amount of enzyme, and the reaction time were set carefully to make the reaction proceed linearly. Reaction was terminated and analyzed by HPLC (12–80% B/(A + B) in 30 min, detection, 230 nm). For the assay with strictosidine and vincoside, a gradient of 20–80% B/(A + B) in 30 min was used. Analysis of galloyl-Glc and gallic acid was done with an isocratic elution of 5% B/(A + B) at 270 nm. Enzyme activity was calculated on the basis of decrease in substrate, except for the assay with galloyl-Glc, flavonoid-Glcs, and pNP-Glcs. Enzyme activities for the flavonoid-Glcs were measured only at 0.2 mm substrate due to their poor solubility. Kinetic parameters (Km and Vmax) were determined from three replicates by the plot [s] versus [s]/v.
Identification of Deglucosylation Products—The reaction mixture (100 μl) containing 0.1 m citrate/0.2 m phosphate buffer (pH 5.0), 1 mm substrate (N-deacetylipecoside, N-deacetylisoipecoside, or ipecoside), and 2 μg of IpeGlu1 enzyme was incubated at 30 °C for 1 h. The reaction under reducing condition was done under the same condition in the presence of 1 mm NaBH3CN. The reaction was terminated by adding 20 μl of 1 n HCl and subjected to LC-MS/MS analysis. The mixture was diluted appropriately before injection. The HPLC separation was done with a gradient of 12–80% B/(A + B) in 30 min at a flow rate of 0.8 ml/min. To identify the molecular ion of the reaction products, mass spectrometry detection was first done by enhanced mass scan (collision energy, 10 V; declustering potential, 30 V), and then the molecular ions detected were further examined by enhanced product ion scan (collision energy, 40 V; declustering potential, 80 V) for their fragmentation pattern.
RESULTS
Isolation of β-Glucosidase cDNA from P. ipecacuanha—The size of cDNA inserts of P. ipecacuanha in the λ-library was checked by PCR. Clones having ∼500- to 2500-bp-long inserts were selected for sequencing. As a result, a total of 1050 single-pass sequenced expressed sequence tags were obtained. The homology search using the BLASTX algorithm revealed that the expressed sequence tag sequence obtained from the clone ph0212-60 was similar to known β-glucosidases in alkaloid biosynthesis, raucaffricine β-glucosidase, and strictosidine β-glucosidase. Complete sequencing of the ph0212-60 insert revealed that it was a partial clone having an internal stop codon. Following 5′-RACE and reverse transcription-PCR, a full-length β-glucosidase cDNA (Ipecac alkaloid β-glucosidase, Ipeglu1) was isolated (GenBank™ accession no. AB455576).
The nucleotide sequence of Ipeglu1 possessed 12 single nucleotide polymorphisms from the clone ph0212-60 in the overlapped region, and contained no stop codon in the coding region. Ipeglu1 cDNA comprised an open reading frame of 1632 bp encoding a polypeptide of 543 amino acids with a calculated molecular mass of 61.8 kDa and a pI of 6.26. The deduced amino acid sequence of IpeGlu1 exhibited the highest identity (54%) to raucaffricine β-glucosidase of R. serpentina (27), and 48% identity to strictosidine β-glucosidase of R. serpentina (20) and C. roseus (19) (supplemental Fig. S2). Phylogenetic analysis of the IpeGlu1 with other plant family 1 glycosyl hydrolases (Fig. 2) showed the close relationship of IpeGlu1 with raucaffricine β-glucosidase and strictosidine β-glucosidases. Subcellular localization of IpeGlu1 was predicted using the programs SignalP version 3.0 (28) and TargetP version 1.1 (29). Both of them predicted the absence of signal peptides, suggesting the localization of IpeGlu1 is in the cytosol.
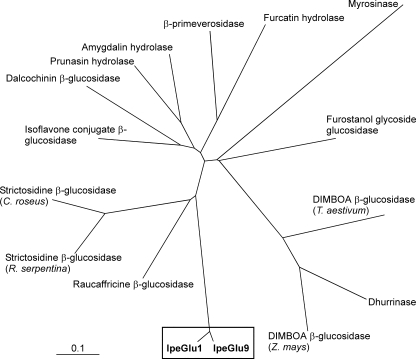
FIGURE 2.
Phylogenetic tree of the Ipecac alkaloid β-glucosidases (IpeGlu1 and IpeGlu9) with plant family 1 glycosyl hydrolases. Full-length amino acid sequences were initially aligned using the ClustalW version 1.83 (clustalw.ddbj.nig.ac.jp/top-j.html). The highly divergent N-terminal region where significant alignment was not constructed was removed, and truncated sequences were aligned again. Calculation was done based on the neighbor-joining method (42), and the tree was visualized with Treeview version 1.66. The amino acid sequences used were IpeGlu1 (P. ipecacuanha, GenBank™ accession no. AB455576, residues 26–543), IpeGlu9 (P. ipecacuanha, AB455584, 26–540), raucaffricine β-glucosidase (R. serpentina, AF149311, 26–540), strictosidine β-glucosidase (R. serpentina, AJ302044, 47–532), strictosidine β-glucosidase (C. roseus, AF112888, 55–555), isoflavone conjugate β-glucosidase (Glycine max, AB259819, 49–514), dalcochinin β-glucosidase (Dalbegia cochinchinensis, AF163097, 49–547), prunasin hydrolase (Prunus serotina, AF411131, 47–542), amygdalin hydrolase (Prunus serotina, U26025, 49–553), β-primeverosidase (Camellia sinensis, AB088027, 43–507), furcatin hydrolase (Viburnum furcatum, AB122081, 78–538), myrosinase (Sinapis alba, X59879, 49–544), furostanol glycoside glucosidase (Costus speciosus, D83177, 100–562), DIMBOA β-glucosidase (Triticum aestivum, AB236422, 82–569), dhurrinase (Sorghum bicolor, U33817, 80–565), and DIMBOA β-glucosidase (Zea mays, U25157, 82–566). The bar corresponds to 10% change.
The presence of two highly identical sequences Ipeglu1 and ph0212-60 made us predict that the Ipecac alkaloid β-glucosidase forms a multigene family. Follow-up cDNA cloning resulted in the isolation of an additional eight glucosidase cDNAs, Ipeglu2–Ipeglu9 (GenBank™ accession nos. AB455577–AB455584). Among them, Ipeglu2–Ipeglu8 encoded polypeptides of 543 amino acids as Ipeglu1, and their amino acid sequences were extremely similar to the IpeGlu1 (99.1–99.8% identities). Ipeglu9 was slightly divergent from the other Ipeglus. It encoded a polypeptide of 540 amino acids and exhibited 93.6% identity to IpeGlu1 (Fig. 2 and supplemental Fig. S2).
Genomic PCR with the primers used for the cloning of Ipeglu2–Ipeglu9 cDNAs also amplified multiple sequences. Among them, exon sequences of the genomic clones-1 (AB455585) and -2 (AB455586) were identical to the Ipeglu3 and Ipeglu5 cDNA sequences, respectively. Both genomic clones comprised 12 exons and 11 introns (supplemental Fig. S3). Exon sequences of other genomic clones were not identical but highly similar to the Ipeglu cDNA sequences. Isolation of the highly identical glucosidase sequences both from cDNA and genomic DNA proved the existence of multiple gene copies for the Ipecac alkaloid β-glucosidase in P. ipecacuanha.
Enzymatic Characterization of Recombinant IpeGlu1—Recombinant IpeGlu1 enzyme was efficiently expressed in a soluble protein fraction in E. coli PlusS strain, although >50% of the expressed protein was precipitated as an inclusion body (data not shown). N-terminal His-tagged IpeGlu1 enzyme was purified to homogeneity from the soluble protein fraction by metal chelation chromatography (Fig. 3).
FIGURE 3.
SDS-PAGE of the recombinant IpeGlu1 enzyme expressed in E. coli. Proteins were separated on 10% SDS-PAGE and stained with Coomassie Brilliant Blue G-250. Lane M, molecular size marker; lane 1, crude extract from the E. coli-expressing IpeGlu1; lane 2, column run through in metal chelation chromatography; lanes 3–5, stepwise elution of the protein from the metal-resin with buffers containing imidazole at 5 mm (lane 3), 50 mm (lane 4), and 200 mm (lane 5).
Optimum pH of the IpeGlu1 enzyme was determined to be 5.0 for both N-deacetylisoipecoside and N-deacetylipecoside. When N-deacetylipecoside was used as substrate, spontaneous lactamization forming demethylalangiside was observed at pH ≥ 6.5. >95% of the substrate was converted to the lactam in 10 min at pH 8.5. In contrast, lactamization of N-deacetylisoipecoside hardly occurred under the same condition. This difference is attributable to the spatial distance between nucleophilic N-atom and the carbonyl group of methyl ester. Optimum temperature of the IpeGlu1 was 55–60 °C, which was similar to those reported for strictosidine β-glucosidases from R. serpentina (50 °C) (20) and C. roseus (stable up to 50 °C) (30).
Effects of metal cations on the IpeGlu1 activity were examined (Table 2), including Cu2+ that was reported to inhibit strictosidine β-glucosidase activity by 90% in R. serpentina (20) and 50% in C. roseus (30). In the presence of 1 mm Cu2+, IpeGlu1 activity was inhibited by 50%. Among the other metal cations, Fe2+ and Zn2+ notably inhibited the IpeGlu1 activity. EDTA and 2-mercaptoethanol did not affect the activity.
Substrate Specificity of IpeGlu1—To examine the substrate specificity of the IpeGlu1 enzyme, kinetic analysis was performed using the 30 compounds listed in Table 1 and supplemental Fig. S1. Although the IpeGlu1 sequence showed the highest identity to raucaffricine β-glucosidase, IpeGlu1 did not accept raucaffricine as substrate. We expected that IpeGlu1 would preferentially deglucosylate N-deacetylisoipecoside rather than N-deacetylipecoside on the basis of a proposed biosynthetic pathway of emetine and cephaeline (Fig. 1). Although the kcat value for N-deacetylisoipecoside (86.3 s–1) was three times higher than that for N-deacetylipecoside (28.2 s–1), the Km value for N-deacetylisoipecoside (5.8 mm) was 18 times larger than that for N-deacetylipecoside (0.32 mm), which diminished the catalytic efficiency for N-deacetylisoipecoside to 16.9% of that for N-deacetylipecoside (Table 1). The best substrate among the compounds tested was ipecoside (Km = 0.1 mm, kcat = 51.6 s–1), for which the catalytic efficiency was 6-fold higher than that for N-deacetylipecoside.
O-Methyl derivatives of N-deacetylisoipecoside and N-deacetylipecoside were also tested. Catalytic efficiencies for 6-O-methyl derivatives were comparable to those for non-methylated substrates in both 1α(S)- and 1β(R)-configurations. On the other hand, 7-O-methylation and 6,7-O,O-dimethylation remarkably lowered the catalytic efficiencies mainly due to an enormous increment in the Km values. When the catalytic efficiencies were compared between 1α(S)- and 1β(R)-epimers, 1β(R)-epimers were always hydrolyzed more efficiently than 1α(S)-epimers regardless of the position of O-methyl group.
IpeGlu1 did not accept the lactams, demethylisoalangiside and demethylalangiside, as substrate. IpeGlu1 hydrolyzed strictosidine and vincoside, which possess a monoterpenoid-indole skeleton. Catalytic efficiency for vincoside (3β(R)-epimer) was 2-fold higher than that for strictosidine (3α(S)-epimer). Their lactams were not accepted as substrate. Although secologanin, loganin, galloyl-Glc, flavonoids (except quercetin-3-O-Glc), and pNP-Glcs were hydrolyzed by IpeGlu1, the activities for those substrates were quite lower than those for Ipecac alkaloid glucosides, such as ipecoside, N-deacetylipecoside, and N-deacetylisoipecoside.
Enzyme Activity of IpeGlu Isozymes—To check the enzyme activity of additionally isolated β-glucosidases, IpeGlu2, IpeGlu3 and IpeGlu9 were representatively expressed in E. coli. They were expressed as efficiently as IpeGlu1 and purified as performed for IpeGlu1. Their enzyme activities measured using N-deacetylipecoside and N-deacetylisoipecoside as substrate were slightly higher (∼1.3- to 2-fold) than IpeGlu1 activity (Table 3). All IpeGlu enzymes tested exhibited ∼4-fold higher activities for N-deacetylipecoside than for N-deacetylisoipecoside.
TABLE 3.
Enzyme activity of the four isozymes of Ipecac alkaloid β-glucosidase
Each glucosidase expressed in E. coli was purified, and enzyme activity was measured using N-deacetylipecoside (1 mm) and N-deacetylisoipecoside (1 mm) as substrate. Activity is shown as the percent ratio of the specific activity to that of the IpeGlu1 at 1 mm N-deacetylipecoside (21.4 μmol/min/mg of protein).
|
Glucosidase |
Relative activity |
|
|---|---|---|
| N-Deacetylipecoside | N-Deacetylisoipecoside | |
| % | ||
| IpeGlu1 | 100 | 24 |
| IpeGlu2 | 126 | 35 |
| IpeGlu3 | 193 | 41 |
| IpeGlu9 | 205 | 53 |
Identification of Deglucosylation Products—HPLC analyses of the enzyme reaction mixture revealed that the deglucosylation of N-deacetylisoipecoside, N-deacetylipecoside, and ipecoside by IpeGlu1 gave multiple products (supplemental Fig. S4, A–C). LC-MS/MS analysis of the reaction mixture of N-deacetylisoipecoside showed that the most reaction products detected by UV (supplemental Fig. S4A) had a molecular ion m/z of 344 (supplemental Fig. S4D). In addition, other reaction products were found to have m/z 362, m/z 667, or m/z 687 as molecular ion, of which m/z 362 corresponds to the mass of immediate deglucosylation product and m/z 687 appears to be a dimer of the reaction products of molecular ion m/z 344. Likewise, LC-MS/MS analysis of the reaction mixture of N-deacetylipecoside showed that the main products had m/z 344 as molecular ion (supplemental Fig. S4E). In addition, reaction products having m/z 362 as molecular ion were detected as in the reaction mixture with N-deacetylisoipecoside. The molecular ion of the two main peaks in the reaction mixture of ipecoside (supplemental Fig. S4C) were m/z 404 (supplemental Fig. S4F), which corresponds to the mass of the immediate deglucosylation product of ipecoside. In the reaction mixture of ipecoside, other ions were not detected. This showed that the occurrence of the products of molecular ion m/z 344 in the reaction mixtures of N-deacetylisoipecoside and N-deacetylipecoside is related to the nucleophilicity of the N-atom. A possible mechanism is as follows: the hemiacetal aglycon ([M+H]+ = m/z 362, Fig. 4A (1)) forms a dialdehyde by ring opening, and the N-atom attacks the carbolyl group of the aldehyde, followed by dehydration to form an iminium cation ([M]+ = m/z 344, Fig. 4A (6)), which is considered to tautomerize to the enamine form, 7.
FIGURE 4.
Plausible reaction scheme of the aglycon formed by IpeGlu1 from N-deacetylisoipecoside and N-deacetylipecoside under normal (A) and reduced (B) conditions. The molecular ion detectable in LC-MS/MS analysis is shown below each structure.
Presence of multiple peaks having m/z 344 as molecular ion (supplemental Fig. S4, D and E) suggested further conversion of 6 and 7. The fragmentation pattern of the molecular ion m/z 344 obtained by LC-MS/MS revealed that there are at least two different molecules; one gave the fragment ion m/z 326, which is formed by the dehydration of the protonated enol hydroxy group (e.g. 7), but the other one did not (data not shown). Accordingly, the formation of the ring-closed enamine 9 is the most probable, which is the counterpart to cathenamine that is formed by deglucosylation of strictosidine (20, 31).
To prove the transition scheme shown in Fig. 4A, the enzyme reaction was performed in the presence of 1 mm NaBH3CN to reduce the iminium cation and possibly the aldehyde. HPLC analyses of the reaction mixture with N-deacetylisoipecoside (supplemental Fig. S5A) and N-deacetylipecoside (supplemental Fig. S5B) detected one and two peaks, respectively. LC-MS/MS analysis revealed that the molecular ion of these products is m/z 346 (supplemental Fig. S5, C and D). In addition, a product having m/z 348 as molecular ion was also detected in both reactions, although its ion intensity was much lower than m/z 346. supplemental Fig. S5E shows the fragmentation pattern of molecular ions m/z 346 and m/z 348 in the reaction mixture with N-deacetylisoipecoside detected by LC-MS/MS analysis. The molecular ion m/z 346 did not give the fragment ion m/z 328, which is formed by dehydration of a protonated enol hydroxy group, showing that it is 13 formed by the reduction of ring-closed enamine 9 (Fig. 4B). Molecular ion m/z 348 is considered to be 11 formed by the reduction of 6 at the imino group and the keto form aldehyde (Fig. 4B).
LC-MS/MS analysis of the reaction mixture with N-deacetylipecoside (supplemental Fig. S5F) revealed the molecular ion m/z 346 at a retention time of 8.4 min gives fragment ion m/z 328, showing that it possesses an enol hydroxy group. In contrast, the m/z 346 at a retention time of 10.3 min did not give a fragment ion m/z 328, and it exhibited the same fragmentation pattern as the molecular ion m/z 346 in supplemental Fig. S5E. These results indicated that the front peak is 10 formed by the reduction of ring-opened iminiun cation 6, and the rear peak is 13 formed by the reduction of enamine 9 (Fig. 4B). The fragmentation pattern of the molecular ion m/z 348 (supplemental Fig. S5F) was the same as that in the N-deacetylisoipecoside reaction (supplemental Fig. S5E), showing that it is 11 formed by further reduction of 10 (Fig. 4B).
DISCUSSION
We have isolated full-length cDNAs encoding Ipecac alkaloid β-glucosidase from cultured tissues of P. ipecacuanha, which efficiently accumulated emetine, cephaeline, and ipecoside (data not shown). IpeGlu1 exhibited many enzymatic properties in common with strictosidine β-glucosidase with respect to optimum pH, optimum temperature, and inhibition by Cu2+, as well as the loss of activity toward lactamized substrates and raucaffricine. However, the stereospecificity for their substrates was totally different. In contrast to the strictosidine β-glucosidase that reacts with 3α(S)-epimer (strictosidine) but not with 3β(R)-epimer (vincoside), IpeGlu1 accepted both 1α(S) (N-deacetylisoipecoside) and 1β(R) (N-deacetylipecoside) epimers as substrate with preference of 1β(R)-epimer to 1α(S)-epimer. Crystal structure of the strictosidine β-glucosidase in complex with the substrate strictosidine demonstrated that the aglycon moiety is surrounded by residues Phe-221, Trp-388, Gly-386, Met-275, Thr-210, and Met-297 (32). The IpeGlu1, however, does not have these residues at the corresponding position, except for Trp-394 and Gly-391, which correspond to Trp-388 and Gly-386 of the strictosidine β-glucosidase (supplemental Fig. S2). Although the aglycon moiety differs between the indole and isoquinoline skeletons, difference in those residues may change the stereospecificity for their substrates. Structure elucidation of the IpeGlu1 enzyme and comparison of the substrate binding pocket with strictosidine β-glucosidase would uncover the mechanism of loose stereospecificity of the IpeGlu1 in its substrate recognition.
It has been shown that the 1α(S)-epimer N-deacetylisoipecoside is the intermediate for emetine biosynthesis in both Psychotria (Cephaelis in the literature) and Alangium (5, 6). In Alangium, the related alkaloid tubulosine, which has a β-carboline skeleton instead of the second tetrahydroisoquinoline moiety of emetine, has also been demonstrated to be synthesized from N-deacetylisoipecoside (33, 34). In contrast, the 1β(R)-epimer N-deacetylipecoside is the precursor mainly of ipecoside and alangiside, the alkaloidal glucosides having a β-configuration. Occurrence of both epimers in Ipecac alkaloid biosynthesis appears to justify the loss of stereospecificity of IpeGlu1. In fact, the stereospecificity of strictosidine β-glucosidase is considered to be due to the formation of only strictosidine by the enzymatic condensation of tryptamine and secologanin. If so, deglucosylation of the alkaloidal glucosides with a β-configuration should play an important physiological role, although it has been considered that the formation of ipecoside (Psychotria) and alangiside (Alangium) functions to deactivate N-deacetylipecoside and they are metabolically inert (6). This seems to be the case for alangiside, because IpeGlu1 did not accept lactam glucosides as substrate (Table 1). Considering that ipecoside is accumulated in Psychotria regardless of the presence of IpeGlu1, which exhibited the highest catalytic efficiency toward ipecoside, they must be localized in distinct subcellular compartments. It has been demonstrated that strictosidine is synthesized and stored in the vacuole (35, 36). Likewise, N-deacetylipecoside and N-deacetylisoipecoside should be localized in the vacuole. On the other hand, IpeGlu1 was predicted to be localized in cytosol due to lack of sorting signals like strictosidine β-glucosidase in R. serpentina, whereas the enzyme is associated with the endoplasmic reticulum in C. roseus (19, 30). It is widely accepted that β-glucosides, as represented by benzoxazinone- and cyanogenic glucosides, are functioning as a sink of toxic aglycons, which are released by corresponding β-glucosidase existing in distinct subcellular compartments, when cells are disrupted by wounding and infection (37, 38). Because ipecoside aglycon was not further converted by spontaneous reactions (supplemental Fig. S4F), it would have another physiological role rather than an intermediate of unknown alkaloid biosynthesis. Involvement of deglucosylated products of strictosidine in the defense against pathogens has been suggested (39). Supposedly ipecoside aglycon may also serve as a defensive chemical in P. ipecacuanha. This implies the functional differentiation of the 1α(S)- and 1β(R)-epimers generated by condensation of dopamine and secologanin: 1α(S)-epimer N-deacetylisoipecoside is transported outside the vacuole, deglucosylated by IpeGlu1, and introduced into emetine biosynthesis, whereas the 1β(R)-epimer N-deacetylipecoside is immediately acetylated to form ipecoside in the vacuole as defense-related glucoside that is deglucosylated by IpeGlu1. IpeGlu1 is potentially involved in both scenarios (Fig. 5).
FIGURE 5.
Overview of the involvement of IpeGlu1 in Ipecac alkaloid biosynthesis on the basis of substrate specificity of the IpeGlu1.
The kinetic analysis of IpeGlu1 revealed that neither 7-O-methyl nor 6,7-O,O-dimethyl derivatives of N-deacetylisoipecoside and N-deacetylipecoside are good substrates for IpeGlu1 (Table 1). Therefore, we can exclude the possibility of 7-O-methylation and 6,7-O,O-dimethylation of the substrates prior to deglucosylation by IpeGlu1 (Fig. 5). Occurrence of 6-O-methylipecoside and 7-O-methylipecoside in P. ipecacuanha (2) suggests the existence of methyltransferases that act on Ipecac alkaloid glucosides. For the biosynthesis of laudanine in opium poppy (Papaver somniferum), the 6-hydroxy group of tetrahydroisoquinoline skeleton is methylated at the first step and 7-hydroxy group is methylated at the last step, where each reaction is catalyzed by regiospecific 6-O-methyltransferase and 7-O-methyltransferase, respectively (40). Similarly, 6-O-methylation in the early step of the emetine biosynthesis is also plausible (Fig. 5), because the IpeGlu1 deglucosylated 6-O-methyl-N-deacetylisoipecoside as efficiently as non-methylated substrate (Table 1). As an alternative way for the formation of 6-O-methyl-N-deacetylisoipecoside, condensation of 4-hydroxy-3-methoxyphenethylamine (=3-O-methyldopamine) with secologanin might be possible. As described under “Experimental Procedures,” however, non-enzymatic condensation did not occur. Although this appears to be because a methoxy group at the meta-position having electron-withdrawing effect renders ring closure of the Schiff base generated by the condensation with secologanin unlikely, we cannot exclude the possibility of the reaction being caused by a corresponding enzyme.
Deglucosylation of N-deacetylisoipecoside by IpeGlu1 gave multiple products having a molecular ion m/z 344, as well as m/z 362, on LC-MS/MS analysis (supplemental Fig. S4D). This showed that the aglycon ([M+H]+ = m/z 362) is immediately dehydrated and further converted to other forms due to its high reactivity. It has been demonstrated that strictosidine aglycon is converted to cathenamine via 4,21-dehydrocorynantheine aldehyde (18, 20, 31). In an analogous manner, it was found that the enamine 9 (Fig. 4A) is formed via the iminium cation 6 from the aglycon of N-deacetylisoipecoside. Although formation of the enamine 9 from the aglycon of the 1β(R)-epimer, N-deacetylipecoside, was also detected, deglucosylation of N-deacetylipecoside does not seem to occur in vivo, because N-deacetylipecoside is immediately acetylated to form ipecoside in P. ipecacuanha as described above. This is supported by the fact that a non-glucosidic Ipecac alkaloid with β-configuration has not been found in nature so far.
In terpenoid-indole alkaloid biosynthesis in C. roseus, reduction of cathenamine occurred enzymatically when tryptamine and secologanin were incubated with crude enzyme in the presence of NADPH or NADH, suggesting the involvement of an oxidoreductase that utilizes NADPH or NADH (18, 41). A similar reaction would also occur in emetine biosynthesis not only on the enamine 9, the counterpart of cathenamine, to generate 13, but also on 6/7 to generate 10. Formation of protoemetine (Fig. 1) from 10 possibly proceeds via reduction of the alkene, hydrolysis of the methyl ester, decarboxylation, and tautomerization to the aldehyde. In addition to these reactions, reactions leading from protoemetine to emetine also need to be elucidated in a future study.
Supplementary Material
Acknowledgments
We greatly thank Dr. Peter Spiteller (Technische Universität München, Germany) for measuring 1H NMR and nuclear Overhauser effect spectroscopy spectra. We appreciate the gifts of raucaffricine, arbutin, and authentic sample of vincoside from Dr. Joachim Stöckigt (Johannes Gutenberg-Universität, Germany). We thank Dr. Thomas Vogt (Leibniz Institute of Plant Biochemistry, Germany) for his generous gifts of flavonoid-glucosides. We are grateful to Dr. Meinhart H. Zenk (Donald Danforth Plant Science Center) for providing ipecoside, secologanin, loganin, galloyl-Glc, and esculin, and for invaluable advice and discussion. We also thank the Tissue Culture Facility of Donald Danforth Plant Science Center for maintaining the shoot and root cultures of P. ipecacuanha.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AB455576–AB455586.
Footnotes
The abbreviations used are: RACE, rapid amplification of cDNA ends; HPLC, high-performance liquid chromatography; LC-MS/MS, liquid chromatography-tandem mass spectrometry; pNP, p-nitrophenyl.
References
- 1.Janot, M.-M. (1953) in The Alkaloids (Manske, R. H. F., and Holmes, H. L., eds) Vol. 3, pp. 363–394, Academic Press, New York [Google Scholar]
- 2.Fujii, T., and Ohba, M. (1998) in The Alkaloids (Cordel, G. A., ed) Vol. 51, pp. 271–321, Academic Press, New York [Google Scholar]
- 3.Battersby, A. R., Burnett, A. R., and Parsons, P. G. (1969) J. Chem. Soc. C 1969 1187–1192 [Google Scholar]
- 4.Battersby, A. R., and Parry, R. J. (1971) Chem. Commun. 1971 901–902 [Google Scholar]
- 5.Nagakura, N., Höfle, G., and Zenk, M. H. (1978) J. Chem. Soc. Chem. Commun. 1978 896–898 [Google Scholar]
- 6.Nagakura, N., Höfle, G., Coggiola, D., and Zenk, M. H. (1978) Planta Med. 34 381–389 [Google Scholar]
- 7.De-Eknamkul, W., Ounaroon, A., Tanahashi, T., Kutchan, T. M., and Zenk, M. H. (1997) Phytochemistry 45 477–484 [Google Scholar]
- 8.De-Eknamkul, W., Suttipanta, N., and Kutchan, T. M. (2000) Phytochemistry 55 177–181 [DOI] [PubMed] [Google Scholar]
- 9.Hampp, N., and Zenk, M. H. (1988) Phytochemistry 27 3811–3815 [Google Scholar]
- 10.Bracher, D., and Kutchan, T. M. (1992) Arch. Biochem. Biophys. 294 717–723 [DOI] [PubMed] [Google Scholar]
- 11.Treimer, J. F., and Zenk, M. H. (1979) Eur. J. Biochem. 101 225–233 [DOI] [PubMed] [Google Scholar]
- 12.Pfitzner, U., and Zenk, M. H. (1989) Planta Med. 55 525–530 [DOI] [PubMed] [Google Scholar]
- 13.DeWaal, A., Meijer, A. H., and Verpoorte, R. (1995) Biochem. J. 306 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevens, L. H., Giroud, C., Pennings, E. J. M., and Verpoorte, R. (1993) Phytochemistry 33 99–106 [Google Scholar]
- 15.Yamazaki, Y., Sudo, H., Yamazaki, M., Aimi, N., and Saito, K. (2003) Plant Cell Physiol. 44 395–403 [DOI] [PubMed] [Google Scholar]
- 16.Stöckigt, J., and Zenk, M. H. (1977) J. Chem. Soc. Chem. Commun. 1977 646–648 [Google Scholar]
- 17.Rueffer, M., Nagakura, N., and Zenk, M. H. (1978) Tetrahedron Lett. 19 1593–1596 [Google Scholar]
- 18.Stöckigt, J., and Zenk, M. H. (1977) FEBS Lett. 79 233–237 [Google Scholar]
- 19.Geerlings, A., Ibanñez, M. M.-L., Memelink, J., van der Heijden, R., and Verpoorte, R. (2000) J. Biol. Chem. 275 3051–3056 [DOI] [PubMed] [Google Scholar]
- 20.Gerasimenko, I., Sheludko, Y., Ma, X., and Stöckigt, J. (2002) Eur. J. Biochem. 269 2204–2213 [DOI] [PubMed] [Google Scholar]
- 21.Linsmaier, E. M., and Skoog, F. (1965) Physiol. Plant. 18 100–127 [Google Scholar]
- 22.Chomczynski, P., and Sacchi, N. (1987) Anal. Biochem. 162 156–159 [DOI] [PubMed] [Google Scholar]
- 23.Bradford, M. M. (1976) Anal. Biochem. 72 248–254 [DOI] [PubMed] [Google Scholar]
- 24.Itoh, A., Tanahashi, T., and Nagakura, N. (1995) Nat. Prod. 58 1228–1239 [Google Scholar]
- 25.Shoeb, A., Raj, K., Kapil, R. S., and Popli, S. P. (1975) J. Chem. Soc. Perkin Trans. 1 1975 1245–1248 [PubMed] [Google Scholar]
- 26.Battersby, A. R., Burnett, A. R., and Parsons, P. G. (1969) J. Chem. Soc. C 1969 1193–1200 [Google Scholar]
- 27.Warzecha, H., Gerasimenko, I., Kutchan, T. M., and Stöckigt, J. (2000) Phytochemistry 54 657–666 [DOI] [PubMed] [Google Scholar]
- 28.Bendtsen, J. D., Nielsen, H., von Heijne, G., and Brunak, S. (2004) J. Mol. Biol. 340 783–795 [DOI] [PubMed] [Google Scholar]
- 29.Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000) J. Mol. Biol. 300 1005–1016 [DOI] [PubMed] [Google Scholar]
- 30.Luijendijk, T. J. C., Stevens, L. H., and Verpoorte, R. (1998) Plant Physiol. Biochem. 36 419–425 [Google Scholar]
- 31.Stöckigt, J., Husson, H. P., Kan-Fan, C., and Zenk, M. H. (1977) J. Chem. Soc. Chem. Commun. 1977 164–166 [Google Scholar]
- 32.Barleben, L., Panjikar, S., Ruppert, M., Koepke, J., and Stöckigt, J. (2007) Plant Cell 19 2886–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhakuni, D. S., Jain, S., and Chaturvedi, R. (1983) J. Chem. Soc. Perkin Trans. 1 1983 1949–1983 [Google Scholar]
- 34.Jain, S., Sinha, A., and Bhakuni, D. S. (2002) Phytochemistry 60 853–859 [DOI] [PubMed] [Google Scholar]
- 35.McKnight, T. D., Bergey, D. R., Burnett, R. J., and Nessler, C. L. (1991) Planta 185 148–152 [DOI] [PubMed] [Google Scholar]
- 36.Stevens, L. H., Blom, T. J. M., and Verpoorte, R. (1993) Plant Cell Rep. 12 573–576 [DOI] [PubMed] [Google Scholar]
- 37.Niemeyer, H. M. (1988) Phytochemistry 27 3349–3358 [Google Scholar]
- 38.Poulton, J. E. (1990) Plant Physiol. 94 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luijendijk, T. J. C., van der Meijden, E., and Verpoorte, R. (1996) J. Chem. Ecol. 22 1355–1366 [DOI] [PubMed] [Google Scholar]
- 40.Ounaroon, A., Decker, G., Schmidt, J., Lottspeich, F., and Kutchan, T. M. (2003) Plant J. 36 808–819 [DOI] [PubMed] [Google Scholar]
- 41.Stöckigt, J., Treimer, J., and Zenk, M. H. (1976) FEBS Lett. 70 267–270 [DOI] [PubMed] [Google Scholar]
- 42.Saitou, N., and Nei, M. (1987) Mol. Biol. Evol. 4 406–425 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.