Abstract
Prostasin (also called channel activating protease-1 (CAP1)) is an extracellular serine protease implicated in the modulation of fluid and electrolyte regulation via proteolysis of the epithelial sodium channel. Several disease states, particularly hypertension, can be affected by modulation of epithelial sodium channel activity. Thus, understanding the biochemical function of prostasin and developing specific agents to inhibit its activity could have a significant impact on a widespread disease. We report the expression of the prostasin proenzyme in Escherichia coli as insoluble inclusion bodies, refolding and activating via proteolytic removal of the N-terminal propeptide. The refolded and activated enzyme was shown to be pure and monomeric, with kinetic characteristics very similar to prostasin expressed from eukaryotic systems. Active prostasin was crystallized, and the structure was determined to 1.45Å resolution. These apoprotein crystals were soaked with nafamostat, allowing the structure of the inhibited acyl-enzyme intermediate structure to be determined to 2.0Å resolution. Comparison of the inhibited and apoprotein forms of prostasin suggest a mechanism of regulation through stabilization of a loop which interferes with substrate recognition.
Prostasin (also known as channel activating protease-1 (CAP1)) is a serine protease originally isolated from seminal fluid as a secreted protein (1). It is widely expressed in mammalian epithelial tissue as a 40-kDa glycosylphosphatidylinositol (GPI)-anchored protein (1–3). Prostasin is implicated in the regulation of sodium and fluid levels via proteolysis of the epithelial sodium channel (ENaC)3 γ subunit (4–9).
ENaC performs an essential function in several epithelial tissues, including the colon, kidney, and lung (5, 7, 9, 10). In these tissues sodium is primarily transported across the apical membrane via ENaC, and fluid transport across the membrane is highly responsive to sodium concentration. Inappropriate function of ENaC leads to misregulation of blood pressure in humans (11, 12). The same ENaC mutations which lead to hypertension in humans give rise to defects in lung fluid clearance in mice (11), suggesting that proper ENaC function may also be important in other diseases where sodium and fluid homeostasis is severely disrupted, for example cystic fibrosis and diarrhea (13, 14).
Although ENaC activity can be regulated via transcription and translation, for example in response to the hormone aldosterone (11), proteolysis of ENaC can lead to channel activation (9), and inhibition of this process might provide a direct and specific method to modulate ENaC activity for therapeutic benefit. Cleavage of an inhibitory peptide in the ENaCγ subunit by prostasin has been shown to stimulate sodium transport by 2–3-fold in cell-based experiments (4, 5, 7–10). Other proteases implicated to activate ENaC in model systems include furin, mCAP-2, mCAP-3, and TMSP-1 (transmembrane serine protease 1) (5, 8–11). Some of these proteases require activation by yet other proteases (15, 16), so clarity regarding which enzymes may represent appropriate targets for reducing ENaC activity has yet to be achieved. Highly selective small-molecule inhibitors of proteases thought to modulate ENaC will aid in understanding the roles of different proteases in channel function and may also provide a therapeutic benefit for hypertension and other diseases, e.g. cystic fibrosis (12–14). Although there are numerous agents for the treatment of hypertension, many individuals require multiple medications to achieve adequate control (17), and the condition continues to exact a high cost to society, and so it seems that additional modes of treatment remain desirable.
Prostasin belongs to the classical serine protease family, with homology to trypsin, chymotrypsin, and kallikrein, and has a trypsin-like substrate specificity (1, 2, 18). In common with these, enzyme activation of prostasin occurs via cleavage of the pro-protein to produce a light chain and a heavy chain that are disulfide-linked. Prostasin has some features unusual in serine proteases, such as a high degree of sensitivity to monovalent and divalent cations, which may relate to the prostasin role in ENaC regulation (18). However, to date there have been no reports of potent and selective small molecule inhibitors of prostasin. High resolution structural studies have been used to significant advantage in the design of selective protease inhibitors (19–22). Availability of the three-dimensional structure of prostasin could offer a significant advantage in the design of prostasin inhibitors.
Previous studies have shown that active prostasin can be expressed in a recombinant baculovirus system and purified to homogeneity, enabling more extensive biochemical studies (18). We report the expression of prostasin in Escherichia coli and the refolding, proteolytic activation, and purification to yield pure and fully active enzyme. Expression in E. coli is novel and can allow for rapid generation of site-directed mutants to allow the biochemical determination of structure-function relationships. We describe the high resolution crystal structure of the protease domain of this enzyme, which may provide valuable information in the design or optimization of specific small-molecule inhibitors.
EXPERIMENTAL PROCEDURES
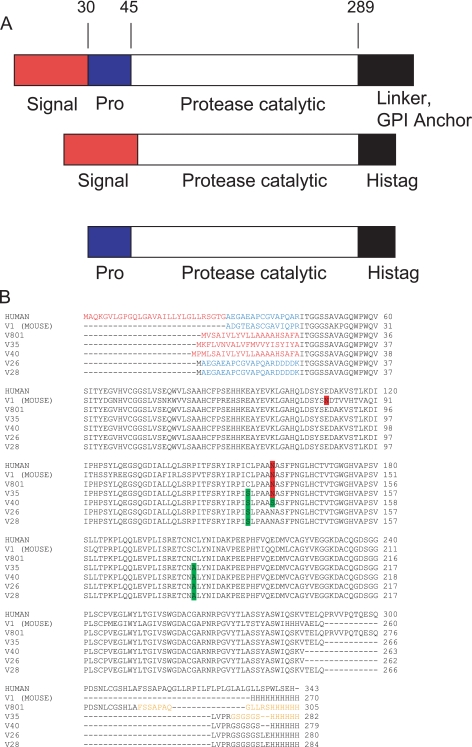
Construction of Prostasin Variants—The native sequence (accession number NM_002773) and all expression constructs are illustrated in Fig. 1. In all constructs the C-terminal glycosylphosphatidylinositol anchor domain and preceding linker were replaced with a His6 tag to aid in purification. Cysteines 154 and 203 were not expected to be involved in intradomain disulfide bonds and were mutated to serine and alanine (18). In the case of baculovirus variant 40, the N-linked glycosylation site was also removed via site-directed mutagenesis. For baculovirus expression constructs, the native signal sequence and propeptide were replaced with an insect cell signal sequence (melittin, GP64, or GP67) to generate the native N terminus of the mature protein, as the propeptide is not required for activity (18). Bacterial expression constructs contained the original native propeptide sequence at the N terminus but with the addition of an enterokinase recognition sequence to allow specific cleavage to generate the active enzyme. Detailed methods for the expression of prostasin in both baculovirus and E. coli are provided in the supplemental Experimental Procedures.
FIGURE 1.
A, map of the prostasin domain structure for the natural human sequence (top) and proteins expressed in insect cells (middle) and bacteria (bottom) showing the signal sequence, propeptide, protease domain, and C-terminal region. B, alignment of prostasin variants. Residues in red represent signal sequences cleaved during expression. The first variant is the full wild-type human sequence, whereas the second variant 1 is recombinant mouse prostasin as purchased from R&D Systems. Residues in blue represent the propeptide sequence either cleaved during expression (mouse) or in vitro (variants 26 and 28). Residues in orange were removed before evaluation of kinetic values. Residues highlighted in red represent N-linked glycosylation sites. Residues highlighted in green represent introduced site-directed mutations, as described under “Experimental Procedures.”
Refolding and Purification of Prostasin from E. coli—Prostasin variants 26 and 28 (Fig. 1) were solubilized from inclusion bodies in 8 m urea with 0.1 m Tris/HCl, pH 8.0, and 2 mm dithiothreitol after extensive washing. Solubilized protein was bound to nickel-nitrilotriacetic acid Superflow resin (Qiagen) and eluted with 0.3 m imidazole. The urea-solubilized material was then refolded in a buffer containing 1 m l-arginine, 0.1 m Tris/HCl, 5 mm reduced glutathione, and 0.5 mm oxidized glutathione, pH 8.0, at a final concentration of 10 mg/liter. After diafiltration, protein was purified via Ni(II) affinity and anion exchange chromatography. Eluted fractions were evaluated via SDS-PAGE and mass spectrometry (MS), and fractions in which prostasin was detected by MS were then pooled for further processing. Prostasin zymogen was converted to the active form by addition of enterokinase (EKMax, Invitrogen) at a final concentration of 2 units/ml (7.5 units/mg prostasin) in the presence of 0.5 mm reduced glutathione. The resultant cleavage reaction was maintained at 4 °C for 48 h and monitored by MS and SDS-PAGE. After completion of cleavage as determined by MS, the reaction was incubated at 4 °C overnight after the addition of 1 mm oxidized glutathione. Active prostasin was purified from enterokinase and misfolded prostasin via Ni(II) affinity and anion exchange chromatography. Fractions were selected based on activity, then pooled to give the final purified prostasin for further characterization and crystallography. Further details of the purification are found in the supplemental Experimental Procedures.
Enzyme Assay—Enzyme assays were performed at ambient temperature (22–24 °C) in 0.1 m Tris/HCl, pH 9, with 0.1% CHAPS using a peptide substrate, Ac-KHYR-7-amino-4-methylcoumarin (Anaspec) at 50 μm (18). Data were collected using a BMGLabTech Fluostar Optima reader, with excitation and emission filter wavelengths of 340 and 450 nm, respectively. Measurements of Km were made by varying the concentration of substrate from 10–250 μm, whereas enzyme concentration was held constant at 19 nm. Absolute quantitation of product was determined by comparison to an independent 7-amino-4-methylcoumarin standard at concentrations from 2 nm to 1.7 μm.
Size Exclusion Chromatography (SEC)—Analytical SEC measurements were made using a Superdex 200 5/150 column (GE Healthcare) equilibrated in 50 mm potassium phosphate, pH 6.8, and 0.3 m NaCl at a flow rate of 0.15 ml/min with injections of 0.5–5 μg of protein. Retention times were converted to apparent molecular masses using mixed protein standards (thyroglobulin, gamma-globulin, ovalbumin, and myoglobin) (Bio-Rad).
MS—Whole-protein mass measurements were made by binding samples (0.1–2 μg) to a reverse-phase protein trap column (Michrom), where they were desalted by washing with 2% acetonitrile, 0.01% trifluoroacetic acid and eluted with a solution of 64% acetonitrile, 0.01% trifluoroacetic acid into an electrospray mass spectrometer (LTQ, Thermo). The resultant spectra were deconvoluted using Promass (Novatia) to yield the whole protein mass. For free cysteine determination, samples were treated with 50 mm iodoacetamide at room temperature for 30 min before analysis. For active site determination, prostasin was incubated with 5 μm nafamostat mesylate (BioMol International) for 5 min in 50 mm Tris/HCl, pH 8.5, with a final NaCl concentration of 0.2 m or less before mass spectroscopic analysis (23).
Protein Crystallization—Prostasin variant 26, comprising human prostasin residues 45–285 with the mutations C154S and C203A, a C-terminal thrombin-cleavage site, and His6 tag (Fig. 1B), and variant 28, comprising human prostasin residues 45–289 with the same mutations and C terminus, were concentrated to a final concentration of 12–14 mg/ml as determined by the Bradford assay (Bio-Rad) in 50 mm Tris/HCl, pH 8.5, 0.1 m NaCl. Crystals of both variants were obtained by the vapor diffusion method. Initial crystals were obtained in hanging drops containing 1 μl of protein mixed with 1 μl of reservoir solution consisting of 100 mm Bis-Tris, pH 6.3, and 30% polyethylene glycol 3350. Diffraction quality crystals formed as a result of streak seeding into hanging drops containing 1 μl of protein mixed with 1 μl of reservoir solution consisting of 100 mm Bis-Tris, pH 5.8, and 21–26% polyethylene glycol 3350. Crystals were cryoprotected for data collection in a solution containing 100 mm Bis-Tris, pH 6.5, 20% polyethylene glycol 3350, 100 mm NaCl, and 20% ethylene glycol.
Protein Structure Determination—In-house diffraction data were collected with Rigaku x-ray sources using R-Axis image plate detectors. Data were processed and scaled with HKL2000 (24). Initial phases were obtained by molecular replacement with Phaser (25) using the model of human plasma kallikrein (PDB code 2ANW) (26). The models for variant 28 apoprotein, and nafamostat-inhibited acyl-enzyme intermediate were built and improved with iterative cycles of manual rebuilding in Coot and MIFit followed by refinement with Refmac (27, 28). There was sufficient density to build the model continuously from residues Ile-45 to Gln-289, which encodes all of the native, mature protein residues. Nafamostat was introduced by soaking crystals of apoprotein in a buffer containing 1 mm nafamostat, 100 mm Bis-Tris, pH 5.8, 100 mm NaCl, and 30% polyethylene glycol 3350. Structural alignments were performed using LSQMAN (29). Figures were generated using PyMOL (30). Model geometry was verified using Procheck (31).
RESULTS AND DISCUSSION
Expression of Prostasin Variants—Initially, prostasin variants were expressed by secretion from insect cells, as used by Shipway et al. (18) to examine the catalytic properties of prostasin. Although the expression levels for the glycosylated prostasin variants 801 and 35 (Fig. 1B) were acceptable (0.4–1.0 mg/liter) and the purified proteins were well behaved in solution and could be readily concentrated to 20 mg/ml, they did not form crystals using a broad screen of conditions. Prostasin variant 40, which lacks the N-linked glycosylation site, expressed to much lower levels in this system (<0.1 mg/liter).
Expression in E. coli was examined in two strains: BL21(DE3) and Origami-2. In BL21(DE3), prostasin variants 26 and 28 expressed at 50–100 mg/liter as insoluble inclusion bodies. Although Origami-2 cells possess an oxidizing intracellular environment sometimes helpful in disulfide formation, as needed for prostasin, expression of prostasin variants 26 and 28 in these cells gave mainly insoluble protein at 1–5 mg/liter. Accordingly, prostasin variants 26 and 28 were expressed in BL21(DE3) cells and refolded in vitro.
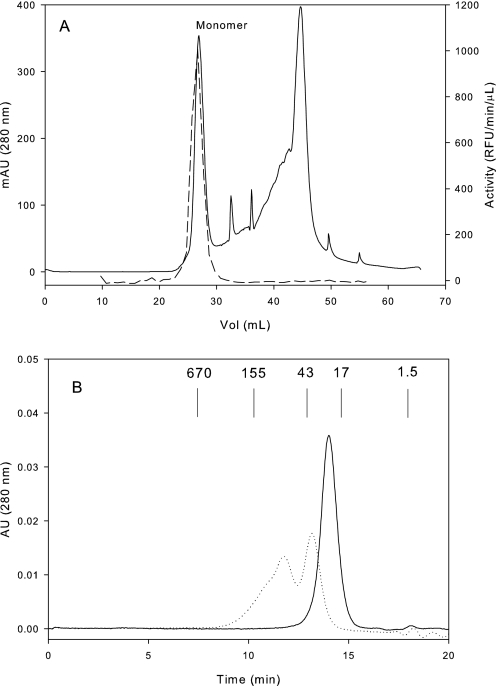
Refolding of Prostasin—The primary challenge to devising a refolding protocol for prostasin was evaluating the product for the correctness of its fold. Although enzymatic activity would normally be the preferred measure of success, prostasin was expressed in the inactive zymogen form to ensure generation of a native N terminus. Enterokinase, the enzyme used to cleave the propeptide, was active in the prostasin assay, making interpretation of activity assays from mixtures containing both enzymes problematic. Although the amount of soluble protein captured on Ni(II) columns was of some guidance (Table 1, buffer additive conditions: none, 2 m urea, and 0.4 m l-arginine), the majority of the protein in these mixtures seemed to be misfolded aggregates. The best tool in these circumstances to judge the quality of refolded zymogen was MS after reverse-phase chromatography. Misfolded proteins are adsorbed much more strongly to the reverse-phase surface than well folded proteins (32, 33), whereas MS ensures accurate identification of the protein species eluted from the column. Purification of the solubilized protein on Ni(II) affinity media was performed to simplify the mass spectrum observed after refolding, making the analysis more consistent and semiquantitative. Using these tools, small-scale refolding experiments were carried out to find optimal conditions, detailed in Table 1. In scaled-up refolds, although the yields of soluble protein after refolding and the first chromatographic step were quite reasonable, the pool still contained substantial amounts of misfolded protein. Prostasin zymogen was heterogeneous by both anion exchange chromatography (supplemental Fig. 1) and analytical SEC, which showed a mixture of monomer with an apparent size of 37 kDa, dimer at ∼ 70 kDa, and larger species (Fig. 2B, dashed line).
TABLE 1.
Results of refolding conditions Shown are refolding results for prostasin variant 28. Yields are expressed as total protein (determined by the Bradford assay) per liter of refold buffer either from pooled fractions after the first chromatographic step or for the final purified protein. Mass spectroscopic signal was assessed on a qualitative basis for different conditions, each with a nominal 1-μg protein injection. All conditions used a buffer containing 0.1 m Tris/HCl, pH 8.0, 5 mm reduced glutathione, and 0.5 mm oxidized glutathione. Similar results were observed for variant 26.
|
Condition | ||||||
|---|---|---|---|---|---|---|
| Buffer additive | None | 2 m Urea | 0.4 m l-Arginine | 0.4 m l-Arginine | 0.4 m l-Arginine | 1.0 m l-Arginine |
| Protein purified prior to refold | No | No | No | Yes | Yes | Yes |
|
Protein concentration in refold (mg/liter)
|
10
|
10
|
10
|
10
|
5
|
10
|
|
Refold results | ||||||
| 1st column yield (mg/liter) | <0.1 | 0.1 | 1 | 1.5 | 2 | 4 |
| MS signal | ND | ND | ND | + | ++ | +++ |
| Final yield (mg/liter) | ND | 0.01 | 0.04 | 0.05 | 0.05 | 0.15a |
ND, not determined
denotes the conditions used in subsequent studies
FIGURE 2.
Chromatographic traces of refolded prostasin variant 28. A, anion-exchange chromatography of prostasin after activation. The peak corresponding to active, monomeric prostasin is marked “monomer.” Superimposed with a dashed line are the results of activity assays of the fractions from this column. B, size exclusion chromatography of the zymogen (dashed line) and activated prostasin (solid line). Molecular weights in kDa of the standards used are shown on the chart at their elution positions. mAU, milliabsorbance units.
Cleavage of the zymogen with enterokinase was carried out in the presence of reduced glutathione to allow for disulfide shuffling, as additional MS experiments suggested that the zymogen had not completely achieved the native disulfide configuration (supplemental Table 1), similar to the behavior observed for trypsinogen (34). Cleavage was rapid and completely specific when monitored by MS, whereas SDS-PAGE continued to show that some of the protein was resistant to cleavage, which we hypothesized was misfolded.
After cleavage, prostasin was purified on two additional columns. The Ni(II) affinity column was used primarily to remove enterokinase, after which prostasin could be tracked and quantified via activity assay. The final anion exchange step proved to be the most important in removing misfolded species. Active prostasin eluted as the first species from the column at a considerably lower ionic strength than either the zymogen or aggregated species (Fig. 2A), and activity coincided only with this first peak (Fig. 2A, dashed line). In contrast to the zymogen, MS of the fractions from the anion exchange column showed that >95% of the MS-active species coincided with the first peak. Analytical SEC and SDS-PAGE of later fractions from the anion-exchange column showed that these contained prostasin in an inactive, aggregated state (data not shown). Although both variants 26 and 28 could be refolded to yield active protein, the final yield of active protein was only 1–2% as the bulk of the protein formed inactive misfolded species and was removed on the final chromatographic step.
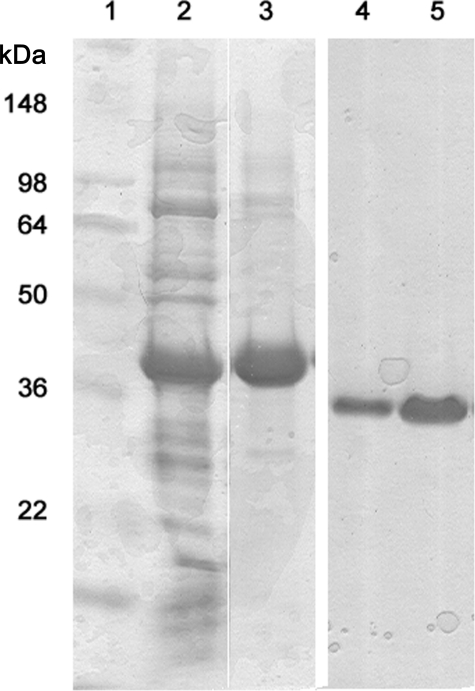
Characterization of the Final Refolded Protein—Refolded prostasin was of high purity as shown in the gel (Fig. 3). SEC is often used to profile refolded protein, as it readily distinguishes lower-order species from the aggregates that result from misfolding. As seen in Fig. 2B, purified refolded prostasin eluted as a single peak, corresponding to a molecular mass of 24 kDa, identical to that seen for prostasin variants 801 and 35 (supplemental Table 2). Given that the cleaved propeptide has a molecular mass of 2 kDa, the difference between monomeric zymogen, which elutes from SEC with an apparent mass of 37 kDa (actual mass 30 kDa) and the active protein (actual mass 28 kDa), is remarkable. The active protein is more compact in terms of hydrodynamic radius, which may be related to its tendency to adopt a native disulfide configuration (34).
FIGURE 3.
Coomassie-stained reduced prostasin 28 examined by 14% Tris-glycine SDS-PAGE. Lane 1, molecular weight markers. Lane 2, washed and urea-solubilized inclusion bodies of prostasin. Lane 3, Ni(II)-purified, urea-solubilized prostasin proenzyme. Lanes 4 and 5, pure, active prostasin at a 1.30 and 5.0 μg load. Lanes 1, 2, and 3 are from the same gel, but intermediate lanes were omitted; lanes 4 and 5 are from a different gel which was aligned by the molecular weight markers.
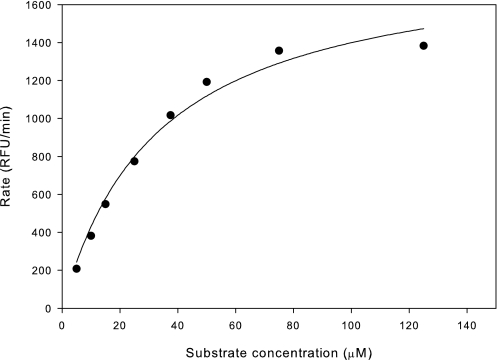
A more demanding measure of the quality of the refolded protein is a comparison of its kinetic constants with that of similar variants folded in eukaryotic cells. Initial reaction rates were measured across a range of substrate concentrations, and an example of the resulting data is shown in Fig. 4 along with the nonlinear curve fit used to obtain Km and kcat. As seen in Table 2, the kcat and kcat/Km for the refolded prostasin variants 26 and 28 are extremely similar to those seen for the analogous variant 40 expressed in insect cells and only slightly lower than seen for the glycosylated variants 801 and 35. Although glycosylation may be important for efficient expression of this protein in eukaryotes, it has no significant effect on enzymatic function in our assays.
FIGURE 4.
Kinetics of hydrolysis of Ac-KHYR-7-amino-4-methylcoumarin by prostasin 28. Rates at each substrate concentration were determined from a linear fit of 8 time points taken over 12 min. The plotted points represent an average of three independent determinations. The line shown represents a nonlinear fit of the Michaelis-Menten equation. RFU, relative fluorescent unit.
TABLE 2.
Kinetic constants for prostasin variants Rate constants were measured for the hydrolysis of Ac-KHYR-7-amino-4-methyl coumarin at room temperature. Mouse prostasin was used as purchased from R&D Systems. Variants 801, 35, and 40 were expressed in insect cells and purified to homogeneity as described in the supplemental Experimental Procedures.
| Variant | Host cell | Glycosylated | KM | kcat | kcat/KM |
|---|---|---|---|---|---|
| μm | s–1 | s–1m–1 × 103 | |||
| 1 | Mouse | Yes | 49.3 ± 8.0 | 0.27 ± 0.04 | 5.4 ± 0.03 |
| 801 | Insect (Sf21) | Yes | 33.5 ± 2.6 | 0.23 ± 0.01 | 7.0 ± 0.85 |
| 35 | Insect (Sf21) | Yes | 43.0 ± 3.5 | 0.17 ± 0.01 | 3.9 ± 0.05 |
| 40 | Insect (Sf21) | No | 40.4 ± 2.4 | 0.13 ± 0.02 | 3.1 ± 0.19 |
| 28 | Bacterial (E. Coli) | No | 34.3 ± 1.8 | 0.14 ± 0.01 | 3.9 ± 0.13 |
| 26 | Bacterial (E. Coli) | No | 38.8 ± 2.0 | 0.12 ± 0.01 | 3.2 ± 0.31 |
Mass spectrometry can offer insights into protein covalent structure complementary to those seen via other analytical measures. Refolded prostasin 26 and 28 give clean mass spectra with only one species seen after deconvolution and excellent correspondence to the predicted mass as seen in Table 3 and supplemental Fig. 2. Treatment of either variant 26 or 28 with iodoacetamide showed no changes in the observed molecular mass (Table 3), indicating that all 8 cysteine residues form intramolecular disulfides. MS can also be used as an active site titrant. Treatment of either variant with nafamostat before mass measurement shows a complete shift of the mass observed to a species at +161 Da from the original mass, indicating that all of the protein observed via MS is capable of conversion to the expected acyl-enzyme intermediate.
TABLE 3.
Mass spectroscopic results for refolded and purified prostasin variants
|
Prostasin variant |
||
|---|---|---|
| 28 | 26 | |
| Da | ||
| Theoretical mass | 28,351 | 27,879 |
| Observed mass | 28,350 | 27,879 |
| Observed mass + iodoacetamide | 28,350 | 27,879 |
| Observed mass + nafamostat | 28,510 | 28,040 |
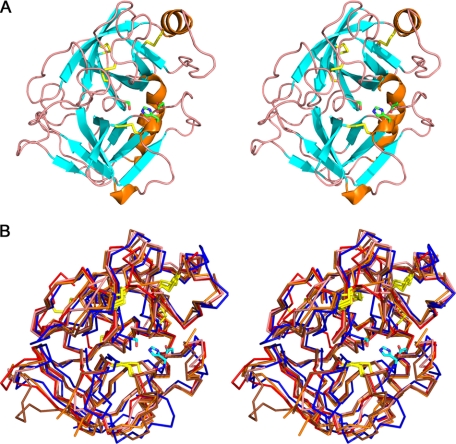
The Structure of Prostasin—The crystal structure of the protease domain of prostasin is structurally similar to other serine proteases, consisting of two β barrel-like subdomains with four conserved disulfide bonds (Fig. 5, Table 4). The two subdomains are separated by a cleft that contains the active site. In prostasin, the catalytic triad is formed by His-85, Asp-134, and Ser-238 (Fig. 5A), corresponding to chymotrypsin residues His-57, Asp-102, and Ser-195. The proximity of the hydroxyl group of Ser-238 to the imidazole ring of His-85 and the hydrogen bond between Oδ of Asp-134 and Nδ of His-85 suggest that the catalytic mechanism observed in other serine proteases is conserved in prostasin.
FIGURE 5.
A, stereoview of the ribbon diagram depicting the prostasin protease domain. The structure is colored by secondary structural elements with β strands in cyan, α helices in orange, and loops in pink. The structure is oriented to show the two β-sheet subunits which form the protease domain. The four disulfide bonds are shown in yellow. The catalytic triad, made of His-85, Asp-134, and Ser-238, are shown as sticks. B, Cα traces of overlays of the protease domains of prostasin, chymotrypsin, trypsin, elastase, and kallikrein. The structure-based alignments are shown with prostasin in blue, chymotrypsin in red (PDB code 4CHA) (45), trypsin in pink (PDB code 2PTN) (46), elastase in orange (PDB code 3EST) (47), and kallikrein in brown (PDB code 2ANW) (26). The disulfide bonds are shown in yellow. The core of the domain is highly conserved in the proximity of the active site. As a representative, the catalytic triad of prostasin is shown.
TABLE 4.
Crystallographic statistics Values in parenthesis are for the highest resolution bin.
| Unbound apoprotein | Nafamostat-inhibited complex | |
|---|---|---|
| Resolution (Å) | 50-1.45 (1.50-1.45) | 50-2.0 (2.07-2.0) |
| Completeness (%) | 99.3 (96.9) | 99.9 (99.8) |
| Average redundancy | 4.2 (3.4) | 4.3 (4.3) |
| Rsym (%) | 5.3 (33.0) | 8.1 (33.7) |
| 〈I/σI〉 | 21.8 (3.6) | 17.6 (4.0) |
| Dimensions of P212121 cell | ||
| a (Å) | 53.0 | 53.5 |
| b (Å) | 58.4 | 54.8 |
| c (Å) | 85.4 | 83.3 |
| α = β = γ (°) | 90 | 90 |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.006 | 0.008 |
| Bond angles (°) | 1.036 | 1.164 |
| No. of protein atoms (non-hydrogen) | 1885 | 1871 |
| No. waters and ions | 408 | 270 |
| Mean B-value of all atoms | 17.35 | 18.04 |
| No. refl. total/no. refl. free set | 42240/2253 | 16236/870 |
| Rwork/Rfree (%) | 19.0/21.3 | 17.3/22.5 |
In comparison with other serine proteases with known structures, the core of the domain is highly conserved, whereas the loops are less conserved (Fig. 5B). The sequence identity between the protease domain of prostasin and those of chymotrypsin, trypsin, elastase, and kallikrein are 37, 38, 32, and 39%, respectively. The root mean square deviation (Å) between the Cα atoms of prostasin and chymotrypsin, trypsin, elastase, and kallikrein was 1.057, 1.084, 1.060, and 0.979, respectively. The four disulfide bonds in prostasin were conserved in the other serine proteases. Prostasin Cys-154, which was mutated to Ser to aid crystallization, is in the same position as Cys-122 in chymotrypsin that forms a disulfide bond with Cys-1 in the N-terminal light chain. Analogous to Cys-1 of chymotrypsin, Cys-37 is present in the N-terminal propeptide fragment of prostasin. A second cysteine, Cys-203, was mutated to Ala, also to aid crystallization. This cysteine is not conserved among serine proteases. Prostasin has a total of 12 cysteines, so Cys-203 may form a sixth disulfide bond with Cys-306 in the C-terminal region. Because the C-terminal region is glycosylphosphatidylinositol-anchored to the cell, it is possible that formation of a sixth disulfide bond may orient the active site toward the surface of the cell to facilitate the co-localization of the catalytic site to its substrate.
Despite the overall similarity to other serine proteases, a comparison of the prostasin structure with other known protease structures of high sequence identity reveals differences in residues that occupy key substrate specificity-determining positions. The closest known structures were chosen by a Blast search of the Protein Data Bank data base using prostasin as the query sequence (35) using an E-score cutoff of E-45. Using the structure of chymotrypsin complexed to bovine pancreatic trypsin inhibitor (PDB code 1T7C) (36) as a model for substrate binding, 22 residues of prostasin appear to form the peptide binding cleft. Of the residues that form the peptide binding cleft, five residues appear to be poorly conserved: Glu-129, Gly-130, Pro-214, His-215, and Gln-235. From the model, Glu-129 appears to interact with the substrate at the P4 position; this is consistent with previous data that suggest a prostasin preference for basic residues in the P4 position (18). Gly-130, Pro-214, and His-215 appear to line the S3 subsite, with the carbonyl oxygens of Gly-130 and Pro-214 oriented toward the substrate. This subsite can accommodate a long residue, but the carbonyl oxygens may restrict the charge in the P3 position. This would support a P3 preference toward histidine, lysine, or arginine which was found by library screening (18). Gln-235 appears to be adjacent to the P1′ and P2′ positions, but it is difficult to determine whether it affects specificity (37). By using only a sequence alignment, the protease hepsin is identical to prostasin in four of the five poorly conserved residues, suggesting that hepsin might have a similar substrate specificity as prostasin. However, a structural comparison shows that the loops that contain these residues are different in lengths and would not form the same interactions with substrate.
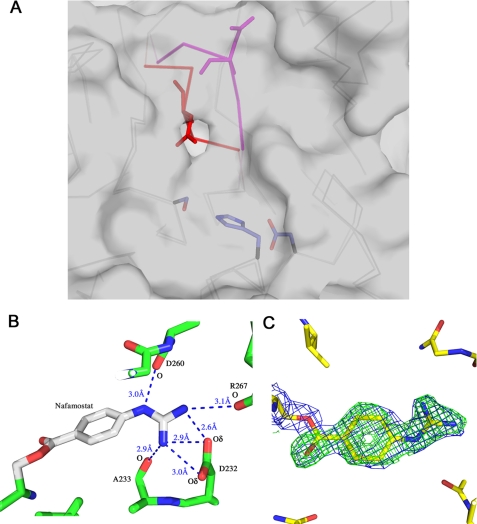
Adjacent to the catalytic triad in serine proteases, there is a pocket that binds and confers specificity for the P1 position of substrates. In the structure of the prostasin apoprotein, this pocket is occluded by a loop containing residues 258–262 (Fig. 6A). A visual inspection of >1200 serine protease structures reveals that blockage of the S1 site is not unique to prostasin but has been found in other serine proteases such as prostate kallikrein (PDB code1GVZ) (38), granzyme K (PDB code 1MZA) (39), and α1-tryptase (PDB codes 1LTO and 2F9N) (40, 41) and either Na+ free or mutant forms of thrombin (PDB codes 2AFQ, 1RD3, and 1TQ0) (42–44). All of these enzymes are described as having reduced catalytic activity; in the case of thrombin, enzyme forms which show the occluded pocket are severely catalytically impaired compared with non-occluded enzyme forms (42–44), presumably because this loop position interferes with substrate binding. To better understand how prostasin recognizes substrate and to facilitate structure-based inhibitor design, the structure of prostasin was solved bound to the inhibitor nafamostat (23). Nafamostat mesylate acts as a slow substrate of many serine proteases; it reacts to form an acyl-enzyme intermediate, with a covalent bond between the catalytic serine and the resulting 4-guanidinobenzoic acid followed by a slower deacylation step (23). The structure of the acyl-enzyme intermediate of prostasin shows that the loop shifts to reveal the S1 site (Fig. 6A). Movement of the loop is tethered on the ends by Trp-258 and the disulfide bonded Cys-262 (chymotrypsin residues Trp-215 and Cys-220). Upon inhibitor binding, the Cα of Asp-260 (chymotrypsin Ser-217) shifts by 5.4 Å. This accompanies a rotation of the side chain away from the active site, shifting the carboxylate oxygens by ∼10 Å. In the apoprotein structure of prostasin, Asp-260 makes hydrogen bonds to a network of water molecules adjacent to the catalytic serine.
FIGURE 6.
A, Cα traces of the prostasin apoprotein and nafamostat-inhibited structures showing the shift of the loop over the S1 substrate site. The apoprotein form is shown in dark gray with the loop shown in red. The nafamostat-inhibited structure is shown in light gray with the loop shown in purple. Asp-260 is shown in stick representation. The Cα atom of Asp-260 shifts by 5.4 Å upon binding to nafamostat. The side chain of Asp-260, which coordinates a network of hydrogen bonded water molecules near the catalytic serine, swings away from the active site upon binding. The catalytic residues are shown for reference. The surface of the nafamostat-bound form of prostasin is illustrated, indicating the location of the S1 site as a cavity near the center. B, hydrogen bonds made by bound nafamostat. The terminal nitrogens of the covalently attached 4-guanidinobenzoic acid form hydrogen bonds with the carboxyl oxygens of Asp-232. Additional hydrogen bonds are formed with the carbonyl oxygens of Ala-233 and Arg-267. The carbonyl oxygen of Asp-260 also makes an additional hydrogen bond with the guanidinium group. C, the electron density maps from the nafamostat-inhibited acyl-enzyme intermediate crystal. Shown in green is the difference density (Fo - Fc) map contoured at 3σ, calculated in the absence of bound 4-guanidinobenzoic acid. The final electron density map (2Fo - Fc), contoured at 1σ, is shown in blue. The density for the guanidinium group is well defined, illustrating the conformational constraints resulting from the hydrogen bonds made to the protein.
For chymotrypsin-like serine proteases, the specificity for the P1 position of the substrate is primarily determined by a residue at the bottom of the S1 pocket (analogous to Ser-189 in chymotrypsin). In prostasin the equivalent residue is Asp-232, which is conserved in proteases characteristic of the trypsin family. This aspartate confers specificity toward lysine and arginine in the P1 position of substrates. In the structure of the 4-guanidinobenzoate ester adduct of prostasin, the terminal guanidinium group of the inhibitor forms hydrogen bonds with both carboxylate oxygens of Asp-232 (Fig. 6B). Additional hydrogen bonds to the inhibitor are provided by the carbonyl oxygens of Ala-233, Asp-260, and Arg-267. As shown in Fig. 6C, the electron density surrounding nafamostat is clearly defined, suggesting that the rotational flexibility of the guanidinium group is constrained by multiple hydrogen bonds. Shipway et al. (18) have extensively characterized prostasin substrate specificity. They have found that prostasin prefers an arginine or lysine in the P1 position. The structure of the acyl-enzyme intermediate suggests that an arginine may be preferred over lysine in the P1 position. The guanidinium moiety of nafamostat makes numerous hydrogen bonds to prostasin, and the arginine Nε may similarly hydrogen-bond to the carbonyl oxygen of Asp-260.
Shipway et al. (18) also have found that metal ions regulate prostasin activity, and divalent cations show more potent inhibition. Based on the movement of the loop from comparisons between the prostasin structures in this study, one may speculate that a mechanism for the previously described divalent cation-mediated regulation may be to affect the energetic favorability for the loop conformation in which the loop moves to block or expose the S1 pocket.
The prostasin structures presented here allow a detailed understanding of substrate specificity that was not available by just sequence alignment. Substrates can be modeled to determine key interactions that determine specificity as well as providing a basis for designing inhibitors. The presence of the blocked S1 subsite in the apoprotein structure reveals a potential mechanism for regulating prostasin activity. The structure of prostasin opens up the opportunity to develop specific inhibitors and chemical probes to further understand and regulate its role in controlling sodium channels in normal and disease states.
Supplementary Material
Acknowledgments
While this manuscript was under revision an article describing inhibitor-bound crystals of prostasin became available electronically (Tully, D. C., Vidal, A., Chatterjee, A. K., Williams, J. A., Roberts, M. J., Petrassi, H. M., Spraggon, G., Bursulaya, B., Pacoma, R., Shipway, A., Schumacher, A. M., Danahaym, H., and Harris, J. L. (2008) Bioorg. Med. Chem. Lett., in press) (PDB codes 3E16 and 3E0P).
The atomic coordinates and structure factors (codes 3DFJ and 3DFL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Tables 1 and 2, and Figs. 1–3.
Footnotes
The abbreviations used are: ENaC, epithelial sodium channel; MS, mass spectrometry; SEC, size-exclusion chromatography; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; Bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1.Yu, J. X., Chao, L., and Chao, J. (1994) J. Biol. Chem. 269 18843-18848 [PubMed] [Google Scholar]
- 2.Yu, J. X., Chao, L., and Chao, J. (1995) J. Biol. Chem. 270 13483-13489 [DOI] [PubMed] [Google Scholar]
- 3.Chen, L. M., Skinner, M. L., Kauffman, S. W., Chao, J., Chao, L., Thaler, C. D., and Chai, K. X. (2001) J. Biol. Chem. 276 21434-21442 [DOI] [PubMed] [Google Scholar]
- 4.Tong, Z., Illek, B., Bhagwandin, V. J., Verghese, G. M., and Caughey, G. H. (2004) Am. J. Physiol. Lung Cell. Mol. Physiol. 287 928-935 [DOI] [PubMed] [Google Scholar]
- 5.Bruns, J. B., Carattino, M. D., Sheng, S., Maarouf, A. B., Weisz, O. A., Pilewski, J. M., Hughey, R. P., and Kleyman, T. R. (2007) J. Biol. Chem. 282 6153-6160 [DOI] [PubMed] [Google Scholar]
- 6.Andreasen, D., Vuagniaux, G., Fowler-Jaeger, N., Hummler, E., and Rossier, B. C. (2006) J. Am. Soc. Nephrol. 17 968-976 [DOI] [PubMed] [Google Scholar]
- 7.Vuagniaux, G., Vallet, V., Jaeger, N. F., Pfister, C., Bens, M., Farman, N., Courtois-Coutry, N., Vandewalle, A., Rossier, B. C., and Hummler, E. (2000) J. Am. Soc. Nephrol. 11 828-834 [DOI] [PubMed] [Google Scholar]
- 8.Vuagniaux, G., Vallet, V., Jaeger, N. F., Hummler, E., and Rossier, B. C. (2002) J. Gen. Physiol. 120 191-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallet, V., Chraibi, A., Gaeggeler, H. P., Horisberger, J. D., and Rossier, B. C. (1997) Nature 389 607-610 [DOI] [PubMed] [Google Scholar]
- 10.Adachi, M., Kitamura, K., Miyoshi, T., Narikiyo, T., Iwashita, K., Shiraishi, N., Nonoguchi, H., and Tomita, K. (2001) J. Am. Soc. Nephrol. 12 1114-1121 [DOI] [PubMed] [Google Scholar]
- 11.Garty, H., and Palmer, L. G. (1997) Physiol. Rev. 77 359-396 [DOI] [PubMed] [Google Scholar]
- 12.Snyder, P. M. (2005) Endocrinology 146 5079-5085 [DOI] [PubMed] [Google Scholar]
- 13.Donaldson, S. H., and Boucher, R. C. (2007) Chest 132 1631-1636 [DOI] [PubMed] [Google Scholar]
- 14.Greig, E. R., Boot-Handford, R. P., Mani, V., and Sandle, G. I. (2004) J. Pathol. 204 84-92 [DOI] [PubMed] [Google Scholar]
- 15.List, K., Hobson, J. P., Molinolo, A., and Bugge, T. H. (2007) J. Cell. Physiol. 213 237-245 [DOI] [PubMed] [Google Scholar]
- 16.Netzel-Arnett, S., Currie, B. M., Szabo, R., Lin, C. Y., Chen, L. M., Chai, K. X., Antalis, T. M., Bugge, T. H., and List, K. (2006) J. Biol. Chem. 281 32941-32945 [DOI] [PubMed] [Google Scholar]
- 17.Chobanian, A. V., Bakris, G. L., Black, H. R., Cushman, W. C., Green, L. A., Izzo, J. L., Jr., Jones, D. W., Materson, B. J., Oparil, S., Wright, J. T., Jr., and Roccella, E. J. (2003) J. Am. Med. Assoc. 289 2560-2572 [DOI] [PubMed] [Google Scholar]
- 18.Shipway, A., Danahay, H., Williams, J. A., Tully, D. C., Backes, B. J., and Harris, J. L. (2004) Biochem. Biophys. Res. Commun. 324 953-963 [DOI] [PubMed] [Google Scholar]
- 19.Congreve, M., Murray, C. W., and Blundell, T. L. (2005) Drug Discov. Today 10 895-907 [DOI] [PubMed] [Google Scholar]
- 20.Abbenante, G., and Fairlie, D. P. (2005) Med. Chem. 1 71-104 [DOI] [PubMed] [Google Scholar]
- 21.Leung, D., Abbenante, G., and Fairlie, D. P. (2000) J. Med. Chem. 43 305-341 [DOI] [PubMed] [Google Scholar]
- 22.Williams, S. P., Kuyper, L. F., and Pearce, K. H. (2005) Curr. Opin. Chem. Biol. 9 371-380 [DOI] [PubMed] [Google Scholar]
- 23.Ramjee, M. K., Henderson, I. M., McLoughlin, S. B., and Padova, A. (2000) Thromb. Res. 98 559-569 [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 25.McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C., and Read, R. J. (2005) Acta Crystallogr. D Biol. Crystallogr. 61 458-464 [DOI] [PubMed] [Google Scholar]
- 26.Tang, J., Yu, C. L., Williams, S. R., Springman, E., Jeffery, D., Sprengeler, P. A., Estevez, A., Sampang, J., Shrader, W., Spencer, J., Young, W., McGrath, M., and Katz, B. A. (2005) J. Biol. Chem. 280 41077-41089 [DOI] [PubMed] [Google Scholar]
- 27.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 28.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 29.Kleywegt, G. J., and Read, R. J. (1997) Structure 5 1557-1569 [DOI] [PubMed] [Google Scholar]
- 30.DeLano, W. L. (2002) The Pymol Molecular Graphics System Delano Scientific, Palo Alto, CA
- 31.Laskowski, R. A., Moss, D. S., and Thornton, J. M. (1993) J. Mol. Biol. 231 1049-1067 [DOI] [PubMed] [Google Scholar]
- 32.Hearn, M. T. W. (2002) in HPLC of Biological Macromolecules (Gooding, K. M., and Regnier, F. E., eds) pp. 99-246, Marcel Dekker, Inc., New York
- 33.Corran, P. H. (1989) in HPLC of Macromolecules: A Practical Approach (Oliver, R. W. A., ed) pp. 127-156, IRL Press at Oxford University Press, Oxford
- 34.al-Obeidi, A. M., and Light, A. (1988) J. Biol. Chem. 263 8642-8645 [PubMed] [Google Scholar]
- 35.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 25 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czapinska, H., Helland, R., Smalas, A. O., and Otlewski, J. (2004) J. Mol. Biol. 344 1005-1020 [DOI] [PubMed] [Google Scholar]
- 37.Herter, S., Piper, D. E., Aaron, W., Gabriele, T., Cutler, G., Cao, P., Bhatt, A. S., Choe, Y., Craik, C. S., Walker, N., Meininger, D., Hoey, T., and Austin, R. J. (2005) Biochem. J. 390 125-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carvalho, A. L., Sanz, L., Barettino, D., Romero, A., Calvete, J. J., and Romao, M. J. (2002) J. Mol. Biol. 322 325-337 [DOI] [PubMed] [Google Scholar]
- 39.Hink-Schauer, C., Estebanez-Perpina, E., Wilharm, E., Fuentes-Prior, P., Klinkert, W., Bode, W., and Jenne, D. E. (2002) J. Biol. Chem. 277 50923-50933 [DOI] [PubMed] [Google Scholar]
- 40.Rohr, K. B., Selwood, T., Marquardt, U., Huber, R., Schechter, N. M., Bode, W., and Than, M. E. (2006) J. Mol. Biol. 357 195-209 [DOI] [PubMed] [Google Scholar]
- 41.Marquardt, U., Zettl, F., Huber, R., Bode, W., and Sommerhoff, C. (2002) J. Mol. Biol. 321 491-502 [DOI] [PubMed] [Google Scholar]
- 42.Pineda, A. O., Chen, Z. W., Caccia, S., Cantwell, A. M., Savvides, S. N., Waksman, G., Mathews, F. S., and Di Cera, E. (2004) J. Biol. Chem. 279 39824-39828 [DOI] [PubMed] [Google Scholar]
- 43.Johnson, D. J., Adams, T. E., Li, W., and Huntington, J. A. (2005) Biochem. J. 392 21-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carter, W. J., Myles, T., Gibbs, C. S., Leung, L. L., and Huntington, J. A. (2004) J. Biol. Chem. 279 26387-26394 [DOI] [PubMed] [Google Scholar]
- 45.Tsukada, H., and Blow, D. M. (1985) J. Mol. Biol. 184 703-711 [DOI] [PubMed] [Google Scholar]
- 46.Fehlhammer, H., and Bode, W. (1975) J. Mol. Biol. 98 683-692 [DOI] [PubMed] [Google Scholar]
- 47.Meyer, E., Cole, G., Radhakrishnan, R., and Epp, O. (1988) Acta Crystallogr. B 44 26-38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.