Abstract
ATP drives the conformational change of the group II chaperonin from the open lid substrate-binding conformation to the closed lid conformation to encapsulate an unfolded protein in the central cavity. The detailed mechanism of this conformational change remains unknown. To elucidate the intra-ring cooperative action of subunits for the conformational change, we constructed Thermococcus chaperonin complexes containing mutant subunits in an ordered manner and examined their folding and conformational change abilities. Chaperonin complexes containing wild-type subunits and mutant subunits with impaired ATP-dependent conformational change ability or ATP hydrolysis activity, one by one, exhibited high protein refolding ability. The effects of the mutant subunits correlate with the number and order in the ring. In contrast, the use of a mutant lacking helical protrusion severely affected the function. Interestingly, these mutant chaperonin complexes also exhibited ATP-dependent conformational changes as demonstrated by small angle x-ray scattering, protease digestion, and changes in fluorescence of the fluorophore attached to the tip of the helical protrusion. However, their conformational change is likely to be transient. They captured denatured proteins even in the presence of ATP, whereas addition of ATP impaired the ability of the wild-type chaperonin to protect citrate synthase from thermal aggregation. These results suggest that ATP binding/hydrolysis causes the independent conformational change of the subunit, and further conformational change for the complete closure of the lid is induced and stabilized by the interaction between helical protrusions.
Chaperonins, a ubiquitous class of molecular chaperones, are double-ring assemblies of about 60-kDa subunits. Each ring has a large central cavity in which a non-native protein can undergo productive folding in an ATP-dependent manner (1, 2). Chaperonin complexes change their conformations to close their chamber and encapsulate the bound substrate, providing a protected environment for protein folding. Opening and closing of the folding chamber is controlled by a conformational cycle driven by ATP binding and hydrolysis. Chaperonins are divided in two groups as follows: group I and group II chaperonins. Group I chaperonins are found in bacteria and endosymbiotic organelles (mitochondria and chloroplasts), and subsets of bacterially related homologs in methanogens (3). On the other hand, group II chaperonins are found in archaea (known as thermosome) (4) and the eukaryotic cytosol (chaperonin-containing t-complex polypeptide-1 or TCP-1 ring complex) (1, 5). Whereas group I chaperonins are homo-oligomeric, eukaryotic and archaeal group II chaperonins are generally hetero-oligomeric (6). All chaperonins share a similar subunit architecture consisting of three distinct domains as follows: an ATP-binding equatorial domain, a distal apical domain harboring the polypeptide-binding sites, and an intermediate hinge domain (7, 8). The most striking structural difference between the two groups is the lid of the chaperonin cavity. The co-chaperonin (GroES in Escherichia coli) acts as a detachable lid for group I chaperonins by forming a heptameric ring-shaped structure. It interacts with one or both GroEL rings in an ATP-dependent fashion, thereby sealing the cavity from the outside (1, 2, 9). In contrast, group II chaperonins have a built-in lid, which is composed of an extension of the apical domain called the helical protrusion (7, 8, 10). The helical protrusion plays the equivalent role of GroES, sealing off the central cavity of the chaperonin complex (11–13). ATP binding triggers the conformational change of archaeal group II chaperonins (11, 14). ATP binding, mimicked by the nonhydrolysable analog AMP-PNP,7 also induced the conformational change of chaperonin-containing t-complex polypeptide-1 to close off the cavity (15). There are also reports on the requirements of ATP hydrolysis for the conformational transitions of group II chaperonins (13, 16). The discrepancy between these results is likely to be caused by some experimental procedures. It might be plausible that the conformational change occurs at the transition state of ATP hydrolysis.
The helical protrusion is not required for substrate binding but is indispensable for ATP-dependent conformational transitions and protein folding (16, 17). Similar to the group I chaperonin (16, 18–20), both archaeal and eukaryotic group II chaperonins demonstrate positive and negative cooperative action with ATP within a ring and between the two rings, respectively (16, 18, 21, 22). The allosteric conformational changes in eukaryotic group II chaperonin are found to spread around the ring in a sequential manner, whereas allosteric transitions in the group I chaperonin GroEL occur in a concerted manner (18, 23, 24). However, despite these findings, the detailed mechanism of this cooperative action in ATP binding/hydrolysis or between helical protrusions for the ATP-dependent conformational changes of group II chaperonins remains obscure. To elucidate these issues, we investigated the details of the intraring cooperativity for ATP-dependent conformational change using a hyperthermophilic archaeal group II chaperonin from Thermococcus sp. strain KS-1 (25). Previously, group I chaperonins composed of various combinations of wild-type and mutated subunits were constructed by covalent assembly of seven subunits (26). Similar to this construction, we constructed chaperonin complexes composed of wild-type and mutant subunits. Analyzing these chaperonin complexes, we demonstrated that ATP binding/hydrolysis causes the independent conformational change of the subunit, and further conformational change for the complete closure of the lid is induced by the interaction between helical protrusions.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, Reagents, and Protein—E. coli strains used in this study were DH5α for plasmid preparation and BL21 Star (DE3) (Invitrogen) for protein expression. KOD-plus-DNA polymerase and restriction endonucleases were obtained from Toyobo Co. Ltd. (Osaka, Japan) and Takara Bio Inc. (Shiga, Japan), respectively. ATP, ADP, and thermolysin were purchased from Wako Pure Chemical Industries (Osaka, Japan). Citrate synthase from porcine heart and Thermoplasma acidophilum were obtained from Sigma. The ammonium sulfate suspension of CS from porcine heart was desalted on an NAP-5 column (GE Healthcare), as described previously (27), before use. ADP was treated with hexokinase (Hoffmann-La Roche) plus glucose to remove contaminating ATP before use as described previously (28). The concentrations of chaperonins were determined with the Bio-Rad protein assay (Bio-Rad) using bovine serum albumin as the standard, and they are expressed as molar concentrations of hexadecamer.
Construction and Purification of Covalently Linked Chaperonins—To express covalently linked chaperonins, three types of fragments containing a wild-type or mutant chaperonin subunit gene were prepared by PCR. The initiation fragment, type “a” fragment, encodes a chaperonin subunit with initiation codon but without termination codon, which contains the NdeI and BamHI digestion sites. It was obtained using the primer pair (forward A, 5′-CAT-ATG-GCA-CAG-CTT-AGT-GGA-CAG-CCG-G-3′, and reverse A, 5′-CGG-GAT-CCA-TTA-CCA-CGA-GGA-TAC-ATG-CCC-ATT-CCG-CCG-GG-3′). Forward A primer contains the N-terminal sequence with an initiation codon in the NdeI site. Reverse A primer corresponds to the C-terminal sequence without the translation termination codon and contains the sequence for the thrombin digestion site and BamHI digestion site. The intermediate fragment, type “b” fragment, is the connecting fragment lacking both initiation codon and termination codon. It is obtained by PCR using the primer pair (forward B, 5′-GAA-GAT-CTG-CAC-AGC-TTA-GTG-GAC-3′, and reverse A). Forward B primer corresponds to the N-terminal sequence with a replacement for the insertion of BglII site and removal of the initiation codon. The terminal fragment, type “c” fragment, is for the C-terminal subunit, which is amplified with the primer pair (forward B and reverse B, 5′-GGA-TCC-TCA-CAT-GCC-CAT-GTC-CAT-TCC-GCC-3′). Reverse B primer contains the C-terminal sequence with termination codon and BamHI site.
The amplified fragments were subcloned into the pT7Blue T-vector (Novagen, Madison, WI) and their sequences were verified. The inserted DNA fragments were excised with NdeI/BamHI (type a) or BglII/BamHI (type b and c) and then purified for further use. For construction of two-, four-, and eight-subunit linked chaperonin complexes, the fragments were inserted into the NdeI/BamHI site of pET9a (Novagen) in the order of a–c, a–b–b–c, and a–b–b–b–b–b–b–c, respectively. At first, the a fragment was inserted into the NdeI/BamHI site of pET9a. Then the type b or type c BglII/BamHI fragment was ligated into the BamHI site. After the confirmation of the orientation of the gene, the next fragment was inserted into the BamHI site, which is located at the C terminus of the last subunit. Insertion of the fragment continues until the insertion of the type c fragment.
All constructed chaperonin complexes were purified as described previously (11). Subsequently, the purified chaperonins were incubated with thrombin protease (100 units/mg, GE Healthcare) at 22 °C for 40 h to digest the linker sequences. After digestion, thrombin was removed by size exclusion chromatography.
Protein Folding Assay—The GFP employed in this study is a thermostable mutant used in assays performed at 60 °C (29). It was expressed and purified as described previously (27, 30). GFP (5 μm) was denatured in TKM buffer (50 mm Tris-HCl, pH 7.5, 100 mm KCl, and 25 mm MgCl2) containing 5 mm dithiothreitol and 0.1 m HCl at room temperature, and it was diluted 100-fold in the preincubated TKM folding buffer with or without chaperonin (100 nm) at 60 °C for 10 min. Nucleotide was added to the mixture to a final concentration of 1 mm 5 min after dilution of denatured GFP. The fluorescence of GFP at 510 nm with excitation at 396 nm was continuously monitored for 20 min using a spectrofluorometer (FP-6500, Jasco, Tokyo, Japan). The reaction mixtures were continuously stirred at 60 °C throughout the assays. As a control, native GFP was diluted in the folding buffer without chaperonin. The fluorescence intensity of native GFP was taken as 100%.
CS from T. acidophilum was subjected to a refolding assay at 50 °C. T. acidophilum CS (19.8 μm, as a monomer) was denatured in 50 mm HEPES-KOH, pH 7.5, containing 6 m guanidine hydrochloride and 5 mm dithiothreitol for 60 min at 50 °C and then diluted 60-fold in the dilution buffer (50 mm HEPES-KOH, pH 7.5, 50 mm MgCl2, and 300 mm KCl) in the absence or presence of chaperonins (0.5 μm). The refolding reactions were conducted for 60 min at 50 °C. 1 mm ATP was added to the mixture 10 min after the dilution. After the refolding reactions, aliquots were removed from the mixture, and the recovered enzyme activity was assayed as described by Furutani et al. (31). The activity of enzyme refolded by wild-type chaperonin was taken as 100%.
ATPase Activity Measurement—ATPase activities were measured at 60 °C in TKM buffer containing 1 mm ATP and 50 μg/ml chaperonin. The amount of Pi produced was measured using the malachite green assay as described previously (30).
SAXS Measurement—The SAXS experiments were performed with beamline 15A at the Photon Factory of the High Energy Accelerator Research Organization, Tsukuba, Japan. Measurements were obtained with protein concentrations of 5 mg/ml in TNM buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, and 25 mm MgCl2) at 60 °C. Before data collection, the protein solutions were incubated with or without nucleotides (1 mm) at 60 °C. Scattering patterns were recorded by a CCD-based x-ray detector, which consisted of a beryllium-windowed x-ray image intensifier (Be-XRII) (V5445P-MOD, Hamamatsu Photonics, Hamamatsu, Japan), an optical lens, a CCD image sensor, and a data acquisition system (C7300, Hamamatsu Photonics), as described previously (32, 33). The experimental details and the analyses of the scattering data were as described (32). Pair distribution (P(r)) functions were calculated using the GNOM program (34). The values for radius of gyration (Rg) and maximum particle distance (Dmax) were derived from the P(r) function.
Protease Sensitivity Assay—Chaperonin (100 nm) was incubated with or without nucleotide (1 mm) at 60 °C in TKM buffer under continuous mixing. Digestion with thermolysin (1 ng/μl) was carried out for 10 min at 60 °C. Proteins in the reaction mixture were precipitated by the addition of trichloroacetic acid and then analyzed on 10% SDS gels. Gels were stained with Coomassie Brilliant Blue R-250.
Fluorescence Intensity Assay—The chaperonin complexes were labeled with fluorescein 5-maleimide (Invitrogen). The fluorescence spectra were measured at 60 °C with Jasco FP-6500 spectrofluorometer. Chaperonin complexes (50 nm) in TKM buffer were preincubated with or without nucleotide (1 mm) at 60 °C. The excitation wavelength was set at 493 nm, and emission was recorded at wavelengths ranging from 500 to 650 nm. All fluorescence intensity spectra were independently analyzed three times.
Fluorescence Polarization Assay—The chaperonin complexes prepared for fluorescence intensity assay were labeled with tetramethylrhodamine-5-maleimide (TMR, Invitrogen). This assay was measured at 60 °C with a Jasco FP-6500 spectrofluorometer equipped with a fluorescence polarization unit (Jasco). TMR-labeled chaperonin complexes (50 nm) in TKM buffer were preincubated with or without nucleotide (1 mm) at 60 °C. The excitation wavelength was set at 552 nm, and the emission was set at 570 nm, which is the maximum of emission intensity of TMR. Fluorescence polarization of TMR-labeled chaperonins was analyzed by a time course assay for 60 s. All fluorescence polarization assays were independently carried out three times.
Thermal Aggregation Measurement—Thermal aggregation of CS from the porcine heart was monitored by measuring light scattering at 500 nm with a spectrofluorometer (FP-6500) for 20 min at 50 °C. Native CS was diluted to a final concentration of 100 nm (as a monomer) in TNM buffer with or without chaperonin (25 nm) and 1 mm nucleotide. The reaction mixtures were preincubated for 10 min at 50 °C and continuously stirred throughout the measurement.
RESULTS
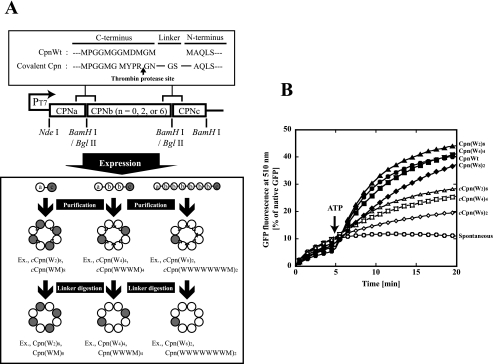
Construction of Chaperonin Complexes Composed of Wild-type and Mutated Subunits in the Defined Order—Thermococcus sp. strain KS-1 chaperonin is a double-ring complex that contains eight subunits per ring. The crystal structure of Thermococcus sp. strain KS-1 chaperonin has shown that the resolvable N-terminal Val-9 residue of one subunit and C-terminal Ala-526 residue of the next subunit are closely located in the central cavity (8). Previously, we have constructed two- or four-subunit covalently linked Thermococcus sp. strain KS-1 chaperonins by connecting the N-terminal Val-9 residue to the C-terminal Ala-526 residue. Although these chaperonins formed proper double-ring structures capable of capturing unfolded proteins and ATPase activity, they lacked ATP-dependent protein folding activity (35). We attributed the loss of folding activity to the decreased flexibility of the N and C termini subunits necessary for the ATP-dependent conformational change. We then tried to obtain active chaperonin complexes by specific digestion of the linker sequence. The C-terminal amino acid sequence of the wild-type subunit was substituted with the thrombin recognized sequence (MYPR↓GN), and then two, four, and eight subunits were covalently connected with flexible linkers (Gly-Ser) (Fig. 1A). These covalently linked chaperonins were expressed and purified in the same manner as the wild type (11). The chaperonin complexes composed of covalently linked dimers, tetramers, and octamers were designated as cCpn(W2)8, cCpn(W4)4, and cCpn(W8)2, respectively. cCpn(W2)8 designates a connected (c) chaperonin hexadecameric complex composed of eight doubly connected wild-type subunits. They were then subjected to thrombin digestion at 22 °C for 40 h, and then the thrombin was removed by size exclusion chromatography. Size exclusion chromatography analysis showed that the prepared linker-digested chaperonin complexes (named as Cpn(W2)8, Cpn(W4)4, and Cpn(W8)2, respectively) retained proper oligomeric structures similar to that of the wild-type chaperonin, CpnWT (data not shown). By SDS-PAGE, cCpn(W2)8 and cCpn(W4)4 appeared as bands of two and four times the size of a chaperonin subunit. They changed to bands of the size corresponding to the chaperonin subunit monomer after thrombin digestion (data not shown). In contrast, cCpn(W8)2 did not appear as a single band of eight times the size of the single subunit, but after thrombin digestion, a single band corresponding to the molecular weight for one chaperonin subunit was detected. The cCpn(W8)2 was likely digested around the thrombin digestion site by the endogenous proteases in E. coli to various sizes of connected chaperonins. GFP folding activities of all covalently linked chaperonins were lower than that of CpnWT. This activity was recovered to the same level as that of the wild-type by thrombin digestion. Cpn(W8)2 also showed protein folding activity, but it was lower than the others (Fig. 1B).
FIGURE 1.
Expression and characterization of covalently linked chaperonin complexes. A, construction of expression plasmids for covalently linked chaperonins. Only a single ring of chaperonin complexes is shown. The chaperonin wild-type subunit, mutant subunit, mutant subunit with G65C replacement, mutant subunit with D64A/D393A replacements, and mutant subunit with helical protrusion deletions were designated as “W,” “M,” “C,” “A,” and “H”. The chaperonin complex was named as follows. The chaperonin hexadecameric complex constructed with the connection of one wild-type and one mutant subunit is named as cCpn(WM)8 and Cpn(WM)8 before and after thrombin digestion, respectively. The details for construction were described under “Experimental Procedures.” Single-ring chaperonin icons are shown as the structural image of the chaperonin complexes. Open and closed circles represent wild-type and mutant subunits, respectively. B, GFP folding assay for chaperonin complexes. The folding mixture was incubated at 60 °C as described under “Experimental Procedures.” The recovery of GFP fluorescence was continuously monitored at 510 nm. At 0 min, acid-denatured GFP (5 μm) was diluted 100-fold in the folding buffer with or without 100 nm wild-type and mutant chaperonins (open circles, spontaneous (without chaperonin); closed circles, CpnWT; open triangles, cCpn(W2)8; closed triangles, Cpn(W2)8; open squares, cCpn(W4)4; closed squares, Cpn(W4)4; open diamonds, cCpn(W8)2; closed diamonds, Cpn(W8)2). Five minutes after dilution, 1 mm ATP was added. The amount recovered is expressed as a percentage of the fluorescence intensity of native GFP.
Using the two- and four-subunit connection system, we constructed various chaperonins composed of wild-type and mutant subunits in the defined order to investigate the molecular mechanism of the cooperative actions between subunits with regard to protein folding activity as well as the ATP-dependent conformational change of group II chaperonins (Fig. 1A). We used the mutants impaired in ATP-dependent conformational change ability (G65C) (designated as C) and ATP hydrolysis activity (D64A/D393A) (designated as A), and also the helical protrusion-deleted mutant Δ245–276 (Δhelical) (designated as H). The ATP hydrolysis activity of the homo-oligomer of G65C, CpnG65C, was almost twice of CpnWT, but conformational change was not induced by the ATP binding signal (30). The homo-oligomer of D64A/D393A, CpnD64A/D393A, showed only trace ATP hydrolysis activity and lacked protein folding ability. The homo-oligomer of Δ245–276, CpnΔhelical, did not show any ATP-dependent conformational change and was also deficient in protein folding activity in spite of the fact that it had ATPase activity and was capable of capturing an unfolded protein (17).
Connected chaperonin complexes containing wild-type and mutant subunits were constructed and then subjected to thrombin digestion. The chaperonin complex containing wild-type and G65C one by one, which was designated as Cpn(WC)8, was prepared from the connected complex of the wild-type and G65C. The subscript 8 designates that 8 units of WC (wild-type and G65C) are contained in a hexadecameric chaperonin complex. The complex containing W and C one by one in the same manner prepared from the four connected complexes was named Cpn(WCWC)4. The other chaperonin complexes composed of wild-type and mutant subunits were designated in the same manner.
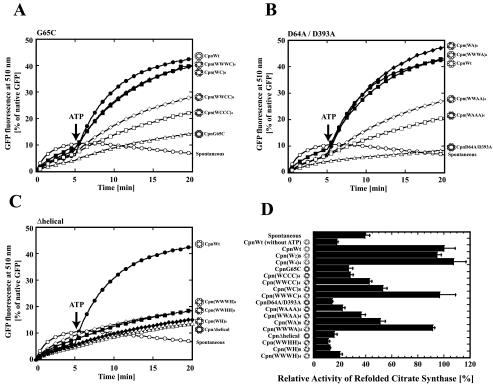
Functional Characterization of Chaperonin Hetero-complexes Containing G65C or D64A/D393A—To examine cooperative action in ATP binding and hydrolysis between subunits in the ring of group II chaperonins, Cpn(WC)8, Cpn(WWCC)4, Cpn(WCCC)4, Cpn(WWWC)4, Cpn(WA)8, Cpn(WWAA)4, Cpn(WAAA)4, and Cpn(WWWA)4 were constructed and characterized. The protein folding activities of these chaperonins were assessed using GFP (27 kDa) and citrate synthase from T. acidophilum (42 kDa) as substrates. The CpnWT arrests spontaneous folding of GFP and refolds it in an ATP-dependent manner, whereas CpnG65C and CpnD64A/D393A cannot refold GFP (Fig. 2, A and B). According to the increase of the number of wild-type subunits in a ring, GFP refolding activity of the chaperonin complexes increased. Although chaperonin complexes containing only two wild-type subunits in a ring (Cpn(WCCC)4 and Cpn(WAAA)4) exhibited marginal GFP folding activity, Cpn(WWWC)4 and Cpn(WWWA)4 possessed almost the same GFP folding activity as the wild-type complex (CpnWT). Interestingly, the order of the mutant subunits also affected the refolding activity. The chaperonin complexes containing the wild-type and mutant subunits alternately (Cpn(WC)8 and Cpn(WA)8) showed higher GFP refolding activity than the mutant complexes containing two mutant subunits adjacently (Cpn(WWCC)4 and Cpn(WWAA)4). Similar results were observed in the T. acidophilum CS folding assay (Fig. 2D). Consequently, these results indicate that mutant subunits (G65C and D64A/D393A), which are impaired in ATP-dependent conformational change activity, can participate in protein folding in response to the effects of contiguous wild-type subunits.
FIGURE 2.
Protein refolding activities of chaperonin complexes composed of wild-type and mutant subunits. GFP folding assay of chaperonin complexes composed of wild-type and G65C (A), wild-type and D64A/D393A (B), or wild-type and Δhelical (C) subunits. This experiment was carried out as described in Fig. 1B. Open circles, spontaneous (without chaperonin); closed circles, CpnWT; open triangles, homo-oligomer of the mutant; closed squares, Cpn(WWWM)4; closed diamonds, Cpn(WM)8; open diamonds, Cpn(WWMM)4; open squares, Cpn(WMMM)4. M designates the mutant subunit. D, thermoplasma citrate synthase refolding assay of the designed chaperonin complexes. The activities of T. acidophilum CS refolded spontaneously or by the wild-type and mutant chaperonin complexes are shown. The folding mixture was incubated at 50 °C as described under the “Experimental Procedures.” Denatured T. acidophilum CS (19.8 μm, monomer) was diluted 60-fold in dilution buffer in the absence or presence of 0.5 μm chaperonins. The refolding reactions were performed for 60 min at 50 °C, and recovered enzyme activity was measured at the indicated time points. Two millimolar ATP was added 10 min after the dilution. The reaction was terminated at 70 min, and the activity of refolded enzyme was measured. The activity of the enzyme refolded by wild-type chaperonin was taken as 100% (n = 3). Wild-type and mutant subunit are shown as open and closed circles in the structural images as Fig. 1A.
We then compared the ATPase activities of Cpn(WC)8 and Cpn(WA)8 with those of CpnWT, CpnG65C, and CpnD64A/D393A. The relative ATP hydrolysis activities of CpnG65C, Cpn(WC)8, CpnD64A/D393A, and Cpn(WA)8 compared with CpnWT were 226, 148, 3, and 51%, respectively. Thus, the ATPase activities of Cpn(WC)8 and Cpn(WA)8 were almost half that of the wild-type and the corresponding mutant homo-oligomers, suggesting that ATP hydrolysis of subunits is independent among subunits.
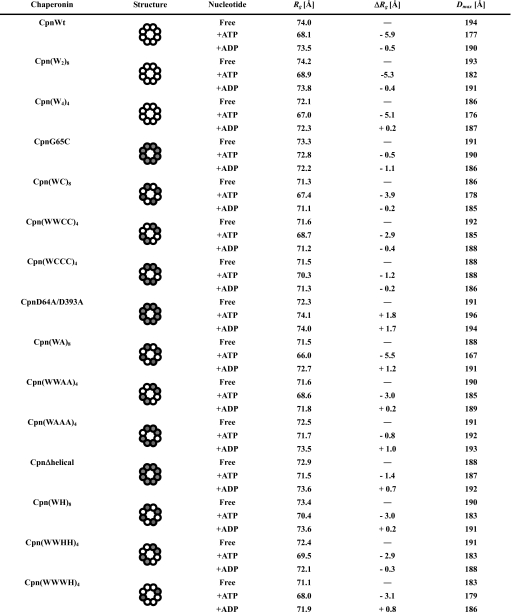
ATP-dependent Conformational Change of Chaperonin Hetero-complexes Containing G65C and D64A/D393A—Protein folding by group II chaperonins tightly correlates with ATP-dependent conformational changes. We investigated the ATP-dependent conformational changes of the mutant complexes by means of small angle x-ray scattering (SAXS). SAXS gives the size of the molecule by two structural parameters, radius of gyration (Rg) and the maximum particle distance (Rg). Rg indicates the mean particle size, and Dmax is the maximum particle distance or the maximal intramolecular distance. Although group II chaperonins form the open conformation in the absence of ATP, they change to the closed conformation by ATP binding or hydrolysis. CpnWT, Cpn(W2)8, and Cpn(W4)4 showed ATP-dependent change in Dmax and also Rg (Table 1). ATP-dependent changes in Rg were estimated to be -5.9, -5.3, and -5.1 Å (Table 1). In contrast, CpnG65C and CpnD64A/D393A did not exhibit significant changes in Dmax and Rg (Table 1). We then examined the conformational change in chaperonin complexes containing mutants (Cpn(WC)8, Cpn(WWCC)4, Cpn(WCCC)4, Cpn(WA)8, Cpn(WWAA)4, and Cpn(WAAA)4) (Table 1). Among them, only Cpn(WC)8 and Cpn(WA)8 exhibited a large ATP-dependent conformational change with Rg changes of -3.9 and -5.5 Å (Table 1), consistent with the protein folding activities. Cpn(WWCC)4 and Cpn(WWAA)4 showed minimal ATP-dependent Rg changes, reflecting their small protein folding activities. The ATP-dependent Rg changes of the mutant complexes decreased according to the increase in the number of mutant subunits in the ring (Table 1). These results demonstrate that the protein folding activities of the chaperonin complexes correlate with their abilities for the conformational change. The conformational change of the mutant subunit is likely to be induced by the action of the contiguous wild-type subunits in the hetero-oligomer.
TABLE 1.
Structural parameters of chaperonin complexes composed of wild-type and mutant subunits determined by small angle x-ray scattering experiments Wild-type and mutant subunits are shown as open and closed circles in the structural images. Error values of Δ radius of gyration (ΔRg) and maximum particle distance (Dmax) are estimated to be under ±0.5 Å and ±5%, respectively.
Requirement of All Helical Protrusions in the Ring for Protein Folding Function—The crystal structure of group II chaperonins illustrates that helical protrusions are located in close contact in the closed conformation (7, 8). We hypothesized that the ATP-dependent allosteric transitions in a ring are caused by the interaction between helical protrusions. To investigate this inter-subunit communication, we characterized various chaperonin complexes composed of wild-type and Δhelical, Cpn(WH)8, Cpn(WWHH)4, and Cpn(WWWH)4.
Although all these chaperonin complexes prevented spontaneous folding of the substrate proteins, they did not exhibit ATP-dependent folding activity for GFP and T. acidophilum CS (Fig. 2, C and D). Even the existence of only one Δhelical subunit in a ring severely affected protein folding activity (data not shown). Therefore, the helical protrusion has the essential role in ATP-dependent conformational change for protein folding.
Then the ATP-dependent transitions of chaperonin complexes containing helical protrusion deletion mutants were analyzed by SAXS. Unexpectedly, these chaperonin complexes showed ATP-dependent changes of Dmax and Rg (Table 1). Curiously, all chaperonin complexes containing Δhelical subunits exhibited almost the same ATP-dependent change of Rg values, about -3.0 Å, irrespective of the number or order of Δhelical subunits in a ring (Table 1). It is speculated that ATP induces the conformational change in the chaperonin complexes containing Δhelical subunits, which is likely to be insufficient for protein folding.
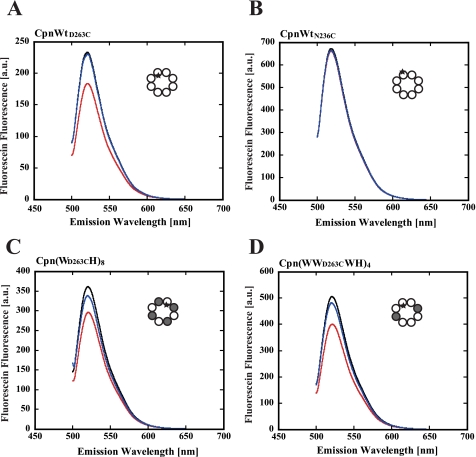
Fluorescence Analysis of the ATP-dependent Conformational Change of Chaperonin Complexes Containing Helical Protrusion Deletion Mutants—To examine whether Cpn(WH)8 and Cpn(WWWH)4 really change their conformations in an ATP-dependent manner, the effect of ATP on the fluorescence of fluorescein attached to the tip of the helical protrusion was examined. To introduce fluorescein at the specific site, Cys-366, the only one cysteine existing in the wild-type subunit, was changed to serine. Because the mutation did not affect the function of the chaperonin, subunits containing C366S mutation were used in the experiments using fluorophore-labeled chaperonins. CpnWT, Cpn(WH)8, and Cpn(WWWH)4 chaperonin complexes were prepared using the mutant subunit D263C to introduce a cysteine residue at the tip of the helical protrusion (CpnWTD263C, Cpn(WD263CH)8, and Cpn(WWD263CWH)4, respectively) (supplemental Fig. 1), and they were labeled with fluorescein. The labeling ratio was less than one fluorescein per one oligomer to minimize the effects of the attached fluorescein on the conformational change. Fluorescein-labeled CpnWTD263C (homo-oligomer of WTD263C) showed an ATP-dependent change in fluorescence intensity (Fig. 3A). The fluorescence peak at 521 nm decreased about 21% by the addition of ATP, but it was not affected by ADP. To examine if the change reflects the closure of the lid, another mutant, WTN236C, which has cysteine at the outside bottom of the helical protrusion (supplemental Fig. 1) was prepared. ATP and ADP did not affect the fluorescence of the fluorescein-labeled CpnWTN236C (homo-oligomer of WTN236C) (Fig. 3B). Thus, the observed fluorescence change in CpnWTD263C should reflect the environmental change of the fluorescein attached on the tip of helical protrusion. Consistent with the results of SAXS, ATP-dependent changes in fluorescence were observed for fluorescein-labeled Cpn(WD263CH)8 and Cpn(WWD263CWH)4 (Fig. 3, C and D). Therefore, we concluded that Cpn(WH)8 and Cpn(WWWH)4 change their conformation from the open lid to the closed lid state in an ATP-dependent manner.
FIGURE 3.
Fluorescence spectra change of fluorescein-labeled chaperonin complexes. The chaperonin complexes were labeled with fluorescein, and their fluorescence spectra were measured by excitation at 493 nm. A, fluorescein-labeled CpnWTD263C (homo-oligomer of WD263C); B, fluorescein-labeled CpnWTN236C (homo-oligomer of WN236C); C, fluorescein-labeled Cpn(WD263CH)8; and D, fluorescein-labeled Cpn(WWD263CWH)4. The experimental details are described under the “Experimental Procedures.” Black line, without addition of nucleotides; red line, with ATP; blue line, with ADP. Wild-type subunits, mutant subunits, and fluorophores are shown as open circles, closed circles, and asterisks in the structural images as Fig. 1A.
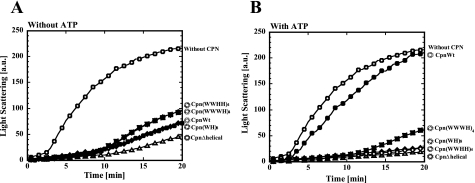
Closure of the Lid of Chaperonin Complexes Containing Helical Protrusion Mutants by ATP Is Transient—The closed conformation of Cpn(WH)8 and Cpn(WWWH)4 formed in the presence of ATP was examined by their ability to protect porcine heart CS from thermal aggregation (Fig. 4). In the absence of ATP, CpnWT protected CS from thermal aggregation. Similar effects were observed for CpnΔhelical, Cpn(WH)8, Cpn(WWHH)4, and (WWHH)4, and Cpn(WWWH)4 were maintained even in the presence of ATP like CpnΔhelical. Thus, the closure of the lid of Cpn(WH)8, Cpn(WWHH)4, and Cpn(WWWH)4 seems to be incomplete. Considering the reduced ATP-dependent Rg changes, they were likely to be in the equilibrium between the open and closed conformations. The slight increase in protective ability in the presence of ATP is likely to reflect the transient closure of the lid, which results in an increase in overall affinity with the unfolded CS.
FIGURE 4.
Effects of chaperonin complexes on thermal aggregation of CS in the presence and absence of ATP. CS from the porcine heart was incubated at 50 °C for denaturation in the presence of chaperonin complexes (open circles, without chaperonin; closed circles, CpnWT; open triangles, CpnΔhelical; closed squares, Cpn(WWWH)4; closed diamonds, Cpn(WH)8; open diamonds, Cpn(WWHH)4) without (A) or with ATP (B) under potassium-free conditions. The experimental details are described under the “Experimental Procedures.” a.u., arbitrary units. Wild-type and mutant subunit are shown as open and closed circles in the structural images as Fig. 1A.
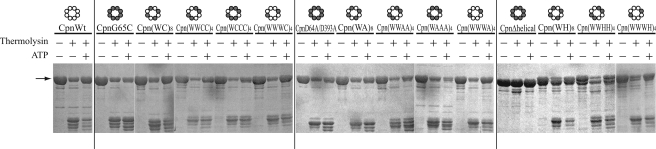
Conformation Analysis by Protease Digestion—Next, we analyzed the ATP-dependent dynamic motion of helical protrusions by thermolysin digestion (Fig. 5). The helical protrusions are digested by thermolysin in the open lid state, whereas those in the closed lid state are resistant to protease digestion. In our previous study (11), the addition of ATP protected CpnWT from digestion by thermolysin, corresponding to the results obtained for SAXS.
FIGURE 5.
Protease digestion assay for the conformational change of chaperonin complexes. Chaperonin incubated with or without ATP (1 mm) was exposed to thermolysin (1 ng/μl) and then analyzed by SDS-PAGE. The arrow indicates the band for the chaperonin subunit. The experimental details are described under the “Experimental Procedures.” Wild-type and mutant subunit are shown as open and closed circles in the structural images as in Fig. 1A.
Among chaperonin complexes composed of wild-type and G65C subunits, Cpn(WC)8 and Cpn(WWWC)4 were protected from digestion in the presence of ATP. Partial protection was also observed for Cpn(WWCC)4 and Cpn(WCCC)4. These results correlate well with the results obtained for SAXS. Helical protrusions of CpnD64A/D393A were not protected from thermolysin even in the presence of ATP, which is likely to be consistent with their deficiency in protein folding and conformational change. The effect of ATP was marginal for the complex containing D64A/D393A, irrespective of their protein folding ability and ATP-dependent conformational change observed by SAXS. The difference between G65C and D64A/D393A is ATP hydrolysis activity. Although ATP hydrolysis of G65C does not induce conformational change required for the closure of the lid, it might induce some movement or mobility of the helical protrusion that correlates with its tolerance against thermolysin.
CpnΔhelical subunit was not digested by thermolysin because the digestion site is located in the helical protrusion. Helical protrusions of Cpn(WH)8, Cpn(WWHH)4, and Cpn(WWWH)4 were protected from protease digestion, reflecting their ATP-dependent conformational change.
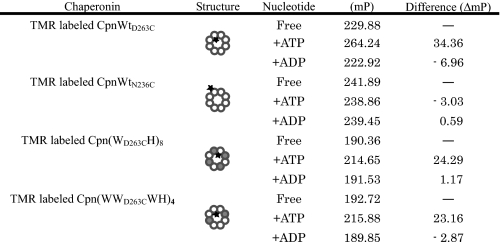
Analysis of the Mobility of the Helical Protrusion by Fluorescence Polarization—We then examined the effects of ATP on the fluorescence polarization (FP) of the fluorophore attached to the tip of the helical protrusion (Table 2). The FP correlates with the mobility of the molecules. As the mobility increases, the FP value decreases. The chaperonin complexes were labeled in the same fashion used for the detection of conformational change by fluorescence intensity assay. TMR was used instead of fluorescein because it gave a higher and more stable FP. The FP of TMR-labeled CpnWTD263C in the presence of ATP was much higher than the value obtained in the absence of ATP. In contrast, ATP had almost no effect on the FP of TMR-labeled CpnWTN236C. The ATP-dependent change in FP of TMR-labeled CpnWTD263C should reflect the lower mobility of the helical protrusion in the closed conformation than that in the open conformation. FPs of TMR-labeled Cpn(WD263CH)8 and Cpn(WWD263CWH)4 were smaller than that of CpnWTD263C, which is likely to correlate with high mobility of the helical protrusions because of the reduced number of helical protrusions in the ring. Addition of ATP affected the FPs, but the increments of FPs of Cpn(WD263CH)8 and Cpn(WWD263CWH)4 were smaller than that of CpnWTD263C (Table 2). It is reasonable to think that the closed conformations of Cpn(WD263CH)8 and Cpn(WWD263CWH)4 are relatively unstable and mobile compared with that of CpnWTD263C.
TABLE 2.
Fluorescence polarization parameters of chaperonin complexes composed of wild-type and mutant subunits Wild-type subunit, mutant subunit, and fluorophore are shown as open circles, closed circles and asterisks in the structural image. Fluorescence polarization analysis was performed as described under “Experimental Procedures.” The polarization value, a ratio of light intensities, is expressed in millipolarization units, mP (1 polarization unit = 1000 milliunits). Error values of fluorescence polarization are estimated to be under ±4.19 mP (n = 12).
DISCUSSION
Arrangement of Subunits of Covalently Subunit-linked Chaperonin Complexes—Based on the visible N and C terminus of subunits within a chaperonin ring, covalently linked subunits seemed to be arranged within a chaperonin ring. However, it is also possible that two covalently linked subunits are arranged in both rings. Although such subunit arrangement is not possible for four-subunit linked complexes, we examined the configuration of Cpn(WH)8 by electron microscopy (supplemental Fig. 2). End-on view images of CpnWT and CpnΔhelical subunits show 8-fold symmetry. In contrast, we found 4-fold symmetrical particles (in the interior of the cavity) in Cpn(WH)8 but hardly any in CpnWT. Although such particles were also observed in CpnΔhelical, very few were present. Thus, we concluded that wild-type and Δhelical subunits are arranged alternately in the ring in Cpn(WH)8. Although Cpn(WC)8 and Cpn(WA)8 were not analyzed by electron microscopy, the results support the idea that the subunits are also arranged alternately in the ring.
Mobility of N and C Termini Are Important for the Function of Group II Chaperonins—GFP folding activities of all covalently linked chaperonins were lower than that of CpnWT (Fig. 1B). These activities were recovered to the same level as that of CpnWT by thrombin digestion. We attributed the loss of folding activity to a decrease in the flexibility of the N and C termini of subunits necessary for ATP-dependent conformational change. It is likely that the conformational change required for function is restrained by the linkage. It is also possible that the inter-ring communication, which is indispensable for the chaperonin function, is hampered by the linkage. Recently, we showed that the C-terminal segments of the Thermococcus sp. strain KS1 chaperonin subunits exhibit higher mobility by NMR (36). Recent studies have also shown the importance of C-terminal regions for the protein folding activity of GroEL (37–39).
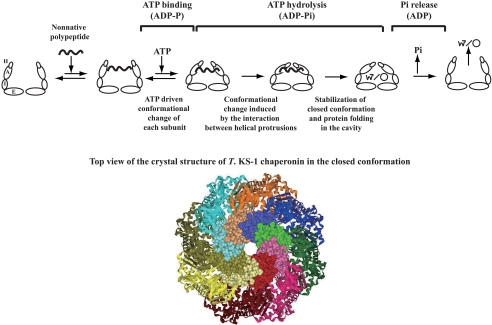
Model for the ATP-dependent Conformational Change of Group II Chaperonins—Thermococcus sp. strain KS-1 chaperonin changes its conformation in an ATP-dependent manner. ATP binding/hydrolysis induces the conformational change of each subunit and closure of the lid. The existence of mutant subunits in the ring affects the conformational change as well as protein folding activities of the complexes. The effects of the ATP-dependent conformational change deficient mutant (G65C) and the ATP hydrolysis deficient mutant (D64A/D393A) were relatively small. The chaperonin complexes containing the wild-type and mutant subunit (G65C or D64A/D393A) one by one (Cpn(WC)8 and Cpn(WA)8) could refold GFP in almost the same manner as the wild type. Folding activity was rescued by the presence of wild-type subunits in the ring. The degree of the rescue effects of wild-type subunits changes with their arrangement in a ring. Complexes containing the wild-type and mutant subunit alternately (Cpn(WC)8 and Cpn(WA)8) showed higher folding activity and a larger conformational change than those containing the mutant subunit contiguously (Cpn(WWCC)4 and Cpn(WWAA)4). In contrast, the effect of the helical protrusion deletion mutant was large. Even Cpn(WWWH)4, which contains only two Δhelical subunits per ring, could not refold denatured proteins. The chaperonin complex containing only one Δhelical subunit or two contiguous Δhelical subunits in a ring exhibited marginal folding activities (data not shown). Unexpectedly, Cpn(WH)8 and Cpn(WWWH)4 could change their conformations to the closed ones in the presence of ATP.
Our results seem to contradict the idea that group II chaperonins mediate protein folding via an ATP-dependent conformational change, from the open conformation to the closed conformation. Although the protein folding activity-deficient complexes, Cpn(WH)8 and Cpn(WWWH)4, showed a reduced change of Rg in response to ATP, some chaperonin complexes containing ATP-dependent conformational change deficient or ATP hydrolysis deficient mutants with ATP-dependent protein folding activities exhibit similar or smaller ATP-dependent Rg changes. This contradiction can be explained based on the model for the ATP-dependent conformational change (Fig. 6). ATP binding/hydrolysis induces the conformational change of each subunit. This conformational change seems to be independent and does not occur in the cooperative manner. The ATP-induced conformational change makes the helical protrusion to get close to each other as observed in the change of fluorescence of the fluorophore attached on the tip of helical protrusion. However, the conformation at this stage is not enough for protein folding or seems to be too transient. For protein folding in the cavity, further conformational change is required, which is induced by the interaction between helical protrusions. When helical protrusions are missing in the ring, the conformational change is impaired. The fact that only one or two deletions severely affected the function of the chaperonin complex suggests the cooperative action of the helical protrusion in the ring. The interaction among eight helical protrusions at the lid of the cavity is likely to be important for stabilization of the closed conformation (Fig. 6). Probably, the chaperonin complex in the closed conformation is likely to be more stable than that in the open conformation. Therefore, ATP hydrolysis energy might be required for the conformational change from the closed conformation to the open conformation.
FIGURE 6.
Model for the ATP-dependent conformational change in group II chaperonins. Only one ring is shown. A, I, E, and H represent the apical domain, intermediate domain, equatorial domain, and the helical protrusion, respectively. The top view of the crystal structure of Thermococcus sp. strain KS-1 (T. KS-1) chaperonin in the closed conformation is shown with the helical protrusions in the CPK model.
The conformational change observed in the chaperonin complex containing the Δhelical subunits seems to be transient because the complexes retained their ability to protect CS from thermal aggregation. It is likely that they are in the dynamic equilibrium between the open and closed conformation. The reduced conformational change in SAXS analysis is likely to reflect the change in equilibrium between the open and closed conformations.
Chaperonin complexes containing G65C or D64A/D393A exhibited conformational change and protein folding activity according to the number and order of mutant subunits. Although the ATP-dependent conformation of Cpn(WC)8 was also observed in the thermolysin digestion assay, only partial protection was observed in Cpn(WA)8. The G65C and D64A/D393A differed in their ATP hydrolysis ability. It is likely that G65C was able to exhibit a partial conformational change by ATP hydrolysis, which was observed in the previous study. Thus, the ATP-dependent conformational change ability of D64A/D393A should be smaller than G65C. As a result, the protease sensitivity of Cpn(WA)8 should be explained by the slow conformational change. Thus, it is likely that the conformational change of the mutant subunit is induced by contiguous wild-type subunits in the ring through the interaction between helical protrusions. The attained final conformation should be same as that of CpnWT.
The binding of ATP and cochaperonin GroES to GroEL triggers a major conformational change, creating an enlarged chamber into which the bound nonnative polypeptide is released. Our results show that closure of the built-in lid following ATP binding/hydrolysis also plays an important role in the conformational change of the protein folding chamber in the group II chaperonin.
Supplementary Material
Acknowledgments
We thank Dr. Akashi Ohtaki (Tokyo University of Agriculture and Technology) for technical advice. We thank the beam line (BL-15A) scientists of the Photon Factory for their help with data collection (2006G396).
This work is a part of the 21st Century Center of Excellence Program of the “Future Nano-Materials” Research and Education Project supported by the Ministry of Education, Science, Sports, Culture, and Technology through Tokyo University of Agriculture and Technology and supported in part by Grants-in-aids for Scientific Research 17028013, 19370038, and 20059013 and a grant from the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Science, Sports, and Culture of Japan (to M. Y.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: AMP-PNP, adenosine 5′-(β,γ-imino)triphosphate; CS, citrate synthase; GFP, green fluorescent protein; SAXS, small angle X-ray scattering; FP, fluorescence polarization; TMR, tetramethylrhodamine.
References
- 1.Hartl, F. U., and Hayer-Hartl, M. (2002) Science 295 1852-1858 [DOI] [PubMed] [Google Scholar]
- 2.Bukau, B., and Horwich, A. L. (1998) Cell 92 351-366 [DOI] [PubMed] [Google Scholar]
- 3.Klunker, D., Haas, B., Hirtreiter, A., Figueiredo, L., Naylor, D. J., Pfeifer, G., Muller, V., Deppenmeier, U., Gottschalk, G., Hartl, F. U., and Hayer-Hartl, M. (2003) J. Biol. Chem. 278 33256-33267 [DOI] [PubMed] [Google Scholar]
- 4.Kagawa, H. K., Osipiuk, J., Maltsev, N., Overbeek, R., Quaite-Randall, E., Joachimiak, A., and Trent, J. D. (1995) J. Mol. Biol. 253 712-725 [DOI] [PubMed] [Google Scholar]
- 5.Gutsche, I., Essen, L. O., and Baumeister, W. (1999) J. Mol. Biol. 293 295-312 [DOI] [PubMed] [Google Scholar]
- 6.Spiess, C., Meyer, A. S., Reissmann, S., and Frydman, J. (2004) Trends Cell Biol. 14 598-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ditzel, L., Lowe, J., Stock, D., Stetter, K. O., Huber, H., Huber, R., and Steinbacher, S. (1998) Cell 93 125-138 [DOI] [PubMed] [Google Scholar]
- 8.Shomura, Y., Yoshida, T., Iizuka, R., Maruyama, T., Yohda, M., and Miki, K. (2004) J. Mol. Biol. 335 1265-1278 [DOI] [PubMed] [Google Scholar]
- 9.Azem, A., Diamant, S., Kessel, M., Weiss, C., and Goloubinoff, P. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 12021-12025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klumpp, M., Baumeister, W., and Essen, L. O. (1997) Cell 91 263-270 [DOI] [PubMed] [Google Scholar]
- 11.Iizuka, R., Yoshida, T., Shomura, Y., Miki, K., Maruyama, T., Odaka, M., and Yohda, M. (2003) J. Biol. Chem. 278 44959-44965 [DOI] [PubMed] [Google Scholar]
- 12.Llorca, O., Smyth, M. G., Carrascosa, J. L., Willison, K. R., Radermacher, M., Steinbacher, S., and Valpuesta, J. M. (1999) Nat. Struct. Biol. 6 639-642 [DOI] [PubMed] [Google Scholar]
- 13.Meyer, A. S., Gillespie, J. R., Walther, D., Millet, I. S., Doniach, S., and Frydman, J. (2003) Cell 113 369-381 [DOI] [PubMed] [Google Scholar]
- 14.Gutsche, I., Holzinger, J., Rauh, N., Baumeister, W., and May, R. P. (2001) J. Struct. Biol. 135 139-146 [DOI] [PubMed] [Google Scholar]
- 15.Llorca, O., Martin-Benito, J., Grantham, J., Ritco-Vonsovici, M., Willison, K. R., Carrascosa, J. L., and Valpuesta, J. M. (2001) EMBO J. 20 4065-4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reissmann, S., Parnot, C., Booth, C. R., Chiu, W., and Frydman, J. (2007) Nat. Struct. Mol. Biol. 14 432-440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iizuka, R., So, S., Inobe, T., Yoshida, T., Zako, T., Kuwajima, K., and Yohda, M. (2004) J. Biol. Chem. 279 18834-18839 [DOI] [PubMed] [Google Scholar]
- 18.Kafri, G., and Horovitz, A. (2003) J. Mol. Biol. 326 981-987 [DOI] [PubMed] [Google Scholar]
- 19.Yifrach, O., and Horovitz, A. (1998) Biochemistry 37 7083-7088 [DOI] [PubMed] [Google Scholar]
- 20.Swain, J. F., and Gierasch, L. M. (2006) Curr. Opin. Struct. Biol. 16 102-108 [DOI] [PubMed] [Google Scholar]
- 21.Kafri, G., Willison, K. R., and Horovitz, A. (2001) Protein Sci. 10 445-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bigotti, M. G., and Clarke, A. R. (2005) J. Mol. Biol. 348 13-26 [DOI] [PubMed] [Google Scholar]
- 23.Rivenzon-Segal, D., Wolf, S. G., Shimon, L., Willison, K. R., and Horovitz, A. (2005) Nat. Struct. Mol. Biol. 12 233-237 [DOI] [PubMed] [Google Scholar]
- 24.Lin, P., and Sherman, F. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 10780-10785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida, T., Yohda, M., Iida, T., Maruyama, T., Taguchi, H., Yazaki, K., Ohta, T., Odaka, M., Endo, I., and Kagawa, Y. (1997) J. Mol. Biol. 273 635-645 [DOI] [PubMed] [Google Scholar]
- 26.Farr, G. W., Furtak, K., Rowland, M. B., Ranson, N. A., Saibil, H. R., Kirchhausen, T., and Horwich, A. L. (2000) Cell 100 561-573 [DOI] [PubMed] [Google Scholar]
- 27.Iizuka, R., Yoshida, T., Ishii, N., Zako, T., Takahashi, K., Maki, K., Inobe, T., Kuwajima, K., and Yohda, M. (2005) J. Biol. Chem. 280 40375-40383 [DOI] [PubMed] [Google Scholar]
- 28.Motojima, F., and Yoshida, M. (2003) J. Biol. Chem. 278 26648-26654 [DOI] [PubMed] [Google Scholar]
- 29.Yoshida, T., Kawaguchi, R., Taguchi, H., Yoshida, M., Yasunaga, T., Wakabayashi, T., Yohda, M., and Maruyama, T. (2002) J. Mol. Biol. 315 73-85 [DOI] [PubMed] [Google Scholar]
- 30.Iizuka, R., Yoshida, T., Maruyama, T., Shomura, Y., Miki, K., and Yohda, M. (2001) Biochem. Biophys. Res. Commun. 289 1118-1124 [DOI] [PubMed] [Google Scholar]
- 31.Furutani, M., Iida, T., Yoshida, T., and Maruyama, T. (1998) J. Biol. Chem. 273 28399-28407 [DOI] [PubMed] [Google Scholar]
- 32.Arai, M., Ito, K., Inobe, T., Nakao, M., Maki, K., Kamagata, K., Kihara, H., Amemiya, Y., and Kuwajima, K. (2002) J. Mol. Biol. 321 121-132 [DOI] [PubMed] [Google Scholar]
- 33.Arai, M., Kataoka, M., Kuwajima, K., Matthews, C. R., and Iwakura, M. (2003) J. Mol. Biol. 329 779-791 [DOI] [PubMed] [Google Scholar]
- 34.Semenyuk, A. V., and Svergun, D. I. (1991) J. Appl. Crystallogr. 24 537-540 [Google Scholar]
- 35.Furutani, M., Hata, J., Shomura, Y., Itami, K., Yoshida, T., Izumoto, Y., Togi, A., Ideno, A., Yasunaga, T., Miki, K., and Maruyama, T. (2005) Protein Sci. 14 341-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurimoto, E., Nishi, Y., Yamaguchi, Y., Zako, T., Iizuka, R., Ide, N., Yohda, M., and Kato, K. (2008) Proteins 70 1257-1263 [DOI] [PubMed] [Google Scholar]
- 37.Machida, K., Kono-Okada, A., Hongo, K., Mizobata, T., and Kawata, Y. (2008) J. Biol. Chem. 283 6886-6896 [DOI] [PubMed] [Google Scholar]
- 38.Tang, Y. C., Chang, H. C., Roeben, A., Wischnewski, D., Wischnewski, N., Kerner, M. J., Hartl, F. U., and Hayer-Hartl, M. (2006) Cell 125 903-914 [DOI] [PubMed] [Google Scholar]
- 39.Farr, G. W., Fenton, W. A., and Horwich, A. L. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5342-5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.