Abstract
The O-GlcNAc post-translational modification is considered to act as a sensor of nutrient flux through the hexosamine biosynthetic pathway. A cornerstone of this hypothesis is that global elevation of protein O-GlcNAc levels, typically induced with the non-selective O-GlcNAcase inhibitor PUGNAc (O-(2-acetamido-2-deoxy-d-glycopyranosylidene) amino-N-phenylcarbamate), causes insulin resistance in adipocytes. Here we address the potential link between elevated O-GlcNAc and insulin resistance by using a potent and selective inhibitor of O-GlcNAcase (NButGT (1,2-dideoxy-2′-propyl-α-d-glucopyranoso-[2,1-d]-Δ2′-thiazoline), 1200-fold selectivity). A comparison of the structures of a bacterial homologue of O-GlcNAcase in complex with PUGNAc or NButGT reveals that these inhibitors bind to the same region of the active site, underscoring the competitive nature of their inhibition of O-GlcNAcase and the molecular basis of selectivity. Treating 3T3-L1 adipocytes with NButGT induces rapid increases in global O-GlcNAc levels, but strikingly, NButGT treatment does not replicate the insulin desensitizing effects of the non-selective O-GlcNAcase inhibitor PUGNAc. Consistent with these observations, NButGT also does not recapitulate the impaired insulin-mediated phosphorylation of Akt that is induced by treatment with PUGNAc. Collectively, these results suggest that increases in global levels of O-GlcNAc-modified proteins of cultured adipocytes do not, on their own, cause insulin resistance.
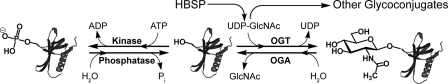
A variety of studies using different methods have demonstrated that increased flux through the hexosamine biosynthetic pathway causes insulin resistance in adipocytes (1, 2). Efforts to uncover the molecular causes of this insulin resistance have focused on the involvement of the end product of the hexosamine biosynthetic pathway; uridine 5′-diphospho-N-acetyl-α-d-glucosamine (UDP-GlcNAc) (3, 4), which is used as a building block for many glycoconjugates (Fig. 1) (5–7). The role of UDP-GlcNAc as a substrate for the enzyme uridine diphospho-N-acetylglucosamine:polypeptidyl-β-N-acetylglucosaminyltransferase (OGT)5 has received particular attention as this enzyme catalyzes the post-translational glycosylation of serine and threonine residues of nucleocytoplasmic proteins in a manner reminiscent of phosphorylation (8) (Fig. 1). Analogous to phosphorylation, residues that are modified by these 2-acetamido-2-deoxy-β-d-glycopyranose (O-GlcNAc) residues can be unmasked by a glycoside hydrolase (family GH84 in the CAZy classification (9)) known as O-GlcNAcase (10). Furthermore, the addition and removal of O-GlcNAc can occur many times during the lifetime of a target protein. Indeed, proteins that are O-GlcNAc-modified have also been found in some cases to be targets of phosphorylation (8), implicating this modification in a variety of processes including transcriptional regulation (11) and cellular signaling (12). A leading hypothesis is that O-GlcNAc, thus, serves as a key molecular sensing mechanism of increased flux through the hexosamine biosynthetic pathway and that increased global O-GlcNAc levels may induce insulin resistance (2, 8, 12).
FIGURE 1.
The dynamic nature of the O-GlcNAc post translational modification. A small percentage of glucose entering the cell is shunted down the hexosamine biosynthetic pathway (HBSP) to form UDP-GlcNAc. This species then acts as a substrate for various glycosyltransferases within the cell to form a wide variety of glycoconjugates. OGT, a nucleocytoplasmic glycosyltransferase, catalyzes the transfer of GlcNAc from UDP-GlcNAc to specific serine or threonine residues of targeted proteins. This process is dynamic, as a glycoside hydrolase termed O-GlcNAcase (OGA) cleaves off GlcNAc to return proteins to their unmodified state. A dynamic interplay between O-GlcNAc and phosphorylation is thought to give rise to high level of control over protein function.
Several observations support O-GlcNAc playing a role in insulin resistance and glucohomeostasis. First, levels of UDP-GlcNAc have been found to vary with nutrient availability (4, 13), a finding that has significance for O-GlcNAc since OGT does not appear to display traditional Michaelis-Menten kinetics and responds to a wide range of UDP-GlcNAc concentrations (14). Consistent with this finding, O-GlcNAc levels vary directly with glucose availability (15, 16). Second, decreased sensitivity to insulin-mediated glucose uptake correlates with increased cellular O-GlcNAc levels in both muscle and fat cells (12, 17–19). Additionally, a number of proteins involved in the insulin signaling cascade such as Akt (20), glycogen synthase kinase 3β (12), insulin receptor substrate-1 (21), and glucose transporter isoform 4 (18) are targets of the O-GlcNAc modification. Very recently, it was shown that insulin stimulates OGT enzymatic activity and promotes its redistribution within 3T3-L1 adipocytes (22).
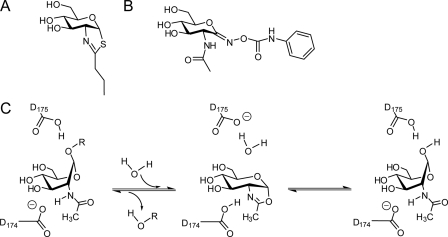
In an effort to provide useful chemical tools for investigating the biological role of O-GlcNAc, we have generated selective inhibitors of O-GlcNAcase. These inhibitors include 1,2-dideoxy-2′-propyl-α-d-glucopyranoso-[2,1-d]-Δ2′-thiazoline (NButGT) (Fig. 2A), the first nanomolar inhibitor of eukaryotic O-GlcNAcases that selectively targets this enzyme over the functionally related lysosomal β-hexosaminidases (1200-fold selective at pH 6.5) (23). This selectivity stems from the mechanism-based design of the inhibitors and structural elaboration of the thiazoline ring to increase its bulk. Indeed, glycoside hydrolase family 20 β-hexosaminidases, such as the lysosomal β-hexosaminidases, display a constrained active site structure around the 2-acetamido moiety (24), whereas GH84 O-GlcNAcases have a large pocket at this position (25, 26), revealing the structural basis for selective inhibition. As part of our ongoing research program into understanding the biological roles of O-GlcNAc, we were stimulated by existing studies, which report the insulin desensitizing effects caused by increasing global O-GlcNAc levels using the O-GlcNAcase inhibitor O-(2-acetamido-2-deoxy-d-glycopyranosylidene) amino-N-phenylcarbamate (PUGNAc) (Fig. 2B), to examine the effect of selective O-GlcNAcase inhibitors.
FIGURE 2.
Catalytic mechanism and inhibitors of O-GlcNAcase. A, structure of the selective, transition state mimic, NButGT. B, structure of the non-selective inhibitor PUGNAc. C, O-GlcNAcase uses a catalytic mechanism involving substrate-assisted catalysis. In this mechanism the carbonyl oxygen on the 2-acetamido group of the substrate acts as the nucleophile to form an oxazoline intermediate. Two important residues coordinate these events; Asp-174 acts to polarize the 2-acetamido group for attack, and Asp-175 serves as general acid catalyst to facilitate departure of the leaving group. In the second step of the mechanism, Asp-175 acts as a general base to aid attack of a water molecule at the anomeric center, facilitating breakdown of the intermediate and liberating GlcNAc and the free hydroxyl group as the products. Residue numbering is for human O-GlcNAcase.
A key motivation for us to generate selective O-GlcNAcase inhibitors was to circumvent the limitations associated with using PUGNAc. PUGNAc is a potent competitive inhibitor of O-GlcNAcase, and indeed, treating cells with this compound results in elevated O-GlcNAc levels stemming from the continuing action of OGT, while O-GlcNAcase function is hindered (27). PUGNAc has, thus, been used in many studies to elevate global O-GlcNAc levels and has thereby been used to implicate O-GlcNAc as a cause of insulin resistance (12, 17, 20, 28). PUGNAc, however, is not a selective inhibitor. PUGNAc also inhibits the lysosomal β-hexosaminidases (23), enzymes that cleave both GlcNAc and GalNAc from a range of glycoconjugates. PUGNAc may also have other targets. Indeed, more recently PUGNAc was also found to inhibit a homologue of the human α-N-acetylglucosaminidase involved in glycosaminoglycan degradation (29). Accordingly, because of these off target effects of PUGNAc, we felt that a more selective inhibitor of O-GlcNAcase would be a useful and welcome tool for investigating the role of O-GlcNAc in a more discriminating manner.
We felt that a clear structural understanding of the similarities and differences between inhibition of O-GlcNAcase by PUGNAc and NButGT would be beneficial. Therefore, we undertook structural studies of the Bacteroides thetaiotaomicron BtGH84 N-acetylglucosaminidase in complex with PUGNAc. BtGH84 is active on O-GlcNAc-modified protein substrates, and the structure of this enzyme is known, revealing that all residues within its active site are conserved in human O-GlcNAcase (25). BtGH84, therefore, provides a valuable model structure with which to probe the molecular basis of O-GlcNAcase inhibition. The results obtained here underscore that NButGT and PUGNAc bind to the same active site and offer a rationale for the competitive mode of inhibition by these inhibitors. The structure also reveals a number of differences in binding between these two inhibitors, clarifying the rationale for the high selectivity of NButGT and offering a clear explanation for the poor selectivity of PUGNAc.
Our first step toward understanding the role of O-GlcNAc in insulin resistance was to examine the effect of selective O-GlcNAcase inhibition in 3T3-L1 adipocytes to enable investigation of the temporal relationship between O-GlcNAc levels and insulin resistance. Our findings, reported here, are surprising. We show the insulin-desensitizing effects of PUGNAc on cultured adipocytes (12), which is a cornerstone of the O-GlcNAc-insulin resistance hypothesis, are not replicated by the selective O-GlcNAcase inhibitor NButGT. This discrepancy between using the non-selective inhibitor PUGNAc and the selective O-GlcNAcase inhibitor NButGT suggests that the promiscuous inhibitory properties of PUGNAc, although not widely appreciated, are a concern. Indeed perturbations in the levels of other glycoconjugates such as N-glycans and glycosphingolipids, which are also biosynthesized from UDP-GlcNAc (Fig. 1), are known to cause insulin resistance (30–32). These results raise some uncertainty about the interpretation of data obtained in cells using the inhibitor PUGNAc. Regardless of the interpretation of PUGNAc-derived data, the results described here collectively suggest that global increases in the levels of O-GlcNAc-modified proteins are not an independent mechanism mediating the development of insulin resistance in cultured adipocytes, although alternative interpretations are discussed.
EXPERIMENTAL PROCEDURES
General—All solutions containing salts and buffers were made from materials obtained from Bioshop unless otherwise noted. The CTD110.6 anti-O-GlcNAc and anti-β-actin antibodies were obtained from Covance and Sigma, respectively, whereas the Akt and phospho-Akt antibodies were obtained from R&D Systems. Secondary antibodies were obtained from Santa Cruz Biotechnology. NButGT and PUGNAc were prepared as reported (23, 33) and verified for purity by NMR spectroscopy. Coordinates for PUGNAc in complex with BTGH84 have been deposited with accession code 2vvs.
Structure Solution and Refinement—BtGH84 was buffer exchanged into 20 mm HEPES, pH 7.5, 300 mm NaCl, and concentrated using a 10-kDa cut off concentration device to a final concentration of ∼11 mg/ml. Crystals were obtained, as described previously for the apoBtGH84 structure (25), from a 1-ml well solution containing 0.3 m ammonium acetate, 13–15% (v/v) polyethylene glycol 3350, 0.1 m MES, pH 6.0, and 20% (v/v) glycerol. Drops were formed from a 1-μl protein solution (11 mg/ml) and 1-μl reservoir solution, with streak seeding from crystals that had formed previously. The complex with PUGNAc (PDB code 2vvs) was obtained by soaking harvested (25% glycerol, 12% polyethylene glycol 3350, 0.3 m ammonium acetate, 0.1 m MES, pH 6.0) crystals for ∼10 min under constant observation with minute quantities of powdered ligand surrounding individual crystals. Crystals were mounted in rayon fiber loops and flash-frozen in liquid nitrogen. X-ray diffraction data were collected at the European Synchrotron Radiation Facility with data processed and scaled using MOSFLM and SCALA from the CCP4 suite (34). Crystals of the BtGH84-PUGNAc complex are in space group C2, with cell dimensions a = 185.6 Å, b = 51.3 Å, c = 81.5 Å, and β = 97 °. There is one molecule of BtGH84 in the asymmetric unit. The structure of BtGH84 in complex with PUGNAc was solved using PHASER (35) with the protein atoms only of a single molecule of apo BtGH84 as the search model (PDB code 2CHO). Manual corrections to the model were made with COOT (36) with refinement cycles incorporating translation, libration, screw-rotation corrections (37) performed with REFMAC (38). Water molecules and ligand were added using COOT with stereochemical target values for the ligand based upon ideal coordinates generated with QUANTA (Accelrys, San Diego Ca). The BtGH84 protein model was built from residue Leu-5 to Pro-592 (except the loop Ser-46—Lys-53). The mobile C-terminal domain (His-593—Lys-716) could not be traced due to poor electron density. Details of data and final structure quality are shown in Table 1.
TABLE 1.
Data collection and refinement statistics for the PUGNAc complex of BtGH84
Outer resolution shell statistics are given in parentheses. r.m.s.d., root mean square deviation.
| BtGH84-PUGNAc complex | |
|---|---|
| Data | |
| Resolution range (Å) | 65-2.2 (2.36-2.24) |
| Rmerge | 0.076 (0.57) |
| 〈I/σI〉 | 14.0 (2.0) |
| Completeness (%) | 99.8 (99.3) |
|
Redundancy
|
4.0 (3.9)
|
| Refinement | |
| Resolution range (Å) | 65-2.2 |
| Rcryst | 0.2 |
| Rfree | 0.24 |
| No. protein atoms | 4727 |
| No. ligand atoms | 25 |
| No. water molecules | 112 |
| r.m.s.d. bonds (Å) | 0.015 |
| r.m.s.d. angles (°) | 1.5 |
| Mean B value (Å2) | |
| Protein atoms | 46 |
| Ligand atoms | 44 |
| Water atoms | 47 |
Cell Culture—3T3-L1 preadipocytes were obtained from Dr. Green (Harvard Medical School). All cells were grown in an incubator with 5% CO2 at a temperature of 37 °C. Preadipocytes were cultured in DMEM (Invitrogen) containing 10% bovine serum (Invitrogen). Two days post-confluence differentiation was initiated by replacing the media with DMEM containing 10% fetal bovine serum (Invitrogen), 120 μg/ml isobutylmethylxanthine (Sigma), 0.4 μg/ml dexamethasone (Sigma), and 1 μg/ml recombinant human insulin (Lilly). Cells were kept in this differentiation medium for 2 days, and then the medium was replaced with DMEM containing 10% fetal bovine serum and further replaced every 2 days after that. Cells were used 10–12 days after differentiation was initiated at a time when >95% of cells displayed characteristic features of adipocytes.
Western Blotting—The procedure was similar to that described previously (23). Fully differentiated 3T3-L1 adipocytes in 6-well plates were treated with either varying doses of PUGNAc or NButGT for varying times. Cells were harvested by adding 400 μl of 1% SDS containing 50 mm β-mercaptoethanol, boiling for 10 min, and using these lysates in subsequent Western blots. Briefly, samples were separated by SDS-PAGE (10% gels), transferred to nitrocellulose membrane (Bio-Rad), blocked for 1 h at room temperature with 1% bovine serum albumin (BSA) in PBS containing 0.1% Tween 20 (PBS-T), and then subsequently probed with the appropriate primary antibody delivered in 1% BSA in PBS-T overnight at 4 °C. Membranes were then extensively washed with PBS-T, blocked again for 30 min with 1% BSA in PBS-T at room temperature, and then probed with the appropriate horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature delivered in 1% BSA in PBS-T. Finally, the membranes were washed extensively and then developed with SuperSignal West Pico chemiluminescence substrate (Pierce).
2-Deoxyglucose (2-DOG) Uptake—All assays took place in 12-well plates (Falcon) with a minimum of six replicates. Cells were treated with PUGNAc or NButGT for 16 h before the start of the protocol at a concentration of 100 μm. Cells were first washed twice with DMEM containing 5 mm glucose (Invitrogen) supplemented with 0.5% BSA (Sigma) and then incubated in this media for 2 h. During this 2-h incubation period, PUGNAc or NButGT was maintained at the same concentration as overnight. Cells were then washed twice in KRP buffer (126 mm NaCl, 4.7 mm KCl, 10 mm NaH2PO4, 0.9 mm MgCl2, 0.9 mm CaCl2, 0.5% BSA, pH 7.4) and incubated in 420 μl of this buffer for 15 min. Subsequently, 30 μl of insulin (Lilly) was added directly to the cultures to yield the required insulin concentration, as required. Also, at this time cytochalasin B (Sigma) was added to some wells to give a concentration of 10 μm to control for glucose transporter-independent uptake. The cells were incubated for another 15 min, and then uptake assays were initiated by the addition of 50 μl of 2-DOG such that the cultures had a final concentration of 100 μm of 2-DOG and a specific activity of 0.5 μCi/ml. After 6 min the uptake was terminated by extensive washing in ice-cold PBS. PBS containing 2% Triton X-100 (500 μl) was then added to each well, and 300 μl from each well was used for scintillation counting. Additional 2-DOG uptake assays were carried out using cells treated with 100 μm NButGT or with vehicle (PBS) alone as described above except that the concentration of insulin was varied from between 0.1 to 10 nm. As well, studies were carried out to evaluate the effect of varying the length of 2-DOG uptake time from 2.5 to 12 min. Last, the concentration of 2-DOG was varied from 0.03 to 10 mm to evaluate the kinetic behavior of 2-DOG transport in cells treated with 100 μm NButGT overnight as compared with control cells.
Analysis of Phospho-Akt Levels—All assays took place in 6-well plates (Falcon) with two replicates. Experiments were also repeated in a second independent trial. Cells were treated with PUGNAc or NButGT for 16 h before the start of the protocol at a concentration of 100 μm. Cells were first washed twice with DMEM containing 5 mm glucose (Invitrogen) supplemented with 0.5% BSA and then incubated in this media for 2 h. During this 2-h incubation period, PUGNAc or NButGT was maintained at the same concentration as overnight. Cells were then washed twice in KRP buffer (126 mm NaCl, 4.7 mm KCl, 10 mm NaH2PO4, 0.9 mm MgCl2, 0.9 mm CaCl2, 0.5% BSA, pH 7.4) and incubated in this buffer for 15 min. Subsequently, the appropriate amount of insulin (0 or 10 nm final concentration) was added directly. The cells were incubated for another 15 min and harvested by adding 400 μl of 1% SDS containing 50 mm β-mercaptoethanol, boiling this for 10 min, and then analyzing these lysates via Western blots. Densitometry was carried out using ImageQuant software (GE Healthcare).
RESULTS AND DISCUSSION
The global elevation of O-GlcNAc levels by PUGNAc forms a cornerstone in the link between global O-GlcNAc levels and insulin resistance and has stimulated considerable efforts in the area (2, 8). Previous studies relying on PUGNAc to elevate O-GlcNAc levels have proposed that elevation of global levels of O-GlcNAc-modified proteins causes insulin resistance in cultured adipocytes (12, 28), primary adipocytes (20), and skeletal muscle studied ex vivo (17). One interesting study by Robinson et al. (39) that is not entirely consistent with the studies making use of PUGNAc showed that a reduction in the levels of O-GlcNAc-modified proteins in 3T3-L1 adipocytes using either OGT knockdown or overexpression of O-GlcNAcase does not relieve the insulin resistance caused by culturing these cells in media containing high glucose. On the basis of these collective observations, we were interested in both revisiting the effects of inhibition of O-GlcNAcase on insulin resistance in 3T3-L1 cells and then examining the temporal relationship between O-GlcNAc levels and insulin resistance. One limitation of earlier studies linking global O-GlcNAc levels and insulin resistance is the reliance on the inhibitor PUGNAc as it was unknown at that time that PUGNAc was not selective for O-GlcNAcase.
PUGNAc (Fig. 2B) is the most widely used inhibitor of O-GlcNAcase; however, this molecule has been recently shown to potently inhibit human lysosomal β-hexosaminidase (KI = 36 nm) (23) in addition to human O-GlcNAcase (KI = 46 nm) (23, 40). One concern about this promiscuity is that inactivation of lysosomal β-hexosaminidases is known to result in the accumulation of gangliosides, which themselves are implicated in the development of insulin resistance (30, 31). More recently, PUGNAc has been shown to inhibit a bacterial family 89 α-N-acetylglucosaminidase that shows high active site similarity to the family 89 human α-N-acetylglucosaminidase known as NAGLU (KI = 6 μm) (29), further highlighting the promiscuous nature of its inhibition. Additionally, PUGNAc causes decreased growth rates in several cultured cell lines (41), a finding that we do not observe with NButGT (data not shown). Therefore, the use of PUGNAc may give rise to disparate physiological effects due to its lack of selectivity or other off-target effects. To circumvent these concerns, which could be profound in animal models in particular, we have developed NButGT (Fig. 2A), a potent (KI = 230 nm) and highly selective inhibitor (KI = 340,000 nm for glycoside hydrolase family 20 β-hexosaminidase) of O-GlcNAcase (23).
O-GlcNAcase is one of a rare group of enzymes that uses a substrate-assisted, “neighboring group” catalytic mechanism in which the carbonyl of the N-acetyl group acts as a nucleophile such that the reaction proceeds via an oxazoline intermediate (Fig. 2C). NAG-thiazoline, which was first synthesized by Knapp et al. (42), is a good mimic of this intermediate and/or a structurally related transition state which confers selectivity on these inhibitors for enzymes that use the substrate-assisted catalytic mechanism. Indeed, recent biochemical analyses have revealed the molecular basis for differences in selectivity between NAG-thiazoline and PUGNAc analogues (43); NButGT is quantifiably a good transition state mimic for the unusual substrate-assisted catalytic mechanism used by O-GlcNAcase. PUGNAc analogues, however, have much lower selectivities (33) because they may not be as good transition state analogues for enzymes using this mechanism (43). With this in mind, a review of the CAZy-glycosidase data base (44) reveals no other obvious human monosaccharide-processing enzymes that use substrate-participation mechanisms and that could be significantly inhibited by a monosaccharide-derived thiazoline; to the best of current knowledge only the two β-hexosaminidases from family glycoside hydrolase family 20 and the O-GlcNAcase from GH84 act on monosaccharide substrates with the neighboring group mechanism. To gain insight into the similarities and differences between inhibition of O-GlcNAcase by PUGNAc and NButGT, we undertook structural studies of a homologue of human O-GlcNAcase, BtGH84 N-acetylglucosaminidase, in complex with PUGNAc and compared the structure with that of the enzyme bound to NButGT (43).
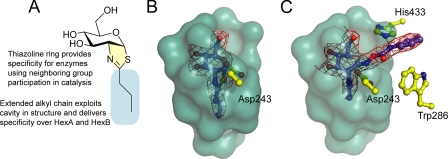
Whereas the thiazoline ring confers selectivity for the lysosomal β-hexosaminidases and O-GlcNAcase over other enzymes, selectivity for O-GlcNAcase over the lysosomal β-hexosaminidases has been achieved by appending substituents to the thiazoline scaffold (23, 45) (Fig. 3A). This alkyl moiety is accommodated in the enlarged acyl binding pocket that is present in human O-GlcNAcase but absent in the more constrained active center of the lysosomal β-hexosaminidases (43). The structure of BtGH84 in complex with NButGT confirmed that this inhibitor occupies a significant volume of the elongated acyl binding cavity to realize selectivity (Fig. 3B). Here we observe that PUGNAc also binds to BtGH84 in the active site, confirming the similar mode of binding by these inhibitors and the competitive nature of their inhibition of O-GlcNAcase. PUGNAc binds with the N-acetyl group below the ring plane, nestled in the acyl binding pocket that is lined by aromatic residues; however, the 2-acetamido group of PUGNAc does not occupy the full extent of this cavity (Fig. 3C). In contrast to NButGT, which interacts only with residues found directly within the active site, the binding of PUGNAc is enhanced by adventitious interactions of its extended N-phenylcarbamate moiety. In the case of BtGH84, these interactions include weak hydrogen bonds from the carbamate nitrogen and carbonyl oxygen of PUGNAc to the enzymic active site residue (Asp-243, BtGH84 numbering) and from the carbamate nitrogen to the side chain of residue His-433 (Fig. 3C). Hydrophobic interactions of the phenyl moiety with the enzyme also likely contribute significantly to binding as LOGNAc, an analogue lacking the phenyl group, is a much worse inhibitor of O-GlcNAcase and glycoside hydrolase family 20 β-hexosaminidases (10). In the case of BtGH84, this group sits above and perpendicular to Trp-286. Highlighting the adventitious binding of PUGNAc, this inhibitor binds with the phenyl group in a different position to the Clostridial GH84 (NagJ) enzyme (26).
FIGURE 3.
Specific and nonspecific O-GlcNAcase inhibitors bound in the active site of O-GlcNAcase. A, the design of the selective inhibitor NButGT includes a thiazoline ring (yellow) that likely mimics the oxazoline intermediate and its closely related transition-state(s), thereby conferring specificity for those enzymes using neighboring group participation in catalysis. Appended to the thiazoline is an n-propyl chain (cyan) designed to fit within the cavity found in the active site of O-GlcNAcase. This cavity is not present in the lysosomal β-hexosaminidases, thus conferring specificity for O-GlcNAcase over those enzymes. B, the three-dimensional structure of the human O-GlcNAcase homologue, BtGH84, in complex with NButGT (43) shows how the n-propyl chain is accommodated within the active center cavity of BtGH84 and human GH84 enzymes. The solvent-accessible surface of the catalytic center is shown in cyan. C, the three-dimensional structure of BtGH84 in complex with the nonspecific inhibitor PUGNAc highlighting the adventitious interactions of the N-phenylcarbamate moiety, notably with Asp-243, His-433, and Trp-286 (discussed under Results and Discussion). Electron density maps where shown are maximum-likelihood weighted 2Fobs – Fcalc syntheses contoured at ∼1σ. Panels B and C were drawn with PyMOL (DeLano Scientific). Residue numbering is for BtGH84.
The potent inhibition by PUGNAc, therefore, stems from two key features. First, it has a trigonal sp2 center at the pseudoanomeric position (C1) that enhances binding through partial mimicry of the transition states of glucosaminidases. This mimicry will occur for all glycoside hydrolases cleaving O-glycosides of GlcNAc irrespective of the catalytic mechanism used. PUGNAc also gains significant binding energy through adventitious interactions between enzyme and the pendent N-phenyl carbamate group. Although these two features lend potency to PUGNAc against O-GlcNAcase, they are features that will also enhance binding to other carbohydrate processing enzymes as underscored by the known lack of specificity.
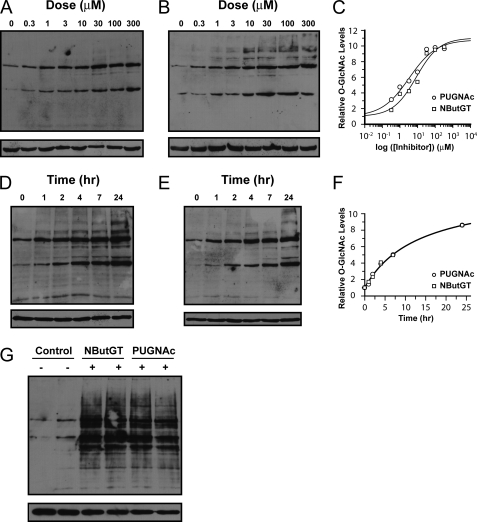
On the basis of its known selectivity and the structural results obtained here, we felt that NButGT was an excellent tool for careful probing of the role of O-GlcNAc in complex physiological settings. More specifically, we initially sought to examine the mechanisms by which increased global O-GlcNAc levels mediate insulin resistance in adipocytes. At the outset of these studies we fully anticipated that the results would lend support to the proposed link between global O-GlcNAc levels and insulin resistance while also providing a powerful chemical tool for future physiological studies. Therefore, as a first step we examined the dose- and time-dependent effect of both PUGNAc and NButGT treatment on O-GlcNAc levels in fully differentiated 3T3-L1 adipocytes. The two inhibitors were first tested over a range of concentrations (300 nm to 300 μm) for 24 h. Both showed a dose-dependent increase in O-GlcNAc levels (Fig. 4, A and B). PUGNAc, however, was somewhat more potent with an EC50 = 3 μm as compared with an EC50 = 8 μm for NButGT (Fig. 4C). Accordingly, PUGNAc acts better at lower concentrations, a finding in keeping with the lower KI value of PUGNAc for O-GlcNAcase. Both PUGNAc and NButGT increased O-GlcNAc levels in a time-dependent manner at a concentration of 100 μm, dramatically increasing global O-GlcNAc levels within 1 h with the maximal increase seen at 24 h (Fig. 4, D and E). The time-dependent increases in O-GlcNAc levels were indistinguishable between the two inhibitors (Fig. 4F), suggesting that under the conditions used both inhibitors act to potently inhibit O-GlcNAcase in differentiated 3T3-L1 cells. This similarity in effect is consistent with both compounds acting as competitive inhibitors and their use at concentrations significantly above their respective KI and EC50 values. Notably, the magnitude of the increase in global O-GlcNAc levels induced by these two inhibitors (10-fold over basal) is identical within error when used at 100 μm after 16 h (Fig. 4G), facilitating a direct comparison of their effects on insulin resistance. Also of note is that the O-GlcNAc banding patterns observed in the PUGNAc- and NButGT-treated cells were equivalent in all cases.
FIGURE 4.
PUGNAc and NButGT produce similar dose- and time-dependent increases in O-GlcNAc modified in 3T3-L1 cells. Fully differentiated 3T3-L1 adipocytes were treated with various doses of NButGT (A) or PUGNAc (B), and cells were harvested after 24 h. C, densitometric analysis of the effect of various doses of NButGT (squares) and PUGNAc (circles) on levels O-GlcNAc modified proteins reveal similar EC50 values with PUGNAc showing a slightly lower value consistent with its lower KI value. Values shown are reported relative to those found for untreated control cells. Cells treated with 100 μm NButGT (D) or PUGNAc (E) display a time-dependent increase in levels of O-GlcNAc modified proteins. F, densitometric analysis of the time-dependent effects of NButGT (squares) and PUGNAc (circles) on levels of O-GlcNAc-modified proteins reveal a near identical effect of these inhibitors. G, direct comparison of 3T3-L1 adipocytes treated for 16 h with 100 μm NButGT or PUGNAc reveals that both inhibitors elevate levels of O-GlcNAc-modified proteins to an equivalent extent over untreated cells. Levels of O-GlcNAc-modified proteins were analyzed by Western blot analysis using anti-O-GlcNAc antibody (CTD110.6) (upper panels) and anti-β-actin to evaluate protein loading (lower panels).
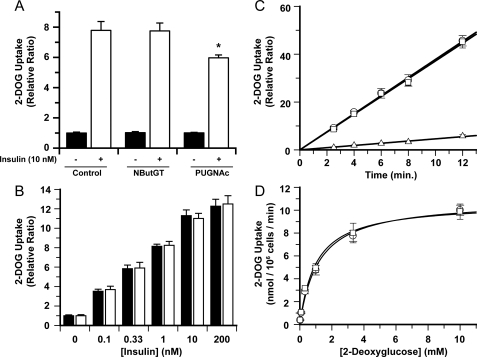
Accordingly, with these results in hand, we turned to carrying out 2-DOG uptake assays in fully differentiated 3T3-L1 adipocytes. On the basis of the time course study and to facilitate comparison with previous studies (12), we incubated fully differentiated cells with 100 μm concentrations of either NButGT or PUGNAc for 16 h. Treating fully differentiated 3T3-L1 adipocytes with the nonselective inhibitor PUGNAc causes a 25% decrease in 2-DOG uptake when stimulated with 10 nm insulin (Fig. 5A), an observation that closely matches earlier reports (12, 28). In contrast, cells treated with NButGT show no decrease in insulin-stimulated 2-DOG uptake (Fig. 5A) despite the significant and essentially indistinguishable increases in O-GlcNAc levels yielded by both inhibitors. To ensure that these assays were carefully executed and to be certain that NButGT does not result in insulin desensitization, three additional variables within the assay were investigated. First, different concentrations of insulin were used to stimulate 2-DOG uptake. As expected there was a dose-dependent increase in insulin-stimulated 2-DOG uptake with an EC50 similar to what has been observed previously (46) (Fig. 5B). However, in contrast to previous observations using PUGNAc (12), no insulin desensitization occurred with NButGT over a broad range of insulin concentrations. Second, the time-dependent uptake of 2-DOG was explored. As demonstrated in Fig. 5C, basal uptake (non-insulin stimulated) as well as cells stimulated with 10 nm insulin displayed linear uptake of 2-DOG for 12 min. Notably, treatment with NButGT resulted in no desensitization over any of the times examined. Third, the effect of different 2-DOG concentrations was investigated. As seen in Fig. 5D, NButGT did not affect the Km (1.2 mm) or Vmax (10 nmol 106 cells–1 min–1) values governing 2-DOG uptake. As well, these values are very similar to values reported previously for these cells, emphasizing that the process being monitored is indeed insulin-sensitive transporter-mediated 2-DOG transport (46).
FIGURE 5.
Selective inhibition of O-GlcNAcase does not replicate the insulin desensitizing effects of PUGNAc. A, insulin sensitivity in fully differentiated 3T3-L1 adipocytes was determined by measuring [1-3H]2-DOG (100 μm, 0.5 μCi/ml) uptake (6-min uptake time) by unstimulated cells (closed bar) or cells stimulated with 10 nm insulin (open bar). Cells were treated with either 100 μm NButGT, PUGNAc, or vehicle (PBS) for 16 h before assay. The asterisk indicates statistical significance from other groups as determined by a t test. B, the effect of different insulin concentrations on 2-DOG uptake (6 min uptake time) in untreated control cells (closed bar) and cells treated with 100 μm NButGT for 16 h before assay (openbar). C, time-dependent 2-DOG uptake. Linear 2-DOG uptake is observed over the indicated time range for unstimulated cells (triangles), untreated control cells stimulated with 10 nm insulin (circles), and NButGT-treated cells (squares). D, effects of variation of 2-DOG concentration on the kinetics of the 2-DOG uptake assay with a 6-min 2-DOG uptake time. Cells treated with (squares) and without (circles) 100 μm NButGT were used in uptake experiments in which the concentration of 2-DOG was varied. For all experiments, a minimum of six replicates (n = 6) were carried out for each condition with data represented as mean ± S.E.
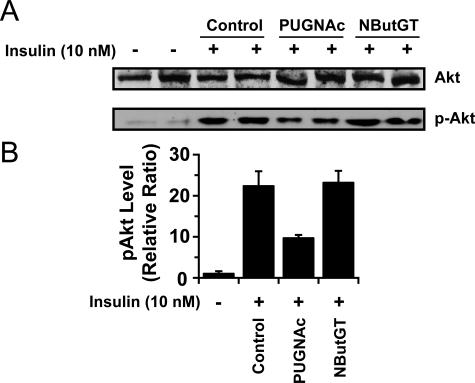
The absence of insulin resistance arising from NButGT treatment, despite global elevations in O-GlcNAc that replicate those seen when using PUGNAc, is extremely surprising. We, therefore, decided to investigate the induction of insulin resistance within these cells by monitoring the insulin-mediated activation of Akt (also known as protein kinase B) by phosphoinositide-dependent kinase-1 phosphorylation at Thr-308. Insulinpromoted phosphorylation at this residue of Akt is known to induce glycogen synthesis and glucose transporter isoform 4 translocation to the cell surface. Interfering with phosphorylation at this residue impairs insulin-stimulated glucose uptake, rendering cells insulin-resistant (for review, see Ref. 47). Accordingly, the pThr-308Akt epitope is considered a useful molecular indicator of insulin sensitivity arising from perturbations in the insulin signaling cascade that compliments glucose uptake assays. Treatment of control cells with insulin resulted in the expected robust activation of Akt as determined by the monitoring the pThr308Akt epitope levels by Western blot (Fig. 6A). PUGNAc-treated cells showed a 2-fold decrease in pThr308Akt levels after treatment with insulin (Fig. 6, A and B), matching previous observations (12, 28) and in accord with the insulin desensitizing effects of this compound. Strikingly, NButGT treatment of cells did not impair insulin-mediated Akt activation; pThr308Akt levels are identical to control cells. The absence of a reduction of phospho-Akt with NButGT contrasts with the results using PUGNAc even though global O-GlcNAc levels are increased to the same extent by both of these inhibitors (Fig. 4G). The absence of impaired Akt activation and robust pThr308Akt levels seen in cells treated with NButGT is entirely consistent with the lack of insulin resistance seen in the glucose uptake assays (Fig. 5). This observation finds good conceptual support from a recent study carried out by an independent group that investigated the effects of global elevation of O-GlcNAc levels using NButGT and PUGNAc on Akt activation in cultured astroglial cells. That study found that NButGT did not affect both pThr308Akt and pSer473Akt levels, whereas PUGNAc resulted in decreased phosphorylation at these residues (48) even as both compounds significantly increased global cellular O-GlcNAc levels.
FIGURE 6.
Selective inhibition of O-GlcNAcase does not result in impairment of AKT phosphorylation. A, the effects of treating fully differentiated 3T3-L1 adipocytes with PUGNAc and NButGT on insulin-mediated Akt activation as measured by monitoring levels of the pThr308Akt epitope (lower panel). pThr308Akt levels were determined after stimulation of fully differentiated 3T3-L1 cells with 10 nm insulin for 15 min. Levels of total Akt were also monitored to ensure equal loading of samples (upper panel). B, densitometric analysis of the effect of PUGNAc and NButGT on levels of insulin-stimulated pThr308Akt levels and levels in control cells. Values are expressed as a ratio of phospho-Akt versus Akt and are relative to cells not stimulated by insulin. Errors are representative of two independent experiments with two replicates in each (n = 4 for each group).
The discrepancy between the effects of PUGNAc and NButGT on insulin resistance that we observe likely arises from one of two possibilities. The first is that PUGNAc has one or more secondary off-target effects in addition to its known ability to inhibit O-GlcNAcase, and it is these secondary effects that induce insulin resistance in 3T3-L1 adipocytes. A second possibility is that NButGT has a secondary off-target effect that coincidentally, and precisely, reverses insulin resistance stemming from the increased global O-GlcNAc levels. Notably, this latter effect would have to occur upstream of Akt activation as pThr308Akt levels are unchanged in NButGT-treated cells as compared with control cells. Although neither scenario can be formally ruled out on the basis of the studies described here, we believe the first is more likely as PUGNAc is known to be promiscuous in its inhibitory tendencies, and the likelihood of an off-target effect of NButGT acting to precisely reverse insulin resistance arising from increased O-GlcNAc levels seems to us improbable. The powerful inhibitory potency of PUGNAc toward lysosomal β-hexosaminidase is intriguing in this context because this enzyme degrades gangliosides, and increases in ganglioside levels have been shown to cause insulin resistance (30, 31). Indeed, the blockage of glycosphingolipid biosynthesis is a validated target for the development of antidiabetogenic agents (49). Therefore, one possibility that may merit future investigation is that perturbed levels of another glycoconjugate arising from impaired processing by an α-or β-N-acetylglucosminidase inhibited by PUGNAc may contribute to insulin resistance. Further studies, however, will be required to define whether the lack of selectivity of PUGNAc toward these glucosaminidases or some entirely adventitious off target effect of PUGNAc gives rise to the insulin resistance observed when using this compound. Regardless, given the results described here, we believe some caution should be exercised when interpreting the results from studies making use of PUGNAc.
The results described here suggest that global increases in O-GlcNAc levels are neither an independent cause of insulin resistance in adipocytes nor do they perturb the insulin signaling cascade at, or above, Akt activation. These studies, however, do not formally rule out the possibility that elevated O-GlcNAc levels do play a role in vivo when established pathways leading to insulin resistance are operative. Further investigation is required to address this question and the role of O-GlcNAc in glucohomeostasis in vivo. These observations were surprising to us given the precedent using PUGNAc and the growing body of evidence using genetic studies that suggests elevated O-GlcNAc levels cause either insulin resistance (50) or perturbed glucohomeostasis (28, 51) in rodents. Recent studies using adenoviral-mediated infection of mouse hepatic tissue with OGT revealed enhanced glucose output in these animals resulting in perturbed glucohomeostasis (28, 51). One interesting observation, however, was that this effect did not appear to be mediated by insulin resistance, as mice that had their β-islets destroyed by streptozotocin showed equivalent perturbations in glucohomeostasis (51). There are obvious differences between using small molecules and genetic methods to study biological phenomena; the two clearly provide complementary strategies, and in some cases one method offers certain benefits over the other. One obvious consideration when using small molecule enzyme inhibitors is their potential lack of selectivity, a concern that is raised in the case of O-GlcNAcase inhibition by PUGNAc by the studies described here. Small molecule inhibitors, however, offer an advantage in that they facilitate the dissection of the catalytic and non-catalytic roles of enzymes because they act to block enzyme function without significantly affecting the levels of the target enzyme within cells, an important point highlighted by a recent investigation using small molecule inhibitors of Cdk7/Kin28 (52). Future investigations examining the effects of global increases of O-GlcNAc levels in vivo using selective small molecule inhibitors of O-GlcNAcase may, therefore, provide insights that are complementary to genetics studies.
Acknowledgments
We thank Dr. K. Stubbs for providing PUGNAc, Dr. D. Tucker for advice about 2-DOG uptake experiments, and Dr. R. Cornell for access to a liquid scintillation counter.
This research was supported by grants from the Canadian Institutes for Health Research and the Natural Sciences and Engineering Research Council of Canada (to D. J. V.) and the Biotechnology and Biological Sciences Research Council (to G. J. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact. The atomic coordinates and structure factors (code 2vvs) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Footnotes
The abbreviations used are: OGT, O-GlcNAc-transferase; O-GlcNAcase, β-N-acetylglucosaminidase; NButGT, 1,2-dideoxy-2′-propyl-α-d-glucopyranoso-[2,1-d]-Δ2′-thiazoline; MES, 2-(N-morpholino)ethanesulfonic acid; PUGNAc, O-(2-acetamido-2-deoxy-d-glycopyranosylidene) amino-N-phenylcarbamate; DMEM, Dulbecco's modified Eagle's medium; PBS, phosphate-buffered saline; BSA, bovine serum albumin; 2-DOG, 2-deoxyglucose.
References
- 1.Traxinger, R. R., and Marshall, S. (1991) J. Biol. Chem. 266 10148–10154 [PubMed] [Google Scholar]
- 2.Buse, M. G. (2006) Am. J. Physiol. Endocrinol. Metab. 290 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossetti, L., Hawkins, M., Chen, W., Gindi, J., and Barzilai, N. (1995) J. Clin. Investig. 96 132–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hawkins, M., Angelov, I., Liu, R., Barzilai, N., and Rossetti, L. (1997) J. Biol. Chem. 272 4889–4895 [DOI] [PubMed] [Google Scholar]
- 5.Kornfeld, R., and Kornfeld, S. (1985) Annu. Rev. Biochem. 54 631–664 [DOI] [PubMed] [Google Scholar]
- 6.Williams, D., Longmore, G., Matta, K. L., and Schachter, H. (1980) J. Biol. Chem. 255 11253–11261 [PubMed] [Google Scholar]
- 7.van Echten, G., and Sandhoff, K. (1993) J. Biol. Chem. 268 5341–5344 [PubMed] [Google Scholar]
- 8.Hart, G. W., Housley, M. P., and Slawson, C. (2007) Nature 446 1017–1022 [DOI] [PubMed] [Google Scholar]
- 9.Henrissat, B., and Bairoch, A. (1996) Biochem. J. 316 695–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, D. L., and Hart, G. W. (1994) J. Biol. Chem. 269 19321–19330 [PubMed] [Google Scholar]
- 11.Jackson, S. P., and Tjian, R. (1988) Cell 55 125–133 [DOI] [PubMed] [Google Scholar]
- 12.Vosseller, K., Wells, L., Lane, M. D., and Hart, G. W. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 5313–5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson, K. A., Weinstein, M. L., Lindenmayer, G. E., and Buse, M. G. (1995) Diabetes 44 1438–1446 [DOI] [PubMed] [Google Scholar]
- 14.Kreppel, L. K., and Hart, G. W. (1999) J. Biol. Chem. 274 32015–32022 [DOI] [PubMed] [Google Scholar]
- 15.Walgren, J. L., Vincent, T. S., Schey, K. L., and Buse, M. G. (2003) Am. J. Physiol. Endocrinol. Metab. 284 424–434 [DOI] [PubMed] [Google Scholar]
- 16.Akimoto, Y., Hart, G. W., Wells, L., Vosseller, K., Yamamoto, K., Munetomo, E., Ohara-Imaizumi, M., Nishiwaki, C., Nagamatsu, S., Hirano, H., and Kawakami, H. (2007) Glycobiology 17 127–140 [DOI] [PubMed] [Google Scholar]
- 17.Arias, E. B., Kim, J., and Cartee, G. D. (2004) Diabetes 53 921–930 [DOI] [PubMed] [Google Scholar]
- 18.Buse, M. G., Robinson, K. A., Marshall, B. A., Hresko, R. C., and Mueckler, M. M. (2002) Am. J. Physiol. Endocrinol. Metab. 283 241–250 [DOI] [PubMed] [Google Scholar]
- 19.Hazel, M., Cooksey, R. C., Jones, D., Parker, G., Neidigh, J. L., Witherbee, B., Gulve, E. A., and McClain, D. A. (2004) Endocrinology 145 2118–2128 [DOI] [PubMed] [Google Scholar]
- 20.Park, S. Y., Ryu, J., and Lee, W. (2005) Exp. Mol. Med. 37 220–229 [DOI] [PubMed] [Google Scholar]
- 21.Ball, L. E., Berkaw, M. N., and Buse, M. G. (2006) Mol. Cell. Proteomics 5 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whelan, S. A., Lane, M. D., and Hart, G. W. (2008) J. Biol. Chem. 283 21411–21417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macauley, M. S., Whitworth, G. E., Debowski, A. W., Chin, D., and Vocadlo, D. J. (2005) J. Biol. Chem. 280 25313–25322 [DOI] [PubMed] [Google Scholar]
- 24.Mark, B. L., Vocadlo, D. J., Knapp, S., Triggs-Raine, B. L., Withers, S. G., and James, M. N. (2001) J. Biol. Chem. 276 10330–10337 [DOI] [PubMed] [Google Scholar]
- 25.Dennis, R. J., Taylor, E. J., Macauley, M. S., Stubbs, K. A., Turkenburg, J. P., Hart, S. J., Black, G. N., Vocadlo, D. J., and Davies, G. J. (2006) Nat. Struct. Mol. Biol. 13 365–371 [DOI] [PubMed] [Google Scholar]
- 26.Rao, F. V., Dorfmueller, H. C., Villa, F., Allwood, M., Eggleston, I. M., and van Aalten, D. M. (2006) EMBO J. 25 1569–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haltiwanger, R. S., Grove, K., and Philipsberg, G. A. (1998) J. Biol. Chem. 273 3611–3617 [DOI] [PubMed] [Google Scholar]
- 28.Yang, X., Ongusaha, P. P., Miles, P. D., Havstad, J. C., Zhang, F., So, W. V., Kudlow, J. E., Michell, R. H., Olefsky, J. M., Field, S. J., and Evans, R. M. (2008) Nature 451 964–969 [DOI] [PubMed] [Google Scholar]
- 29.Ficko-Blean, E., Stubbs, K. A., Nemirovsky, O., Vocadlo, D. J., and Boraston, A. B. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 6560–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nojiri, H., Stroud, M., and Hakomori, S. (1991) J. Biol. Chem. 266 4531–4537 [PubMed] [Google Scholar]
- 31.Sasaki, A., Hata, K., Suzuki, S., Sawada, M., Wada, T., Yamaguchi, K., Obinata, M., Tateno, H., Suzuki, H., and Miyagi, T. (2003) J. Biol. Chem. 278 27896–27902 [DOI] [PubMed] [Google Scholar]
- 32.Ohtsubo, K., Takamatsu, S., Minowa, M. T., Yoshida, A., Takeuchi, M., and Marth, J. D. (2005) Cell 123 1307–1321 [DOI] [PubMed] [Google Scholar]
- 33.Stubbs, K. A., Zhang, N., and Vocadlo, D. J. (2006) Org. Biomol. Chem. 4 839–845 [DOI] [PubMed] [Google Scholar]
- 34.Collaborative Computational Project Number 4 (1994) 50 760–763 [Google Scholar]
- 35.Mccoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C., and Read, R. J. (2007) J. Appl. Crystallogr. 40 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley, J., Knight, C. G., Farndale, R. W., and Barnes, M. J. (2004) J. Mol. Biol. 335 1019–1028 [DOI] [PubMed] [Google Scholar]
- 37.Winn, M. D., Isupov, M. N., and Murshudov, G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57 122–133 [DOI] [PubMed] [Google Scholar]
- 38.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53 240–255 [DOI] [PubMed] [Google Scholar]
- 39.Robinson, K. A., Ball, L. E., and Buse, M. G. (2007) Am. J. Physiol. Endocrinol. Metab. 292 884–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao, Y., Wells, L., Comer, F. I., Parker, G. J., and Hart, G. W. (2001) J. Biol. Chem. 276 9838–9845 [DOI] [PubMed] [Google Scholar]
- 41.Slawson, C., Zachara, N. E., Vosseller, K., Cheung, W. D., Lane, M. D., and Hart, G. W. (2005) J. Biol. Chem. 280 32944–32956 [DOI] [PubMed] [Google Scholar]
- 42.Knapp, S., Vocadlo, D., Gao, Z. N., Kirk, B., Lou, J. P., and Withers, S. G. (1996) J. Am. Chem. Soc. 118 6804–6805 [Google Scholar]
- 43.Whitworth, G. E., Macauley, M. S., Stubbs, K. A., Dennis, R. J., Taylor, E. J., Davies, G. J., Greig, I. R., and Vocadlo, D. J. (2007) J. Am. Chem. Soc. 129 635–644 [DOI] [PubMed] [Google Scholar]
- 44.Henrissat, B., and Davies, G. (1997) Curr. Opin. Struct. Biol. 7 637–644 [DOI] [PubMed] [Google Scholar]
- 45.Yuzwa, S. A., Macauley, M. S., Heinonen, J. E., Shan, X., Dennis, R. J., He, Y., Whitworth, G. E., Stubbs, K. A., McEachern, E. J., Davies, G. J., and Vocadlo, D. J. (2008) Nat. Chem. Biol. 4 483–490 [DOI] [PubMed] [Google Scholar]
- 46.Frost, S. C., and Lane, M. D. (1985) J. Biol. Chem. 260 2646–2652 [PubMed] [Google Scholar]
- 47.Sale, E. M., and Sale, G. J. (2008) Cell. Mol. Life Sci. 65 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matthews, J. A., Belof, J. L., Acevedo-Duncan, M., and Potter, R. L. (2007) Mol. Cell. Biochem. 298 109–123 [DOI] [PubMed] [Google Scholar]
- 49.Zhao, H., Przybylska, M., Wu, I. H., Zhang, J., Siegel, C., Komarnitsky, S., Yew, N. S., and Cheng, S. H. (2007) Diabetes 56 1210–1218 [DOI] [PubMed] [Google Scholar]
- 50.McClain, D. A., Lubas, W. A., Cooksey, R. C., Hazel, M., Parker, G. J., Love, D. C., and Hanover, J. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 10695–10699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dentin, R., Hedrick, S., Xie, J., Yates, J., III, and Montminy, M. (2008) Science 319 1402–1405 [DOI] [PubMed] [Google Scholar]
- 52.Kanin, E. I., Kipp, R. T., Kung, C., Slattery, M., Viale, A., Hahn, S., Shokat, K. M., and Ansari, A. Z. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]