Abstract
Sphingosine kinase 1 (SK1) is an important regulator of cellular signaling that has been implicated in a broad range of cellular processes. Cell exposure to a wide array of growth factors, cytokines, and other cell agonists can result in a rapid and transient increase in SK activity via an activating phosphorylation. We have previously identified extracellular signal-regulated kinases 1 and 2 (ERK1/2) as the kinases responsible for the phosphorylation of human SK1 at Ser225, but the corresponding phosphatase targeting this phosphorylation has remained undefined. Here, we provide data to support a role for protein phosphatase 2A (PP2A) in the deactivation of SK1 through dephosphorylation of phospho-Ser225. The catalytic subunit of PP2A (PP2Ac) was found to interact with SK1 using both GST-pulldown and coimmunoprecipitation analyses. Coexpression of PP2Ac with SK1 resulted in reduced Ser225 phosphorylation of SK1 in human embryonic kidney (HEK293) cells. In vitro phosphatase assays showed that PP2Ac dephosphorylated both recombinant SK1 and a phosphopeptide based on the phospho-Ser225 region of SK1. Finally, both basal and tumor necrosis factor-α-stimulated cellular SK1 activity were regulated by molecular manipulation of PP2Ac activity. Thus, PP2A appears to function as an endogenous regulator of SK1 phosphorylation.
Sphingosine kinase 1 (SK1)2 has emerged as a critical regulator of cellular signaling through the generation of the bioactive phospholipid, sphingosine 1-phosphate (S1P). S1P has been implicated in a broad range of cellular processes, including cell proliferation, apoptosis, calcium homeostasis, angiogenesis, and vascular maturation (1). This diverse range of actions of S1P may be due in part to the ability of this lipid mediator to function as both an intracellular second messenger and a ligand for cell surface receptors (2).
Cellular S1P levels are largely controlled by the actions of sphingosine kinases, which catalyze the formation of S1P from sphingosine (3). Although low cellular levels of S1P are present under basal conditions, growth factor and other agonist stimulation of cells can result in a rapid and transient increase in S1P levels as a direct consequence of SK1 activation (4). Thus, the activation state of SK1 is critical in the control of cellular S1P levels and the cellular effects of this bioactive phospholipid.
A basal level of SK activity has been proposed to play a “housekeeping” role in maintaining relatively low levels of sphingosine and ceramide in the cell (5). In contrast, agonist stimulation can result in a rapid and transient increase in SK1 activity, as a result of an activating phosphorylation. We have previously identified extracellular signal-regulated kinases 1 and 2 (ERK1/2) as the kinases responsible for this phosphorylation, which occurs at Ser225 in human SK1 (6). Interestingly, this phosphorylation results not only in increased SK1 activity, but is also crucial for agonist-induced translocation of SK1 from the cytosol to the plasma membrane (6), and subsequent oncogenic signaling by this enzyme (7).
Phosphorylation of cellular proteins is a reversible process and the transient activation/phosphorylation of SK1 (6, 8) suggests that levels of SK1 phosphorylation at Ser225 are governed by the competing actions of the protein kinases and protein phosphatases that mediate SK1 phosphorylation and dephosphorylation, respectively. As noted above, we have previously identified ERK1/2 as the protein kinases responsible for phosphorylation of SK1 at Ser225 (6). Despite its likely importance in the regulation of SK1, and SK1-mediated cell signaling, the protein phosphatase responsible for dephosphorylation of this residue has, however, remained undefined. In this study we report that protein phosphatase 2A (PP2A) deactivates SK1 through dephosphorylation of phospho-Ser225.
EXPERIMENTAL PROCEDURES
Materials—Protein Phosphatase 1 (PP1) purified from rabbit skeletal muscle was obtained from New England Biolabs (Ipswich, MA). Protein phosphatase 2A1 (PP2A1) purified from bovine kidney, okadaic acid, human recombinant protein phosphatase inhibitor-2 (I-2), and fostriecin were obtained from Calbiochem. Monoclonal anti-FLAG (M2) antibody, anti-HA antibody and p-nitrophenyl phosphate were obtained from Sigma. Anti-His antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-PP2A antibody, recognizing the PP2A catalytic subunit, was obtained from Upstate/Millipore (Temecula, CA). Mouse monoclonal antibody (DM1A) to α-tubulin was obtained from Abcam (Cambridge, UK). The phosphopeptide based on the SK1 phospho-Ser225 region (phospho-SK1 peptide) was synthesized by Auspep (Victoria, Australia) and had the sequence CGSKTPApSPVVVQQ, where pS denotes phospho-Ser. Protein G-Sepharose was obtained from Invitrogen (Carlsbad, CA), and recombinant human tumor necrosis factor-α (TNFα) was from R&D Systems (Minneapolis, MN).
Generation of Expression Constructs—The generation of mammalian expression vectors for FLAG-epitope-tagged wild-type human SK1 and a truncated version of SK1 (SK1ΔCT) lacking 17 residues (368–384) at the C terminus have been previously described (9).
Human PP2A catalytic subunit (PP2Ac) cDNA (GenBank™ accession number NM_002715) was amplified by Pfu DNA polymerase chain reaction from placenta cDNA with the oligonucleotide primers 5′-TAGAATTCCAATGGACGAGAAGGTGTTCAC-3′ and 5′-TAGGTACCTTACAGGAAGTAGTCTGGGGT-3′. The resultant product was cloned into pCMV(HA) (Clontech Laboratories Inc., Mountain View, CA) with EcoRI and KpnI to allow expression of N-terminal hemagglutinin (HA)-tagged PP2Ac in mammalian cells. To generate a glutathione S-transferase (GST)-PP2Ac fusion protein in bacteria, PP2Ac was also subcloned into pGEX4T2 (GE Healthcare, Piscataway, NJ) following digestion with EcoRI and NotI.
Human protein phosphatase 4 catalytic subunit (PP4c) (GenBank™ accession number BC001416) was amplified by Pfu DNA polymerase chain reaction from placenta cDNA with the oligonucleotide primers 5′-TAGAATTCCCATGGCGGAGATCAGCG-3′ and 5′-TAGAATTCTCACAGGAAGTAGTCGGCC-3′. The resultant product was cloned into pCMV(HA) and pGEX4T2, with EcoRI for mammalian and bacterial expression, respectively.
The HA-tagged catalytically inactive versions of PP2Ac (10) and PP4c (11) (PP2AcR89A (human protein phosphatase 2A catalytic subunit with Arg89 → Ala mutation) and PP4cR236L (human protein phosphatase 4 catalytic subunit with Arg236 → Leu mutation), respectively) were generated by QuikChange™ mutagenesis using the primers 5′-GGAGATTATGTTGACGCCGGCTATTATTCAGTTGAA-3′, 5′-TTCAACTGAATAATAGCCGGCGTCAACATAATCTCC-3′ and 5′-CGCAGCCAATGACATCGATATGATCTGCCTTGCCCACCAACTGGT-3′, 5′-ACCAGTTGGTGGGCAAGGCAGATCATATCGATGTCATTGGCTGCG-3′, respectively. DNA sequencing verified the integrity and orientation of all cloned cDNAs.
Cell Culture and Transfection—Human embryonic kidney (HEK293) cells were cultured in Dulbecco's modified Eagle's medium (GIBCO® Invitrogen), containing 10% bovine calf serum (JRH Biosciences, Lenexa, KS), 2 mm glutamine, 0.2% (w/v) sodium bicarbonate, 1 mm HEPES, penicillin (1.2 mg/ml), and streptomycin (1.6 mg/ml). Cells were transiently transfected using Lipofectamine™ 2000 Transfection Reagent (Invitrogen) according to the manufacturer's instructions. Transfected cells were harvested and lysed 24 h post-transfection.
For immunoprecipitations, transfected cells were lysed by sonication in a Bioruptor™ (Diagenode, NY) (4 × 25 s pulses with 25 s breaks, 200 watts) in extraction buffer composed of 50 mm Tris/HCl buffer (pH 7.4) containing 150 mm NaCl, 10% glycerol, 0.05% Triton X-100, 1 mm dithiothreitol, 2 mm Na3VO4, 10 mm NaF, 10 mm β-glycerophosphate, 1 mm EDTA, and protease inhibitors (Complete™, Roche Applied Sciences). Where samples were used for coimmunoprecipitation, transfected cells were lysed in extraction buffer containing no dithiothreitol, and drawn 5 times through a 26.5-gauge needle rather than being sonicated. All lysates were clarified by centrifugation (17,000 × g, 15 min, 4 °C) prior to subsequent analyses. Lysates for use in in vitro phosphatase assays were prepared in a modified format, as described in detail below.
Preparation of GST Fusion Proteins and Pulldown Analyses—GST fusion proteins were prepared and purified as described previously (12), except that after growth and induction with isopropyl 1-thio-β-d-galactopyranoside, bacteria were lysed in phosphate-buffered saline supplemented with protease inhibitors (Complete™). For pulldown analyses, 2 μg of each fusion protein immobilized on glutathione-Sepharose (GE Healthcare, Piscataway, NJ) was incubated with 1 μg of recombinant His-tagged SK1 protein (13) in extraction buffer for 1.5 h at 4 °C with constant mixing. Protein complexes were then washed three times in extraction buffer and bound SK1 resolved by SDS-PAGE and visualized by immunoblotting via its His epitope.
Coimmunoprecipitation and Immunoblotting—For coimmunoprecipitation analyses, lysates from transfected cells were incubated in the presence of the appropriate antibody (anti-FLAG or anti-HA) with mixing for 1.5 h at 4 °C, prior to addition of protein G-Sepharose and further incubation with mixing (1 h, 4 °C). Immunocomplexes were washed three times in extraction buffer prior to SDS-PAGE, transfer of the proteins to nitrocellulose and immunoblotting. Following incubation with primary antibodies and horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG (Pierce) as appropriate, an enhanced chemiluminescence kit (ECL, Amersham Biosciences) was used for detection.
In Vitro Phosphatase Assays—Phosphorylase b was 32P-labeled by phosphorylase kinase to form 32P-phosphorylase a as previously described (14). This labeled phosphorylase a was then used as a substrate to standardize the activities of the commercial PP1 and PP2A phosphatases via scintillation counting of 32P released, such that 1 unit of each protein phosphatase could be included in subsequent experiments. A unit of phosphatase activity is defined as the amount of enzyme required to release 1 nmol of 32P/min from [32P]phosphorylase a.
Assays to detect endogenous phosphatase activity were performed as previously described (15). Briefly, whole cell extracts from cells overexpressing SK1 were incubated at 37 °C in phosphatase buffer (50 mm Tris/HCl (pH 7.0), 5 mm MnCl2, 5 mm MgCl2, 0.1 mm EDTA, 5 mm dithiothreitol, 0.025% Tween 20) in a 10-μl total reaction volume. Phosphatase inhibitors (1 μm I-2, 1.6 μm fostriecin) were added, as specified in individual experiments. Control reactions were carried out in the presence of the relevant vehicle solutions. Reactions were terminated by the addition of 5× Laemmli sample buffer with boiling, and then subjected to SDS-PAGE. Dephosphorylation of SK1 was quantitated by immunoblotting with antibodies recognizing phospho-Ser225 of SK1 (6), with total SK1 levels assessed by anti-FLAG antibodies.
Where overexpressed phosphatases were immunoprecipitated prior to use in in vitro phosphatase assays, transfected HEK293 cells were lysed by sonication in modified extraction buffer containing 50 mm Tris/HCl (pH 7.4), 150 mm NaCl, 10% (v/v) glycerol, 1 mm dithiothreitol, 0.05% (v/v) Triton X-100, and Complete™ protease inhibitors. Lysates were clarified by centrifugation (17,000 × g, 15 min, 4 °C), and overexpressed phosphatases were immunoprecipitated with anti-HA antibody and protein G-Sepharose. Immunoprecipitation complexes were washed three times in wash buffer (50 mm Tris/HCl (pH 7.4), 150 mm NaCl, 10% (v/v) glycerol) prior to use in in vitro phosphatase assays with either p-nitrophenyl phosphate (0.9 mg/ml) or phospho-SK1 peptide (625 μm) substrates, as per the manufacturer's protocols for Ser/Thr assay kit 1 (Upstate Biotechnology Inc., Waltham, MA) and as described previously (16).
Sphingosine Kinase Assays—Sphingosine kinase activity was routinely determined using d-erythro-sphingosine and [γ-32P]ATP as substrates, as described previously (9). A unit of sphingosine kinase activity is defined as the amount of enzyme required to produce 1 pmol of S1P/min.
siRNA Silencing—The siGENOME SMART pool (M-003598) targeting human PP2A catalytic subunit, α isoforms (NM_ 002715) was purchased from Dharmacon (Thermo Fisher Scientific, Lafayette, CO). The Stealth universal negative control siRNA reagent and transfection reagents (Lipofectamine™ 2000 and Lipofectamine™ RNAiMAX) were purchased from Invitrogen. For transfection of siRNA into HEK293 cells, Lipofectamine™ RNAiMAX was used according to the manufacturer's guidelines. For cotransfection of plasmid DNA and siRNA into HEK293 cells, Lipofectamine™ 2000 was used according to the manufacturer's guidelines. siRNAs were used at a final concentration of 10 nm. Cells were harvested 72–96 h post-transfection and lysed as described above.
Cell Fractionation—For analyses of soluble versus membrane localization of SK1, cells were lysed first by sonication as described above, but in extraction buffer containing no Triton X-100. These whole cell extracts were then clarified by centrifugation (17,000 × g, 15 min, 4 °C) yielding the supernatant as the soluble fraction. The cell pellets were then sonicated as above in extraction buffer containing 1% (v/v) Triton X-100, and clarified as above, yielding the supernatant as the membrane fraction.
RESULTS
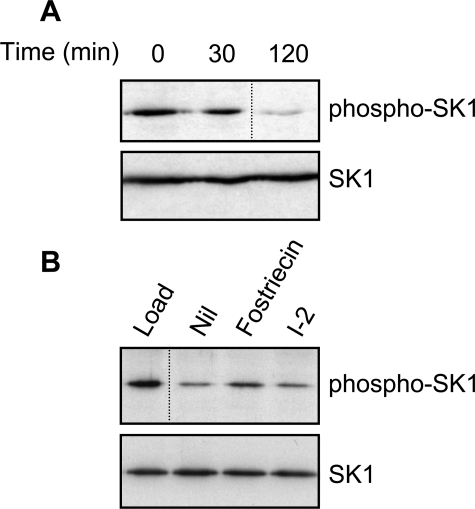
PP2A-family Phosphatases Dephosphorylate SK1—A number of growth factors and cytokines induce a rapid and transient activation of SK1 (4). In the case of TNFα, we have previously shown that this correlated with the rapid phosphorylation and subsequent rapid dephosphorylation of SK1 at Ser225 (6). Because this dephosphorylation and deactivation of SK1 suggested the importance of a protein phosphatase in SK1 regulation, we sought to establish the identity of this enzyme. To do this, we initially exploited the observation that overexpression of SK1 in HEK293 cells results in the detectable phosphorylation of this protein at Ser225 (6). Assay conditions were then established to follow SK1 dephosphorylation in vitro due to endogenous protein phosphatase activity. Specifically, lysates were prepared from cells overexpressing SK1 and incubated under conditions that permitted phosphatase activity. Immunoblot analysis with an antibody that specifically detects phospho-Ser225 of SK1 (6) revealed a noticeable decrease in phospho-SK1 levels over time, while levels of total SK1 protein remained unchanged (Fig. 1A).
FIGURE 1.
Endogenous PP2A-family phosphatases regulate phospho-SK1 levels. A, whole cell extracts from cells overexpressing SK1 were incubated at 37 °C for the indicated times. Dephosphorylation of SK1 was assessed by immunoblotting with an antibody recognizing phospho-SK1 (upper panel), and total SK1 levels were confirmed by immunoblotting with anti-FLAG antibodies (lower panel). B, as in A, but reactions were incubated for 120 min the presence of fostriecin (1.6 μm) or I-2 (1 μm), to assess the relative contributions from PP2A- and PP1-family phosphatases, respectively. Results are representative of three independent experiments. The dividing lines indicate where lanes have been spliced to simplify viewing, but results in each panel are from a single experiment.
Human Ser/Thr-specific phosphatases can be grouped into five broad classes, comprising the PP1, PP2/4/6, PP3, PP5, and PP7 subfamilies (17). However, the PP1 and PP2/4/6 subclasses have been reported to contribute most of the Ser/Thr phosphatase activity in cells (18). PP2A is by far the best-characterized member within the PP2/4/6 subclass, given that its catalytic, structural, and regulatory subunits have been sequenced and identified, and its susceptibility to inhibition by compounds such as okadaic acid and fostriecin is well known (19). Although the related PP4 and PP6 proteins are known to be similarly susceptible to active site inhibitors (20, 21), considerably less is known about their component subunits.
Using the conditions we established to follow SK1 dephosphorylation, we assessed the relative contributions to this activity from endogenous PP1- and PP2A-family phosphatases, given that these are known to constitute the majority of cellular phosphatase activity. In order to distinguish between these phosphatase subclasses, we included I-2 and fostriecin in the assay, which are selective inhibitors of the PP1- and PP2A-family phosphatases, respectively. Fostriecin displayed noticeable inhibition of SK1 dephosphorylation under these conditions, whereas I-2 showed no such effect (Fig. 1B), supporting a role for the PP2A-family phosphatases, but not PP1-family phosphatases, in regulating levels of phospho-SK1.
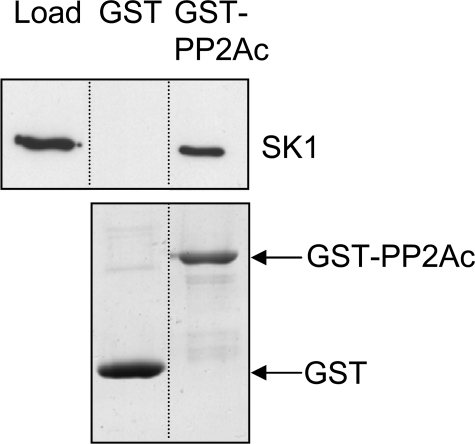
Direct Association of PP2Ac and Recombinant SK1—The previous assays demonstrated that PP2A activity reduced phospho-SK1 levels in cell lysates, but did not demonstrate a direct interaction between the proteins. To address whether PP2A exerted direct or indirect effects on phospho-SK1 levels, we prepared the catalytic subunit of PP2A as a GST fusion protein (GST-PP2Ac) and used this in pulldown analyses with purified recombinant human SK1 generated in insect cells (13). We found that this purified recombinant SK1 specifically associated with GST-PP2Ac, but not GST alone (Fig. 2), indicating a direct interaction between PP2Ac and SK1.
FIGURE 2.
Direct association of SK1 with PP2Ac. Purified recombinant SK1 (1 μg) was included in GST-pulldown assays with either GST alone or GST-PP2Ac (2 μg). Samples were subjected to SDS-PAGE analysis and bound SK1 was detected via its His tag by immunoblotting with an anti-His antibody (upper panel). Coomassie Blue staining was used to confirm the amounts of GST and GST-PP2Ac in each pulldown (lower panel). Results are representative of three independent experiments. The dividing lines indicate where lanes have been spliced to simplify viewing, but results in each panel are from a single experiment.
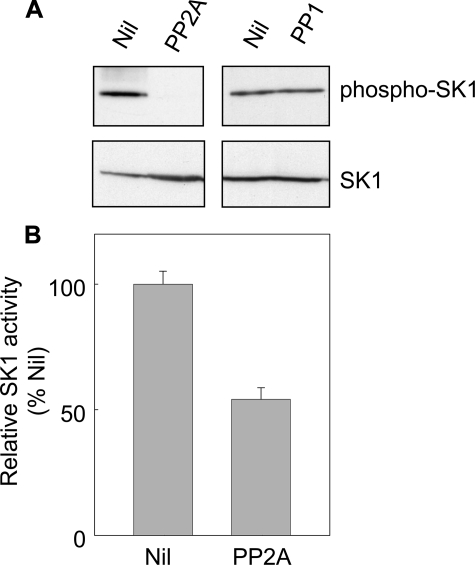
To reinforce the role of PP2A in SK1 dephosphorylation, we examined the ability of commercial preparations of PP1 and PP2A to dephosphorylate purified recombinant SK1 (13), which is known to be phosphorylated at Ser225 (22). Immunoblotting to detect levels of phospho-SK1 indicated that PP2A treatment resulted in substantial SK1 dephosphorylation, while, in contrast, PP1 had no effect on phospho-SK1 levels (Fig. 3A). Furthermore, PP2A-treated recombinant SK1 demonstrated reduced catalytic activity in vitro (Fig. 3B). This is consistent with observations that phosphorylation of SK1 at Ser225 correlates with an increase in the activity of the protein (6), and thus dephosphorylation of this site results in reduced SK1 activity.
FIGURE 3.
PP2A, but not PP1, promoted dephosphorylation of SK1 in vitro. A, purified recombinant SK1 (1 μg) was incubated at 37 °C for 30 min in phosphatase buffer with 1 unit of commercial PP2A or PP1. 32P-labeled phosphorylase a was used as a control substrate to standardize the activities of the commercial PP1 and PP2A phosphatases. One unit of phosphatase activity is defined as the amount required to release 1 nmol of 32P/minute from phosphorylase a. Dephosphorylation of SK1 was assessed by immunoblotting with an antibody recognizing phospho-SK1 (upper panel) and total SK1 levels were confirmed by immunoblotting with an anti-His antibody (lower panel). B, relative catalytic activity of SK1 in the untreated versus PP2A-treated extract. Data are mean (±S.D.) from three independent experiments.
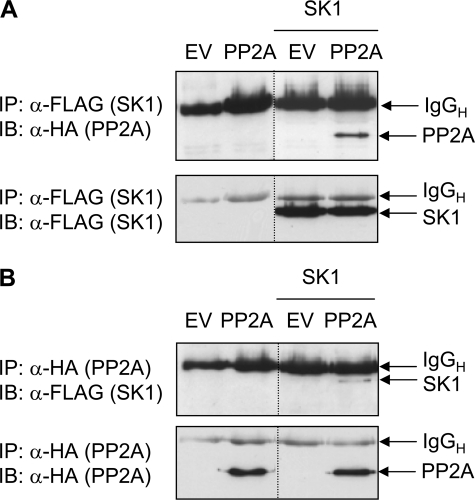
Coimmunoprecipitation of PP2Ac and SK1—The GST pulldown analysis above indicated that protein complexes of SK1 and PP2Ac could form in vitro. Next, we investigated whether similar protein complexes could form within cells by performing coimmunoprecipitation analysis using lysates from HEK293 cells coexpressing HA-tagged PP2Ac and FLAG-tagged SK1. We found that PP2Ac could be detected in SK1 immunoprecipitates (Fig. 4A), and additionally, SK1 was detected in equivalent PP2Ac immunoprecipitates (Fig. 4B). These results indicated that the catalytic subunit of PP2A could interact with SK1 protein not only in vitro, but also in a more physiologically relevant cellular context.
FIGURE 4.
Coimmunoprecipitation of SK1 and PP2Ac. A, HEK293 cells were transfected with empty vector (EV), or vector encoding HA-tagged PP2Ac, in the presence or absence of FLAG-tagged SK1 plasmid. Cell lysates were immunoprecipitated with anti-FLAG antibody to capture SK1 and coimmunoprecipitated PP2Ac was detected by immunoblotting with anti-HA antibody (upper panel). The lower panel indicates the presence of equal amounts of immunoprecipitated SK1. B, lysates were prepared as in A, but immunoprecipitated with anti-HA antibody to capture PP2Ac, and the associated SK1 was detected by immunoblotting with anti-FLAG antibody (upper panel). The lower panel indicates the presence of equal amounts of immunoprecipitated PP2Ac. Results are representative of three independent experiments. The dividing lines indicate where lanes have been spliced to simplify viewing, but results in each panel are from a single experiment performed on the same day. Detected immunoglobulin heavy chain (IgGH) from the immunoprecipitating antibody is indicated.
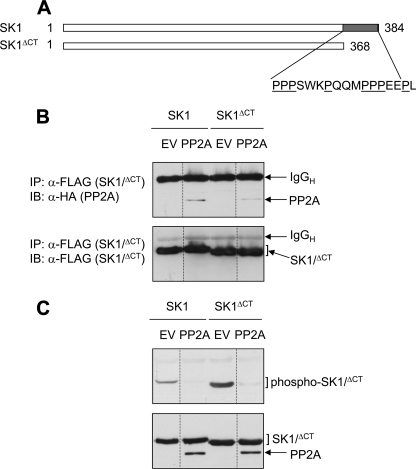
Role of the Pro-rich C Terminus of SK1 in Regulation by PP2Ac—Thus far, our data indicated a direct interaction between SK1 and PP2Ac, but did not highlight the binding interface(s) between these proteins. It seemed most likely that the PP2A catalytic subunit would interact with the phospho-Ser225 motif of SK1, given that its activity was directed toward this region of the protein. However, it seemed likely that other regions of the protein might also participate in the interaction, given previous reports of the interactions of the related PP2A-family member, PP4, and its substrates. The PP4 substrate hematopoietic progenitor kinase 1, an upstream activating kinase in the pathway leading to c-Jun N-terminal kinase activation, binds PP4c through a Pro-rich region in its C terminus (23). Because SK1 also possesses a Pro-rich C-terminal region (13), we sought to investigate its significance for the SK1-PP2Ac interaction. For these studies, HEK293 cells were cotransfected with PP2Ac and a truncated version of SK1, lacking the 17 extreme C-terminal residues, which constitutes the Prorich region (SK1ΔCT) (Fig. 5A). We found that while PP2Ac coimmunoprecipitated with both SK1 and SK1ΔCT, the amount of PP2Ac detected in SK1ΔCT immunoprecipitates was substantially reduced (Fig. 5B). This prompted us to examine whether this reduced interaction with PP2Ac also had implications for the levels of phosphorylated SK1ΔCT in cells. We found that similar to wildtype SK1, coexpression of PP2Ac reduced levels of SK1ΔCT phosphorylated at Ser225 (Fig. 5C). Strikingly, however, in the absence of PP2A coexpression, the basal levels of Ser225 phosphorylation were found to be considerably higher for SK1ΔCT compared with wild-type SK1 (Fig. 5C). In light of the reduced amount of PP2Ac detected in SK1ΔCT immunoprecipitates (Fig. 5B), it is possible that this could result from a reduced ability of this mutant to interact with endogenous PP2A. Taken together, these results suggest that while the Pro-rich C terminus of SK1 is not essential for the interaction with PP2Ac, it may contribute to both the affinity of the PP2Ac-SK1 interaction and also the ability of PP2Ac to dephosphorylate SK1 at Ser225.
FIGURE 5.
Role of the Pro-rich C terminus of SK1 in regulation by PP2Ac. A, comparison of the SK1 and SK1ΔCT sequences, indicating the Pro-rich C-terminal region that is not present in SK1ΔCT. B, HEK293 cells were transfected with vectors encoding either FLAG-tagged SK1 or SK1ΔCT, with either vector encoding HA-tagged PP2Ac or the corresponding empty vector (EV). Cell lysates were immunoprecipitated with anti-FLAG antibody to capture SK1/SK1ΔCT and the presence of associated PP2Ac was detected by immunoblotting with anti-HA antibody (upper panel). The bracket in the lower panel indicates the presence of equal amounts of immunoprecipitated SK1/SK1ΔCT. C, HEK293 cells were transfected as in B, and cell lysates were immunoblotted with an antibody recognizing phospho-SK1 (upper panel). Total SK1/SK1ΔCT and PP2Ac expression levels were confirmed by immunoblotting with anti-FLAG and anti-HA antibodies, respectively (lower panel). Results are representative of three independent experiments. The dividing lines indicate where lanes have been spliced to simplify viewing, but results in each panel are from a single experiment performed on the same day. Detected immunoglobulin heavy chain (IgGH) from the immunoprecipitating antibody is indicated.
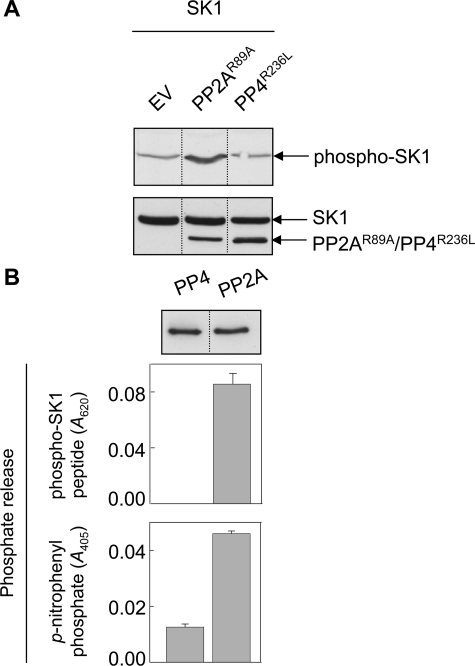
Implications of PP2A Activity for Cellular Phospho-SK1 Levels—Thus far, our overexpression studies strongly suggested that PP2A regulated cellular levels of phospho-SK1. We next performed complementary experiments using a catalytically inactive form of PP2A, given that the use of such a dominant-negative is a more physiological tool than overexpression of an active phosphatase. Thus, we cotransfected HEK293 cells with SK1 and catalytically inactive forms of either PP2Ac, or the related PP4c (PP2AcR89A or PP4cR236L, respectively). Immunoblotting analysis on these cell lysates for phospho-SK1 indicated that PP2AcR89A expression resulted in hyperphosphorylation of SK1 compared with the control cells (Fig. 6A). In contrast, coexpression of PP4cR236L had no effect on cellular phospho-SK1 levels (Fig. 6A). This again supported a role for PP2A in the regulation of SK1 phosphorylation, rather than its related family members, such as PP4. To further confirm this finding, we expressed PP2Ac and the related PP4c in HEK293 cells, immunoprecipitated the active phosphatases, and assessed their activities toward a phosphopeptide substrate based on the phospho-Ser225 region of SK1 (phospho-SK1 peptide) using an in vitro assay measuring phosphate release. PP2Ac dephosphorylated the phospho-SK1 peptide, whereas PP4c had no noticeable activity toward the substrate (Fig. 6B), despite both protein phosphatases displaying activity against a generic substrate, p-nitrophenyl phosphate (Fig. 6B). Notably, even when 3.5-fold more PP4c was used in these assays, dephosphorylation of the phospho-SK1 peptide substrate remained undetectable, while activity toward the pNPP substrate increased proportionately (data not shown). Thus, it appeared that PP2Ac, but not the related PP4c, had the ability to directly dephosphorylate the phospho-SK1 peptide. These results were in keeping with the above coexpression studies, and provided further evidence supporting a role for PP2A, rather than the related PP4, in regulating cellular levels of phospho-SK1.
FIGURE 6.
Regulation of cellular phospho-SK1 levels by PP2A. A, HEK293 cells were transfected with vectors encoding FLAG-tagged SK1 and HA-tagged catalytically inactive mutants of PP2Ac or PP4c (PP2AcR89A or PP4cR236L, respectively), or the corresponding empty vector (EV). Cell lysates were immunoblotted with an antibody recognizing phospho-SK1 (upper panel). Total SK1 and phosphatase expression levels were confirmed by immunoblotting with anti-FLAG and anti-HA antibodies, respectively (lower panel). Results are representative of three independent experiments. B, HEK293 cells were transfected with vectors encoding HA-tagged PP4c or PP2Ac and the phosphatases immunoprecipitated from cell lysates using an anti-HA antibody. Immunoprecipitations with no primary antibody were used as negative controls. Immunoprecipitated phosphatases present in the phosphatase assays were detected by immunoblotting with an anti-HA antibody (representative blot, upper panel). Immunoprecipitated phosphatases were incubated with either phospho-SK1 peptide for 30 min, and reactions were terminated with Malachite Green stop solution prior to reading the absorbance at 620 nm (middle panel), or incubated with p-nitrophenyl phosphate for 45 min prior to reading the absorbance at 405 nm (lower panel). Data are expressed as the phosphate release, relative to the negative controls. Data for the phospho-SK1 peptide substrate are mean (±S.E.) from three independent experiments, and data for the p-nitrophenyl phosphate substrate are mean (±range) from two independent experiments. All analyses in each experiment were performed in duplicate. The dividing lines indicate where lanes have been spliced to simplify viewing, but results in each panel are from a single experiment.
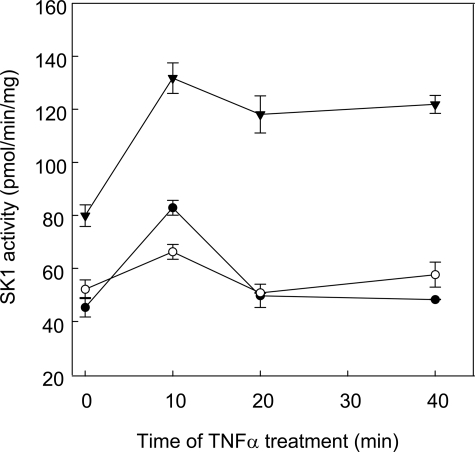
PP2A Mediates Deactivation of SK1 following TNFα Stimulation—Although our findings indicated a role for PP2A activity in SK1 regulation, all cellular experiments thus far relied on ectopic expression of SK1 and PP2Ac. We next chose to investigate the potential for PP2A to participate in the regulation of SK1 phosphorylation in a more physiological setting. It has been previously shown that endogenous SK1 activity and phospho-SK1 levels are transiently elevated in response to TNFα stimulation, peaking at 10 min and rapidly decreasing thereafter (6, 8, 24). Because total SK1 levels remained constant during this time (6), it appeared likely that PP2A may be responsible for this deactivation process. To examine this, we transfected HEK293 cells with either PP2Ac, or the catalytically inactive PP2Ac mutant (PP2AcR89A), prior to stimulating cells with TNFα and assessing endogenous SK1 activity in the cell lysates. As expected, empty vector control cells showed a transient increase in SK1 activity, peaking at 10 min and returning to baseline by 20–40 min (Fig. 7). Cells overexpressing wild-type PP2Ac showed a similar transient increase in SK1 activity, but the increase in TNFα-induced SK1 activity was markedly reduced (Fig. 7). Strikingly, cells transfected with catalytically inactive PP2AcR89A showed an elevated level of basal SK1 activity, and the TNFα-stimulated SK1 activity peaked at 10 min, but remained at elevated levels at 20–40 min (Fig. 7). Together these findings indicated that PP2A activity could indeed regulate endogenous SK1, and provided further support for PP2A as an endogenous regulator of SK1 dephosphorylation and deactivation.
FIGURE 7.
PP2A mediates deactivation of endogenous SK1 activity following TNFα stimulation. HEK293 cells were transfected with vectors encoding HA-tagged PP2Ac (○), catalytically-inactive PP2AcR89A (▾), or the corresponding empty vector (EV) (•). The cells were stimulated with TNFα (2 ng/ml) 24 h after transfection, for 0, 10, 20, or 40 min, harvested and analyzed for endogenous SK1 activity. Data are mean (±range) of duplicate determinations from a single experiment, and are representative of results obtained in two independent experiments.
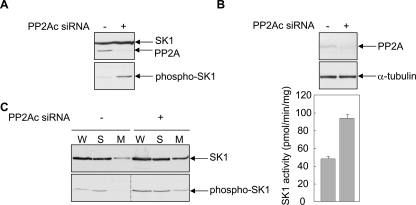
Silencing PP2Ac Expression Increases SK1 Phosphorylation, Activity, and Membrane Localization—To further examine the role of PP2A in SK1 regulation, we used siRNA directed toward the PP2A catalytic subunit to investigate the effects on SK1 phosphorylation, activity and the proportion of SK1 detected at cellular membranes. We first used overexpression of PP2Ac and SK1, to assess knockdown of PP2Ac expression by the siRNA, and because relatively low levels of endogenous SK1 expression hinder the detection of endogenous phospho-SK1. Thus, HEK293 cells were cotransfected with vectors encoding SK1, PP2Ac, and either PP2Ac siRNA or the equivalent control siRNA. Immunoblotting of the cell lysates indicated that reduced PP2Ac expression resulted in a marked increase in phospho-SK1 without altering levels of total SK1 (Fig. 8A). We next assessed the effects of silencing endogenous PP2Ac expression on endogenous SK1 activity in cells. PP2Ac siRNA markedly reduced endogenous PP2Ac expression, which resulted in an approximate 2-fold increase in endogenous SK1 activity relative to cells transfected with the control siRNA (Fig. 8B). Notably, this increase in SK1 activity was similar to the effect of overexpressing the catalytically inactive PP2AcR89A mutant (Fig. 7).
FIGURE 8.
Silencing PP2Ac expression increases SK1 phosphorylation, activity and the proportion of membrane-localized SK1. A, HEK293 cells were transfected with vectors encoding FLAG-tagged SK1 and HA-tagged PP2Ac, in the presence of siRNA targeting the PP2A catalytic subunit (+) or a negative control siRNA (-). Total SK1 and PP2Ac expression levels in the cell lysates were assessed by immunoblotting with anti-FLAG and anti-HA antibodies, respectively (upper panel) and levels of phosphorylated SK1 were assessed using an antibody recognizing phospho-SK1 (lower panel). Results are representative of triplicate transfections. B, HEK293 cells were transfected with either siRNA targeting the PP2A catalytic subunit (+), or a negative control siRNA (-). Cell lysates were immunoblotted with an antibody recognizing the PP2A catalytic subunit (upper panel), and equal protein loading was confirmed using an antibody recognizing α-tubulin (middle panel). Results are representative of triplicate transfections. The lower panel shows the mean (±range) for duplicate determinations of endogenous SK1 activity measured in the cell lysates and is representative of duplicate transfections. C, HEK293 cells were transfected with a vector encoding FLAG-tagged SK1, in the presence of siRNA targeting the PP2A catalytic subunit (+) or a negative control siRNA (-). Whole cell extracts (W), soluble fractions (S), and membrane-enriched fractions (M) were prepared from the transfected cells, which were then assessed for levels of total SK1 (upper panel) and phospho-SK1 (lower panel), by immunoblotting with anti-FLAG and anti-phospho-SK1 antibodies, respectively. Results are representative of three experiments. The dividing lines indicate where lanes have been spliced to simplify viewing, but results in each panel are from a single experiment.
Because phosphorylation of SK1 regulates its association with the plasma membrane (6, 22) we then further assessed whether silencing PP2Ac expression altered the proportion of soluble- versus membrane-localized SK1. Immunoblot analysis of soluble and membrane fractions from cells cotransfected with SK1 and either the control or PP2Ac siRNA demonstrated an increased amount of total SK1 in the membrane fraction following PP2Ac silencing (Fig. 8C). Furthermore, although PP2Ac silencing increased phospho-SK1 levels in the whole cell extracts, the proportion of membrane-bound phospho-SK1 was also substantially increased (Fig. 8C). Thus, endogenous PP2A activity appears important for SK1 phosphorylation, activity, and subcellular localization, further supporting an important role for PP2A as an endogenous regulator of SK1.
DISCUSSION
SK1 has been implicated in tumorigenesis, and we have previously demonstrated that phosphorylation of SK1 at Ser225 by ERK1/2 is essential for its oncogenic potential (7). However, phosphorylation status is often a balance between protein kinase and protein phosphatase activity, and to date, the corresponding Ser/Thr phosphatase responsible for dephosphorylation of SK1 has remained undefined. Here, we show through various lines of evidence that PP2A is the phosphatase responsible for SK1 dephosphorylation and deactivation.
SK1 and PP2A have been separately implicated in the regulation of cell survival, proliferation, and tumor formation, but our current work highlights an important link between these proteins. This is especially true when considering therapeutic strategies to combat oncogenesis and tumor formation. SK1 expression is elevated in several naturally occurring tumors (25, 26) and specific SK inhibitors have been reported to significantly reduce tumor growth in vivo in mice (25) and sensitize many tumor cells to chemotherapeutics (27). PP2A has also been implicated in the control of cell growth and is targeted by viral proteins that promote tumorigenesis (19, 28–30). Additionally, studies have indicated a decreased expression of selective PP2A subunits (Aα, Aβ, and B′/B56γ) in lung, colorectal, breast, and brain cancer and various cancer cell lines (31), and also shown that reduced expression of the PP2A catalytic subunit in prostate cancer significantly correlated with tumor stage (32). When considered separately, either elevated SK1 expression and activity, or decreased PP2A function, biases cells toward the pro-survival, pro-proliferative phenotype characteristic of tumors. However, in light of our current observations that PP2A functions as a regulator of SK1, it is feasible that the interplay between these proteins could contribute to disease pathogenesis.
The actions of PP2A are likely to be complex, given its ability to interact with a broad range of transcription factors, cell cycle regulators, and protein kinases (19). Furthermore, its physiological role is somewhat confounding, given that it can oppose the pro-growth signals associated with Ser/Thr phosphorylation (33–35), but also promote cell growth and survival (18, 36). This apparent discrepancy could be resolved by considering that specific PP2A holoenzymes in distinct subcellular locations have restricted substrate specificity and thus mediate specific subsets of cellular effects, along with the fact that PP2A activity can result in activation of some substrates and inactivation of others (35, 37–40). The importance of subcellular localization for PP2A activity is highlighted by studies in ovarian cancer cells, where cytosolic PP2A was observed to translocate to the plasma membrane in response to the gonadotropin-releasing hormone antagonist, cetrorelix, and contribute to the induction of apoptosis in these cells (41). Furthermore, membrane-associated PP2A had a lower Km for substrate, whereas cytosolic PP2A had a higher maximal velocity (42, 43). Thus, in future studies it will be of interest to determine whether PP2A regulates SK1 at multiple cellular locations. This is particularly relevant in light of our previous findings showing that phosphorylation of SK1 results in its translocation from the cytoplasm to the plasma membrane (6) and that this drives its oncogenic signaling (7).
SK1 plays a critical role in regulating the dynamic balance between prosurvival S1P and its proapoptotic precursors, sphingosine, and ceramide, a “sphingolipid rheostat” that controls cell fate (44). Notably, ceramide has been reported to activate heterotrimeric PP2A (45), which has been implicated in promoting apoptosis via dephosphorylation of Bcl2 (46, 47) and Bad (48). Given that the “sphingolipid rheostat” is likely to be tightly regulated, our current findings that PP2A dephosphorylates and deactivates SK1 appear somewhat counterintuitive. Tight regulation of the “sphingolipid rheostat” may imply that high levels of cellular ceramide should enhance, rather than reduce, its metabolism. We could speculate, however, that this may be a mechanism to enhance apoptotic efficiency, with the ceramide-stimulated PP2A activity deactivating SK1 as part of a positive feedback mechanism. This would shift the “sphingolipid rheostat” away from S1P and even further toward ceramide and cell death.
FTY720 (2-amino-2-[2-(4-octylphenyl)ethyl]-1,3-propanediol hydrochloride) is an immunomodulator that is structurally similar to sphingosine, and like ceramide, can also activate PP2A. Although FTY720 can be phosphorylated by sphingosine kinase 2 (49, 50) to form an S1P analog with activity at Gi protein-coupled S1P receptors (51, 52), these events are not required for PP2A activation. FTY720 can activate purified PP2A via a direct interaction (53) and activate cellular PP2A to effectively antagonize leukemogenesis in both in vitro and in vivo disease models (53, 54). Notably, FTY720 may also contribute to leukemia therapy via direct inhibition of SK1 (54). Again, the results of our current study allow us to hypothesize that PP2A-mediated deactivation of SK1 may also contribute to the antitumor effects of FTY720.
In summary, this study has identified PP2A as an important regulator of SK1. Further characterization of the interaction between these proteins might facilitate the discovery of specific drugs targeting the cellular effects of this protein-protein interaction. Furthermore, clarification of the roles that these proteins play in tumorigenesis may assist in the development of more effective therapeutic strategies to treat disease.
This work was supported by the Fay Fuller Foundation, a Senior Research Fellowship from the Cancer Council of South Australia (to Y. K.-G.), and the National Health and Medical Research Council of Australia through a Peter Doherty Postdoctoral Research Fellowship 430909 (to R. K. B.), a Senior Research Fellowship 508098 (to S. M. P.), and Project Grant 453512. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SK1, sphingosine kinase 1; ERK1/2, extracellular signal-regulated kinases 1 and 2; PP, protein phosphatase; PP2Ac, protein phosphatase 2A catalytic subunit; HEK293, human embryonic kidney cells; TNFα, tumor necrosis factor-α; S1P, sphingosine 1-phosphate; I-2, human recombinant protein phosphatase inhibitor-2; HA, hemagglutinin; GST, glutathione S-transferase; IgGH, immunoglobulin heavy chain.
References
- 1.Hannun, Y. A., and Obeid, L. M. (2008) Nat. Rev. Mol. Cell. Biol. 9 139-150 [DOI] [PubMed] [Google Scholar]
- 2.Spiegel, S., and Kolesnick, R. (2002) Leukemia 16 1596-1602 [DOI] [PubMed] [Google Scholar]
- 3.Leclercq, T. M., and Pitson, S. M. (2006) IUBMB Life 58 467-472 [DOI] [PubMed] [Google Scholar]
- 4.Pitson, S. M., Moretti, P. A., Zebol, J. R., Xia, P., Gamble, J. R., Vadas, M. A., D'Andrea, R. J., and Wattenberg, B. W. (2000) J. Biol. Chem. 275 33945-33950 [DOI] [PubMed] [Google Scholar]
- 5.Wattenberg, B. W., Pitson, S. M., and Raben, D. M. (2006) J. Lipid Res. 47 1128-1139 [DOI] [PubMed] [Google Scholar]
- 6.Pitson, S. M., Moretti, P. A., Zebol, J. R., Lynn, H. E., Xia, P., Vadas, M. A., and Wattenberg, B. W. (2003) EMBO J. 22 5491-5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pitson, S. M., Xia, P., Leclercq, T. M., Moretti, P. A., Zebol, J. R., Lynn, H. E., Wattenberg, B. W., and Vadas, M. A. (2005) J. Exp. Med. 201 49-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia, P., Wang, L., Gamble, J. R., and Vadas, M. A. (1999) J. Biol. Chem. 274 34499-34505 [DOI] [PubMed] [Google Scholar]
- 9.Pitson, S. M., D'Andrea, R. J., Vandeleur, L., Moretti, P. A., Xia, P., Gamble, J. R., Vadas, M. A., and Wattenberg, B. W. (2000) Biochem. J. 350 429-441 [PMC free article] [PubMed] [Google Scholar]
- 10.Ogris, E., Mudrak, I., Mak, E., Gibson, D., and Pallas, D. C. (1999) J. Virol. 73 7390-7398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu, M. C., Tang-Oxley, Q., Qiu, W. R., Wang, Y. P., Mihindukulasuriya, K. A., Afshar, R., and Tan, T. H. (1998) J. Biol. Chem. 273 33561-33565 [DOI] [PubMed] [Google Scholar]
- 12.Sutherland, C. M., Moretti, P. A., Hewitt, N. M., Bagley, C. J., Vadas, M. A., and Pitson, S. M. (2006) J. Biol. Chem. 281 11693-11701 [DOI] [PubMed] [Google Scholar]
- 13.Pitson, S. M., Moretti, P. A., Zebol, J. R., Zareie, R., Derian, C. K., Darrow, A. L., Qi, J., D'Andrea, R. J., Bagley, C. J., Vadas, M. A., and Wattenberg, B. W. (2002) J. Biol. Chem. 277 49545-49553 [DOI] [PubMed] [Google Scholar]
- 14.Tung, H. Y., Pelech, S., Fisher, M. J., Pogson, C. I., and Cohen, P. (1985) Eur. J. Biochem. 149 305-313 [DOI] [PubMed] [Google Scholar]
- 15.Mayer-Jaekel, R. E., Ohkura, H., Ferrigno, P., Andjelkovic, N., Shiomi, K., Uemura, T., Glover, D. M., and Hemmings, B. A. (1994) J. Cell Sci. 107 2609-2616 [DOI] [PubMed] [Google Scholar]
- 16.Zhou, G., Mihindukulasuriya, K. A., MacCorkle-Chosnek, R. A., Van Hooser, A., Hu, M. C., Brinkley, B. R., and Tan, T. H. (2002) J. Biol. Chem. 277 6391-6398 [DOI] [PubMed] [Google Scholar]
- 17.Cohen, P. T., Philp, A., and Vazquez-Martin, C. (2005) FEBS Lett. 579 3278-3286 [DOI] [PubMed] [Google Scholar]
- 18.Strack, S., Cribbs, J. T., and Gomez, L. (2004) J. Biol. Chem. 279 47732-47739 [DOI] [PubMed] [Google Scholar]
- 19.Janssens, V., and Goris, J. (2001) Biochem. J. 353 417-439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prickett, T. D., and Brautigan, D. L. (2006) J. Biol. Chem. 281 30503-30511 [DOI] [PubMed] [Google Scholar]
- 21.Brewis, N. D., Street, A. J., Prescott, A. R., and Cohen, P. T. (1993) EMBO J. 12 987-996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stahelin, R. V., Hwang, J. H., Kim, J. H., Park, Z. Y., Johnson, K. R., Obeid, L. M., and Cho, W. (2005) J. Biol. Chem. 280 43030-43038 [DOI] [PubMed] [Google Scholar]
- 23.Zhou, G., Boomer, J. S., and Tan, T. H. (2004) J. Biol. Chem. 279 49551-49561 [DOI] [PubMed] [Google Scholar]
- 24.Xia, P., Wang, L., Moretti, P. A., Albanese, N., Chai, F., Pitson, S. M., D'Andrea, R. J., Gamble, J. R., and Vadas, M. A. (2002) J. Biol. Chem. 277 7996-8003 [DOI] [PubMed] [Google Scholar]
- 25.French, K. J., Schrecengost, R. S., Lee, B. D., Zhuang, Y., Smith, S. N., Eberly, J. L., Yun, J. K., and Smith, C. D. (2003) Cancer Res. 63 5962-5969 [PubMed] [Google Scholar]
- 26.Kawamori, T., Osta, W., Johnson, K. R., Pettus, B. J., Bielawski, J., Tanaka, T., Wargovich, M. J., Reddy, B. S., Hannun, Y. A., Obeid, L. M., and Zhou, D. (2006) FASEB J. 20 386-388 [DOI] [PubMed] [Google Scholar]
- 27.Pchejetski, D., Golzio, M., Bonhoure, E., Calvet, C., Doumerc, N., Garcia, V., Mazerolles, C., Rischmann, P., Teissie, J., Malavaud, B., and Cuvillier, O. (2005) Cancer Res. 65 11667-11675 [DOI] [PubMed] [Google Scholar]
- 28.Mumby, M. (1995) Semin. Cancer Biol. 6 229-237 [DOI] [PubMed] [Google Scholar]
- 29.Hahn, W. C., Dessain, S. K., Brooks, M. W., King, J. E., Elenbaas, B., Sabatini, D. M., DeCaprio, J. A., and Weinberg, R. A. (2002) Mol. Cell. Biol. 22 2111-2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu, J., Boyapati, A., and Rundell, K. (2001) Virology 290 192-198 [DOI] [PubMed] [Google Scholar]
- 31.Sontag, J. M., and Sontag, E. (2006) Cell Mol. Life Sci. 63 2979-2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh, A. P., Bafna, S., Chaudhary, K., Venkatraman, G., Smith, L., Eudy, J. D., Johansson, S. L., Lin, M. F., and Batra, S. K. (2008) Cancer Lett. 259 28-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mumby, M. C., and Walter, G. (1993) Physiol. Rev. 73 673-699 [DOI] [PubMed] [Google Scholar]
- 34.Schonthal, A. H. (1998) Front Biosci. 3 D1262-D1273 [DOI] [PubMed] [Google Scholar]
- 35.Schonthal, A. H. (2001) Cancer Lett. 170 1-13 [DOI] [PubMed] [Google Scholar]
- 36.Silverstein, A. M., Barrow, C. A., Davis, A. J., and Mumby, M. C. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 4221-4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomez, N., and Cohen, P. (1991) Nature. 353 170-173 [DOI] [PubMed] [Google Scholar]
- 38.Mueller, P. R., Coleman, T. R., and Dunphy, W. G. (1995) Mol. Biol. Cell 6 119-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cegielska, A., Gietzen, K. F., Rivers, A., and Virshup, D. M. (1998) J. Biol. Chem. 273 1357-1364 [DOI] [PubMed] [Google Scholar]
- 40.Ricciarelli, R., and Azzi, A. (1998) Arch. Biochem. Biophys. 355 197-200 [DOI] [PubMed] [Google Scholar]
- 41.Imai, A., Sugiyama, M., Furui, T., and Tamaya, T. (2006) J. Obstet. Gynaecol. 26 37-41 [DOI] [PubMed] [Google Scholar]
- 42.Sugiyama, M., Imai, A., Furui, T., and Tamaya, T. (2003) Am. J. Obstet. Gynecol. 189 1666-1669 [DOI] [PubMed] [Google Scholar]
- 43.Sugiyama, M., Imai, A., Furui, T., and Tamaya, T. (2003) Oncol. Rep. 10 1885-1889 [PubMed] [Google Scholar]
- 44.Cuvillier, O., Pirianov, G., Kleuser, B., Vanek, P. G., Coso, O. A., Gutkind, S., and Spiegel, S. (1996) Nature 381 800-803 [DOI] [PubMed] [Google Scholar]
- 45.Dobrowsky, R. T., Kamibayashi, C., Mumby, M. C., and Hannun, Y. A. (1993) J. Biol. Chem. 268 15523-15530 [PubMed] [Google Scholar]
- 46.Ruvolo, P. P., Clark, W., Mumby, M., Gao, F., and May, W. S. (2002) J. Biol. Chem. 277 22847-22852 [DOI] [PubMed] [Google Scholar]
- 47.Ruvolo, P. P., Deng, X., Ito, T., Carr, B. K., and May, W. S. (1999) J. Biol. Chem. 274 20296-20300 [DOI] [PubMed] [Google Scholar]
- 48.Xin, M., and Deng, X. (2006) J. Biol. Chem. 281 18859-18867 [DOI] [PubMed] [Google Scholar]
- 49.Paugh, S. W., Payne, S. G., Barbour, S. E., Milstien, S., and Spiegel, S. (2003) FEBS Lett. 554 189-193 [DOI] [PubMed] [Google Scholar]
- 50.Kharel, Y., Lee, S., Snyder, A. H., Sheasley-O'neill, S. L., Morris, M. A., Setiady, Y., Zhu, R., Zigler, M. A., Burcin, T. L., Ley, K., Tung, K. S., Engelhard, V. H., Macdonald, T. L., Pearson-White, S., and Lynch, K. R. (2005) J. Biol. Chem. 280 36865-36872 [DOI] [PubMed] [Google Scholar]
- 51.Brinkmann, V., Davis, M. D., Heise, C. E., Albert, R., Cottens, S., Hof, R., Bruns, C., Prieschl, E., Baumruker, T., Hiestand, P., Foster, C. A., Zollinger, M., and Lynch, K. R. (2002) J. Biol. Chem. 277 21453-21457 [DOI] [PubMed] [Google Scholar]
- 52.Mandala, S., Hajdu, R., Bergstrom, J., Quackenbush, E., Xie, J., Milligan, J., Thornton, R., Shei, G. J., Card, D., Keohane, C., Rosenbach, M., Hale, J., Lynch, C. L., Rupprecht, K., Parsons, W., and Rosen, H. (2002) Science 296 346-349 [DOI] [PubMed] [Google Scholar]
- 53.Matsuoka, Y., Nagahara, Y., Ikekita, M., and Shinomiya, T. (2003) Br. J. Pharmacol. 138 1303-1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neviani, P., Santhanam, R., Oaks, J. J., Eiring, A. M., Notari, M., Blaser, B. W., Liu, S., Trotta, R., Muthusamy, N., Gambacorti-Passerini, C., Druker, B. J., Cortes, J., Marcucci, G., Chen, C. S., Verrills, N. M., Roy, D. C., Caligiuri, M. A., Bloomfield, C. D., Byrd, J. C., and Perrotti, D. (2007) J. Clin. Investig. 117 2408-2421 [DOI] [PMC free article] [PubMed] [Google Scholar]