Abstract
FE65 is an adaptor protein that binds to and forms a transcriptionally active complex with the γ-secretase-derived amyloid precursor protein (APP) intracellular domain. The regulatory mechanisms of FE65-APP-mediated transcription are still not clear. In this report, we demonstrate that Dexras1, a Ras family small G protein, binds to FE65 PTB2 domain and potently suppresses the FE65-APP-mediated transcription. The suppression is not via competition for binding of FE65 between Dexras1 and APP because the two proteins can simultaneously bind to the FE65 PTB2 domain. Phosphorylation of FE65 tyrosine 547 within the PTB2 domain has been shown to enhance FE65-APP-mediated transcription but not to influence binding to APP. Here we find that this phosphorylation event reduces the binding between Dexras1 and FE65. We also demonstrate that Dexras1 inhibits the FE65-APP-mediated transcription of glycogen synthase kinase 3β (GSK3β). Moreover, small interfering RNA knockdown of Dexras1 enhances GSK3β expression and increases phosphorylation of Tau, a GSK3β substrate. Thus, Dexras1 functions as a suppressor of FE65-APP-mediated transcription, and FE65 tyrosine 547 phosphorylation enhances FE65-APP-mediated transcription, at least in part, by modulating the interaction between FE65 and Dexras1. These findings reveal a novel regulatory mechanism for FE65-APP-mediated signaling.
FE65 is an adaptor protein with multiple protein-protein interaction domains including a WW domain and two C-terminal PTB domains (1). It is believed that FE65 functions as a “scaffold” protein to recruit various binding partners together to form a functional complex. In fact, FE65 has been shown to interact with a number of proteins. For example, transcription factors CP2 and Tip60 interact with the FE65 PTB1 (2, 3), c-Abl tyrosine kinase and Mena are FE65 WW domain binding partners (4, 5) and the nucleosome assembly factor SET binds FE65 (6). Of great interest, the Alzheimer disease amyloid precursor protein (APP)2 has been shown to interact with the FE65 PTB2 domain (7–10).
APP is a ubiquitously expressed type I integral transmembrane protein with a large ectodomain and a short intracellular domain (11, 12). The functions of APP are not properly understood. However, APP is known to be processed first by either α-or β-secretase and then by γ-secretase. Cleavage of APP by β- and γ-secretases results in the generation of the 4-kDa β-amyloid peptide (Aβ). Aggregation of Aβ to form neuritic plaques in brains is a pathological hallmark of Alzheimer disease (for reviews, see Refs. 1 and 13). FE65 has been shown to modulate the production of Aβ (14–16). In addition to Aβ generation, cleavage of the APP by γ-secretase releases the APP intracellular domain (AICD). Importantly, AICD has been shown to translocate to the nucleus as a complex with FE65 and the FE65·AICD complex strongly stimulates transcription of a GAL4-dependent reporter system (3, 17–20). However, the control mechanism(s) of FE65-AICD nuclear transcription is currently unclear.
Nuclear transcription can be regulated by various means including hormonal control (for reviews, see Refs. 21–23). Glucocorticoids are steroid hormones and are known to be involved in control of gene transcription (for reviews, see Refs. 24 and 25). There is evidence to suggest that Alzheimer disease is linked to abnormal functions of glucocorticoids (26–35). Thus, glucocorticoid-regulated genes may contribute to some aspects of Alzheimer disease. Dexras1 is a member of Ras family small G protein that is induced by dexamethasone (an analogue of glucocorticoid) and contains an extended C terminus that is found to interact with the PTB domain of CAPON (36, 37), an adaptor protein that interacts with neuronal nitric-oxide synthase. Dexras1 is widely expressed in various brain regions with high levels in the cerebellum and hippocampus, which is similar to the distribution pattern of FE65 in the brain (37–39). As Dexras1 has been shown to interact with PTB domain bearing protein and FE65 contains two PTB domains, these observations led us to investigate if Dexras1 and FE65 interact. In this study, we demonstrate that Dexras1 is an FE65 PTB2 domain interacting protein and that this interaction is regulated by phosphorylation of FE65 Tyr547 within the PTB2. Moreover, both Dexras1 and FE65 are found in the nucleus and FE65-APP-mediated transcription is significant repressed by Dexras1. Our data reveals a novel regulatory mechanism for the FE65-APP-mediated transcription.
EXPERIMENTAL PROCEDURES
All the experiments were performed at least three times with similar results.
Cell Culture and Transfection—CHO, HEK293 and SH-SY5Y cells were cultured as described previously (40, 41). Primary rat cortical neurons were obtained from E18 rat embryos and cultured on either culture plates or glass coverslips (for indirect immunofluorescence staining) coated with poly-d-lysine in Neurobasal medium and B27 supplement (Invitrogen) containing 100 units/ml penicillin, 100 mg/ml streptomycin (Invitrogen), and 2 mm glutamine (Invitrogen). Neurons were cultured for 7 days prior to analyses. For plasmid transfections, CHO cells were transfected using Lipofectamine (Invitrogen), HEK293 and SHSY5Y cells were transfected using FuGENE 6 (Roche), and rat cortical neurons were transfected with Lipofectamine 2000 (Invitrogen). The transfection efficiency for neurons was ∼5%, which was determined by counting the number of green fluorescent protein-transfected cells in pilot experiments and is consistent with other studies from our laboratory. siRNA knockdown was performed using human Dexras1 and control non-targeting siRNAs (Dharmacon). siRNAs were transfected using Lipofectamine 2000 (Invitrogen). Efficiency of knockdown was determined using semi-quantitative PCR and immunoblotting (see below).
Antibodies—Antibodies were as follows: anti-myc 9B11 (Cell Signaling Technology), Dexras1 (Santa Cruz and Abcam), tubulin DM1A (Sigma), c-Jun (Santa Cruz), anti-GST (Sigma), c-Abl (Santa Cruz), total Tau (DAKO), and anti-FLAG antibodies (Sigma). Anti-FE65, anti-APP, and anti-phospho-Tau PHF-1 were as described (42, 43).
Plasmids—Mammalian expression constructs for myc-tagged FE65, APP695, and GAL4-AICD were as described (42, 44). Wild-type Tau 2N4R construct was as described (45). Full-length APP into which the GAL4 DNA binding domain was inserted after the APP transmembrane domain (pMst-APP) and myc-tagged FE65 constructs, with either PTB1 or PTB2 deleted were as described (3). The pCMV-GST-Dexras1 mammalian expression construct was as described (37). FLAG-tagged Dexras1 wild-type and A178V mutant were generated by subcloning the corresponding full-length cDNA isolated either from pcDNA3.1/His-Dexras1 or pcDNA3.1/His-Dexras1(A178V) (46) into pCMV-Tag2 (Stratagene). GAL4UAS-dependent firefly luciferase reporter pFR Luc and transfection efficiency vector Renilla luciferase phRL-TK plasmids were obtained from Stratagene and Promega, respectively. Human GSK3β promoter luciferase reporter construct (GSK3β promoter Luc) was generated by subcloning the promoter fragment isolated from the p-2090CAT GSK3β promoter construct into pGL3 (47).
Protein Binding Assays—For mammalian GST fusion protein binding assays, CHO cells were transfected with GST + FE65, GST-Dexras1 + FE65, GST + APP, GST-Dexras1 + APP, GST + FE65 + APP, or GST-Dexras1 + FE65 + APP. Cells were harvested in ice-cold cell “lysis buffer.” This comprised 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, and Complete™ proteinase inhibitor (Roche) and was used in a number of procedures below where it is also termed lysis buffer. Following lysis, cells were cleared by centrifugation at 14,000 × g. The cell lysates were incubated with glutathione-Sepharose at 4 °C for 1 h. The captured proteins were then isolated by boiling in SDS-PAGE sample buffer and analyzed by SDS-PAGE and immunoblotting all as described (48, 49).
The bacterial GST-Dexras1 expression construct was created by subcloning the full-length Dexras1 cDNA isolated from pCMV-GST-Dexras1 into pGEX-5X2 (GE Healthcare). GST-Dexras1 fusion protein was expressed in Escherichia coli BL21 and captured by glutathione-Sepharose 4B according to the manufacturer's instructions (GE Healthcare). GST or GST-Dexras1 “baits” were used in pull-down assays from FE65-transfected cell lysates. The captured proteins were analyzed by SDS-PAGE and Western blotting.
For immunoprecipitation, CHO cells transfected either with myc-tagged FE65 + FLAG-tagged Dexras1 or Dexras1 were harvested in ice-cold lysis buffer as described above. Myc-tagged FE65 was immunoprecipitated from cell lysates using 9B11 anti-myc antibody for 16 h at 4 °C. The antibody was captured by protein G-Sepharose (Sigma) for 2 h at 4 °C and the immunoprecipitates were washed 3 times with ice-cold lysis buffer. Proteins in the immunoprecipitates were analyzed by SDS-PAGE and Western blotting. To detect the tripartite complex of Dexras1·FE65·APP, CHO cells were transfected either with Dexras1 + APP or Dexras1 + FE65 + APP. FLAG-tagged Dexras1 was immunoprecipitated using M2 monoclonal anti-FLAG antibody and the proteins in immunoprecipitates were detected as described above. Endogenous FE65/Dexras1 interaction was determined by immunoprecipitation of FE65 from rat brain lysate. Rat brain lysate was prepared by homogenizing a fresh rat brain in ice-cold lysis buffer and then cleared by centrifugation as described above. FE65 and Dexras1 in the immunoprecipitate were detected by a rabbit anti-FE65 polyclonal antibody and a goat anti-Dexras1 antibody, respectively. Signal intensities on immunoblots were quantified by pixel densitometry using a Bio-Rad GS710 imager with Quantity 1 software as previously described (50).
Indirect Immunofluorescence Staining—Transfected HEK293 cells and 7-day-old rat cortical neurons cultured on glass coverslips were fixed in 4% paraformaldehyde for 10 min followed by permeabilization with 0.1% Triton X-100 in phosphate-buffered saline for 20 min and blocked by blocking solution containing 5% fetal bovine serum in phosphate-buffered saline. The cells were then stained with primary antibodies diluted in blocking solution. Myc-tagged FE65 was detected either by an FE65 polyclonal antibody (44) or 9B11 monoclonal antibody. Dexras1 was detected using M2 anti-FLAG antibody (Sigma) or a goat anti-dexras1 antibody (Santa Cruz). Goat anti-rabbit, goat anti-mouse, and rabbit anti-goat Ig coupled with Alexa Fluor 488 or Alexa Fluor 568 (Molecular Probes) were used to visualize the primary antibodies by confocal microscopy (Zeiss). Nuclei were stained by 4′,6-diamidino-2-phenylindole (Sigma).
Subcelluar Fractionation—Subcelluar fractionation of cells was performed as described (51) and the purity of the different fractions determined by probing with fraction-specific markers (tubulin and c-Jun).
FE65-APP/Dexras1 Competition Assay—For competition assays, CHO cells were transfected with FE65 + APP and increasing amounts of Dexras1 DNA, or with FE65 + Dexras1 with increasing amounts of APP DNA. Transfections were balanced with pCIneoCAT DNA (vector containing the chloramphenicol acetyltransferase gene) such that all cells received the same total amounts of plasmid. Cell lysates were prepared by scraping cells into ice-cold lysis buffer comprising 50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100 supplemented with protease inhibitors (Complete, Roche Molecular Biochemicals) and then cleared by centrifugation at 14,000 × g for 10 min at 4 °C. Cell lysates were then incubated for 16 h with the 9B11 anti-myc monoclonal antibody against the myc tag of FE65 at 4 °C. The antibody was captured by protein G-Sepharose (Sigma) for 2 h at 4 °C and the immunoprecipitates were washed 3 times with ice-cold lysis buffer. The proteins in the immunoprecipitates were analyzed by SDS-PAGE and Western blotting.
GAL4-APP and GSK3β Promoter Luciferase Reporter Assays—Luciferase assays for GAL4 APP reporter and GSK3β promoter reporter transactivation were performed by a Dual-Glo luciferase assay system (Promega). In the GAL4 reporter assay, cells were transfected with the relevant constructs together with pFR-Luc and phRL-TK. In the GSK3β promoter assay, cells were transfected with the relevant constructs together with the GSK3β promoter reporter (GSK3β promoter Luc) and phRL-TK. phRL-TK, which expresses the Renilla luciferase, was used as a control to quantify transfection efficiency. Cells were harvested in Dual-Glo luciferase substrate at 48 h post-transfection. The firefly luciferase activities produced by pFR-Luc and GSK3β promoter Luc were measured by a luminometer (Wallace). Then, the Renilla luciferase activities produced by the phRL-TK were assayed by adding an equal volume of Dual-Glo Stop&Glo substrate (comprising the stop solution for firefly luciferase and substrate for Renilla luciferase) and analyzed by the luminometer. The firefly luciferase activity was normalized to the corresponding Renilla luciferase activity. For all reporter gene assays, statistical analyses were performed using analysis of variance tests. Significance is indicated between different treatments as * (p < 0.05), ** (p < 0.005), *** (p < 0.001). Error bars shown are standard deviations.
Semi-quantitative Polymerase Chain Reaction Analyses—Total RNA was isolated from transfected HEK293 cells by TRIzol reagent (Invitrogen) and was reverse transcribed into first strand cDNA in the presence of oligo(dT) primer by using the First Strand cDNA synthesis kit for reverse transcriptase-PCR (Roche). Amplification of Dexras1 was performed by using the following two primers:(5′-AGCCGAGGGTGGATTTATCT-3′ and 5′-AACCCGGAATCACAGACAAG-3′. PCR of GSK3β and glyceraldehyde-3-phosphate dehydrogenase were performed as described previously (52).
RESULTS
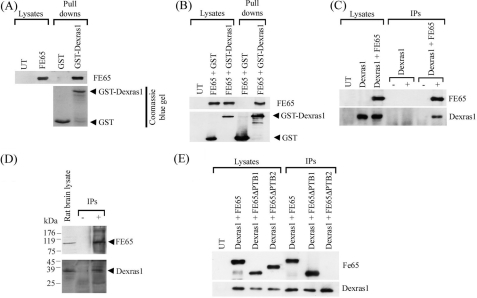
Dexras1 Is an FE65 Interacting Protein—Dexras1 is a dexamethasone (an analogue of glucocorticoid)-induced protein and is highly expressed in brain. To explore the possibility that FE65 and Dexras1 might interact, we first tested whether FE65 and Dexras1 interact in bacterial GST fusion protein binding assays using GST or GST-Dexras1 expressed from E. coli as baits to pull down FE65 from transfected CHO cell lysate. Probing the protein pulled down by the baits revealed that FE65 bound to the GST-Dexras1 bait but not GST (Fig. 1A).
FIGURE 1.
FE65 interacts with Dexras1 via its PTB2 domain. A, E. coli expressed GST and GST-Dexras1 were used as bait in pull-down assays from FE65-transfected cells. FE65 was detected using the myc sequence engineered to the C terminus of FE65. B, FE65 was either co-transfected to cells with mammalian expression constructs for GST or GST-Dexras1. GST and GST-Dexras1 were captured from the cell lysates by glutathione-Sepharose 4B and FE65 bound to the complex was detected as above. C, immunoprecipitations were performed from cells either transfected with Dexras1 or Dexras1 + FE65. FE65 was immunoprecipitated by myc antibody 9B11. Immunoprecipitated FE65 was detected by a rabbit anti-FE65 antibody and co-immunoprecipitated Dexras1 was detected by a rabbit anti-FLAG antibody to the FLAG sequence placed at the N terminus of Dexras1. (–) and (+) refer to the absence or presence of 9B11 in the immunoprecipitations. D, endogenous FE65·Dexras1 complex was detected by immunoprecipitating FE65 from rat brain lysate and probing for Dexras1. (–) and (+) refer to the absence or presence of anti-FE65 antibody in the immunoprecipitations with minus (–) indicating non-immune serum. Under these conditions APP co-immunoprecipitates with FE65 (data not shown). E, FLAG-Dexras1 was co-transfected with myc-tagged FE65, FE65ΔPTB1, or FE65ΔPTB2 into CHO cells. FLAG-Dexras1 was immunoprecipitated and detected by an anti-FLAG antibody, and FE65 in the immunoprecipitates was detected by using the myc tag.
To confirm FE65 and Dexras1 interaction in a mammalian system, FE65 was transfected into CHO cells either with GST or GST-Dexras1. GST or GST-Dexras1 was pulled down by using glutathione-Sepharose from the transfected cell lysate. Again, probing the pulled down protein complex revealed that FE65 was present in GST-Dexars1 + FE65 co-transfected cells but not in GST + FE65 co-transfected cells (Fig. 1B).
We next tested FE65 and Dexras1 interaction by co-immunoprecipitation. FLAG-tagged Dexras1 was transfected to CHO cells either alone or with myc-tagged FE65. FE65 was immunoprecipitated using an anti-myc antibody. Immunoblotting showed that Dexras1 was co-immunoprecipitated with FE65 in Dexras1 + FE65 co-transfected cells but not in Dexras1 only transfected cells (Fig. 1C). The existence of an endogenous FE65·Dexras1 complex was confirmed by co-immunoprecipitation of the endogenous proteins from rat brain (Fig. 1D).
Dexras1 has been demonstrated to bind to other PTB domain containing protein (37). To inquire which FE65 PTB domain mediates the binding of Dexras1, we used full-length FE65 and FE65 constructs with either PTB1 or PTB2 deleted in co-immunoprecipitation assays. We found that only full-length FE65 and FE65ΔPTB1, but not FE65ΔPTB2, could co-immunoprecipitate with Dexras1 (Fig. 1E). This indicates that the FE65 PTB2 domain is required for the binding of Dexras1.
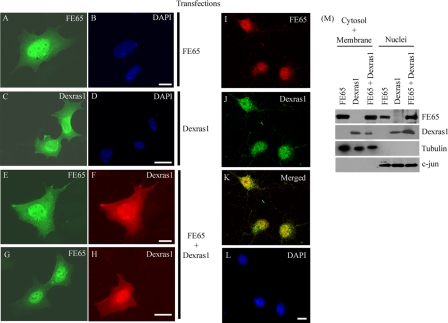
Dexras1 and FE65 Colocalize in Nuclei—To interact in cells, FE65 and Dexras1 must be localized in the same cellular compartments. We therefore examined the localization of FE65 and Dexras1 in transfected HEK293 cells. When transfected alone, FE65 was present in both nuclei and cytoplasm as reported previously (3, 53, 54) (Fig. 2A). Dexras1 was expressed in both the cytoplasm (particularly in perinuclear regions) and nuclei when transfected alone (Fig. 2C). However, when FE65 and Dexras1 were co-transfected, the amount of Dexras1 in the nuclei was increased dramatically (Fig. 2, F and H). There was no noticeable difference in the distribution of FE65 in the presence or absence of Dexras1 (Fig. 2, A, E, and G). To confirm these distributions, we prepared nuclear and cytosolic/membrane fractions from the transfected cells and probed for the presence of Dexras1 and FE65 by immunoblotting. Both FE65 and Dexras1 were present in the cytosol/membrane and nuclear fractions but transfection of FE65 induced a marked increase in nuclear and a corresponding decrease in cytosol/membrane Dexras1 (Fig. 2M). A minor increase in the proportion of nuclear FE65 was also detected in the presence of Dexras1 in these assays although this could not be detected in the non-quantified immunocytochemical assays. Finally, we studied the localization of endogenous FE65 and Dexras1 in primary cortical neurons by confocal microscopy. FE65 and Dexras1 co-localized within the nuclei of these cells (Fig. 2, I–L). Together, these findings indicate that FE65 and Dexras1 are colocalized in cells and one possibility is that FE65 may function to translocate Dexras1 from the cytosol to the nucleus.
FIGURE 2.
FE65 co-localizes with Dexras1 to the nucleus. Immunofluorescent staining of HEK293 cells transfected either with FE65 (A and B), Dexras1 (C and D) or FE65 + Dexras1 (E–H). A, E, and G, labeled for FE65; C, F, and H, labeled for Dexras1. Confocal imaging show endogenous FE65 (I) and Dexras1 (J) colocalize in the nucleus of rat cortical neurons (K, overlaid image). B, D, and L, labeled for nucleus by 4′,6-diamidino-2-phenylindole (DAPI); scale bars are 10 μm. M, subcellular fractionation of FE65, Dexras1, and FE65 + Dexras1-transfected HEK293 cells. The combined cytoplasmic + membrane and nuclear fractions were probed with antibodies for FE65, Dexras1, and subcellular fraction specific markers (tubulin and c-jun).
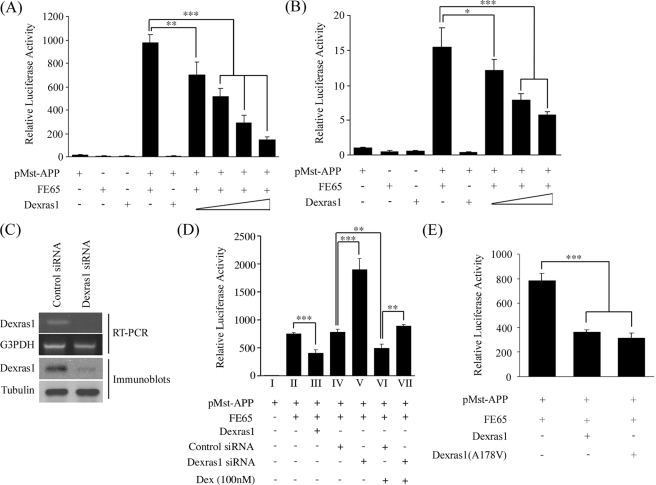
Dexras1 Inhibits FE65-APP-mediated Transcription—γ-Secretase-cleaved AICD has been shown to translocate to the nucleus with FE65 and the complex has been reported to participate in transcription events using GAL4-dependent reporter gene assays (3, 17–19). The finding that Dexras1 interacts and colocalizes with FE65 in nuclei prompted us to investigate the role of Dexras1 in FE65-AICD signaling. We used a previously described GAL4-dependent reporter system that involves monitoring the transcriptional activity of the APP-GAL4 DNA binding domain fusion genes using a GAL4UAS-luciferease reporter. GAL4-APP transcription was strongly stimulated by transfection of FE65. Dexras1 alone did not cause any transactivation. However, the FE65-stimulated transcription was repressed by co-transfection with Dexras1 in a dose-dependent manner in both HEK293 cells (Fig. 3A) and primary cortical neurons (Fig. 3B). A similar effect was also observed in SHSY5Y neuroblastoma cells (data not shown).
FIGURE 3.
Dexras1 represses FE65-APP-dependent transcription. HEK293 cells (A) and cortical neurons (B) were transfected with the constructs indicated. Transcription induced by a fusion gene comprising the GAL4 DNA binding domain fused to the full-length APP (pMst-APP) is stimulated by FE65. The FE65-stimulated transcription was repressed by Dexras1 in a dose-dependent manner. The amounts of Dexras1 DNA transfected in A were 0.05, 0.1, 0.2 and 0.4 μg, and in B were 0.25, 0.5, and 1 μg. C, siRNA knockdown of Dexras1 mRNA and protein in HEK293 cells. D, FE65-stimulated transcription is increased by siRNA knockdown of Dexras1. FE65-APP transcription was also repressed by dexamethasone (Dex), which induces the expression of Dexras1 (36, 55, 56). The inhibitory effect of dexamethasone on FE65-APP signaling was partially abrogated by siRNA knockdown of Dexras1. E, Dexras1(A178V) mutant, in which the guanyl nucleotide-binding pocket is interrupted, also inhibits FE65-APP transcription. A, B, D, and E, n ≥ 12.
To determine whether inhibition of Dexras1 expression might increase FE65-dependent transcriptional activity, we reduced Dexras1 expression by transfection of siRNAs in HEK293 cells. Dexras1 but not control siRNAs led to a decrease in Dexras1 mRNA and protein levels (Fig. 3C). This in turn stimulated FE65-dependent transcription (Fig. 3D, cf. histograms IV versus V). Thus, overexpression and inhibition of Dexras1 lead to complementary changes in FE65-mediated gene transcription.
We also treated HEK293 cells transfected with FE65-APP GAL4-dependent reporter constructs with dexamethasone because Dexras1 was first identified as a dexamethasone-induced protein (36, 55, 56). After dexamethasone treatment, FE65-APP signaling was inhibited, which is in line with the result we observed from transfection of Dexras1 (Fig. 3D, histograms IV versus VI). To determine whether this dexamethasone-induced inhibition of FE65-APP signaling involved Dexras1, we monitored the effect of dexamethasone in cells in which Dexras1 expression was reduced with siRNA. Treatment with Dexras1 siRNA partially rescued the effect of dexamethasone on FE65-APP signaling although it did not increase signaling to the level seen in cells treated with Dexras1 siRNAs alone (Fig. 3D, cf. histograms V–VII). Thus dexamethasone inhibits FE65-APP signaling and at least part of this effect is mediated by Dexras1.
The function of Dexras1 in signaling is still not fully understood. However, Dexras1 has been shown to act as a guanyl nucleotide exchange factor (46, 57). To investigate whether the effect of Dexras1 on FE65-APP-mediated transcription involved guanyl nucleotide exchange, we used a constitutively active Dexras1(A178V) mutant (46) in the FE65-APP-mediated transcription assay. As with wild-type Dexras1, the Dexras1(A178V) mutant also repressed the FE65-APP-mediated transcription and to a similar magnitude (Fig. 3E). This suggests that guanyl nucleotide exchange by Dexras1 is not required for the inhibition of the FE65-APP transactivation.
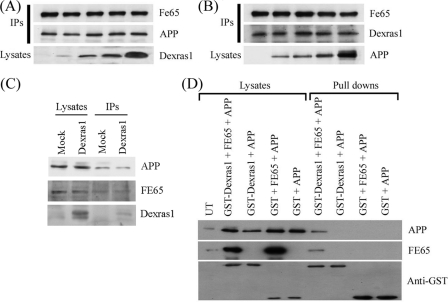
Dexras1 and APP Do Not Compete for FE65, but Dexras1·FE65·APP Forms a Tripartite Complex—Because both Dexras1 and APP interact with the FE65 PTB2 domain, the repression that was observed in the FE65-APP transactivation assays in the presence of Dexras1 may due to competition between APP and Dexras1 for FE65. To test this hypothesis, we transfected cells with the same amount of FE65 + APP DNA along with increasing amounts of Dexras1 DNA. FE65 was immunoprecipitated from the cell lysates and the amounts of APP coimmunoprecipitated were determined. The amounts of APP in the immunoprecipitates remained the same when the amount of Dexras1 was increasing (Fig. 4A). Similarly, increasing the amount of APP did not have a noticeable effect on Dexras1 binding to FE65 (Fig. 4B). We also tested if overexpression of Dexras1 affected the interaction of endogenous FE65 and APP. Again, the amounts of FE65 and APP remained the same in immunoprecipitates from cells transfected with or without Dexras1 (Fig. 4C). We conclude that Dexras1 and APP do not compete for FE65 at least under these experimental conditions.
FIGURE 4.
Dexras1 and APP do not compete for FE65 but Dexras1·FE65·APP form a tripartite complex. A, FE65 and APP were co-transfected into HEK293 cells with increasing amounts of Dexras1 DNA (0, 1, 2, 4, and 8 μg). FE65 was co-immunoprecipitated from the transfected cell lysates by an anti-myc antibody. The amounts of FE65 and APP in the immunoprecipitates and Dexras1 in the lysates were detected using rabbit polyclonal antibodies as appropriate. B, FE65 and Dexras1 were co-transfected with increasing amounts of APP DNA to cells (0, 1, 2, 4, and 8 μg). FE65 was co-immunoprecipitated from the transfected cell lysates as above. The amounts of FE65 and Dexras1 in the immunoprecipitates and APP in the lysates were detected by appropriate rabbit polyclonal antibodies. C, HEK293 cells were transfected either with or without Dexras1. APP was immunoprecipitated from the cell lysates by an anti-APP antibody. The amounts of FE65 and APP in the immunoprecipitates were determined by probing of blots with antibodies to FE65 and APP, respectively. D, APP was either co-transfected to cells with mammalian expression constructs for GST or GST-Dexras1 in the presence or absence of FE65. GST and GST-Dexras1 were captured from the cell lysates by glutathione-Sepharose 4B. APP and FE65 bound to the complex were detected by immunoblotting as described above. GST and GST-Dexras1 were detected using anti-GST antibody. GST-Dexras1 pulled down APP only in the presence of co-transfected FE65.
Several reports have demonstrated that certain PTB domains are able to bind two ligands simultaneously (58, 59). This prompted us to examine whether Dexras1 and APP bind simultaneously to the FE65 PTB2 domain. We co-transfected CHO cells with either GST-Dexras1 + APP or GST-Dexras1 + FE65 + APP; for controls, we transfected cells with GST + APP and GST + FE65 + APP. GST-Dexras1 was pulled down from the transfected cell lysates by glutathione-Sepharose 4B and the samples probed for the presence of APP and FE65. A strong signal of APP was found in the pull-down complex only in the presence of FE65 (Fig. 4D). Long exposure of the blot revealed a faint APP band in GST-Dexras1 + APP pull-down and this was probably due to the presence of endogenous FE65 in CHO cells. Thus, APP and Dexras1 can bind simultaneously to the FE65 PTB2 domain to form a tripartite complex.
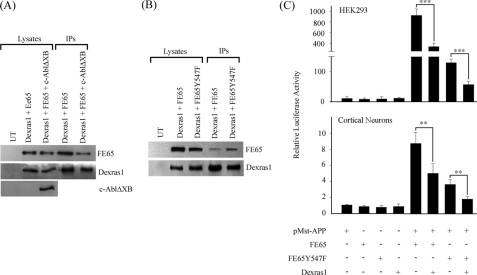
Phosphorylation of Tyrosine 547 of FE65 Reduces Dexras1 and FE65 Interaction—We have previously demonstrated that the tyrosine kinase c-Abl phosphorylates Tyr547 of FE65, which is located within its PTB2 domain, and that this does not affect the binding of APP to FE65 (42). Here, we have shown that Dexras1 also binds to the FE65 PTB2 domain. Therefore we investigated if phosphorylation of FE65 Tyr547 modulates Dexras1 binding. We co-transfected Dexras1 + FE65 either with or without c-Abl. Dexras1 was pulled down from the cell lysates and the amount of bound FE65 was determined. The amount of FE65 bound to Dexras1 was significantly reduced in the presence of c-Abl (Fig. 5A). In a complementary assay, we used an FE65 mutant in which Tyr547 was mutated to phenylalanine (FE65-Y547F) to preclude phosphorylation. We found that the FE65-Y547F bound more to Dexras1 than the wild-type FE65 (Fig. 5B). These findings suggest that phosphorylation on Tyr547 of FE65 reduces its ability to interact with Dexras1.
FIGURE 5.
FE65 and Dexras1 interaction is reduced by FE65 Tyr547 phosphorylation and the transactivation ability of FE65 is diminished by Y547F mutation. A, Dexras1 was co-transfected either with FE65 or FE65 + c-AblΔXB into HEK293 cells. Dexras1 was immunoprecipitated (IP) from the lysates and the amount of bound FE65 was determined by immunoblotting. B, Dexras1 was co-transfected with either FE65 or FE65-Y547F mutant (to preclude phosphorylation) into HEK293 cells. Dexras1 was immunoprecipitated from the lysates as described and the amount of bound FE65 was determined by immunoblotting. C, HEK293 cells and cortical neurons were transfected with the constructs indicated in the FE65-APP transcription assays. The ability of FE65-Y547F to stimulate transcription is significantly lower than wild-type FE65. The transcription was further suppressed by co-transfection of Dexras1 (n ≥ 12).
The effect of FE65 Tyr547 phosphorylation on FE65-APP signaling was studied by using the APP-GAL4 transcription assay. In both HEK293 cells and cortical neurons, FE65-Y547F still stimulated transcription but this was significantly lower than the wild-type FE65 (Fig. 5C). This is similar to previous observations in other cultured cell types (42). Dexras1 reduced the transcription further in both FE65 wild-type and FE65-Y547F co-transfected cells (Fig. 5C). Moreover, these inhibitory effects of Dexras1 on FE65 wild-type as compared with FE65-Y547F were ∼8-fold in the HEK293 cells and 3-fold in neurons (histograms 6 versus 8 in Fig. 5C). This 8-fold reduction in the HEK293 cells was similar to the difference observed in binding of FE65 wild-type versus FE65-Y547F to Dexras1 in this cell type. Quantification of the FE65 signals in the immunoprecipitation assays revealed a 6–8-fold reduction (depending upon experiment n = 3) in binding of FE65 wild-type compared with FE65-Y547F (Fig. 5B). Together, these findings suggest that phosphorylation of FE65 Tyr547 reduces binding of Dexras1 to FE65 and can thereby regulate FE65-APP transactivation.
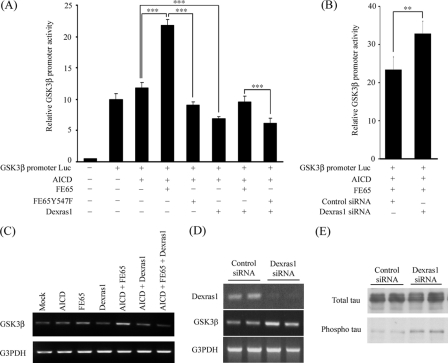
Dexras1 Suppresses the FE65-APP-mediated Activation of GSK3β Promoter and mRNA and Alters Tau Phosphorylation Status—Several genes have been proposed as targets for FE65-APP signaling. There has been some controversy over how many of the proposed gene targets are physiologically relevant (60, 61). GSK3β is one of the FE65-APP-regulated genes that has been reported by several different groups (42, 54, 62, 63). We therefore determined if Dexras1 modulates FE65-APP-mediated GSK3β promoter activity. Similar to other reports, GSK3β promoter activity was strongly enhanced in the presence of AICD and FE65; the FE65 stimulatory effect was lost with FE65-Y547F (Fig. 6A). However, FE65-AICD-mediated activation of the GSK3β promoter was inhibited by Dexras1 (Fig. 6A). We also studied the effect of siRNA knockdown of Dexras1 expression on GSK3β promoter activity. GSK3β promoter activity was significantly stimulated following Dexras1 but not control siRNA treatment (Fig. 6B). Thus, transfection and inhibition of Dexras1 expression induce complementary changes in the AICD-FE65-dependent effects on GSK3β promoter activity.
FIGURE 6.
Dexras1 represses FE65-AICD dependent transcription of the human GSK3β promoter to reduce GSK3β mRNA levels. A, HEK293 cells were transfected with a human GSK3β promoter reporter (GSK3β promoter Luc) and the constructs as indicated. The GSK3β promoter activity was strongly stimulated by FE65 and AICD. However, the FE65-AICD stimulation of the GSK3β promoter was inhibited by Dexras1. The FE65-Y547F mutant did not stimulate the GSK3β promoter (n ≥ 12). B, HEK293 cells were transfected with GSK3β promoter Luc, AICD, FE65, and the siRNA as indicated. Knockdown of Dexras1 enhanced GSK3β promoter activity (n = 5). C, reverse transcriptase-PCR analyses of GSK3β (upper panel) and glyceraldehyde-3-phosphate dehydrogenase (G3PDH)(lower panel) mRNA from cells transfected with the indicated constructs. D, HEK293 cells were transfected with either non-targeting control or Dexras1 siRNA. The amounts of Dexras1, GSK3β, and glyceraldehyde-3-phosphate dehydrogenase in the samples were analyzed by reverse transcriptase-PCR. E, HEK293 cells were transfected with human Tau and either with non-targeting control or Dexras1 siRNA. Total and phosphorylated Tau were detected by phospho-independent and phosphorylation specific (PHF-1) Tau antibodies, respectively.
We next tested if Dexras1 influences expression of endogenous GSK3β mRNA. Increased levels of GSK3β mRNA were observed in cells transfected with FE65 and AICD (Fig. 6C). However, the effect of FE65 and AICD on endogenous GSK3β mRNA was suppressed following co-transfection with Dexras1 (Fig. 6C). In contrast, knockdown of Dexras1 enhanced the expression of GSK3β (Fig. 6D). Together, these findings strongly suggest that Dexras1 is a repressor of FE65-APP-mediated transcription and this includes FE65-AICD transcriptional effects on the GSK3β gene.
Tau is a substrate of GSK3β, and hyperphosphorylation of Tau is observed in Alzheimer disease. We therefore tested if Dexras1 affects Tau phosphorylation. Cells were cotransfected with human Tau and either with non-targeting control or Dexras1 siRNAs. Probing of the samples with PHF-1 antibody that detects Tau phosphorylated on serines 396 and 404, which are two known GSK3β-targeted sites in Tau, revealed that Tau phosphorylation was increased in Dexras1 knockdown cells (Fig. 6E). Thus, Dexras1 stimulates GSK3β gene expression and this induces increased phosphorylation of Tau, a known GSK3β substrate.
DISCUSSION
In the present study, we have shown that FE65 associates with Dexras1. The interaction between FE65 and Dexras1 was confirmed by various biochemical assays. We have also demonstrated that Dexras1 binds to the FE65 PTB2 domain. The PTB domain was initially discovered as a protein-protein interaction domain that bound to tyrosine-phosphorylated NPXY motifs (for review, see Ref. 64). However, Dexras1 does not possess an NPXY sequence. Increasing evidence indicates that PTB domains have more diverse binding specificity. For example, Numb-associated kinase interacts with the Numb PTB domain through a non-NPTY sequence (65–67).
Dexras1 is a member of the Ras superfamily of GTPases and was originally identified as a dexamethasone-inducible gene (36, 55, 56). Although Dexras1 is a GTPase, it possesses several differences from other Ras members. With an extended C terminus, the molecular mass of Dexras1 is about 34 kDa, which is significantly higher than the 22-kDa typical for Ras proteins (66). The extended C terminus has previously been shown to interact with another PTB domain containing protein, CAPON (37). Additionally, Dexras1 has been found in both cytosol and membrane, whereas most other Ras proteins are membrane associated (37). Here, we further demonstrate that a significant proportion of Dexras1 is located in the nuclei. This observation is in line with the sequence analysis that Dexras1 contains a bipartite nuclear localization signal (amino acids 207–224) that is required for targeting proteins to the nucleus. Among more than 100 known small G proteins, Dexras1 is only the third to be found in the nucleus. Interestingly, when Dexras1 and FE65 were co-expressed in cells, a large proportion of Dexras1 was shifted from the cytosol to nucleus. Therefore, FE65 may play a role in translocation of Dexras1 from the cytosol to nucleus. Nevertheless, the differences between Dexras1 and other Ras members suggests that Dexras1 is involved in a broad range of biological events. In fact, we have demonstrated here that Dexras1 plays a direct regulatory role in FE65-APP-mediated transcription.
FE65 and AICD have been shown to form a transcription stimulation complex for a GAL4-dependent reporter (3, 42). In the present study, we have shown that Dexras1 binds to and co-localizes with FE65 in the nucleus. These observations prompted us to investigate the role of Dexras1 in FE65-APP nuclear signaling. Dexras1 strongly repressed FE65-APP-mediated transcription in GAL4-APP reporter and GSK3β promoter assays using various cell types. Interestingly, Telese and colleagues (6) demonstrated that FE65 mutants with their PTB domains deleted enhance transactivation. However, the mechanism(s) by which FE65 PTB domains inhibit transactivation is not fully understood. Our finding provides one possible mechanism as Dexras1 binds to the FE65 PTB2 domain and inhibits transcription. We initially expected that Dexras1 might inhibit the FE65-AICD-mediated transcription by competing with AICD for binding to FE65 PTB2 domain. However, we did not observe Dexras1 and APP competing for FE65 in our competition assays. Another possibility for this repression is that Dexras1 retains FE65 in the cytosol. However, our immunostaining results do not support this notion as the subcellular distribution of FE65 was not noticeably altered by Dexras1.
Unexpectedly, our pull-down assays showed that Dexras1·FE65·APP could form a tripartite complex. However, a previous study showed that X11β PTB interacts with Alcadein and APP at the same time (58). Additionally, Stolt and colleagues (59) demonstrated that the adaptor protein Disable-1 binds to two ligands simultaneously by structurally distinct binding sites within its PTB domain. These findings suggest that certain PTB domains could accommodate two binding partners at the same time. In this report, we found that Tyr547 phosphorylation within the FE65 PTB2 domain only influences the binding of Dexras1, but not APP (42). This may suggest that Dexras1 and APP bind to different regions within the FE65 PTB2 domain.
One mechanism to regulate protein-protein interaction is by phosphorylation. For example, the interaction between FE65 and APP can be regulated by phosphorylation of APP threonine 668 (16). In a previous study, we showed by mass spectrometry that the Tyr547 resides within the FE65 PTB2 domain is a phosphorylated residue. The FE65 Tyr547 phosphorylation does not influence the interaction between FE65 and APP but stimulates FE65-APP-mediated transcription (42). Here, we have demonstrated that FE65-APP-mediated transcription was significantly repressed when the Y547F mutation (mimicking permanent dephosphorylation) was introduced to FE65. This observation is in line with our previous finding that phosphorylation of FE65 Tyr547 enhances the FE65-APP-mediated transcription (42). Because Dexras1 preferentially binds to the nonphosphorylated FE65 (at Tyr647), one explanation for the enhancement of FE65-APP-mediated transcription is the amount of Dexras1 that bound to FE65 is reduced by phosphorylation of FE65 Tyr547.
Of interest was our finding that Dexras1 inhibited FE65-mediated transcription without an apparent effect on binding of FE65 to APP. The precise mechanisms whereby FE65 and APP interact and stimulate transcription are far from clear. One suggestion is that the FE65 WW and PTB2 domains bind to one another such that FE65 is in a closed configuration that is refractory to its transcriptional ability. Binding of APP and other “factors” then induces FE65 to adopt an open conformation that is more permissible to transcription (68). The identity of these factors is not known but suggestions include one or more membrane-associated proteins, kinases (because FE65 is a phosphorylated on multiple residues (42, 69)), or some special lipids (68). There is also evidence that once FE65 is activated in this way, then binding to AICD is dispensable for its transcriptional activity (68). In this context, it is therefore not surprising that the inhibitory effect of Dexras1 might not involve an effect on the FE65-APP interaction. Indeed, it is even possible that Dexras1 represents one of these previously described factors but that its release from FE65 stabilizes the open conformation that is more permissible to transcription. Clearly, identifying the full complement of FE65 interacting proteins and factors in future studies will help resolve this issue.
In summary, we have identified Dexras1 as a novel interacting partner of FE65, and their interaction suppresses the FE65-APP-mediated transcription. We have also demonstrated that the FE65-Dexras1 interaction is regulated by FE65 Tyr547 phosphorylation. FE65 is a phosphoprotein, although, the role of FE65 phosphorylation is not well understood. Aberrant protein phosphorylation has been implicated in the pathogenesis of several neurodegenerative diseases. Defective FE65 phosphorylation might disrupt the regulatory processes of FE65-APP signaling including FE65-Dexras1 interaction. This could lead to mistranscription of potential FE65-APP-regulated genes such as BACE, GSK3β, and APP, which are closely linked to the pathogenesis of Alzheimer disease.
Acknowledgments
We thank Richard Dorin for mammalian expression Dexras1 constructs, Solomon Snyder for GST-Dexras1 construct, Thomas Südhof for FE65 deletion constructs, and Peter Davies for PHF-1 antibody.
This work was supported by the Research Grant Council Hong Kong, a Chinese University of Hong Kong direct grant scheme, the Wellcome Trust, United Kingdom Medical Research Council, Biotechnology and Biological Sciences Research Council, European Union NeuroNE, the Alzheimer Society, the Alzheimer Research Trust, and the Alzheimer Association. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: APP, amyloid precursor protein; AICD, amyloid precursor protein intracellular domain; Aβ, β-amyloid peptide; GSK3β, glycogen synthase kinase 3β; PTB domain, phosphotyrosine binding domain; CHO, Chinese hamster ovary; HEK, human embryonic kidney; GST, glutathione S-transferase; siRNA, small interfering RNA.
References
- 1.McLoughlin, D. M., and Miller, C. C. (2008) J. Neurosci. Res. 86 744–754 [DOI] [PubMed] [Google Scholar]
- 2.Zambrano, N., Minopoli, G., de Candia, P., and Russo, T. (1998) J. Biol. Chem. 273 20128–20133 [DOI] [PubMed] [Google Scholar]
- 3.Cao, X., and Sudhof, T. C. (2001) Science 293 115–120 [DOI] [PubMed] [Google Scholar]
- 4.Zambrano, N., Bruni, P., Minopoli, G., Mosca, R., Molino, D., Russo, C., Schettini, G., Sudol, M., and Russo, T. (2001) J. Biol. Chem. 276 19787–19792 [DOI] [PubMed] [Google Scholar]
- 5.Ermekova, K. S., Zambrano, N., Linn, H., Minopoli, G., Gertler, F., Russo, T., and Sudol, M. (1997) J. Biol. Chem. 272 32869–32877 [DOI] [PubMed] [Google Scholar]
- 6.Telese, F., Bruni, P., Donizetti, A., Gianni, D., D'Ambrosio, C., Scaloni, A., Zambrano, N., Rosenfeld, M. G., and Russo, T. (2005) EMBO Rep 6 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zambrano, N., Buxbaum, J. D., Minopoli, G., Fiore, F., De Candia, P., De Renzis, S., Faraonio, R., Sabo, S., Cheetham, J., Sudol, M., and Russo, T. (1997) J. Biol. Chem. 272 6399–6405 [DOI] [PubMed] [Google Scholar]
- 8.McLoughlin, D. M., and Miller, C. C. (1996) FEBS Lett. 397 197–200 [DOI] [PubMed] [Google Scholar]
- 9.Borg, J. P., Ooi, J., Levy, E., and Margolis, B. (1996) Mol. Cell. Biol. 16 6229–6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiore, F., Zambrano, N., Minopoli, G., Donini, V., Duilio, A., and Russo, T. (1995) J. Biol. Chem. 270 30853–30856 [DOI] [PubMed] [Google Scholar]
- 11.Kang, J., Lemaire, H. G., Unterbeck, A., Salbaum, J. M., Masters, C. L., Grzeschik, K. H., Multhaup, G., Beyreuther, K., and Muller-Hill, B. (1987) Nature 325 733–736 [DOI] [PubMed] [Google Scholar]
- 12.Tanzi, R. E., Gusella, J. F., Watkins, P. C., Bruns, G. A., St. George-Hyslop, P., Van Keuren, M. L., Patterson, D., Pagan, S., Kurnit, D. M., and Neve, R. L. (1987) Science 235 880–884 [DOI] [PubMed] [Google Scholar]
- 13.Selkoe, D. J. (2000) Ann. N. Y. Acad. Sci. 924 17–25 [DOI] [PubMed] [Google Scholar]
- 14.Santiard-Baron, D., Langui, D., Delehedde, M., Delatour, B., Schombert, B., Touchet, N., Tremp, G., Paul, M. F., Blanchard, V., Sergeant, N., Delacourte, A., Duyckaerts, C., Pradier, L., and Mercken, L. (2005) J. Neurochem. 93 330–338 [DOI] [PubMed] [Google Scholar]
- 15.Sabo, S. L., Lanier, L. M., Ikin, A. F., Khorkova, O., Sahasrabudhe, S., Greengard, P., and Buxbaum, J. D. (1999) J. Biol. Chem. 274 7952–7957 [DOI] [PubMed] [Google Scholar]
- 16.Ando, K., Iijima, K. I., Elliott, J. I., Kirino, Y., and Suzuki, T. (2001) J. Biol. Chem. 276 40353–40361 [DOI] [PubMed] [Google Scholar]
- 17.Kimberly, W. T., Zheng, J. B., Guenette, S. Y., and Selkoe, D. J. (2001) J. Biol. Chem. 276 40288–40292 [DOI] [PubMed] [Google Scholar]
- 18.Baek, S. H., Ohgi, K. A., Rose, D. W., Koo, E. H., Glass, C. K., and Rosenfeld, M. G. (2002) Cell 110 55–67 [DOI] [PubMed] [Google Scholar]
- 19.Scheinfeld, M. H., Ghersi, E., Davies, P., and D'Adamio, L. (2003) J. Biol. Chem. 278 42058–42063 [DOI] [PubMed] [Google Scholar]
- 20.Kinoshita, A., Whelan, C. M., Smith, C. J., Berezovska, O., and Hyman, B. T. (2002) J. Neurochem. 82 839–847 [DOI] [PubMed] [Google Scholar]
- 21.Costello, L. C., and Franklin, R. B. (2002) Horm. Metab. Res. 34 417–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amanatullah, D. F., Zafonte, B. T., and Pestell, R. G. (2002) Minerva Endocrinol. 27 7–20 [PubMed] [Google Scholar]
- 23.Morrison, R. F., and Farmer, S. R. (2000) J. Nutr. 130 3116S-3121S [DOI] [PubMed] [Google Scholar]
- 24.De Martino, M. U., Alesci, S., Chrousos, G. P., and Kino, T. (2004) Ann. N. Y. Acad. Sci. 1024 72–85 [DOI] [PubMed] [Google Scholar]
- 25.Klein-Hitpass, L., Schwerk, C., Kahmann, S., and Vassen, L. (1998) J. Mol. Med. 76 490–496 [DOI] [PubMed] [Google Scholar]
- 26.Touma, C., Ambree, O., Gortz, N., Keyvani, K., Lewejohann, L., Palme, R., Paulus, W., Schwarze-Eicker, K., and Sachser, N. (2004) Neurobiol. Aging 25 893–904 [DOI] [PubMed] [Google Scholar]
- 27.Swaab, D. F., Raadsheer, F. C., Endert, E., Hofman, M. A., Kamphorst, W., and Ravid, R. (1994) J. Neuroendocrinol. 6 681–687 [DOI] [PubMed] [Google Scholar]
- 28.Hoschl, C., and Hajek, T. (2001) Eur. Arch. Psychiatry Clin. Neurosci. 251 Suppl. 2, II81–II88 [DOI] [PubMed] [Google Scholar]
- 29.Maeda, K., Tanimoto, K., Terada, T., Shintani, T., and Kakigi, T. (1991) Neurobiol. Aging 12 161–163 [DOI] [PubMed] [Google Scholar]
- 30.Pascualy, M., Petrie, E. C., Brodkin, K., Peskind, E. R., Wilkinson, C. W., and Raskind, M. A. (2000) Biol. Psychiatry 48 247–254 [DOI] [PubMed] [Google Scholar]
- 31.Peskind, E. R., Wilkinson, C. W., Petrie, E. C., Schellenberg, G. D., and Raskind, M. A. (2001) Neurology 56 1094–1098 [DOI] [PubMed] [Google Scholar]
- 32.Giubilei, F., Patacchioli, F. R., Antonini, G., Sepe Monti, M., Tisei, P., Bastianello, S., Monnazzi, P., and Angelucci, L. (2001) J. Neurosci. Res. 66 262–265 [DOI] [PubMed] [Google Scholar]
- 33.Dai, J., Buijs, R., and Swaab, D. (2004) Br. J. Pharmacol. 143 606–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulstad, J. J., McMillan, P. J., Leverenz, J. B., Cook, D. G., Green, P. S., Peskind, E. R., Wilkinson, C. W., Farris, W., Mehta, P. D., and Craft, S. (2005) J. Neuropathol. Exp. Neurol. 64 139–146 [DOI] [PubMed] [Google Scholar]
- 35.Green, K. N., Billings, L. M., Roozendaal, B., McGaugh, J. L., and LaFerla, F. M. (2006) J. Neurosci. 26 9047–9056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kemppainen, R. J., and Behrend, E. N. (1998) J. Biol. Chem. 273 3129–3131 [DOI] [PubMed] [Google Scholar]
- 37.Fang, M., Jaffrey, S. R., Sawa, A., Ye, K., Luo, X., and Snyder, S. H. (2000) Neuron 28 183–193 [DOI] [PubMed] [Google Scholar]
- 38.Bressler, S. L., Gray, M. D., Sopher, B. L., Hu, Q., Hearn, M. G., Pham, D. G., Dinulos, M. B., Fukuchi, K., Sisodia, S. S., Miller, M. A., Disteche, C. M., and Martin, G. M. (1996) Hum. Mol. Genet. 5 1589–1598 [DOI] [PubMed] [Google Scholar]
- 39.Kesavapany, S., Banner, S. J., Lau, K. F., Shaw, C. E., Miller, C. C., Cooper, J. D., and McLoughlin, D. M. (2002) Neuroscience 115 951–960 [DOI] [PubMed] [Google Scholar]
- 40.Brownlees, J., Yates, A., Bajaj, N. P., Davis, D., Anderton, B. H., Leigh, P. N., Shaw, C. E., and Miller, C. C. (2000) J. Cell Sci. 113 401–407 [DOI] [PubMed] [Google Scholar]
- 41.Kesavapany, S., Lau, K. F., McLoughlin, D. M., Brownlees, J., Ackerley, S., Leigh, P. N., Shaw, C. E., and Miller, C. C. (2001) Eur. J. Neurosci. 13 241–247 [PubMed] [Google Scholar]
- 42.Perkinton, M. S., Standen, C. L., Lau, K. F., Kesavapany, S., Byers, H. L., Ward, M., McLoughlin, D. M., and Miller, C. C. (2004) J. Biol. Chem. 279 22084–22091 [DOI] [PubMed] [Google Scholar]
- 43.Jicha, G. A., Lane, E., Vincent, I., Otvos, L., Jr., Hoffmann, R., and Davies, P. (1997) J. Neurochem. 69 2087–2095 [DOI] [PubMed] [Google Scholar]
- 44.Lau, K. F., McLoughlin, D. M., Standen, C. L., Irving, N. G., and Miller, C. C. (2000) Neuroreport 11 3607–3610 [DOI] [PubMed] [Google Scholar]
- 45.Lovestone, S., Reynolds, C. H., Latimer, D., Davis, D. R., Anderton, B. H., Gallo, J. M., Hanger, D., Mulot, S., Marquardt, B., Stabel, S., Woodgett, J. R., and Miller, C. C. (1994) Curr. Biol. 4 1077–1086 [DOI] [PubMed] [Google Scholar]
- 46.Graham, T. E., Key, T. A., Kilpatrick, K., and Dorin, R. I. (2001) Endocrinology 142 2631–2640 [DOI] [PubMed] [Google Scholar]
- 47.Lau, K. F., Miller, C. C., Anderton, B. H., and Shaw, P. C. (1999) Genomics 60 121–128 [DOI] [PubMed] [Google Scholar]
- 48.Lau, K. F., McLoughlin, D. M., Standen, C., and Miller, C. C. (2000) Mol. Cell. Neurosci. 16 557–565 [DOI] [PubMed] [Google Scholar]
- 49.McLoughlin, D. M., Standen, C. L., Lau, K. F., Ackerley, S., Bartnikas, T. P., Gitlin, J. D., and Miller, C. C. (2001) J. Biol. Chem. 276 9303–9307 [DOI] [PubMed] [Google Scholar]
- 50.Lau, K. F., Howlett, D. R., Kesavapany, S., Standen, C. L., Dingwall, C., McLoughlin, D. M., and Miller, C. C. (2002) Mol. Cell. Neurosci. 20 13–20 [DOI] [PubMed] [Google Scholar]
- 51.Thomas, J. E., Smith, M., Rubinfeld, B., Gutowski, M., Beckmann, R. P., and Polakis, P. (1996) J. Biol. Chem. 271 28630–28635 [DOI] [PubMed] [Google Scholar]
- 52.Lau, K. F., Miller, C. C., Anderton, B. H., and Shaw, P. C. (1999) J. Pept. Res. 54 85–91 [DOI] [PubMed] [Google Scholar]
- 53.Minopoli, G., de Candia, P., Bonetti, A., Faraonio, R., Zambrano, N., and Russo, T. (2001) J. Biol. Chem. 276 6545–6550 [DOI] [PubMed] [Google Scholar]
- 54.von Rotz, R. C., Kohli, B. M., Bosset, J., Meier, M., Suzuki, T., Nitsch, R. M., and Konietzko, U. (2004) J. Cell Sci. 117 4435–4448 [DOI] [PubMed] [Google Scholar]
- 55.Brogan, M. D., Behrend, E. N., and Kemppainen, R. J. (2001) Neuroendocrinology 74 244–250 [DOI] [PubMed] [Google Scholar]
- 56.Tu, Y., and Wu, C. (1999) Biochim. Biophys. Acta 1489 452–456 [DOI] [PubMed] [Google Scholar]
- 57.Cismowski, M. J., Ma, C., Ribas, C., Xie, X., Spruyt, M., Lizano, J. S., Lanier, S. M., and Duzic, E. (2000) J. Biol. Chem. 275 23421–23424 [DOI] [PubMed] [Google Scholar]
- 58.Araki, Y., Tomita, S., Yamaguchi, H., Miyagi, N., Sumioka, A., Kirino, Y., and Suzuki, T. (2003) J. Biol. Chem. 278 49448–49458 [DOI] [PubMed] [Google Scholar]
- 59.Stolt, P. C., Vardar, D., and Blacklow, S. C. (2004) Biochemistry 43 10979–10987 [DOI] [PubMed] [Google Scholar]
- 60.Hass, M. R., and Yankner, B. A. (2005) J. Biol. Chem. 280 36895–36904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hebert, S. S., Serneels, L., Tolia, A., Craessaerts, K., Derks, C., Filippov, M. A., Muller, U., and De Strooper, B. (2006) EMBO Rep. 7 739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim, H. S., Kim, E. M., Lee, J. P., Park, C. H., Kim, S., Seo, J. H., Chang, K. A., Yu, E., Jeong, S. J., Chong, Y. H., and Suh, Y. H. (2003) FASEB J. 17 1951–1953 [DOI] [PubMed] [Google Scholar]
- 63.Chang, K. A., Kim, H. S., Ha, T. Y., Ha, J. W., Shin, K. Y., Jeong, Y. H., Lee, J. P., Park, C. H., Kim, S., Baik, T. K., and Suh, Y. H. (2006) Mol. Cell. Biol. 26 4327–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uhlik, M. T., Temple, B., Bencharit, S., Kimple, A. J., Siderovski, D. P., and Johnson, G. L. (2005) J. Mol. Biol. 345 1–20 [DOI] [PubMed] [Google Scholar]
- 65.Chien, C. T., Wang, S., Rothenberg, M., Jan, L. Y., and Jan, Y. N. (1998) Mol. Cell. Biol. 18 598–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li, S. C., Zwahlen, C., Vincent, S. J., McGlade, C. J., Kay, L. E., Pawson, T., and Forman-Kay, J. D. (1998) Nat. Struct. Biol. 5 1075–1083 [DOI] [PubMed] [Google Scholar]
- 67.Zwahlen, C., Li, S. C., Kay, L. E., Pawson, T., and Forman-Kay, J. D. (2000) EMBO J. 19 1505–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao, X., and Sudhof, T. C. (2004) J. Biol. Chem. 279 24601–24611 [DOI] [PubMed] [Google Scholar]
- 69.Standen, C. L., Perkinton, M. S., Byers, H. L., Kesavapany, S., Lau, K. F., Ward, M., McLoughlin, D., and Miller, C. C. (2003) Mol. Cell. Neurosci. 24 851–857 [DOI] [PubMed] [Google Scholar]