Abstract
Cells sense nutrients present in the extracellular environment and modulate the activities of intracellular signaling systems in response to nutrient availability. This study demonstrates that RalA and its activator RalGDS participate in nutrient sensing and are indispensable for activation of mammalian target of rapamycin complex 1 (mTORC1) induced by extracellular nutrients. Knockdown of RalA or RalGDS abolished amino acid- and glucose-induced mTORC1 activation, as judged by phosphorylation of S6 kinase and eukaryotic translation initiation factor 4E-binding protein 1. The amount of GTP-bound RalA increased in response to increased amino acid availability. In addition, RalA knockdown suppressed Rheb-induced S6 kinase phosphorylation, and the constitutively active form of RalA induced mTORC1 activation in the absence of Rheb. These results collectively suggest that RalGDS and RalA act downstream of Rheb and that RalA activation is a crucial step in nutrient-induced mTORC1 activation.
Nutrients such as amino acids and carbohydrates are available in the extracellular space and are vital for cell homeostasis. Cells are able to detect extracellular nutrients and modulate their intracellular signaling systems in response to changes in nutrient levels. Recent studies have shown that mammalian target of rapamycin complex 1 (mTORC1)2 functions as a critical intracellular component of the nutrient-sensing system and governs many aspects of cellular responses to changing nutrient levels (1-5). Signaling cascades initiated by growth factors and leading to mTORC1 activation have been extensively characterized. Growth factor stimulation results primarily in the phosphorylation of TSC2, a GTPase-activating protein (GAP) for Rheb GTPase, in a phosphoinositide 3-kinase- and protein kinase B (PKB)-dependent manner, followed by inactivation of TSC2 GAP activity (6-9). It has been proposed that the decreased GAP activity allows Rheb to stay in an “active” GTP-bound form, leading to mTORC1 activation. In contrast to the mechanism of the growth factor-initiated mTORC1 activation, little is known about the mechanism(s) by which mTORC1 activity is regulated by amino acid availability (10-16). Recent studies have shown that calmodulin and hVps34 play a role in the amino acid-sensing system (12) and that Rag GTPases escort mTORC1 to sites where they can activate the kinase (17, 18). However, most of the amino acid-sensing signaling machinery, including the amino acid sensors themselves, is unknown, and the mechanism(s) underlying the nutrient-sensing process remains to be identified.
RalA and RalB GTPases belong to the Ras superfamily and are known to participate in multiple cellular processes through binding to diverse effector proteins. RalA effectors described to date include Ral-binding protein 1, Sec5, Exo84, filamin, and ZO-1-associated nucleic acid-binding protein (19-22); these interactions are indicative of crucial roles for RalA in cell migration, membrane dynamics, and transcriptional regulation. RalB is highly homologous to RalA and shares an identical effector domain with RalA. However, these two proteins are believed to have distinct functions in cells (23-25); RalB participates in apoptotic signaling, whereas RalA is predominantly involved in proliferation-related signaling, particularly anchorage-independent proliferation (26, 27). In addition, an accumulating body of evidence shows that both RalA and RalGDS, an activator of RalA, are crucial for Ras-induced oncogenic transformation of cells and that Ras in fact binds to RalGDS and induces RalA activation (23, 24, 26, 28). These observations suggest that the RalGDS-RalA signaling unit plays a role in cell proliferation; however, how RalA controls cell proliferation and participates in oncogenic transformation remains unknown.
In this report, we show that RalA is activated in response to extracellular amino acids and that both RalGDS and RalA are indispensable for nutrient-induced mTORC1 activation. The RalGDS-RalA signaling unit functions downstream of Rheb and participates selectively in the nutrient-sensing system.
MATERIALS AND METHODS
Cell Culture and Transfection—HeLa cells (ATCC CCL-2) were maintained as described previously (29). Amino acid-free RPMI1640 medium (30) and glucose-free RPMI1640 medium (Invitrogen) were used for amino acid and glucose deprivation, respectively. Amino acid and glucose stimulation were performed by the addition of normal RPMI1640 medium and RPMI1640 medium containing 25 mm glucose, respectively. Serum starvation was carried out in RPMI1640 medium containing 0.1% fatty acid-free bovine serum albumin (Sigma). Transfection of plasmid DNA was carried out using either FuGENE HD (Roche Applied Science) or Lipofectamine 2000 (Invitrogen). Lipofectamine RNAi Max (Invitrogen) was used for transfection of siRNA; a mixed siRNA mixture was used unless otherwise specified. Transfections were performed according to the manufacturers' protocols.
Plasmids and siRNAs—SMARTpool siRNA against human Rheb was purchased from Dharmacon. Other siRNAs (Stealth RNAi), including negative control siRNA, were purchased from Invitrogen: RALA RNAi 1 (HSS109033), RALA RNAi 3 (HSS109035), RALB RNAi 1-3 (HSS109036 to -109038), RALGDS RNAi 1 (HSS109039), and RALGDS RNAi 2 (HSS109040). Full-length human Rheb (NP_005605, amino acids 1-184) cDNA was amplified from HeLa cell cDNA by PCR and cloned into the EcoRI site of pCAGGS (31) to generate Rheb/pCAGGS. Rheb mutations S16H (tct to cat) and S20N (tcc to aat) were introduced by site-directed mutagenesis using a PCR-based strategy. Full-length human RalA (NP_005393, amino acids 1-206) cDNA was amplified from HeLa cell cDNA by PCR with the addition of Myc tag sequence at the 5′-end, followed by cloning into the EcoRI/SalI site of pCMV5 (gift from David W. Russell) to generate Myc-RalA/pCMV5. The Q72L mutation (cag to ctc) was introduced by site-directed mutagenesis employing a PCR-based strategy. The coding region of Sec5-RBD was amplified from the Sec5-RBD/pGEX vector (32) (gift of Dr. Hisanori Horiuchi; Kyoto University) and tagged with a FLAG sequence at the 5′-end by PCR, followed by cloning into the BamHI/EcoRI site of pEF1-MycHisA (Invitrogen) to generate Sec5-RBD/pEF1-FLAG. PrimerSTAR DNA polymerase (Takara) was used for all PCR amplifications, and sequences were confirmed using the BigDye Terminator method (Applied Biosystems).
Immunoblot—Immunoblot analyses were conducted as described previously (29). Antibodies used were anti-phospho-p70S6K (recognizes Thr412 of the α1 isoform, which corresponds to Thr389 of the α2 isoform; Upstate Biotechnology), anti-p70S6K (Epitomics), anti-RalA (R&D Systems), anti-RalA/B (R&D Systems), anti-Akt H136 (Santa Cruz Biotechnology), anti-phospho-4EBP1 (Ser65) (Cell Signaling Technology), anti-4EBP1 (Bethyl Laboratories), anti-Rheb (Abcam), anti-α-tubulin (Zymed Laboratories Inc.), and anti-DDDDK-Tag (MBL). The relative intensities of immunoreactive phospho-S6K and S6K bands were measured using the Image J Java applet. Briefly, the signal intensity of a selected band or corresponding neighboring control area was measured. After the control value was subtracted from each experimental value as background, the ratio of phospho-S6K to S6K signal was determined. The calculated pS6K/S6K values were normalized to that of the control experiment, and the normalized values were represented in each figure. Typical images from repeated experiments are represented in each figure.
Reverse Transcription-PCR—RNA was prepared from cultured cells using TRIzol reagent (Invitrogen) and reverse-transcribed into cDNA using Moloney murine leukemia virus reverse transcriptase (Takara). In order to confirm the RNAi against RalGDS, PCR was conducted in a 10-μl reaction mixture using 0.1 μg of cDNA as template and Blend-Taq polymerase (Toyobo). RalGDS primer sequences were 5′-agtcggccctgaacctttat-3′ (forward) and 5′-tccggaggttgacagaaatc-3′ (reverse); actin primer sequences were 5′-ggagaaaatctggcaccacacct-3′ (forward) and 5′-aggaaggaaggctggaagagtg-3′ (reverse).
RalA Activation Assay—The assay was essentially conducted as described previously (32) with minor modifications. Briefly, HeLa cells were lysed in lysis buffer consisting of 25 mm sodium phosphate (pH 7.5), 78 mm KCl, 1% Nonidet P-40, 1 mm MgCl2, 0.2 mm CaCl2, 1 mm dithiothreitol, 0.1 μm microcystin LR, and protease inhibitor mixture (Nacalai Tesque). After removal of insoluble materials, the lysate was incubated with glutathione-Sepharose beads (GE Healthcare) for 10 min. Beads were removed by centrifugation, and then glutathione beads that were conjugated to glutathione S-transferase-fused Sec5-RBD protein were added to the cleared lysate and incubated for 30 min to allow GTP-bound RalA to bind to the beads. Beads were washed with lysis buffer, and bound proteins and lysate were subjected to immunoblot analysis. Band intensity of the RalA blot was measured using Image J, as described under “Immunoblot.” Typical images from repeated experiments are represented. All Sepharose beads were pretreated with 5% bovine serum albumin prior to use to reduce nonspecific binding. All procedures were carried out at 4 °C unless otherwise specified. Glutathione S-transferase-fused Sec5-RBD protein was prepared as described previously (32), with the exception that Escherichia coli strain BL21(DE3) Codon Plus RP (Clontech) was used.
RESULTS AND DISCUSSION
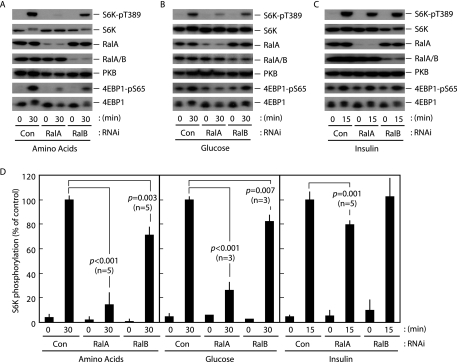
RalA Is an Indispensable Component of the Nutrient-sensing System—Cell proliferation is a dynamic event that requires highly coordinated multiple cellular processes, such as DNA replication and protein synthesis, which are controlled by growth factor-initiated growth/proliferation signals and nutrient-sensing signals. mTORC1 is a critical component in the organization of these signaling systems to regulate cell proliferation (1-5). In order to understand the biological significance of Ral GTPases in the mTORC1-driven signaling systems, we knocked down Ral GTPases using conventional siRNA technology and then analyzed the effects on growth factor- and nutrient-induced mTORC1 activation in HeLa cells (Fig. 1). We successfully achieved RNAi-mediated suppression of both RalA and RalB as assayed by immunoblot analysis using anti-RalA and anti-RalA/B antibodies, respectively (Fig. 1). It is of note that the anti-RalA/B antibody used in this study (lot WIS01) recognizes both RalA and RalB; however, the antibody shows ∼10 times higher affinity for RalB than for RalA (according to the manufacturer's data sheet). We monitored phosphorylation of human p70S6K at Thr389 as a measure of mTORC1 activity throughout this study, since the Thr389 residue has been identified as a direct mTORC1 phosphorylation site (33-35). When cells were incubated in amino acid-depleted medium, phosphorylation of Thr389 was reduced to an almost undetectable level within 2 h. Thr389 phosphorylation became detectable 5-10 min after the readdition of amino acids and reached a plateau within 60 min in HeLa cells (data not shown). RalA knockdown robustly (∼90%) inhibited S6K phosphorylation induced by the addition of amino acids, whereas knockdown of RalB showed a relatively low level (∼30%) of suppression of S6K phosphorylation (Fig. 1, A and D). A similar effect was observed when two individual siRNAs were used to knock down RalA (supplemental Fig. 1), indicating that the inhibition of mTORC1 activation was not due to an “off-target” effect of the RNAi. This conclusion was further supported by the rescue experiment in which an RNAi-resistant RalA mutant, bearing silent mutations at codons 150 and 151, restored Thr389 phosphorylation under conditions of endogenous RalA knockdown (supplemental Fig. 2).
FIGURE 1.
Ral GTPase knockdown inhibits nutrient-induced mTORC1 activation. HeLa cells were transfected with siRNAs against the indicated targets. After 2 days in culture, cells were deprived of amino acids (A), glucose (B), or serum (C) for 2 h. Cells were then exposed to amino acids (A), glucose (B), or 2 μg/ml bovine insulin (C) for the indicated times, followed by immunoblot analysis. D, quantification of the phospho-S6K (normalized to total S6K) immunoblots shown in A-C, as described under “Materials and Methods.” Values represent the mean ± S.D. of 3-5 independent experiments. Calculated p values (Student's t test) are presented.
Knockdown of either RalA or RalB also showed inhibitory effects on glucose-induced S6K phosphorylation that were of similar magnitude to those observed for amino acid-induced inhibition (Fig. 1, B and D), suggesting that both the amino acid- and glucose-sensing systems share the Ral GTPase-dependent core signaling machinery. In contrast, when cells were stimulated by insulin, which is known to activate mTORC1 in a phosphoinositide 3-kinase/PKB-dependent manner, RalA knockdown resulted in partial (∼20%) inhibition of S6K phosphorylation, and RalB knockdown resulted in no inhibition (Fig. 1, C and D). It is noteworthy that incubation of the cells in the presence of insulin for 15 min resulted in a level of S6K phosphorylation that was almost comparable with that observed in a 30-min incubation with either amino acids or glucose. This partial inhibition caused by the RalA knockdown might be a reflection of the requirement for a nutrient signal for growth factor-induced mTORC1 activation as described previously (1). In addition, the effects of RalA and RalB knockdown on the phosphorylation of 4EBP1, another well characterized readout for mTORC1 activation, were very similar to their effects on S6K phosphorylation (Fig. 1, A-C). These results suggest that the nutrient-sensing system employs Ral GTPases to activate mTORC1 and that RalA is an indispensable component, whereas the phosphoinositide 3-kinase/PKB-dependent mTORC1 activation pathway does not depend to a significant extent on Ral GTPases.
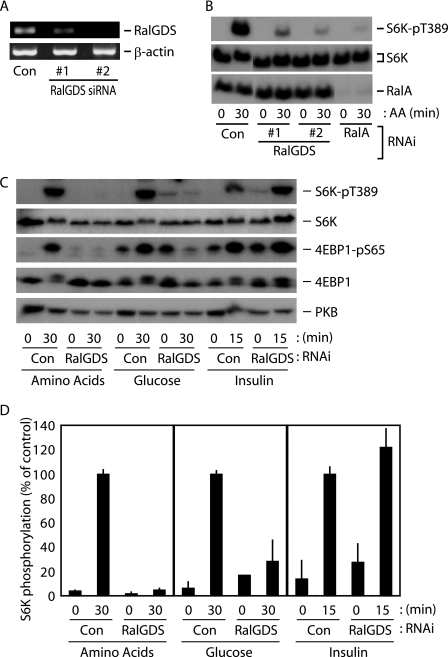
The Nutrient-sensing System Requires the RalGDS-RalA Signaling Unit—RalGDS is a well characterized upstream activator of the Ral GTPases in context of its relevance for cell proliferation. We therefore used RNAi to inactivate RalGDS in HeLa cells in order to analyze its role in the amino acid-sensing system. Unfortunately, we were unable to confirm the knockdown using commercially available anti-RalGDS antibodies to detect endogenous RalGDS; however, two distinct siRNAs against RalGDS successfully reduced expression of the RalGDS transcript (Fig. 2A). Cells transfected with these siRNAs showed reduced S6K phosphorylation at Thr389 upon the addition of amino acids to the medium (Fig. 2B). Because Ral GTPases appear to preferentially participate in the nutrient-induced mTORC1 activation pathway, we next tested whether the RalGDS contribution was selective for the nutrient-sensing system. As shown in Fig. 2, C and D, results of the RalGDS knockdown were fundamentally consistent with those observed for RalA knockdown (i.e. robust inhibition of amino acid- and glucose-induced mTORC1 activation). These findings strongly suggest that RalGDS participates in the nutrient-sensing system, presumably as an activator of Ral GTPases. In contrast, insulin-induced mTORC1 activation was not inhibited by RalGDS knockdown (Fig. 2, C and D), indicating that insulin induces mTORC1 activation in a RalGDS-independent manner. Hao et al. (36) recently reported that RalGDS forms a complex with PKB, a critical component in growth factor-regulated mTORC1 activation, and promotes PKB activation in breast cancer cell lines. We are not able to completely explain the conflict between those findings and the results of this study or the discrepancy in the requirement for RalA and RalGDS in insulin-induced mTORC1 activation. However, taken together, these results suggest that RalGDS has more diverse effects on signaling systems than RalA and potentially provides a feedback mechanism for RalA action under some conditions.
FIGURE 2.
RalGDS plays an indispensable role in nutrient-induced mTORC1 activation. A and B, HeLa cells were transfected with siRNAs, as indicated. After 2 days in culture, cells were deprived of amino acids for 2 h and then exposed to amino acids for the indicated times, followed by immunoblot analysis (B). The original unprocessed gel image is presented in supplemental Fig. 6. The parallel experiment to confirm RalGDS knockdown was also performed by reverse transcription-PCR analysis (A). C, HeLa cells were transfected with siRNAs, as indicated. After 2 days in culture, cells were deprived of amino acids, glucose, or serum for 2 h. Cells were then exposed to amino acids, glucose, or 2 μg/ml bovine insulin for the indicated times, followed by immunoblot analysis. D, quantification of the phospho-S6K (normalized to total S6K) immunoblots shown in C, as described under “Materials and Methods.” Values represent the mean ± S.D. of 3-5 independent experiments.
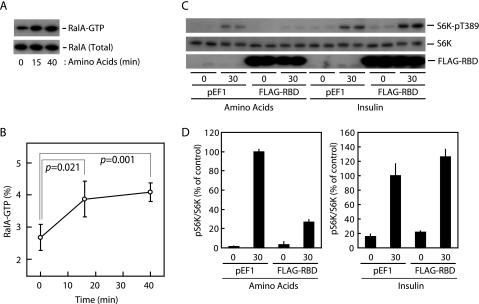
RalA Is Activated in Response to Amino Acid Availability—The observation that knockdown of either RalA or RalGDS, which facilitates GDP-GTP exchange of RalA, inhibited the amino acid-sensing system prompted us to ask whether RalA was activated in response to amino acid availability. We assayed RalA activation in cells using the Sec5-RBD protein, which selectively binds to the GTP-bound form of Ral GTPases and, therefore, can be used as an “active” Ral GTPase probe (32). Fig. 3, A and B, show that levels of the active, GTP-bound form of RalA increased following a 15-min incubation of cells with amino acids, reaching a plateau within 40 min. In contrast, RalB activation was not observed under these conditions (data not shown). In addition, overexpression of Sec5-RBD, which may inhibit RalA-dependent signals by binding toward active RalA, clearly suppressed amino acid-induced Thr389 phosphorylation of S6K (Fig. 3, C and D). In contrast, insulin-induced Thr389 phosphorylation was not inhibited by Sec5-RBD overexpression (Fig. 3, C and D). On the basis of these results, we conclude that nutrient-induced activation of RalA is a critical step for the activation of mTORC1.
FIGURE 3.
RalA is activated in response to amino acid availability, and the activation is required for nutrient-induced mTORC1 activation. A and B, HeLa cells were deprived of amino acids for 2 h, followed by exposure to amino acids for the indicated times. The RalA activation assay was then performed as described under “Materials and Methods.” Values represent the mean ± S.D. of four independent experiments. Calculated p values (Student's t test) are presented. C, HeLa cells were transfected with the DNA construct as indicated. After 1 day in culture, cells were deprived of amino acids or serum for 2 h. Cells were then exposed to amino acids or 2 μg/ml bovine insulin for the indicated times followed by immunoblot analysis. D, quantification of the phospho-S6K (normalized to total S6K) immunoblots shown in C, as described under “Materials and Methods.” Values represent the mean ± S.D. of 3-5 independent experiments.
RalA Functions as a Downstream Component of Rheb—Rheb is the small GTPase that plays a crucial role in the mTORC1 activation machinery. Although the action of Rheb remains controversial, an accumulating body of evidence places Rheb upstream of and in proximity to mTORC1 (11, 37, 38). Our question arising here is whether RalA acts upstream or downstream of Rheb in the nutrient-sensing system, because RalA may exert its effect on diverse processes, such as cytoskeletal reorganization and membrane dynamics (19-22), which potentially influence the nutrient uptake process. In fact, a Ral effector, the exocyst complex, is involved in vesicle transport (20, 39, 40); Exo70, an exocyst component, plays a role in targeting of the glucose transporter 4 to the plasma membrane (41, 42). Therefore, we first investigated the effect of RalA and RalGDS knockdown on nutrient uptake and confirmed that neither knockdown inhibited uptake of amino acids or glucose (supplemental Fig. 3).
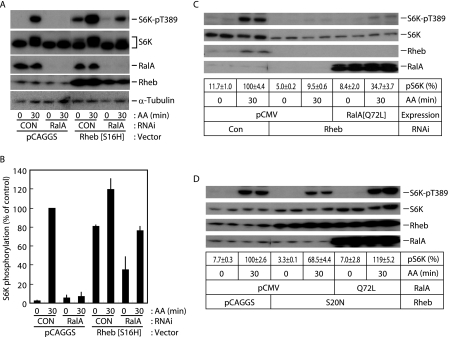
Ectopic expression of the hyperactive Rheb mutant S16H resulted in a robust increase in S6K phosphorylation (Fig. 4, A and B), consistent with a previous report (43). RalA knockdown resulted in significant (40-50%) suppression of S6K phosphorylation in S16H-Rheb-expressing cells (Fig. 4, A and B), raising the possibility that RalA functions downstream of Rheb. In order to explore this possibility, we next asked whether RalA expression induced mTORC1 activation in the absence of Rheb. Knockdown of Rheb abolished amino acid-induced mTORC1 activation, and expression of the constitutively active Q72L-RalA mutant partially restored mTORC1 activation (Fig. 4C). The dominant negative S20N-Rheb mutant also inhibited amino acid-induced mTORC1 activation by ∼30%, and Q72L-RalA restored the activity (Fig. 4D). In addition, it should be noted that neither RalA nor RalGDS knockdown altered the “activation” (guanine nucleotide-binding) status of Rheb (supplemental Fig. 4). These observations collectively suggest that RalA primarily functions as a downstream component of Rheb in the amino acid-sensing system and that RalA requires Rheb to achieve full activation of mTORC1.
FIGURE 4.
RalA functions downstream of Rheb. A, HeLa cells were transfected with control siRNA or siRNA against RalA. After 1 day in culture, cells were further transfected with empty pCAGGS or Rheb[S16H]/pCAGGGS plasmid DNA. After 1 day in culture, cells were deprived of amino acids for 2 h. Cells were then exposed to amino acids for the indicated times, followed by immunoblot analysis. B, quantification of the phospho-S6K (normalized to total S6K) immunoblot shown in A, as described under “Materials and Methods.” Bars represent differences between two independent experiments. C, HeLa cells were transfected with control siRNA or siRNA against Rheb. After 1 day in culture, cells were further transfected with empty pCMV5 or RalA[Q72L]/pCMV5 plasmid DNA. After 1 day in culture, cells were deprived of amino acids for 2 h. Cells were then exposed to amino acids for the indicated times, followed by immunoblot analysis. Quantification of the phospho-S6K (normalized to total S6K) immunoblot was performed as described under “Materials and Methods.” The first row of lane description in the figure represents averages of and differences between duplicate determinations in a typical study. D, HeLa cells were transfected with equal amounts of RalA expression construct (pCMV5 empty vector or RalA[Q72L]/pCMV5) and Rheb expression construct (pCAGGS empty vector or Rheb[S20N]/pCAGGS). After 1 day in culture, cells were deprived of amino acids for 2 h. Cells were then exposed to amino acids for the indicated times followed by immunoblot analysis. Quantification of the phospho-S6K (normalized to total S6K) immunoblot was performed as described under “Materials and Methods.” The first row of lane description in the figure represents averages of and differences between duplicate determinations in a typical study. Original unprocessed gel images are presented in supplemental Fig. 6.
Cross-talk Occurs between RalA and Rheb in mTORC1 Activation—We then asked how RalA transduces the signal from Rheb to mTORC1. One plausible mechanism is direct activation of mTORC1 by RalA; however, we were unable to detect direct activation of immunopurified mTORC1 by recombinant RalA protein (data not shown). Further, we were not able to detect direct association of either RalA or RalGDS with mTORC1 (data not shown). These results indicate that RalA alone would not be sufficient to induce mTORC1 activation, and another factor(s) must be required. This hypothesis is supported by the observation that Q72L-RalA did not fully restore S6K phosphorylation in cells when Rheb was depleted (Fig. 4C). Taken together, these results suggest that RalA promotes the action of Rheb to activate mTORC1 in response to extracellular nutrients, although further study will be required to unravel the underlying biochemistry (Fig. 5).
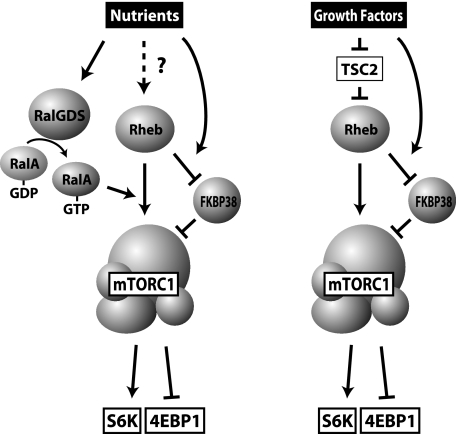
FIGURE 5.
Model for the action of RalGDS and RalA. Extracellular nutrients activate RalGDS to promote guanine nucleotide exchange of RalA. Activated RalA then facilitates the action of Rheb to activate mTORC1 in a FKBP38-independent manner. In contrast, growth factor stimulation activates Rheb primarily through the inactivation of TSC2. Rheb then activates mTORC1 in a RalA-independent manner.
Rheb was shown to bind directly to and activate mTORC1 (11, 38) (Fig. 5). In addition, a recent report (37) demonstrated that Rheb binds to FKBP38, an endogenous inhibitor of mTORC1, and sequesters it from mTORC1 (Fig. 5). Therefore, we next asked whether RalA acts on FKBP38 to regulate mTORC1 activity. To address this question, we first tested the interaction between ectopically expressed Myc-RalA and FLAG-FKBP38; however, we were unable to detect the interaction of these molecules in HeLa cells (supplemental Fig. 5). In addition, it should be noted that we could not reproduce the inhibitory effect of FKBP38 on mTORC1 activity. Both S6K and 4EBP1 phosphorylation remained unaffected by overexpression of FKBP38 (supplemental Fig. 5). Further, knockdown of FKBP38 affected neither S6K nor 4EBP1 phosphorylation (supplemental Fig. 5). These observations are supported by a recent report (44), which argued that FKBP38 did not inhibit mTORC1 signaling. Taking these results together, we conclude that RalA participates in mTORC1 regulation independently to FKBP38 (Fig. 5).
Growth factor signaling cascades also include Rheb as an indispensable regulator of mTORC1 activity. However, in contrast to the pathway employed by nutrient-sensing systems, growth factor-induced mTORC1 activation employs TSC2, a Rheb-GAP, as a critical mediator (45, 46). Since the precise mechanism by which cells discriminate between nutrient- and growth factor-induced Rheb signals remains unknown, it would be difficult to incorporate a specific mechanism for the action of RalA in mTORC1 activation into a simple scheme. However, our observation that growth factor-induced mTORC1 activation does not require RalA (Fig. 1) suggests that Rheb, which is activated as a result of growth factor stimulation and TSC2-inactivation, drives a pathway distinct from the nutrient-dependent pathway to activate mTORC1. S16H-Rheb might activate both RalA-dependent and RalA-independent signals, presumably due to its hyperactive characteristics (43), providing an explanation for the observation that depletion of RalA did not result in complete inhibition of S6K phosphorylation in S16H-Rheb-expressing cells (Fig. 4, A and B).
Biological Significance of RalA as a Component of the Nutrient-sensing System—RalGDS and RalA are known to be potential oncogenic factors that are required for Ras-induced transformation of cells. A number of studies have indicated that these two molecules are essential but not necessarily sufficient for the transformation (23, 24, 26, 28); however, the exact reason for the RalGDS-RalA requirement has been poorly understood to date. This study has demonstrated that the RalGDS-RalA signaling unit plays a critical role in nutrient-sensing systems that drive mTORC1 activation in the regulation of diverse intracellular processes. In order to achieve accelerated proliferation, a hallmark of transformed cells, cells require large amounts of extracellular nutrients, such as amino acids for protein synthesis and carbohydrates as a major energy source. In addition, networks of nutrient-initiated intracellular processes, such as protein synthesis and ATP production, should be up-regulated simultaneously; however, up-regulation of these systems alone may not directly induce transformation. Intriguingly, these characteristics also apply to the requirement for RalA and RalGDS in cell proliferation and oncogenic transformation. Although further studies will be required to confirm the biological significance of RalGDS and RalA in vivo, our findings on the participation of RalGDS-RalA in the mTORC1 activation pathway provide novel insight into roles of RalGDS and RalA in cell proliferation and oncogenic transformation.
Supplementary Material
Acknowledgments
We are grateful to Dr. Takehana of Ajinomoto Co., Inc. for providing amino acid-free medium and to Drs. Ryutaro Shirakawa, Mitsunori Kawato, and Hisanori Horiuchi (Kyoto University) for providing the Sec5-RBD/pGEX vector and for invaluable discussions on the RalA activation assay.
This work was supported by Grants-in-Aid for Scientific Research 18057028, 20054024, 19370058, and 19657044 from the Japanese Ministry of Education, Culture, Sports, Science, and Technology. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1-6.
Footnotes
The abbreviations used are: mTORC1, mammalian target of rapamycin complex 1; 4EBP1, eukaryotic translation initiation factor 4E-binding protein 1; GAP, GTPase-activating protein; PKB, protein kinase B; RBD, Ras-binding domain; S6K, S6 kinase; siRNA, small interfering RNA; RNAi, RNA interference.
References
- 1.Avruch, J., Hara, K., Lin, Y., Liu, M., Long, X., Ortiz-Vega, S., and Yonezawa, K. (2006) Oncogene 25 6361-6372 [DOI] [PubMed] [Google Scholar]
- 2.Corradetti, M. N., and Guan, K. L. (2006) Oncogene 25 6347-6360 [DOI] [PubMed] [Google Scholar]
- 3.Dann, S. G., and Thomas, G. (2006) FEBS Lett. 580 2821-2829 [DOI] [PubMed] [Google Scholar]
- 4.Guertin, D. A., and Sabatini, D. M. (2007) Cancer Cell 12 9-22 [DOI] [PubMed] [Google Scholar]
- 5.Wullschleger, S., Loewith, R., and Hall, M. N. (2006) Cell 124 471-484 [DOI] [PubMed] [Google Scholar]
- 6.Inoki, K., Li, Y., Zhu, T., Wu, J., and Guan, K. L. (2002) Nat. Cell Biol. 4 648-657 [DOI] [PubMed] [Google Scholar]
- 7.Potter, C. J., Pedraza, L. G., and Xu, T. (2002) Nat. Cell Biol. 4 658-665 [DOI] [PubMed] [Google Scholar]
- 8.Castro, A. F., Rebhun, J. F., Clark, G. J., and Quilliam, L. A. (2003) J. Biol. Chem. 278 32493-32496 [DOI] [PubMed] [Google Scholar]
- 9.Zhang, Y., Gao, X., Saucedo, L. J., Ru, B., Edgar, B. A., and Pan, D. (2003) Nat. Cell Biol. 5 578-581 [DOI] [PubMed] [Google Scholar]
- 10.Roccio, M., Bos, J. L., and Zwartkruis, F. J. (2006) Oncogene 25 657-664 [DOI] [PubMed] [Google Scholar]
- 11.Long, X., Lin, Y., Ortiz-Vega, S., Yonezawa, K., and Avruch, J. (2005) Curr. Biol. 15 702-713 [DOI] [PubMed] [Google Scholar]
- 12.Gulati, P., Gaspers, L. D., Dann, S. G., Joaquin, M., Nobukuni, T., Natt, F., Kozma, S. C., Thomas, A. P., and Thomas, G. (2008) Cell Metab. 7 456-465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahara, T., Hara, K., Yonezawa, K., Sorimachi, H., and Maeda, T. (2006) J. Biol. Chem. 281 28605-28614 [DOI] [PubMed] [Google Scholar]
- 14.Hara, K., Yonezawa, K., Weng, Q. P., Kozlowski, M. T., Belham, C., and Avruch, J. (1998) J. Biol. Chem. 273 14484-14494 [DOI] [PubMed] [Google Scholar]
- 15.Smith, E. M., Finn, S. G., Tee, A. R., Browne, G. J., and Proud, C. G. (2005) J. Biol. Chem. 280 18717-18727 [DOI] [PubMed] [Google Scholar]
- 16.Nobukuni, T., Joaquin, M., Roccio, M., Dann, S. G., Kim, S. Y., Gulati, P., Byfield, M. P., Backer, J. M., Natt, F., Bos, J. L., Zwartkruis, F. J., and Thomas, G. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 14238-14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, E., Goraksha-Hicks, P., Li, L., Neufeld, T. P., and Guan, K. L. (2008) Nat. Cell Biol. 10 935-945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sancak, Y., Peterson, T. R., Shaul, Y. D., Lindquist, R. A., Thoreen, C. C., Bar-Peled, L., and Sabatini, D. M. (2008) Science 320 1496-1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cantor, S. B., Urano, T., and Feig, L. A. (1995) Mol. Cell. Biol. 15 4578-4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moskalenko, S., Henry, D. O., Rosse, C., Mirey, G., Camonis, J. H., and White, M. A. (2002) Nat. Cell Biol. 4 66-72 [DOI] [PubMed] [Google Scholar]
- 21.Ohta, Y., Suzuki, N., Nakamura, S., Hartwig, J. H., and Stossel, T. P. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 2122-2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frankel, P., Aronheim, A., Kavanagh, E., Balda, M. S., Matter, K., Bunney, T. D., and Marshall, C. J. (2005) EMBO J. 24 54-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim, K. H., Baines, A. T., Fiordalisi, J. J., Shipitsin, M., Feig, L. A., Cox, A. D., Der, C. J., and Counter, C. M. (2005) Cancer Cell 7 533-545 [DOI] [PubMed] [Google Scholar]
- 24.Lim, K. H., O'Hayer, K., Adam, S. J., Kendall, S. D., Campbell, P. M., Der, C. J., and Counter, C. M. (2006) Curr. Biol. 16 2385-2394 [DOI] [PubMed] [Google Scholar]
- 25.Shipitsin, M., and Feig, L. A. (2004) Mol. Cell. Biol. 24 5746-5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chien, Y., and White, M. A. (2003) EMBO Rep. 4 800-806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bodemann, B. O., and White, M. A. (2008) Nat. Rev. Cancer 8 133-140 [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez-Garcia, A., Pritchard, C. A., Paterson, H. F., Mavria, G., Stamp, G., and Marshall, C. J. (2005) Cancer Cell 7 219-226 [DOI] [PubMed] [Google Scholar]
- 29.Okahara, F., Itoh, K., Nakagawara, A., Murakami, M., Kanaho, Y., and Maehama, T. (2006) Mol. Biol. Cell 17 4888-4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ijichi, C., Matsumura, T., Tsuji, T., and Eto, Y. (2003) Biochem. Biophys. Res. Commun. 303 59-64 [DOI] [PubMed] [Google Scholar]
- 31.Niwa, H., Yamamura, K., and Miyazaki, J. (1991) Gene (Amst.) 108 193-199 [DOI] [PubMed] [Google Scholar]
- 32.Kawato, M., Shirakawa, R., Kondo, H., Higashi, T., Ikeda, T., Okawa, K., Fukai, S., Nureki, O., Kita, T., and Horiuchi, H. (2008) J. Biol. Chem. 283 166-174 [DOI] [PubMed] [Google Scholar]
- 33.Moser, B. A., Dennis, P. B., Pullen, N., Pearson, R. B., Williamson, N. A., Wettenhall, R. E., Kozma, S. C., and Thomas, G. (1997) Mol. Cell. Biol. 17 5648-5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romanelli, A., Dreisbach, V. C., and Blenis, J. (2002) J. Biol. Chem. 277 40281-40289 [DOI] [PubMed] [Google Scholar]
- 35.Saitoh, M., Pullen, N., Brennan, P., Cantrell, D., Dennis, P. B., and Thomas, G. (2002) J. Biol. Chem. 277 20104-20112 [DOI] [PubMed] [Google Scholar]
- 36.Hao, Y., Wong, R., and Feig, L. A. (2008) Mol. Cell. Biol. 28 2851-2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai, X., Ma, D., Liu, A., Shen, X., Wang, Q. J., Liu, Y., and Jiang, Y. (2007) Science 318 977-980 [DOI] [PubMed] [Google Scholar]
- 38.Long, X., Ortiz-Vega, S., Lin, Y., and Avruch, J. (2005) J. Biol. Chem. 280 23433-23436 [DOI] [PubMed] [Google Scholar]
- 39.Brymora, A., Valova, V. A., Larsen, M. R., Roufogalis, B. D., and Robinson, P. J. (2001) J. Biol. Chem. 276 29792-29797 [DOI] [PubMed] [Google Scholar]
- 40.Sugihara, K., Asano, S., Tanaka, K., Iwamatsu, A., Okawa, K., and Ohta, Y. (2002) Nat. Cell Biol. 4 73-78 [DOI] [PubMed] [Google Scholar]
- 41.Inoue, M., Chang, L., Hwang, J., Chiang, S. H., and Saltiel, A. R. (2003) Nature 422 629-633 [DOI] [PubMed] [Google Scholar]
- 42.Inoue, M., Chiang, S. H., Chang, L., Chen, X. W., and Saltiel, A. R. (2006) Mol. Biol. Cell 17 2303-2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan, L., Findlay, G. M., Jones, R., Procter, J., Cao, Y., and Lamb, R. F. (2006) J. Biol. Chem. 281 19793-19797 [DOI] [PubMed] [Google Scholar]
- 44.Wang, X., Fonseca, B. D., Tang, H., Liu, R., Elia, A., Clemens, M. J., Bommer, U. A., and Proud, C. G. (2008) J. Biol. Chem. 283 30482-30492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garami, A., Zwartkruis, F. J., Nobukuni, T., Joaquin, M., Roccio, M., Stocker, H., Kozma, S. C., Hafen, E., Bos, J. L., and Thomas, G. (2003) Mol. Cell 11 1457-1466 [DOI] [PubMed] [Google Scholar]
- 46.Inoki, K., Li, Y., Xu, T., and Guan, K. L. (2003) Genes Dev. 17 1829-1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.