Abstract
The chemical composition of Persistent Organic Pollutants (POPs) in the environment is not uniform throughout the world, and these contaminants contain many structurally different lipophilic compounds. In a well-defined study cohort in the Slovak Republic, the POP chemicals present in the peripheral blood of exposed children were chemically analyzed. The chemical analysis data revealed that the relative concentration and profile of structurally different organic pollutants, including polychlorinated biphenyls (PCBs), 2,2’-bis(4-chlorophenyl)-1,1- dichloroethylene (p,p’-DDE), 2,2’-bis(4-chlorophenyl)-1,1,1-trichloro-ethane (p,p’-DDT), hexachlorobenzene (HCB) and β-hexachlorocyclohexane (β-HCH), may vary from individual to individual, even within the same exposure area. These chemicals can be broadly classified into two groups. The first group, the PCB congeners, primarily originated from industrial compounds and their byproducts. The second group of compounds originated from or was commonly used in the agricultural sector (e.g., DDT, HCB). The objective of this study was to examine the effects of the two POP exposure profiles on gene expression. For the study population, we selected prepubertal girls (mean age of 46.2 ± 1.4 months) with high POP concentrations in their blood (> 75% tile of total POP) and classified them in the high ‘PCB’ group when the total PCB concentration was significantly higher than the total concentration of other POP components and in the ‘Other Than PCB’ (OTP) group, when the total PCB concentration was significantly lower than the concentration of the other major POP constituents. A matched control group of girls (< 25% tile of total POP) was selected for comparison purpose (n = 5 per group). Our aims were to determine whether there were any common effects of high POP exposure at a toxicogenomic level and to investigate how exposure may affect physiological functions of the children in two different exposure scenarios. Global gene expression analysis using a microarray (Affymetrix Gene Chip Human genome U133 Plus 2.0 Array) platform was conducted on the total RNA of peripheral blood mononuclear cells from the girls. The results were analyzed by Partek GS, Louis, MI, which identified twelve genes (ATAD2B, BIVM, CD96, CXorf39, CYTH1 ETNK1, FAM13A, HIRA, INO80B, ODG1, RAD23B, and TSGA14) and two unidentified probe sets, as regulated differentially in both the PCB and OTP groups against the control group. The qRT-PCR method was used to validate the microarray results. The Ingenuity Pathway Analysis (IPA) software package identified the possible molecular impairments and disease risks associated with each gene set. Connective tissue disorders, genetic disorders, skeletal muscular disorders and neurological diseases were associated with the 12 common genes. The data therefore identified the potential molecular effects of POP exposure on a genomic level. This report underscores the importance of further study to validate the results in a random population and to evaluate the use of the identified genes as biomarkers for POP exposure.
Keywords: Persistent Organic Pollutants, POP, Gene Environment Interaction, Gene Expression, Human PBMC, IPA Analysis, Disease and Disorders
1. Introduction
Persistent organic pollutant (POP) is a collective term for any organic, lipid-soluble compound that survives physicochemical and biological degradation and persists in the environment. These chemicals often bio-accumulate in fatty tissues and maintain a close equilibrium with the lipid content of the extracellular fluid of the body due to their slow metabolic clearance. The production and uses of many POP compounds, especially organochlorine insecticides and industrial chemicals, have been banned in developed countries. Information about the sources, release, and environmental levels of POPs are yet to be completely understood, particularly in underdeveloped and developing countries (Bouwman 2003). The global transport is common for POP compounds, so the effects of these chemicals on human health are not restricted by national boundaries.
These bioaccumulated POP compounds may be of different chemical structures. Even within a particular group of chemicals, such as polychlorinated biphenyls (PCB), there may be different chemical structures depending on the degree of chlorination and their positional substitution at the biphenyl nucleus.The relative distribution of each POP chemical in the body may be different, depending on several factors, including the source of the contamination, dietary habits, and the timing of the exposure (e.g., pre- or post-natal) (Webster et al., 2004).
Previous epidemiological studies reported an association between POP exposure and higher risks of different diseases (Sergeev and Carpenter 2010), including cancer (Brody et al., 2007; Bencko et al., 2009). In vitro and in vivo animal models have been used to understand the mechanisms of action responsible for the health risks of POP components (Zhu et al., 2009). The structural similarities of these POP chemicals with the naturally occurring metabolites, particularly with steroid and thyroid hormones may induce pseudo-hormonal behavior and may cause imbalances in normal physiological processes through receptor-mediated interactions (Kelce et al., 1995). Additionally, these compounds and their metabolites may also produce oxidative stress (Lin et al., 2009).
Microarray-based gene expression studies conducted in our lab and by other groups have so far used specific POP compound(s) in different combinations to study the effects of the exposure. These studies were mainly conducted in cultured cells, rodents or aquatic models (Dutta et al., 2008; De et al., 2010; Ghosh et al., 2007, 2010, 2011; Lyche et al., 2010). Royland (2008 a,b) investigated the effects of a commercial PCB mixture on the expression of genes related to developmental neurobiology in rats. However, the composition of actual bioaccumulated exposure profiles is different from the composition of the commercial chemical. Moreover, the species differences between rodents and humans also affect the pharmacokinetics of the toxicity. In our recent work, we have established that differences in gene expression due to PCB exposure can help elucidate the molecular mechanisms behind the associated disease processes (Dutta et al., 2011). In actual exposure, different lipophilic organochemicals are present at the same time. The evaluation of the integrated toxicogenomics impact of these compounds is essential to understand the molecular mechanism of the associated disease risk in individuals. The scientists were able to delineate the effects of a cocktail of serum POPs in the Greenlandic Inuit by evaluating receptor interactions in an in vivo set up. They found that the overall effect of the exposure was endocrine disruption in that particular population. (Kruger et al., 2008, Long et al., 2007). Toxicological potential other than endocrine disruption have also been observed for these chemicals (Faroon et al., 2001; Farnandes et al., 2010).
So far, no data are available on the effects of POP on global gene expression or the comparative effects of different POP profiles in humans. A single study has been performed with pooled serum samples to understand the effects of POP exposure on a moderate number of genes (908 genes) using an array-based method. This study detected the down regulation of tumor suppressor genes (Tsai et al., 2008). However, their work did not provide information about the effects of POP exposure on the whole genome, or the effects of variations in the POP profile on gene expression.
In the present study, our goal was to examine the effects of POP exposure on global gene expression and determine how the variation in POP components in actual human exposure may affect gene expression in the children of a well-studied cohort to expand our knowledge about the molecular mechanisms responsible for different POP exposure-related pathophysiological effects.
2.Materials and Methods
2.1 Study subjects
We selected subjects for this study from a unique cohort of mothers and children from the Slovak Republic in Eastern Europe. This cohort has already been explicitly documented for differential pathophysiological effects due to high POP exposure (Jan et al., 2007; Park et al., 2009; Radikova et al., 2008; Trnovec et al., 2010; Ukoprek et al., 2010). Each child was evaluated using standardized clinical assessments. We analyzed only the girls’ microarray results (mean age 46.2 months) because there were few boys for whom both the Affymetrix expression data and blood POP concentrations were available. We classified these subjects into two groups: the ‘high PCB’ group (Group A), for which the total PCB concentration was significantly higher than total concentration of other POP components, and the ‘Other Than PCB’ (OTP) group (Group B), for whom the total PCB concentration was significantly lower than the concentration of other major POP constituents. A matched control group of girls (<25% tile of total POP, Group C) was selected for comparison purpose (n = 5 per group) (Figure 1). This study was approved by the Howard University Institutional Review Board (IRB-07-GSAS-30), and written consent in the approved format was obtained from all study subjects prior to the collection of blood.
Figure 1.
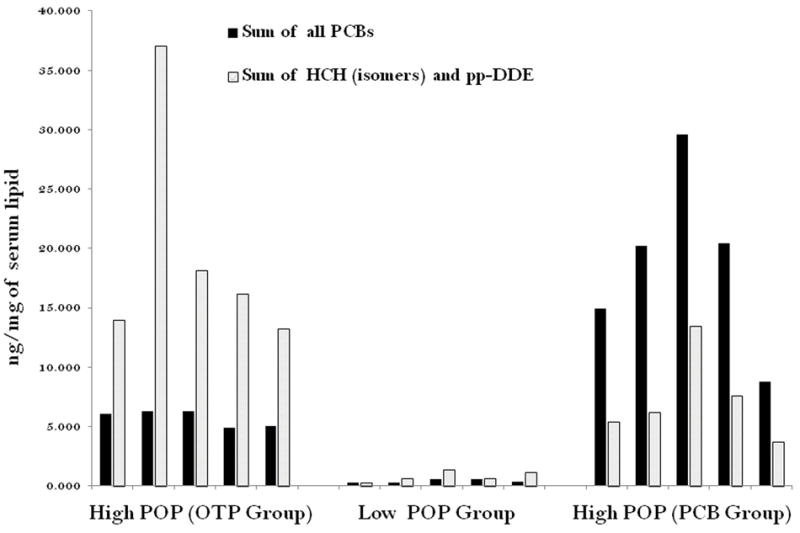
Distribution of POP components in the subjects selected for microarray experiments: High POP (OTP Group) means, high total POP value in their blood (>75 percentile) and Other Than PCB of POP component / PCB > 1; High POP (PCB Group) means high total POP value in their blood (>75 percentile) but PCB / Other Than PCB of POP component > 1. Low POP Group (Control): low total POP value in their blood (< 25 percentile).
2.2 POP analysis
The details of the estimation of polychlorinated biphenyl and pesticide concentrations have been described previously (Ukoprek et al., 2010). The serum concentrations of fifteen PCB congeners (IUPAC numbers 28, 52, 101, 105, 114, 118, 123, 138+163, 153, 156+171, 157, 167, 170, 180 and 189) and the concentrations of p,p’-DDE, 2,2’-bis(4-chlorophenyl)-1,1,1-trichloro-ethane (p,p’-DDT), HCB and β-hexachlorocyclohexane (β-HCH) were determined in serum using a high-resolution gas chromatography device (HP 6890; Agilent, Santa Clara, CA, USA) equipped with a Ni-63 micro-electron capture detector and a 60-m DB-5 capillary column (J&W Scientific, Folsom, CA, USA) (Kocan et al.,1994,2001). For congeners that were below the detection limit the values corresponding to half the limit of detection were used. The detection limit of each PCB congener varied from 3.9 ng/g lipid for PCB-157 to 7.5 ng/g lipid for PCB-52. For organochlorine pesticides, the limits of detection were between 2.8 ng/g for γ-HCH and 7.4 ng/g for β-HCH (Petrk et al.,2006; Kokan et al.,1994). The sum of all 15 individual PCB congeners was calculated as the sum of PCBs including half the limit of detection for nondetected PCBs. The organochlorine concentrations were adjusted to the total lipid level. The total cholesterol, non-esterified cholesterol, phospholipids and triacylglycerol were measured using enzymatic methods (Ukropec et al., 2010; Patric et al., 2006).
2.3 Sample collection, RNA isolation
Whole blood was collected in PAXgene® Blood RNA tubes in Slovakia and dispatched to the Molecular Genetics Laboratory at Howard University, Washington, DC, USA according to the specific protocol of the manufacturer (Qiagen, MD). RNA was extracted from the PAXgene tubes using the PAXgene Blood RNA Kit IVD (Qiagen, Cat. # 762164) according to manufacturer’s instructions. Contaminating DNA was removed using the Ambion DNA-free kit. The RNA content was determined spectrophotometrically on a nanodrop at 230, 260 and 280 nm. RNA quality was also verified with an Agilent bioanalyzer using an RNA 6000 nanochip before the microarray chip hybridization; the RNA was stored at −80 °C.
2.4 Microarray experiments
The microarray analysis was performed at the Center for Genetic Medicine, Children’s National Medical Center, Washington, DC, USA. Expression profiling data was obtained for each sample individually using the GeneChip® Human Genome U133 Plus 2.0 array with standard operating procedures and following the protocols of Zhao and Hoffman (2004) for the quality control procedure. The expression data sets are available at the following link: www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=vtcvzaqsusyysdy&acc=GSE28805.
2.5 Gene expression data analysis
To summarize the information obtained in the microarray analysis and to check data quality, we used Principal Component Analysis (PCA) and plotted the PCA scores. In PCA analysis, the aim was to reduce a complex, multi-dimensional problem to a few understandable components. The plot demonstrated one point per array. Each color represented one experimental factor (grouping). This allows viewing of separations between groups of replicates and from arrays of other groups. In the 3D PCA plot PCA scores (components) are represented in the X,Y, and Z axes and are numbered as PC1,PC2,and PC3, according to their decreasing significance. PC1 is the first axis which account for the most variability in the data. The proportion of variance accounted for by each subsequent axis, PC2 and PC3 (in three dimensional presentations) will sequentially decrease.
To identify differentially expressed gene sets, we analyzed the microarray results using a oneway ANOVA model by Partek GS. Briefly, the raw microarray values from the Affymetrix.cel files were imported into Partek Genomics Suite, version 6.5, release 6.10.0915 (Partek Inc., St. Louis, MI). The probe summarization and probe set normalization were done using the GC RMA algorithm (Wu and Irizarry 2004), which included GC-RMA background correction, quantile normalization, log2 transformation and median polish probe set summarization. We used a one-way ANOVA model and the method of moments (Eisenhart, 1947). Model: Yij = μ + PCB Other Control i+ εij, where Yij represents the jth observation of the ith PCB Other Control; μ is the common effect for the whole experiment. εij represents the random error present in the jth observation on the ith PCB Other Control. The errors εij are assumed to be normally and independently distributed with mean 0 and standard deviation δ for all measurements. The following comparisons were performed: Other than PCB vs. Control; PCB vs. Control.
The actual variation among the groups is expressed using F ratio and presented in the ‘Source of Variation Plot’. Thus, if the null hypothesis is correct that there is no variation between the groups, we expect F to be about 1, whereas large F-ratio indicates a likely significant effect. The bar chart shows the variation contributed by effects across all the test variables (response variables) in the ANOVA model, i.e., X axis represents the factors or interaction in the ANOVA model and Y axis represents the F ratio.
With more than 54,000 probe sets, whole-genome microarray studies are susceptible to type I errors, because they simultaneously test multiple hypotheses. Our goal was to identify a true positive gene set associated with POP exposure. We employed the False Discovery Rate (FDR) correction, the most frequently used method of controlling the type I error. The results yielded the expected proportion of truly null hypotheses among all the rejected null hypotheses (Kaizer et al., 2007) and kept the false positive rate below five percent.
We selected gene sets that represented at least a 1.5-fold change in expression level because at this level of altered gene expression, the biological impact of chemical exposure could be reliably deduced (Guo et al., 2006). This analysis generated two highly significant sets of differentially expressed genes that originated from two different high-POP exposure conditions relative to low POP exposure (FDR < 0.05 and Fold Change > 1.5 or < -1.5).
The differential expression of the genes in POP exposures is presented using ‘Volcano Plot’. It is an effective and easy-to-interpret graph that summarizes both fold-change and level of significance. It is a scatter-plot of the negative log10-transformed p-values against the log2 fold change. Genes with statistically significant differential expression will lie above a horizontal threshold line. Genes with large fold-change values will lie outside a pair of vertical threshold lines. The significant genes will tend to be located in the upper left or upper right parts of the plot (Cui and Churchill, 2003).
The common differentially expressed genes in two types of exposure are represented in Venn diagram. Venn Diagrams are useful for examining similarities and differences in two sets of genes. It is often used in mathematics to show relationships between sets. The overlapping sets of genes representing both the PCB set and OTP set are observed in the intersection of the circles.
2.6 Functional analysis by Ingenuity Pathway Analysis (IPA)
The data mining software Ingenuity Pathway Analysis (IPA) was used for further analysis of the gene lists obtained from the Partek GS analysis. The gene identifiers (called focus molecules) of the lists were mapped to their corresponding gene objects and overlaid onto a global molecular network developed from information contained in the Ingenuity knowledge base. Gene identifiers were defined as value parameters for the analysis and identified the relationship between gene expression changes and related changes in biofunctions that fell into the subcategories of molecular and cellular functions, physiological system development, and function and disease and disorders.
Biofunction analysis identified the biofunctions that were significantly associated with the data set based on the association between the data set and the information in the IPA library. This association was calculated from the ratio of the number of genes from the data set that are associated with the biofunction divided by the total number of molecules that are associated with the function. The probability that each biological function and/or disease assigned to that data set is due to chance alone was calculated using a Right-tailed Fischer’s Exact Test. Overrepresentation of the molecules in a given process was considered to be statistically significant when P < 0.05. The over-represented functional or pathway processes are those which have more focus molecules than expected by chance.
2.7 Validation of the genes by quantitative real time PCR (qRT-PCR)
The genes identified by microarray analysis were validated by qRT-PCR. The sets of forward and reverse primers are given in Supplemental Table 3. The amplification efficiency of the primer pairs are 100±10 as mentioned by Applied Pharmaceuticals user’s guide. Total RNA was DNase treated to eliminate contaminating DNA using a DNA-free kit (AM1906) from Ambion (Austin TX). The reverse transcription reaction was performed with a High Capacity RNA-to cDNA Kit (P/N 4387949) from Applied Biosystems (Foster City, CA) according to the manufacturer’s protocol. The reverse transcription reaction was carried out using Taqman Universal master Mix II (Applied Biosystems, Foster City, CA). All reactions were run in 3 technical replicates. The endogenous control GAPDH (Glyceraldehyde-3-phosphate dehydrogenase) was measured from the same cDNA preparations. All primers were designed to amplify only from cDNA. All quantitative PCR reactions were performed in a 7900 HT instrument. The reactions with Taqman Universal Master Mix II and specific primers were carried out for 10 min at 95°C, to allow for polymerase activation, followed by 40 cycles of 15 sec at 95 °C for denaturation and 1 min at 60 °C for the annealing and extension step. Values from 3 technical replicates for each expression assay were averaged and normalized to the expression level of GAPDH from the same cDNA preparation. Relative differences were calculated using the ΔΔCt method (Livak and Schmittgen, 2001).
3. Results
3.1 The POP concentration in three groups
The relative distribution of the two groups of POP components (PCB and OTP) in each subject is presented in Figure1. The median and mean values and ±SD of the three groups are as follows. Median POP concentration of High PCB Group is 4.8 ng/mg of lipid with a range 1.8-7.4 and the mean (±SD) of this group is 4.5 ± 1.9 ng/mg of serum lipid. Median POP concentration of High POP Group is 3.3 ng/mg of lipid with a range of 2.9-7.5 ng/mg of lipid and the mean (±SD) value of this group is 4.2 ±1.6 ng/mg of serum lipid. The control group has the Median of 0.15 ng/mg of lipid with a range of 0.07-0.4ng/mg of serum lipid and a mean (±SD) value of 0.18 ± 0.1 ng/mg of lipid.
3.2 Global view of changes in gene expression by Principal Component Analysis (PCA)
To determine the actual differences in over all gene expression, between the three groups, if any, we conducted principal component analysis on the microarray data set. Figure 2 indicated the relationships among the groups in the 3D PCA plot. The colors represented red for control, blue for OTP and green for PCB group respectively. The contour of the ellipsoids demonstrated the possible variation within the replicates in each groups, constructed with Partek software. In this microarray experiment PC1 accounted for 34.5%, of the variability in the data followed by the subsequent axis, PC2 (13.2 %) and PC3 (9.22 %). The distribution and clustering of the samples within the PCA models indicated that the OTP and PCB group were mostly separated from control group though a small partial overlap was there. It also indicated that the two different exposure profiles may have two different impacts on gene expression. However, the complete absence of separation of the two high exposure groups from that of control group indicated that the effect of gene expression is modest.
Figure 2.
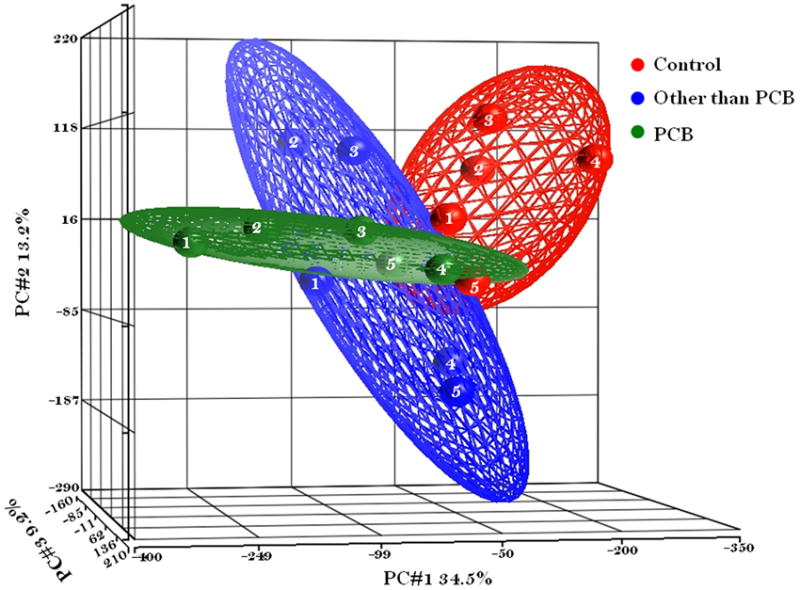
Principle Component Analysis (PCA) mapped scatter plot: The global gene expression profiles of the PBMC cells from Girls exposed to environmental POPs in Slovakia were analyzed by PCA. The figure represented the first three principal components of microarray analysis data (PC1, PC2 and PC3) in X, Y, and Z axis respectively, and demonstrated the expression profile of the three groups of five subjects per group; Other than PCB (Blue # 1-5), PCB (Green # 1-5), and Control (Red # 1-5).
3.3 Identification of differentially expressed genes due to High POP exposure
We identified 38 differentially expressed genes between group A and C, of which 19 were up regulated and 21 were down regulated. The notable genes in this category are APC, ETNK1, and CAPN2 (for the complete list see Supplemental Table 1). With the same statistical stringency, we identified 82 genes that were differentially expressed between group B and C. The up regulated (22) and down regulated (60) gene sets with their location and respective fold change are included in the table. The notable genes in this category are ALCAM, CDK6, and CDK8 (for the complete list see Supplemental Table 2).
The effects of the exposure were tested for the F-ratio between the groups. In an ANOVA model, the F-ratio is the statistic used to test the hypothesis that the effects are real (i.e., the means are significantly different from one another). If the difference between the means is due only to chance, and there are no real effects, then the expected value of the F-ratio would be one (1.00). The variations in the F-ratios for our analysis were 27.32 and 23.30 for OTP vs. the control group and PCB vs. the control group, respectively (Supplemental Figure 3). The volcano plot (Supplemental Figure 4) represents the fold-changes as well as the statistical significance of the difference in terms of the P value of the two gene sets. The red circles represent up regulated (> 1.5-fold) genes and blue circles down regulated (< -1.5-fold) genes.
3.4 Common Genes in the differentially expressed gene sets for two POP profiles
The two POP distribution patterns yielded two different gene sets with partial overlap on a Venn diagram (Figure 3). A large proportion of the genes that were significantly altered were POPprofile specific. The common area in the Venn diagram (Figure 3) represents 14 Affymetrix probe IDs common to two differentially expressed gene sets (Table 1). In two cases, the IDs did not represent any known gene. Among twelve known genes, 10 genes were down-regulated and only 2 genes were up-regulated. The heat map of the 14 gene IDs is shown in Figure 4. The hierarchal clustering pattern of these differentially expressed genes shows the admixture of subjects in the two POP groups, although they are significantly different from the lower exposure group. Table 1 shows the differential expression patterns in the two groups side by side. They changed in the same direction in both groups, and were highly correlated.
Figure 3.
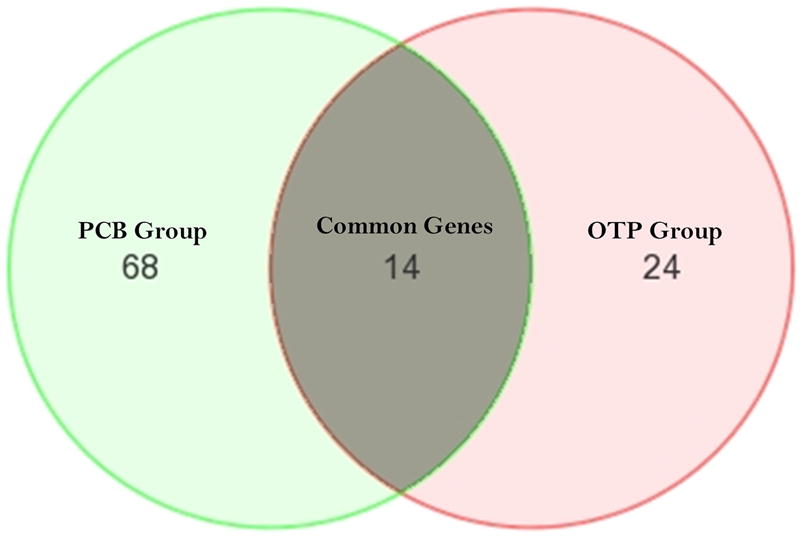
Venn diagram represented the distribution of the two differentially expressed gene sets. 14 genes were common in both gene sets. 24, and 68 genes were differentially expressed exclusively in OTP Group (A), PCB Group (B).
Table 1. The list of differentially expressed gene set common in OTP and PCB group.
(Fold change values are shown in each case. Out of 14 Affimetrx IDs, 12 IDs encoded for known gene product and identified by Ingenuity Knowledge base for functional analysis).
| Probe set ID | Gene Symbol | Gene Title | Fold-Change(OT P vs. Control) | Fold-Change(PCB vs. Control) |
|---|---|---|---|---|
| 237287_at | --- | --- | 1.84638 | 1.80332 |
| 241226_at | --- | --- | 1.69705 | 1.65254 |
| 213387_at | ATAD2B | ATPase family, AAA domain containing 2B | -2.34685 | -2.51108 |
| 233255_s_at | BIVM | basic, immunoglobulin-like variable motif containing | -1.65087 | -1.51936 |
| 1555120_at | CD96 | CD96 molecule | -2.5444 | -2.67458 |
| 227133_at | CXorf39 | chromosome X open reading frame 39 | -2.7657 | -2.53817 |
| 202879_s_at | CYTH1 | cytohesin 1 | 1.65071 | 1.63352 |
| 219017_at | ETNK1 | ethanolamine kinase 1 | -2.91183 | -2.48733 |
| 217047_s_at | FAM13A | family with sequence similarity 13, member A | -1.70893 | -1.64237 |
| 217427_s_at | HIRA | HIR histone cell cycle regulation defective homolog A (S. cerevisiae) | -1.98679 | -2.32324 |
| 222093_s_at | INO80B | INO80 complex subunit B | 1.89113 | 1.72936 |
| 205728_at | ODZ1 | odz, odd Oz/ten-m homolog 1(Drosophila) | -2.4357 | -2.19683 |
| 201222_s_at | RAD23B | RAD23 homolog B (S. cerevisiae) | -2.0173 | -1.84372 |
| 225485_at | TSGA14 | testis specific, 14 | -1.74307 | -1.62052 |
Figure 4.
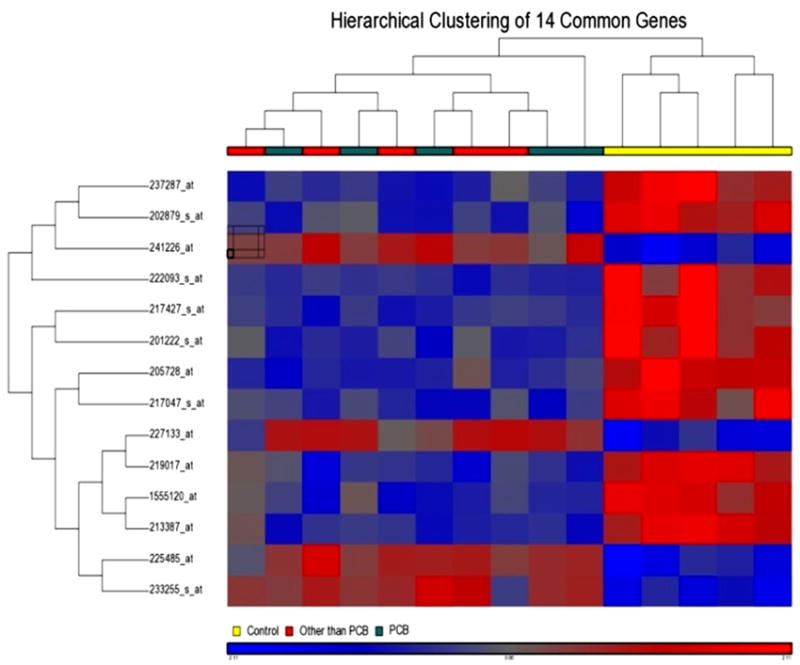
Hierarchical cluster analysis of the differentially expressed common gene set between two exposure groups: Dendrogram displayed the clustering results systematically and color mosaic display provided a visual support for the 14 differentially expressed genes, up regulated, red and down regulated, blue. The legend color for subject groups are: OTP red, PCB green and Control yellow.
3.5 Identification of biological network functions by Ingenuity Pathway Analysis
Of the 38 gene IDs from the differentially expressed gene set in Group A, 31 IDs were successfully mapped to the molecules in the Ingenuity Knowledge Base. The molecules formed 8 different networks (Table 2). The functions associated with network 1, which contains 12 focus molecules, are cellular growth and proliferation, tissue development, DNA replication, recombination, and repair. The biological functions associated with network 2 are cellular growth and proliferation, cell cycle, hepatic system development and function. Network 2 contains six focus molecules. These two networks are interconnected and in total contain 18 focus molecules.
Table 2.
The networks of genes associated with the 38 genes differentially expressed in OTP group.
| ID | Molecules in Network | Score | Focus Molecules | Top Functions |
|---|---|---|---|---|
| 1 | ALPP, APC, ARHGEF4, ATP6V0A2, BUB1, CAPN2, CD96, CD3G, CDKN1A, CTNNBIP1, CYTH1, CYTIP, DDB2, DNM, DNM3, EGF, ERK1/2, FLT3LG, FNBP1, FYN, HIRA, HRAS, IL10, IL12 (complex), INO80B, IPCEF1, PAX7, PRDX6, prostaglandin E2, PVR, RAD23B, SENP2, SPDEF, SUMO1, TNFSF8 (includes EG:944) | 29 | 12 | Cellular Growth and Proliferation, Tissue Development, DNA Replication, Recombination, and Repair |
| 2 | 4-phenylbutyric acid, CDKN1A, CDKN1B, CEBPA, CLDND1, D-glucose, DERA, FABP2, GBE1, GCKR, GH1, GSK3B, HIF1A, Histone h3, HNF1A, HNF4A, INS1, JUN, KLHDC10, KLHL6, MIR30A (includes EG:407029), ORMDL1, PAAF1, PCNP, Pka, PPARD, PSMD1, PTPRN, REG1A, SPATS2L, SRPR, TGFB1, TP53 | 12 | 6 | Cellular Growth and Proliferation, Cell Cycle, Hepatic System Development and Function |
| 3 | ethanolamine kinase, ETNK1, norepinephrine | 3 | 1 | Carbohydrate Metabolism, Cardiovascular System Development and Function, Cell Cycle |
| 4 | ATAD2B, MIR27A (includes EG:407018), MIR27B (includes EG:407019), ZFP36 | 2 | 1 | Cancer, Reproductive System Disease, Cell Cycle |
| 5 | CIITA, GFI1, ZXDA, ZXDC | 2 | 1 | Cell Death, Connective Tissue Development and Function, Gene Expression |
| 6 | FAM13A, MIR124-1 (includes EG:406907), MIR124-2 (includes EG:406908), MIR124-3 (includes EG:406909) | 2 | 1 | Cellular Growth and Proliferation, Tissue Morphology, Cancer |
| 7 | MIR133A1, MIR133A2, MIR133A-1, MIR25 (includes EG:407014), TSGA14 | 2 | 1 | Cardiovascular System Development and Function, Organ Development, Cancer |
| 8 | Ap1 gamma, ARRB1, ARRB2, CLINT1, CLTC, SCYL2, ZFYVE9 | 2 | 1 | Cell Signaling, DNA Replication, Recombination, and Repair, Nucleic Acid Metabolism |
In Group B, 70 gene IDs out of 82 were mapped to the Ingenuity Knowledge Base. These molecules formed 7 different networks (Table 3). The biological functions associated with the three major interconnected networks are cancer, gastrointestinal disease, cellular growth and proliferation for network 1, cancer developmental disorder, genetic disorder for network 2 and cellular development, cellular growth and proliferation, hematological system development and function for network 3. The three interconnected networks in total contain 38 focus molecules. Another two major networks contain eleven and 8 focus molecules.
Table 3.
The networks of genes associated with the 82 genes differentially expressed in PCB group.
| ID | Molecules in Network | Score | Focus Molecules | Top Functions |
|---|---|---|---|---|
| 1 | ACTR5, AKIRIN1, BPTF, CEBPG, CRIPT, DSTN, EED, ERCC3, ERVK6, FN1, GH1, HNF4A, INO80, INO80B, INSIG1, JAZF1, LOC284393, MIR128-1 (includes EG:406915), MIR25 (includes EG:407014), PIN4, PPP2R4, RAD23B, RBBP4, retinoic acid, RFC3, RNASE4, RUVBL2, STAG1, TLK2, TP53, TROVE2, TSGA14, Vegf, WDR12, WRNIP1 | 33 | 16 | Cancer, Gastrointestinal Disease, Cellular Growth and Proliferation |
| 2 | AKR1C14, ALCAM, CCDC80, CCDC92 (includes EG:80212), CD6, CD96, CDK6, CYTH3, EIF5, EIF2S1, ERK1/2, FAM13A, FEZF1, HNRNPC, HRAS, Igh, KRAS, LGALS8, MIR124-1 (includes EG:406907), MIRLET7A (produces hsa-let-7a), NBL1, NOTCH2NL, PCOLCE, Pdgf, PGGT1B, PIF, PRKCZ, PVR, RAP1GDS1, RASGRP1, SMARCA4, STK24, TMEM109, VPS41, ZBTB24 (includes EG:9841) | 21 | 11 | Cancer, Developmental Disorder, Genetic Disorder |
| 3 | ARNT2, ATAD2B, C10ORF26, CD6, CDC42EP3, CLK4, ELOVL6, ERLIN2, FOXO4, FSTL1, GALNT4, GPN1, HEXA, IL12RB1, IL17D, IL17F, IL18BP, IL23A, IL23R, L-tyrosine, MIR205 (includes EG:406988), OVOL1, PDPN, POMC, SLC35A5, SOAT1, SUPT4H1, TGFB1, TLR2/TLR4, TNF, TNFAIP2, USP5, USP25, ZFP36, ZFP36L1 | 21 | 11 | Cellular Development, Cellular Growth and Proliferation, Hematological System Development and Function |
| 4 | AGT, Calmodulin, CCK, CNST, COL4A1, cyclic AMP, CYTH1, CYTH3, DDX3X, DNAJA2, ERRFI1, EWSR1, HMGXB4, HNRNPA1, HNRNPF, HNRNPH1, HNRNPM, IGFBP1, INS1, Lfa-1, LRCH4, MIRLET7B (includes EG:406884), P2RY12, Pde, PTBP1, RFC1, SLC38A9, SUV39H2, TNPO1, TRIM13, TWISTNB, UBE2V2, WASL (includes EG:8976), YBX1, ZCCHC14 | 20 | 11 | Behavior, Cell Cycle, Metabolic Disease |
| 5 | ASF1B, BRD2, CDK8, CDK12, CRSP5, DIO1, ETNK1, GTF2E1, H3F3B, H3F3C, HIRA, HIRIP3, Histone h3, Histone h4, MED4, MED12, MED16, MED17, MED20, MED22, MED23, MED25, MED27, MED30, MED7 (includes EG:9443), MSH5, norepinephrine, PAX7, PNN, RNA polymerase II, TCEA1, THRAP3, TRIM24, UBN1, ZCCHC17 | 14 | 8 | Gene Expression, Cell Signaling, Infection Mechanism |
| 6 | GBGT1, globoside alpha-N-acetylgalactosaminyltransferase, lipid | 2 | 1 | Lipid Metabolism, Small Molecule Biochemistry, Molecular Transport |
| 7 | GTF3C1, POLA2, PRPF38A | 2 | 1 | Gene Expression, Molecular Transport, Protein Trafficking |
3.6 RT-PCR confirmation of selected genes from the arrays
To validate the array data, the gene expression levels of selected genes were measured by quantitative Real Time Polymerase Chain Reaction (qRT-PCR) (Supplemental Figure 1). The selected genes that were significantly changed in two exposure groups were validated by RT-PCR technique. The PCR results indicated the similar effect in expression due to POP exposure, as observed in microarray experiments. The array data tend to have a higher fold change than PCR data.
4. Discussion
Selection of subjects
The selection of the subjects was very important for this study. The girls were selected from a well-defined cohort of children from the Slovak Republic. This eastern part of Slovakia is one of the most POP-contaminated areas due to negligence in the proper disposal of organochemicals from chemical manufacturing units. The details of the cohort have been described previously (Park et al., 2009, 2010; Wimmerova et al., 2010). The POP profiles were dominated by two different classes of chemicals, which represented two different sources of environmental contamination. Chemical analysis of extracellular fluid to identify the accumulated POPs in these children demonstrated that there was variation between the POP profiles in these children (Figure 1). The‘t test’ between the total POP levels in the OTP and PCB groups demonstrated no statistically significant difference (0.94). However, the POP concentration of each of the group was highly significant when compared against the control group. The prior epidemiological study of this cohort indicated that the common POP markers showed a different association pattern with the incidence of certain diseases (Ukopreck et al., 2010). The wide inter-individual variability in exposure (Llop et al., 2010) and the involvement of different known covariant is of major concern. We did not consider the confounding factors like, education, socio-economic conditions, and other epidemiological settings, while elucidating the gene expression results due to the fiscal constrain of microarray experiments. We did not analyze the data on that basis due to inadequate sample size in each category for statistically meaningful analysis result. However, health parameters of the girls and their mothers were within the physiological normal limit at the time of birth. The subjects were also within the physiological normal limit and healthy at the time of drawing the blood. There was no manifestation of disease either diabetes or obesity in these girls which was checked by the physician for any disease.
The pre- and post-natal routes of bioaccumulation are important determinants of the POP exposure profile. Upon lipophilic chemical exposure, the concentration of lipids in extracellular fluid determines the bioavailability and metabolic clearance of these compounds. Therefore, the concentration of lipids is an important factor that can influence the perturbation of physiological functions. We compared the lipid level and did not find any significant difference between the groups. The lipid concentrations of the subjects were in the same range in all three groups. A high correlation between plasma POP concentration and lipid-adjusted POP concentration was observed in these study subjects (Supplemental Figure 2). The free POP level, which has also been reported as being important for some physiological functions, was expected to be proportional to the POPs in the extra cellular fluid in the subjects (Wigle 2008). Similarly, the age and gender were consistent determinants of serum POPs (Jonsson et al., 2005). We included only the girls in this study, mean age of 46.2 ± 1.4 months. There was no significant difference in age between the groups. The chronological age of the girls was below the age of menarche. During this period of life, the gonadal and thyroid hormones maintain a steady level. Therefore, the differential changes in endocrine parameters and their functional effects, if any, are expected to only be due to the differences in POP exposure between the groups. These groups of girls, one with low blood POP levels and another with high blood POP levels, divided into two different exposure profiles and therefore represented a good match for the comparison analysis. The welldefined subjects are one of the major strengths of this study.
Microarray procedure for investigating the toxicogenomic effect of POP exposure
We identified a common set of 14 genes that were differentially expressed in the PBMC cells of the two groups of girls with different POP exposure profiles compared with the girls who had low POP exposure levels (Table 1). This finding suggested that there may be a set of common genes associated with POP exposure, regardless of the profile of the accumulated POPs in an individual subject or the exposure scenario. Our analysis also indicated that this common toxicogenomic response may explain some general functional effects common to POP exposure, and the specific functional manifestation and pathophysiology would depend on the common toxicogenomic perturbation and other, profile-specific genes.
The common differentially expressed genes in the two high POP groups
The common differentially expressed genes in the two groups with high POP levels were mostly downregulated. The direction of the change in each gene was the same in both groups (Table 1). HIRA, a transcriptional regulator gene located in the nucleus, was down-regulated approximately two-fold in both cases. This gene encodes a histone chaperone that preferentially places the variant histone H3.3 in nucleosomes and plays an important role in the formation of the senescence-associated heterochromatin foci. These foci are thought to mediate the irreversible cell cycle changes that occur in senescent cells. Insufficient production of this gene may disrupt normal embryonic development (Lorain et al., 1998). An animal study has provided evidence supporting the observed associations between behavioral and learning disabilities and prenatal exposure to PCBs in humans (Nakagami et al., 2011). Du et al. (2008) showed in an animal model that HIRA may play a role in the development of embryos.
The two other nuclear genes, RAD23B and INO80B, also followed the same expression pattern in these two groups. RAD23B was found to be a component of the protein complex that specifically complements the NER (nucleotide excision repair) defect (Neher et al., 2010). This protein has also been shown to interact with and elevate the nucleotide excision activity of 3- methyladenine-DNA glycosylase (MPG), which is involved in DNA damage recognition in base excision repair. This protein is also involved in other biological processes, including spermatogenesis and proteasomal ubiquitin-dependent protein catabolism. The toxic effects of this gene under stressful environmental conditions, especially oxidative damage induced by different classes of chemical pollutants, have been studied (Valavanidis et al., 2006). It has also been reported that some environmentally persistent organochlorines may alter the expression of tumor suppressor genes that are important in the DNA repair process (Rattenborg et al., 2002). INO80B was another nuclear gene that was upregulated in both groups with high POP levels. Although very little is known about the biological function of this gene, the activator of its nucleolar binding protein, Pim-1, is a proto-oncogene. It has been used as a prognostic marker in prostate cancer, which suggests a link between this group of proteins and prostate carcinogenesis. INO80B induces growth arrest by halting the cell cycle at the G1phase (Miyako et al., 2009; Kuroda et al., 2004). The effect of the POP chemical HCB was studied in detail in NIH 3T3 (mouse) and WS1 (human) embryonic cells. Exposure of both cell types to HCB results in cell membrane damage, a short-term decrease in cell number, increased DNA strand breaks, and a long-term decrease in colony survival. This study demonstrated that relevant environmental concentrations of HCB have significant effects on mammalian embryonic cells in culture (Salmon et al., 2002). Therefore, our results support the in vitro mechanistic findings.
Among the differentially regulated cytoplasmic genes, ETNK1 and TSGA14 were down regulated, and CYTH1 was up regulated in both sets of subjects with high POP levels. The ETNK1 gene encodes a kinase, which is the first enzyme in the phosphatidyl ethanolamine synthesis pathway. This cytosolic enzyme is specific for ethanolamine and exhibits negligible kinase activity on choline. The specific function of TSGA14 is unknown, but a possible link between the gene and a centrosomal function in familial autism has been reported (Korvatska et al., 2011). The protein encoded by CYTH1 gene is a member of the PSCD family and appears to mediate the regulation of protein sorting and membrane trafficking. This gene is highly expressed in natural killer and peripheral T cells and regulates the adhesiveness of integrins at the plasma membrane of lymphocytes. Molecular functions of CYTH1 include ARF guanylnucleotide exchange factor activity and protein binding. These genes were associated with vesicle-mediated transport; regulation of cell adhesion; and regulation of ARF protein signal transduction.
The expression levels of CD96 and ODZ1, which have been tentatively located at the plasma membrane, were also down-regulated in both groups with high POP levels. The protein encoded by the CD96 gene belongs to the immunoglobulin superfamily. This protein is a type I membrane protein and may play a role in the adhesive interactions between activated T and NK cells during the late phase of the immune response. CD96 may also have a role in antigen presentation. C (Opitz trigonocephaly) syndrome can be caused by disruption of the CD96 gene by a missense mutation (Kaname et al., 2007), although the effects of this deficiency have not been discussed in the literature. The protein encoded by the ODZ1 gene belongs to the tenascin family and teneurin subfamily. It is expressed in the neurons and may function as a cellular signal transducer (Zhou et al., 2003). The expression patterns of these genes are not completely consistent with the gene expression analysis of Royland et al. (2008 a, b) in neural tissue of animals after PCB exposure. However, the neurobehavioral effects have also been reported in an epidemiological study on this particular population (Park et al., 2009).
The four other common genes, for which the subcellular localization is not known, were ATAD2B, BIVM, CXorf39 and FAM13A. ATAD2B overexpression has been demonstrated by immunostaining in human breast carcinoma. In tumors, ATAD2B appears to be cytoplasmic or membrane bound, not nuclear. ATAD2B is a phylogenetically conserved nuclear protein expressed during neuronal differentiation and tumorigenesis (Leachman et al., 2010). BIVM possesses virtually no sequence similarity to any currently described protein, making the prediction of a function challenging. It is highly likely that BIVM is essential for some aspect of basic cellular functioning and is expressed in a near-ubiquitous manner. The presence of a CpG island at the 5’ end of BIVM and its wide tissue distribution suggest that it may function as a housekeeping gene (Yoder et al., 2002). CXorf39 (FAM199Y) is an uncharacterized gene. However, FAM13A variants are associated with lung function (Pillai et al., 2010). At present, no known pathophysiology has been associated with these genes in epidemiological studies.
The functional effects of the differentially expressed gene(s)
In our effort to understand the functional consequences of the differential expression induced by high POP exposure, we used IPA analysis software to identify some key mechanisms involved in POP-related toxicity (http://people.mbi.ohio-state.edu/baguda/PathwayAnalysis/ipa_help_manual_5.5_v1.pdf). The IPA Analysis identified the functions and/or diseases that were most significant to the data set. Fischer‘s exact test was used to calculate a p-value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone.
The IPA analysis explained some of the cellular effects and aberrations in pathology commonly associated with POP exposure. The analysis of the gene sets differentially expressed in the OTP and PCB groups indicated some common disease processes, but these were associated with different gene sets. The involvement of different toxicogenomic pathways induced by the POP chemicals has been described in the literature (Sergent et al., 1989; Khan and Thomas 2001; Langer et al., 2008; Mariussen and Fonnum 2006; Sagerup et al., 2009., Park et al., 2009; Ruzzin et al., 2010; Shi et al., 2010; De et al., 2010). The effects of POP exposure can be better understood by studying the networks formed by the two groups of genes. However, the functional effect categorized by Ingenuity as ‘cellular growth and proliferation’ can be associated with three different networks of genes (Network 1, Network 2, Network 3) in the IPA analysis (Table 2) in the gene set from the OTP group (Group B) and two different networks (Network 1, Network 3) in the PCB group (Group B) (Table 3). In the OTP group, the first two of eight networks are interconnected, and in the PCB group, the first three of seven networks are interconnected. The change in one gene in the networks may affect the function of the other connected genes and influence the related biological functions. The IPA analysis of the genes in the PCB group did not include HIRA in any of the three networks in the analysis. This particular gene, along with seven other focus molecules (RTNK1, CDK6MSH1, TRIM24 etc.) formed another network with a different set of functions (Table 3). The IPA analysis of the genes from the OTP group demonstrated that HIRA, along with five other common genes (CD96, HIRA, CYTH1, RAD23A and INO80B), was incorporated in network 1. The second network formed by the genes from the OTP group was interconnected with the first one, through the common relationship of a single gene that was not present in either of the PCB or OTP sets; i.e., it was not significantly affected by high POP exposure. This can be explained by the fact that a particular function, e.g., ‘cellular growth and proliferation’, can involve different gene sets. At the same time a particular gene (HIRA) can be associated with different genes, depending on the cellular conditions perturbed by different chemical exposures. It seems that the combination of altered gene(s) resulting from the specific distribution of the POP chemicals determined the particular phenotypic manifestation and pathophysiology of the exposure.
The functional significance of the differential expression between the two groups
To understand the common effects of POP exposure and to identify a common marker for different exposure scenarios, we compared the disease and disorder biofunctions in IPA for the two gene sets. There were 12 disease and disorder categories common to the two gene sets. However, the associated genes were not the same for the two groups. Some diseases (Table 4) were associated with group-specific gene sets. The inflammatory response, developmental disorders, cardiovascular disorders, and reproductive system diseases were associated with the genes that were differentially expressed in the PCB group (Group B). Respiratory disease was also associated with the same group (Table 4). We also performed the same analysis for the 14 genes regulated in common (Table 5). This analysis identified many important disease and disorder functions. Some of the disease processes associated with POP exposure have been reported in epidemiological association studies (Howsome et al., 2004; Hertz-Picciotto et al., 2008; Brody et al., 2007; Ruzzin et al., 2010; Sagiv et al., 2010; Trnovec et al., 2010; Sergeev and Carpenter, 2010). It has long been indicated that the immunotoxicity is one of the most sensitive endpoint and the human immune system is vulnerable to the effects of environmental contaminants and may have detrimental health effects (Tryphonus 2001). Our result indicated that in both group the differentially expressed genes are highly associated with immunological disease. To understand the specific contribution of the common gene sets to disease, we performed a comparison analysis using the IPA platform, comparing the common gene set with the OTP- and PCB-specific gene sets. We found only four biofunctions that were significantly associated with all three gene sets and independent of the POP profile. These were connective tissue disorders, genetic disorders, neurological diseases, and skeletal and muscular disorders (Figure 5). These four disease and disorder functions may be associated with common functions of the genes in these two POP profiles. This does not necessarily exclude an association with other diseases and disorders reported to be caused by POP-related exposure. One such disease is cancer, which is a controversial topic in PCB/ POP related toxicity (Brody et al., 2007). Our analysis suggested that the genes known to be responsible for cancer differ between the two high POP exposed groups (Table 4). Similarly, immunological disease, which is also a common complication of POP exposure, was associated with two different gene sets (Table 4). A particular gene can also exhibit two different connections in separate groups, as was the case for HIRA, which is a common gene between the two gene sets but did not show the same type of association and biological function in the two exposure scenarios.
Table 4.
Comparison of Significantly associated Disease and Disorder from IPA Analysis associated with two gene sets.
| Disease and Disorder | Gene set from OTB group | Gene set from PCB group |
|---|---|---|
| Hematological Disease | TNFSF8 (includes EG:944), CD3G, PRDX6 | CDK8, CDK6, KRAS |
| Immunological Disease | TNFSF8 (includes EG:944), CD3G, PRDX6 | JAZF1, SUV39H2, TNPO1, WDR27, CDK6, KRAS, CDK8, CNST, LGALS8, TAGAP, P2RY12, MSH5, ALCAM, IL23A, BPTF, TROVE2 |
| Cancer | CAPN2, PRDX6, APC | FOXO4, CDK12, TRIM24, INSIG1, SUV39H2, CDK6, LRCH4, KRAS, SMA4, CLK4, CEBPG, CDK8, DSTN, LGALS8, ALCAM, IL23A, USP25 |
| Connective Tissue Disorders | APC, PRDX6 | IL23A |
| Dermatological Diseases and Conditions | TNFSF8 (includes EG:944), ATP6V0A2 | CDK8, CDK6, KRAS, IL23A |
| Gastrointestinal Disease | APC | DSTN, KRAS, IL23A |
| Genetic Disorder | TNFSF8 (includes EG:944), ATP6V0A2, GBE1, APC | CDK8, INSIG1, LGALS8, CDK6, KRAS, SMA4, IL23A |
| Metabolic Disease | GBE1 | INSIG1, CNST, SUV39H2, LGALS8, WDR27, ALCAM, MSH5, P2RY12, BPTF, TROVE2 |
| Organismal Injury and Abnormalities | PRDX6, APC | P2RY12, KRAS, IL23A |
| Endocrine System Disorders | APC | CNST, SUV39H2, LGALS8, WDR27, ALCAM, MSH5, P2RY12, KRAS, BPTF, TROVE2 |
| Inflammatory Disease | TNFSF8 (includes EG:944) | IL23A |
| Neurological Disease | CAPN2 | CDK6, KRAS, CLK4 |
| Renal and Urological Disease | APC | |
| Inflammatory Response | ALCAM, IL23A | |
| Cardiovascular Disease | TRIM24, INSIG1, P2RY12, IL23A | |
| Developmental Disorder | INSIG1, KRAS | |
| Reproductive System Disease | CDK8, LGALS8, CDK6, KRAS, SMA4, IL23A | |
| Respiratory Disease | KRAS | |
| Hepatic System Disease | INSIG1 | |
| Skeletal and Muscular Disorders | IL23A | |
| Infectious Disease | IL23A |
Table 5.
Significantly associated Disease and Disorder observed in IPA Analysis associated with common twelve genes.
| Category | p-value | Molecules |
|---|---|---|
| Connective Tissue Disorders | 6.04E-04-6.04E-04 | CD96 |
| DNA Replication, Recombination, and Repair | 6.04E-04-1.5E-02 | HIRA, RAD23B |
| Embryonic Development | 6.04E-04-2.33E-02 | HIRA, RAD23B |
| Genetic Disorder | 6.04E-04-6.04E-04 | CD96 |
| Neurological Disease | 6.04E-04-6.04E-04 | CD96 |
| Organismal Development | 6.04E-04-2.33E-02 | HIRA, RAD23B |
| Skeletal and Muscular Disorders | 6.04E-04-6.04E-04 | CD96 |
| Tissue Morphology | 6.04E-04-9.03E-03 | HIRA, INO80B |
| Carbohydrate Metabolism | 1.81E-03-1.81E-03 | ETNK1 |
| Lipid Metabolism | 1.81E-03-1.81E-03 | ETNK1 |
| Small Molecule Biochemistry | 1.81E-03-3.98E-02 | CYTH1, ETNK1, RAD23B |
| Cell Signaling | 2.42E-03-3.98E-02 | CYTH1 |
| Molecular Transport | 2.42E-03-2.42E-03 | CYTH1 |
| Nucleic Acid Metabolism | 2.42E-03-3.98E-02 | CYTH1, RAD23B |
| Cellular Function and Maintenance | 6.63E-03-6.63E-03 | INO80B |
| Cellular Growth and Proliferation | 9.03E-03-9.03E-03 | INO80B |
| Cell-To-Cell Signaling and Interaction | 2.45E-02-3.39E-02 | CYTH1 |
| Nervous System Development and Function | 2.45E-02-2.45E-02 | CYTH1 |
| Tissue Development | 3.34E-02-3.34E-02 | CYTH1 |
| Cardiovascular System Development and Function | 3.75E-02-3.75E-02 | HIRA |
| Organ Development | 3.75E-02-4.39E-02 | HIRA |
| Organ Morphology | 3.75E-02-3.75E-02 | HIRA |
| Reproductive System Development and Function | 4.39E-02-4.39E-02 | HIRA |
| Cell Death | 4.68E-02-4.68E-02 | CD96 |
Figure 5.
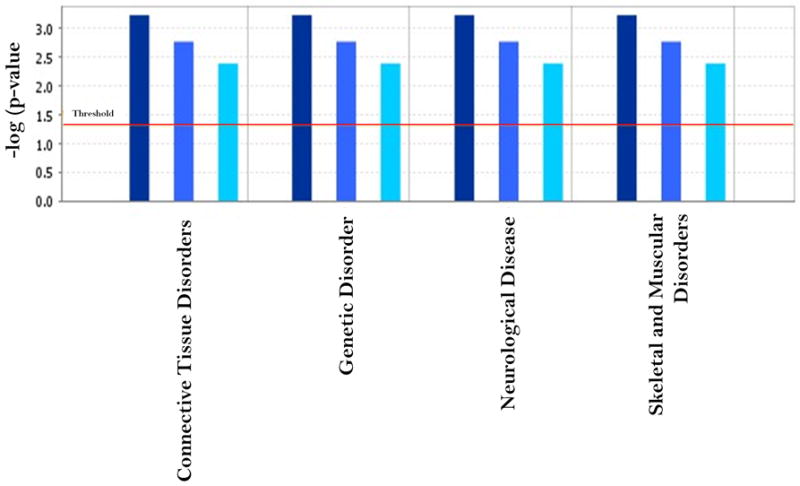
The comparison of significant disease disorder function generated by IPA based on the 14 common genes between the OTP and PCB group and the total differentially expressed gene sets in the two groups. The threshold line in the bar chart represents a p-value of 0.05. Dark blue, blue and light blue represented the common, OTP and PCB group respectively.
The 14 genes altered by the PCB and OTP groups are strikingly similar and this merits emphasis even though both induce many other genes independently. This is important given the similarities of some adverse health effects of PCB and pesticide exposures. The exact POP composition and their individual toxic contribution in disease development are difficult to measure in actual exposure situation. In both of our experimental groups there are chemicals of varied structure. Moreover there are other lipophilic substances which may be migrating selectively in PCB and OTP groups with unknown effects. Even within PCBs which is a mixture of different congeners, quite different patterns of gene induction was observed as in dioxin-like compared to non-dioxin-like congeners (Vezina et al., 2001), so one should expect complexity in POP exposure as well.
While the number of Functions/Pathways/Lists Eligible molecules associated with a given function/pathway is an important measure when calculating the p-value for Functional Analyses, the p-value does not only depend on this number. According to IPA algorithm, the p-value for a given function is calculated by considering: 1) The number of Functions/Pathways/Lists Eligible molecules that participate in that Annotation. 2) The total number of knowledge base molecules known to be associated with that function. 3) The total numbers of Functions/Pathways/Lists Eligible molecules 4) The total number of molecules in the Reference Set of IPA library. The number of genes differentially expressed in the high PCB group was higher than OTP group under same statistical stringency. It may introduce the positive bias in p value in IPA functional analysis in case of PCB group. So the functional effect due to high PCB group may be more pronounced due to the additional number of focus molecule in PCB group. However, we selected an identical cut of value for the fold change and statistical analysis of the microarray data in two cases. We did not consider the effect of other molecule which may have substantial influence in a disease process but did not pass the selection cut off. In actual physiological situation every single alteration in the pathway would count in the resultant disease process.
In the present work, we have utilized one of the most advanced data mining software, an enriched database and a highly sophisticated algorithm (in IPA), to understand the possible overall effect associated with the altered gene expression level due to two different high POP exposure scenarios compared to the low exposure level. A single study like this one is neither a conclusive evidence of the effected bio-functions nor it excludes the possibilities of other physiological impairments, unless validated through other methodological and detailed mechanistic pathway investigations. The present study is the first to its kind to reveal a comparative picture of gene expressions due to different POP exposure profiles from the same exposed population.
5.Conclusion
This study demonstrated a comparative toxicogenomic effect of two distinct POP exposure profiles, where a common set of 14 genes (including 2 hypothetical probes) were differentially expressed, regardless of the POP profile. Functional analysis provided an overview of the molecular mechanisms involved in POP exposure and helped explain the associated disease processes. This information is essential for the identification of a robust biomarker for POP exposure. It will also help to explain the molecular mechanisms responsible for the different pathophysiological effects reported in epidemiological studies. The array data tend to have a higher fold change than PCR data, though each gene demonstrated the same directionality in two methods. The random validation of the gene panel in a larger sample size is needed, and robustness of the expression should also be checked in a different method like RNA sequencing technique.
This result also underscores the importance of a future multicenter global validation study of other POP exposure conditions and the incorporation of different ethnic groups as well. This would increase the study power and the probability of identifying genomic biomarker(s) for POP exposure, regardless of the specific POP constituents in an individual exposure scenario. Complementary to the gene expression study, the analysis of epigenetic components will be required to understand the gene-environment interactions and the underlying disease risk for an individual due to chronic POP exposure.
Supplementary Material
Comparison of the gene expression in microarray and in RT PCR experiments. The RT PCR was ATAD2B, ETNK1, INO80B, HIRA, and RAD23B performed on a subset of differentially expressed genes common in two groups.
Correlation between Total PCB concentration (Y axis in ng/ml of blood) vs. Lipid Adjusted PCB Concentration (X axis in ng/mg of serum lipid) in 45 month children selected for microarray analysis.
The effect of the exposure was tested for F ratio in between the groups. A. High POP (OTP group) vs. Low POP control group and B. High POP (PCB group) vs. Low POP control group.
Volcano plot represents the differential expression, both in terms of fold-changes (x-axis) and the statistical significance of difference (-log10 of p-value, yaxis). The red circles represent up regulated (>1.5 fold) genes and blue circles down regulated (<-1.5 fold) genes. A-Comparison of Control Group, and OTP group. B-Comparison of Control Group and genes in PCB group.
Research Highlights.
A structural effect of POP exposure on gene expression was established.
Common genes of interest to POP exposure were identified.
The associated network of genes involved in POP exposure was identified.
The molecular mechanism of toxicity in POP exposure was provided.
This study will help with the identification of predictive markers in a POP exposure scenario.
Acknowledgments
This study is supported by the 1UO1ES016127-01 from the National Institute of Environmental Health Sciences (NIEHS/NIH). Thanks are also due to the General Clinical Research Center (GCRC) of Howard University for their assistance with the blood collection from healthy donors, as per approved HU IRB # IRB-07- GSAS-30. The contents of this report are solely the responsibility of the authors.
Footnotes
Authors’ contributions PM developed the POP work, performed the statistical analysis of the microarray results, and the functional analysis of the results. He also wrote the manuscript. SG and SZ did the handling of the human subject’s specimens, and the isolation of the RNA for the microarray studies. SG helped with critical revisions on the manuscript. SGM ran the microarray experiments. TT, LP and ES were responsible for the collection of the human subjects’ blood and data. IHP and DS provided the required background data for the human subjects in this work. EH and SKD provided support and direction and revised the manuscript. SKD holds the NIEHS/UO1 grant.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bencko V, Rames J, Ondrusova M, Plesko I, Jurickova L, Trnovec T. Human exposure to polyhalogenated hydrocarbons and incidence of selected malignancies — central European experience. Neoplasma. 2009;56:353–7. doi: 10.4149/neo_2009_04_353. [DOI] [PubMed] [Google Scholar]
- Bouwman H. Persistent organic pollutants. Berlin Heidelberg: © Springer-Verlag; 2003. The Handbook of Environmental Chemistry H. Fiedler. [Google Scholar]
- Brody JG, Moysich KB, Humblet O, Attfield KR, Beehler GP, Rudel RA. Environmental pollutants and breast cancer: epidemiologic studies. Cancer. 2007;109:2667–711. doi: 10.1002/cncr.22655. [DOI] [PubMed] [Google Scholar]
- Cui X, Churchill GA. Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 2003;4:210. doi: 10.1186/gb-2003-4-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Ghosh S, Chatterjee R, Chen YQ, Moses L, Kesari A, Hoffman EP, Dutta SK. PCB congener specific oxidative stress response by microarray analysis using human liver cell line. Environ Int. 2010;36:907–17. doi: 10.1016/j.envint.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du XZ, Zhou L, Zhao HB, Wang YF, Gui JF. Identical sequences but different expression patterns of Hira gene in gynogenetic and gonochoristic crucian carps. Fish Physiol Biochem. 2008;34:175–84. doi: 10.1007/s10695-007-9158-1. [DOI] [PubMed] [Google Scholar]
- Dutta SK, Ghosh S, De S, Hoffman EP. CYP1A1 and MT1K are Congener Specific Biomarker Genes for Liver Diseases Induced by PCBs. Environ Toxicol Pharmacol. 2008;25:218–21. doi: 10.1016/j.etap.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Dutta SK, Mitra PS, Ghosh S, Zang S, Sonneborn D, Hertz-Picciotto I, Trnovec T, Palkovicova L, Sovcikova E, Ghimbovschi S, Hoffman EP. Differential gene expression and functional analysis of PCB-exposed children: Understanding disease and disorder development. Environ Intl. 2011 doi: 10.1016/j.envint.2011.07.008. in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhart C. The assumptions underlying the analysis of variance. Biometrics. 1947;3:1–21. [PubMed] [Google Scholar]
- Faroon O, Jones D, de Rosa C. Effects of polychlorinated biphenyls on the nervous system. Toxicol Ind Health. 2001;16:305–33. doi: 10.1177/074823370001600708. [DOI] [PubMed] [Google Scholar]
- Fernandes EC, Hendriks HS, van Kleef RG, van den Berg M, Westerink RH. Potentiation of the human GABA(A) receptor as a novel mode of action of lower-chlorinated non-dioxin-like PCBs. Environ Sci Technol. 2010;44:2864–9. doi: 10.1021/es902321a. [DOI] [PubMed] [Google Scholar]
- Ghosh S, De S, Dutta SK. Altered protein expressions in chronic PCB-153-induced human liver (HepG2) cells. Int J Toxicol. 2007;26:203–12. doi: 10.1080/10915810701352648. [DOI] [PubMed] [Google Scholar]
- Ghosh S, De S, Chen Y, Sutton DC, Ayorinde FO, Dutta SK. Polychlorinated biphenyls (PCB-153) and (PCB-77) absorption in human liver (HepG2) and kidney (HK2) cells in vitro: PCB levels and cell death. Environ Int. 2010;36:893–900. doi: 10.1016/j.envint.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Zang S, Mitra PS, Ghimbovschi S, Hoffman EP, Dutta SK. Global gene expression and Ingenuity biological functions analysis on PCBs 153 and 138 induced human PBMC in vitro reveals differential mode(s) of action in developing toxicities. Environ Int. 2011;37:838–57. doi: 10.1016/j.envint.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Lobenhofer EK, Wang C, Shippy R, Harris SC, Zhang L, Mei N, et al. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nat Biotechnol. 2006;24:1162–9. doi: 10.1038/nbt1238. [DOI] [PubMed] [Google Scholar]
- Hertz-Picciotto I, Park HY, Dostal M, Kocan A, Trnovec T, Sram R. Prenatal exposures to persistent and non-persistent organic compounds and effects on immune system development. Basic Clin Pharmacol Toxicol. 2008;102:146–54. doi: 10.1111/j.1742-7843.2007.00190.x. [DOI] [PubMed] [Google Scholar]
- Howsam M, Grimalt JO, Guino E, Navarro M, Marti-Rague J, Peinado MA, et al. Organochlorine exposure and colorectal cancer risk. Environ Health Perspect. 2004;112:1460–6. doi: 10.1289/ehp.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan J, Sovcikova E, Kocan A, Wsolova L, Trnovec T. Developmental dental defects in children exposed to PCBs in eastern Slovakia. Chemosphere. 2007;67:S350–4. doi: 10.1016/j.chemosphere.2006.05.148. [DOI] [PubMed] [Google Scholar]
- Jonsson BA, Rylander L, Lindh C, Rignell-Hydbom A, Giwercman A, Toft G, et al. Inter-population variations in concentrations, determinants of and correlations between 2,2’,4,4’,5,5’-hexachlorobiphenyl (CB-153) and 1,1-dichloro-2,2-bis (p-chlorophenyl)-ethylene (p,p’-DDE): a cross-sectional study of 3161 men and women from Inuit and European populations. Environ Health. 2005;4:27. doi: 10.1186/1476-069X-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizer EC, Glaser CL, Chaussabel D, Banchereau J, Pascual V, White PC. Gene expression in peripheral blood mononuclear cells from children with diabetes. J Clin Endocrinol Metab. 2007;92:3705–11. doi: 10.1210/jc.2007-0979. [DOI] [PubMed] [Google Scholar]
- Kaname T, Yanagi K, Chinen Y, Makita Y, Okamoto N, Maehara H, et al. Mutations in CD96, a member of the immunoglobulin superfamily, cause a form of the C (Opitz trigonocephaly) syndrome. Am J Hum Genet. 2007;81:835–41. doi: 10.1086/522014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelce WR, Stone CR, Laws SC, Gray LE, Kemppainen JA, Wilson EM. Persistent DDT metabolite p,p’-DDE is a potent androgen receptor antagonist. Nature. 1995;375:581–5. doi: 10.1038/375581a0. [DOI] [PubMed] [Google Scholar]
- Khan IA, Thomas P. Disruption of neuroendocrine control of luteinizing hormone secretion by aroclor 1254 involves inhibition of hypothalamic tryptophan hydroxylase activity. Biol Reprod. 2001;64:955–64. doi: 10.1095/biolreprod64.3.955. [DOI] [PubMed] [Google Scholar]
- Kocan A, Petrik J, Drobna B, Chovancova J. Levels of PCBs and some organochlorine pesticides in the human population of selected areas of the Slovak Republic. I. Blood. Chemosphere. 1994;29:2315–25. doi: 10.1016/0045-6535(94)90400-6. [DOI] [PubMed] [Google Scholar]
- Korvatska O, Estes A, Munson J, Dawson G, Bekris LM, Kohen R, et al. Mutations in the TSGA14 gene in families with autism spectrum disorders. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:303–11. doi: 10.1002/ajmg.b.31162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger T, Ghisari M, Hjelmborg PS, Deutch B, Bonefeld-Jorgensen EC. Xenohormone transactivities are inversely associated to serum POPs in Inuit. Environ Health. 2008;7:38. doi: 10.1186/1476-069X-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda TS, Maita H, Tabata T, Taira T, Kitaura H, Ariga H, Iguchi-Ariga SM. A novel nucleolar protein, PAPA-1, induces growth arrest as a result of cell cycle arrest at the G1 phase. Gene. 2004;340:83–98. doi: 10.1016/j.gene.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Langer P. Persistent organochlorinated pollutants (PCB, DDE, HCB, dioxins, furans) and the thyroid--review 2008. Endocr Regul. 2008;42:79–104. [PubMed] [Google Scholar]
- Leachman NT, Brellier F, Ferralli J, Chiquet-Ehrismann R, Tucker RP. ATAD2B is a phylogenetically conserved nuclear protein expressed during neuronal differentiation and tumorigenesis. Dev Growth Differ. 2010;52:747–55. doi: 10.1111/j.1440-169X.2010.01211.x. [DOI] [PubMed] [Google Scholar]
- Lin T, Hu Z, Zhang G, Li X, Xu W, Tang J, Li J. Levels and mass burden of DDTs in sediments from fishing harbors: the importance of DDT-containing antifouling paint to the coastal environment of China. Environ Sci Technol. 2009;43:8033–8. doi: 10.1021/es901827b. [DOI] [PubMed] [Google Scholar]
- Lin CH, Huang CL, Chuang MC, Wang YJ, Chen DR, Chen ST, Lin PH. Protective role of estrogen receptor-alpha on lower chlorinated PCB congener-induced DNA damage and repair in human tumoral breast cells. Toxicol Lett. 2009 Jul 10;188(1):11–9. doi: 10.1016/j.toxlet.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Llop S, Ballester F, Vizcaino E, Murcia M, Lopez-Espinosa MJ, Rebagliato M, et al. Concentrations and determinants of organochlorine levels among pregnant women in Eastern Spain. Sci Total Environ. 2010;408:5758–67. doi: 10.1016/j.scitotenv.2010.07.085. [DOI] [PubMed] [Google Scholar]
- Long M, Deutch B, Bonefeld-Jorgensen EC. AhR transcriptional activity in serum of Inuits across Greenlandic districts. Environ Health. 2007;6:32. doi: 10.1186/1476-069X-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorain S, Quivy JP, Monier-Gavelle F, Scamps C, Lecluse Y, Almouzni G, Lipinski M. Core histones and HIRIP3, a novel histone-binding protein, directly interact with WD repeat protein HIRA. Mol Cell Biol. 1998;18:5546–56. doi: 10.1128/mcb.18.9.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyche JL, Nourizadeh-Lillabadi R, Almaas C, Stavik B, Berg V, Skare JU, Alestrom P, Ropstad E. Natural mixtures of persistent organic pollutants (POP) increase weight gain, advance puberty, and induce changes in gene expression associated with steroid hormones and obesity in female zebrafish. J Toxicol Environ Health A. 2010;73:1032–57. doi: 10.1080/15287394.2010.481618. [DOI] [PubMed] [Google Scholar]
- Mariussen E, Fonnum F. Neurochemical targets and behavioral effects of organohalogen compounds: an update. Crit Rev Toxicol. 2006;36:253–89. doi: 10.1080/10408440500534164. [DOI] [PubMed] [Google Scholar]
- Mariussen E, Fonnum F. The effect of polychlorinated biphenyls on the high affinity uptake of the neurotransmitters, dopamine, serotonin, glutamate and GABA, into rat brain synaptosomes. Toxicology. 2001;159:11–21. doi: 10.1016/s0300-483x(00)00374-7. [DOI] [PubMed] [Google Scholar]
- Miyako K, Cobb LJ, Francis M, Huang A, Peng B, Pintar JE, Ariga H. Cohen PPAPA-1 Is a nuclear binding partner of IGFBP-2 and modulates its growth-promoting actions. Mol Endocrinol. 2009;23:169–75. doi: 10.1210/me.2008-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami A, Koyama T, Kawasaki K, Negishi T, Ihara T, Kuroda Y, Yoshikawa Y. Maternal plasma polychlorinated biphenyl levels in cynomolgus monkeys (Macaca fascicularis) affect infant social skills in mother-infant interaction. Dev Psychobiol. 2011;53:79–88. doi: 10.1002/dev.20493. [DOI] [PubMed] [Google Scholar]
- Neher TM, Rechkunova NI, Lavrik OI, Turchi JJ. Photo-cross-linking of XPC-Rad23B to cisplatin-damaged DNA reveals contacts with both strands of the DNA duplex and spans the DNA adduct. Biochemistry. 2010;49:669–78. doi: 10.1021/bi901575h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Linderholm L, Charles MJ, Athanasiadou M, Petrik J, Kocan A, Drobna B, Trnovec T, Bergman A, Hertz-Picciotto I. Polychlorinated biphenyls and their hydroxylated metabolites (OH-PCBS) in pregnant women from eastern Slovakia. Environ Health Perspect. 2007;115:20–7. doi: 10.1289/ehp.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HY, Park JS, Sovcikova E, Kocan A, Linderholm L, Bergman A, Trnovec T, Hertz-Picciotto I. Exposure to hydroxylated polychlorinated biphenyls (OH-PCBs) in the prenatal period and subsequent neurodevelopment in eastern Slovakia. Environ Health Perspect. 2009;117:1600–6. doi: 10.1289/ehp.0900611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HY, Hertz-Picciotto I, Sovcikova E, Kocan A, Drobna B, Trnovec T. Neurodevelopmental toxicity of prenatal polychlorinated biphenyls (PCBs) by chemical structure and activity: a birth cohort study. Environ Health. 2010;9:51. doi: 10.1186/1476-069X-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrik J, Drobna B, Pavuk M, Jursa S, Wimmerova S, Chovancova J. Serum PCBs and organochlorine pesticides in Slovakia: age, gender, and residence as determinants of organochlorine concentrations. Chemosphere. 2006;65:410–418. doi: 10.1016/j.chemosphere.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Pillai SG, Kong X, Edwards LD, Cho MH, Anderson WH, Coxson HO, Lomas DA, Silverman EK. ECLIPSE and ICGN Investigators. Loci identified by genome-wide association studies influence different disease-related phenotypes in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182:1498–505. doi: 10.1164/rccm.201002-0151OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rádiková Ž, Koška J, Kšinantová L, Imrich R, Kočan A, Petrík J, Hučková M, Wsolová L, Langer P, Trnovec T, Šeböková E, Klimeš I. Increased frequency of diabetes and other forms of dysglycemia in the population of specific areas of eastern Slovakia chronically exposed to contamination with polychlorinated biphenyls (PCB) Organohalogen Compounds. 2004;6:3498–502. [Google Scholar]
- Radikova Z, Tajtakova M, Kocan A, Trnovec T, Sebokova E, Klimes I, Langer P. Possible effects of environmental nitrates and toxic organochlorines on human thyroid in highly polluted areas in Slovakia. Thyroid. 2008;18:353–62. doi: 10.1089/thy.2007.0182. [DOI] [PubMed] [Google Scholar]
- Rattenborg T, Gjermandsen I, Bonefeld-Jorgensen EC. Inhibition of E2-induced expression of BRCA1 by persistent organochlorines. Breast Cancer Res. 2002;4:R12. doi: 10.1186/bcr461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royland JE, Kodavanti PR. Gene expression profiles following exposure to a developmental neurotoxicant, Aroclor 1254: pathway analysis for possible mode(s) of action. Toxicol Appl Pharmacol. 2008a;231:179–96. doi: 10.1016/j.taap.2008.04.023. [DOI] [PubMed] [Google Scholar]
- Royland JE, Wu J, Zawia NH, Kodavanti PR. Gene expression profiles in the cerebellum and hippocampus following exposure to a neurotoxicant, Aroclor 1254: developmental effects. Toxicol Appl Pharmacol. 2008b;231:165–78. doi: 10.1016/j.taap.2008.04.022. [DOI] [PubMed] [Google Scholar]
- Ruzzin J, Petersen R, Meugnier E, Madsen L, Lock EJ, Lillefosse H, et al. Persistent organic pollutant exposure leads to insulin resistance syndrome. Environ Health Perspect. 2010;118:465–71. doi: 10.1289/ehp.0901321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylander L, Nilsson-Ehle P, Hagmar L. A simplified precise method for adjusting serum levels of persistent organohalogen pollutants to total serum lipids. Chemosphere. 2006;62:333–6. doi: 10.1016/j.chemosphere.2005.04.107. [DOI] [PubMed] [Google Scholar]
- Sagerup K, Larsen HJ, Skaare JU, Johansen GM, Gabrielsen GW. The toxic effects of multiple persistent organic pollutant exposures on the post-hatch immunity maturation of glaucous gulls. J Toxicol Environ Health A. 2009;72:870–83. doi: 10.1080/15287390902959516. [DOI] [PubMed] [Google Scholar]
- Sagiv SK, Thurston SW, Bellinger DC, Tolbert PE, Altshul LM, Korrick SA. Prenatal organochlorine exposure and behaviors associated with attention deficit hyperactivity disorder in school-aged children. Am J Epidemiol. 2010;171:593–601. doi: 10.1093/aje/kwp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon ML, Madanagopal SG, Blando R, Berner J, Williams K. Effects of hexachlorobenzene on embryonic mammalian cells. Toxicol In Vitro. 2002;16:539–48. doi: 10.1016/s0887-2333(02)00054-1. [DOI] [PubMed] [Google Scholar]
- Sargent L, Roloff B, Meisner L. In vitro chromosome damage due to PCB interactions. Mutat Res. 1989;224:79–88. doi: 10.1016/0165-1218(89)90006-2. [DOI] [PubMed] [Google Scholar]
- Sergeev AV, Carpenter DO. Increased hospitalizations for ischemic stroke with comorbid diabetes and residential proximity to sources of organic pollutants: a 12-year population-based study. Neuroepidemiology. 2010;35:196–201. doi: 10.1159/000316874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YQ, Wang YP, Song Y, Li HW, Liu CJ, Wu ZG, Yang KD. p,p’-DDE induces testicular apoptosis in prepubertal rats via the Fas/FasL pathway. Toxicol Lett. 2010;193:79–85. doi: 10.1016/j.toxlet.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Trnovec T, Sovcikova E, Pavlovcinová G, Jakubikova J, Jusko TA, Hustak M, Jureckova D, Palkovicova L, Kocan A, Drobna B, Lancz K, Wimmerova S. Serum PCB concentrations and cochlear function in 12-year-old children. Environ Sci Technol. 2010;44:2884–9. doi: 10.1021/es901918h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PC, Wang YJ, Tsai JH, Guo YL, Ueng TH, Liu HS, Huang W. Reduced expression of von Hippel-Lindau gene in subjects exposed to polychlorinated biphenyls and dibenzofurans. Environ Res. 2008;108:247–51. doi: 10.1016/j.envres.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Tryphonas H. Approaches to detecting immunotoxic effects of environmental contaminants in humans. Environ Health Perspect. 2001;109(Suppl 6):877–84. doi: 10.1289/ehp.01109s6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukropec J, Radikova Z, Huckova M, Koska J, Kocan A, Sebokova E, Drobna B, Trnovec T, Susienkova K, Labudova V, Gasperikova D, Langer P, Klimes I. High prevalence of prediabetes and diabetes in a population exposed to high levels of an organochlorine cocktail. Diabetologia. 2010;53:899–906. doi: 10.1007/s00125-010-1683-2. [DOI] [PubMed] [Google Scholar]
- Valavanidis A, Vlahogianni T, Dassenakis M, Scoullos M. Molecular biomarkers of oxidative stress in aquatic organisms in relation to toxic environmental pollutants. Ecotoxicol Environ Saf. 2006;64:178–89. doi: 10.1016/j.ecoenv.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Vezina CM, Walker NJ, Olson JR. Subchronic exposure to TCDD, PeCDF, PCB126, and PCB153: effect on hepatic gene expression. Environ Health Perspect. 2004;112:1636–44. doi: 10.1289/txg.7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster E, Mackay D, Di Guardo A, Kane D, Woodfine D. Regional differences in chemical fate model outcome. Chemosphere. 2004;55:1361–76. doi: 10.1016/j.chemosphere.2003.10.061. [DOI] [PubMed] [Google Scholar]
- Wigle DT, Arbuckle TE, Turner MC, Berube A, Yang Q, Liu S, Krewski D. Epidemiologic evidence of relationships between reproductive and child health outcomes and environmental chemical contaminants. J Toxicol Environ Health B Crit Rev. 2008;11:373–517. doi: 10.1080/10937400801921320. [DOI] [PubMed] [Google Scholar]
- Wimmerova S, Lancz K, Tihanyi J, Sovcikova E, Kocan A, Drobna B, Palkovicova L, Jureckova D, Fabisikova A, Conka K, Trnovec T. Half-lives of serum PCB congener concentrations in environmentally exposed early adolescents. Chemosphere. 2011;82:687–91. doi: 10.1016/j.chemosphere.2010.10.099. [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA. Preprocessing of oligonucleotide array data. Nat Biotechnol. 2004;22:656–8. doi: 10.1038/nbt0604-656b. author reply 658. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Hawke NA, Eason DD, Mueller MG, Davids BJ, Gillin FD, Litman GW. BIVM, a novel gene widely distributed among deuterostomes, shares a core sequence with an unusual gene in Giardia lamblia. Genomics. 2002;79:750–5. doi: 10.1006/geno.2002.6768. [DOI] [PubMed] [Google Scholar]
- Zhao P, Hoffman EP. Embryonic myogenesis pathways in muscle regeneration. Dev Dyn. 2004;229:380–92. doi: 10.1002/dvdy.10457. [DOI] [PubMed] [Google Scholar]
- Zhou XH, Brandau O, Feng K, Oohashi T, Ninomiya Y, Rauch U, Fassler R. The murine Ten-m/Odz genes show distinct but overlapping expression patterns during development and in adult brain. Gene Expr Patterns. 2003;3:397–405. doi: 10.1016/s1567-133x(03)00087-5. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Kalen AL, Li L, Lehmler HJ, Robertson LW, Goswami PC, Spitz DR, Aykin-Burns N. Polychlorinated-biphenyl-induced oxidative stress and cytotoxicity can be mitigated by antioxidants after exposure. Free Radic Biol Med. 2009;47:1762–71. doi: 10.1016/j.freeradbiomed.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of the gene expression in microarray and in RT PCR experiments. The RT PCR was ATAD2B, ETNK1, INO80B, HIRA, and RAD23B performed on a subset of differentially expressed genes common in two groups.
Correlation between Total PCB concentration (Y axis in ng/ml of blood) vs. Lipid Adjusted PCB Concentration (X axis in ng/mg of serum lipid) in 45 month children selected for microarray analysis.
The effect of the exposure was tested for F ratio in between the groups. A. High POP (OTP group) vs. Low POP control group and B. High POP (PCB group) vs. Low POP control group.
Volcano plot represents the differential expression, both in terms of fold-changes (x-axis) and the statistical significance of difference (-log10 of p-value, yaxis). The red circles represent up regulated (>1.5 fold) genes and blue circles down regulated (<-1.5 fold) genes. A-Comparison of Control Group, and OTP group. B-Comparison of Control Group and genes in PCB group.


