Abstract
Seasonal epizootics of vector-borne pathogens infecting multiple species are ecologically complex and difficult to forecast. Pathogen transmission potential within the host community is determined by the relative abilities of host species to maintain and transmit the pathogen and by ecological factors influencing contact rates between hosts and vectors. Increasing evidence of strong feeding preferences by a number of vectors suggests that the host community experienced by the pathogen may be very different from the local host community. We developed an empirically informed transmission model for West Nile virus (WNV) in four sites using one vector species (Culex pipiens) and preferred and non-preferred avian hosts. We measured strong feeding preferences for American robins (Turdus migratorius) by Cx. pipiens, quantified as the proportion of Cx. pipiens blood meals from robins in relation to their abundance (feeding index). The model accurately predicted WNV prevalence in Cx. pipiens at three of four sites. Sensitivity analysis revealed feeding preference was the most influential parameter on intensity and timing of peak WNV infection in Cx. pipiens and a threshold feeding index for transmission was identified. Our findings indicate host preference-induced contact heterogeneity is a key mediator of vector-borne pathogen epizootics in multi-species host communities, and should be incorporated into multi-host transmission models.
Keywords: West Nile virus, Culex pipiens, American robin, biodiversity, transmission model
1. Introduction
Zoonotic pathogens cause significant mortality, morbidity and economic loss to human, livestock and wildlife hosts throughout the world, constituting an estimated 75 per cent of emerging infectious diseases [1–3]. The complex dynamics characterizing these systems, combined with costly ecological data collection, make predicting epizootics and spill-over to humans (or livestock) difficult [4]. It is therefore critical to identify the major determinants of pathogen transmission, both to develop tractable models capturing natural transmission dynamics, and to inform surveillance and control efforts in the field.
For vector-borne zoonoses, pathogen transmission potential of a host community is determined by the relative abilities of host species to maintain the pathogen and transmit it to a feeding vector (‘reservoir competence’) and by ecological factors influencing contact rates between hosts and vectors. Previous research has focused on how heterogeneity in reservoir competence among species influenced pathogen persistence, transmission [5] and human disease risk [6]. However, recent developments in molecular methods allow estimation of species-specific vector–host contact rates by identifying the source species from vector blood meals. The potential for vector host preference to influence transmission has been postulated by several empirical studies evaluating mosquito feeding preferences [7–9]. In this study, we addressed the question of whether heterogeneity in contact rates between vectors and hosts influenced transmission dynamics, independently of the relative reservoir competence of the hosts.
The central aims of this study were to quantify vector host-feeding preferences at multiple field sites and determine the extent to which these preferences shape West Nile virus (WNV) transmission dynamics. To accomplish these aims, we developed an empirically informed model using an epidemiological susceptible–infected–recovered (SIR) framework to describe daily enzootic transmission of WNV in a community of multiple avian hosts and a single mosquito vector, Culex pipiens (figure 1). We modelled transmission over 1 year at four field sites in Connecticut (CT, USA; electronic supplementary material, figure S1). We measured and incorporated into our model, seasonal mosquito abundances using data collected with Centres for Disease Control and Prevention (CDC) light traps, bird abundances using data from field surveys, and Cx. pipiens feeding behaviour through DNA analysis to determine the blood meal source [10]. Using sensitivity analysis, we evaluated the influence of all model parameters on two model outcomes, intensity and timing of peak transmission. We found that host-feeding preferences by Cx. pipiens were the most influential determinant for WNV transmission. To our knowledge, our model is the first arbovirus transmission model to be both informed and validated by field data at multiple sites, and thoroughly analysed using robust sensitivity analysis. Our findings demonstrate the critical role vector host preferences play in multi-host vector-borne pathogen systems. From an applied perspective, this model could be extended to determine transmission and emergence of other infectious vector-borne diseases involving multiple host species.
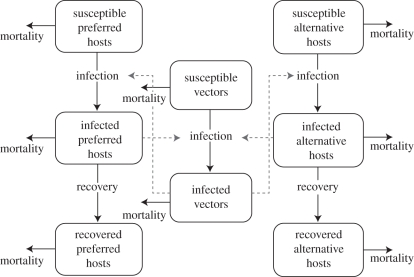
Figure 1.
Flow diagram for the WNV model. Transmission was simulated among two host types (vector-preferred and alternative). See text for model equations and table 1 for list of parameter values and sources.
2. Methods
(a). Field surveys
Four CT field study sites (site A in East Haven, site B in New Haven, site C in West Haven and site D in Stratford) were selected among 91 fixed locations where the CT Agricultural Experiment Station maintains mosquito surveillance traps. These sites have consistently experienced WNV activity in years since its initial introduction and have typical Culex spp. habitat, including waterways, parks, undeveloped wood lots and temporary wetlands in densely populated residential areas [11–14]. A 500 m buffer centred on the traps delineated the sites.
Bird point counts were conducted during the breeding season at 5–11 randomly distributed positions in each site, located at least 200 m apart. Each position was visited once between 22 June and 8 July 2006, and all birds seen or heard within a 50 m radius of a survey position during a 10 min period were recorded according to established methods [15]. Survey results are provided in the electronic supplementary material. The total bird population at each site was estimated as the number of observed birds divided by the area of all survey points (birds per square metre), multiplied by the total area of the site (πr2). No adjustments were made for variation in detection probabilities (see the electronic supplementary material).
CDC-type light traps at the centre of each field site collected host-seeking mosquitoes overnight at least once in every 10 days from 5 June to 26 October. Traps were gathered the following morning and collections were sorted and identified to species with the aid of a stereomicroscope and descriptive keys [16]. Blood-engorged mosquitoes were collected using backpack aspirators. Mosquitoes were collected in plastic mesh-covered containers and transported alive to the laboratory where they were frozen, sorted and identified to species.
3. Laboratory methods
(a). Blood meal analysis
Blood-fed Cx. pipiens were processed individually to determine blood source and WNV infection status. The abdomens of blood-fed mosquitoes were separated from the body under a dissecting microscope, under sterile conditions to avoid cross-contamination. DNA was isolated from the abdominal contents of engorged mosquitoes using DNA-zol BD (Molecular Research Center, Cincinnati, OH, USA) according to established protocol [13,17,18]. Isolated DNA from the mosquito blood meal was screened using a polymerase chain reaction (PCR)-based technique taking advantage of the mitochondrial cytochrome b gene [13,17,18]. PCR products were sequenced at the W. M. Keck Foundation Biotechnology Resource Laboratory (Yale University, New Haven, CT, USA), a core DNA sequencing facility. The source of the blood meal was identified by comparison to the GenBank DNA sequence database (http://www.ncbi.nlm.nih.gov). The performance of the molecular-based assay was validated by isolating DNA from the blood of a number of known vertebrate species, subjecting them to PCR amplification, and sequencing [13].
(b). Feeding index
The ‘feeding index’ assesses the proportion of blood meals from a particular host species in relation to the proportional abundance of that species in the host community [10]. Therefore, a feeding index of 1 indicates opportunistic feeding habits, while a feeding index greater than 1 indicates preferential feeding. Confidence intervals for the feeding index were generated by bootstrapping with 1000 samples (STATA SE 10; Stata Corporation).
(c). Mosquito infection
Blood-fed mosquitoes were tested individually for infection with WNV. The head and thorax of individual mosquitoes were screened for viral infection by inoculating mosquito homogenates onto confluent Vero cell cultures as previously described [14]. RNA was extracted from virus isolates using viral RNA kit (Qiagen, Valencia, CA) and tested for WNV by real-time RT-PCR assays [19]. For estimating infection rate (IR), non blood-fed mosquitoes were pooled by species, collecting site, trap type, and date, and processed as above. The number of mosquitoes per pool ranged from 1 to 50. The IR and its skew-corrected 95% confidence intervals at each site were calculated using the Pooled IR add-in developed for Excel [20].
(d). Model
We constructed a model using a standard SIR epidemiological framework (e.g. [21]) to model enzootic transmission in a community of multiple avian hosts with a single mosquito vector for 1 year. The model incorporates WNV transmission between the vector, Cx. pipiens, and two categories of avian hosts: a primary avian species preferred by the vector, and all other bird species, referred to collectively as alternative avian hosts (figure 1). The resulting model equations (3.1)–(3.12) are given below.
Both primary and alternative avian hosts are further divided into classes of susceptible (S), infected (I) and recovered (R) individuals so the total population size is N = S + I + R. The variables for the preferred population of birds are indicated with subscript p, alternative hosts with subscript a, and vectors with subscript v. We assumed that mosquitoes do not recover from infection with WNV, so only susceptible (Sv) and infected (Iv) classes of vectors were modelled. The model is given by the system of differential equations:
| 3.1 |
| 3.2 |
| 3.3 |
| 3.4 |
| 3.5 |
| 3.6 |
| 3.7 |
| 3.8 |
| 3.9 |
| 3.10 |
| 3.11 |
| 3.12 |
We assumed that bird populations remain constant over the year, with balanced rates of recruitment and loss. To model seasonal dynamics of the mosquito populations, we forced the total number of mosquitoes to follow an empirically derived function for the population over time, r(t) in equation (3.7).
IRs for primary hosts, alternative hosts and vectors are written in terms of the force of infection (λp, λa and λv), respectively; equations (3.10)–(3.12). These equations describe WNV transmission between vectors and hosts, and incorporate the vector biting rate (v), reservoir and vector competence (β1 and β2, respectively), feeding preference (αv) and the number of infected individuals (I).
Model simulations for all four sites used parameter values and sampling ranges derived from published values [22–33](table 1). Further information on model assumptions are described in the electronic supplementary material.
Table 1.
Model parameters. (The rates have units per day.)
| parameter, definition | baseline value | source (s) | sampling distributions | mean (s.d.) |
|---|---|---|---|---|
| b, avian recruitment rate | 0.0014 | 22 | normal | 0.0014 (0.0001) |
| d, background avian death rate | 0.0014 | 23–26 | normal | 0.0014 (0.0001) |
| dv, mosquito mortality rate | 0.10 | imposed | n.a. | n.a. |
| v, biting rate of vectors on hosts | 0.14 | 27, 28 | normal | 0.14 (0.01) |
| αv, feeding index for preferred host | 6.70 | site A | poisson | 6.70 |
| 8.53 | site B | poisson | 8.53 | |
| 24.83 | site C | poisson | 24.83 | |
| 31.89 | site D | poisson | 31.86 | |
| m, trap conversion multiplier | 10.00 | 29 | normal | 10.00 (2.00) |
| β1, vector- to host-transmission rate | 0.44 | 30–32 | normal | 0.465 (0.013) |
| β2, host- to vector-transmission rate | 0.974 | 31, 33 | normal | 0.974 (0.0075) |
| γp, recovery rate of preferred host | 0.195 | 32 | normal | 0.195 (0.005) |
| γa, recovery rate of alternate host | 0.091 | 32 | normal | 0.091 (0.01) |
| dWNV1, WNV deaths of preferred hosts | 0 | 32 | normal | 0.005 (0.0025) |
| dWNV2, WNV deaths of alternate hosts | 0.11 | 32 | normal | 0.11 (0.02) |
(e). Model validation
To validate the model, we compared the model output (daily abundance of infected Cx. pipiens) to the average monthly estimate of infected Cx. pipiens, or Vector Index (VI; described in [17]) which is calculated by the mean monthly Cx. pipiens abundance multiplied by the bias-corrected maximum-likelihood estimate (MLE) IR. A monthly estimate of the VI for each site for June through October 2006 was calculated (electronic supplementary material).
(f). Sensitivity analysis
We conducted sensitivity analyses to explore the model's behaviour and identify the key parameters driving transmission. We used a probabilistic method with Latin Hypercube Sampling (LHS), a type of stratified Monte Carlo sampling [34–36]. Briefly, probability distribution functions were ascribed to each parameter (table 1), representing the degree of belief associated with each quantity. Sample values for each parameter are drawn from their distributions and LHS is used to better cover the whole space of possible combinations of parameter values with a moderate number of samples [34]. The model is run many times using the sampled parameter values to produce sets of model output. This sampling scheme generates input and output distributions useful in assessing model and parameter uncertainties in a ‘global' sense [34,35,37]. The partial rank correlation coefficient (PRCC) is a widely used and powerful test for parameter sensitivity [37] that calculates PRCC values for each input parameter (sampled by the LHS scheme) and model outcome variables. Since the PRCCs show which parameters have the largest influence on model outcomes, this analysis identifies which biological mechanisms are important determinants of intensity and timing of pathogen transmission.
4. Results
(a). Field results
To determine avian species abundances, a total of 489 birds representing 40 species were observed at point count stations in four field sites in CT (sites A–D). To parametrize the model, the abundance of American robins in each site was estimated for the total area of the site (site A = 88, site B = 133, site C = 25 and site D = 67 robins). The number of other bird species was calculated in the same manner (site A = 1613, site B = 1856, site C = 1100 and site D = 2483 birds).
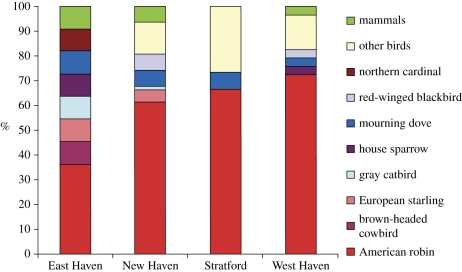
We found that Cx. pipiens fed primarily on birds (139 of 147 blood meals, or 94.56%), and an average of 59.18 per cent (range 36.36–72.41%) of the avian blood meals were derived from American robins (T. migratorius) (figure 2). There were five species that were present in the blood meals but not observed during point counts: black-crowned night heron, Northern flicker, rose-breasted grosbeak, wild turkey and wood thrush. Since the blood meals and counts of non-robin species were pooled, we did not consider it necessary to make specific adjustments to the calculation of the feeding index, assuming that the proportion of blood meals on all other species is representative of the species abundances as estimated by the point counts. Using feeding indices, American robins were the preferred bird host at all sites (αv) >1 (site A = 6.70, CI: 2–18.90; site B = 8.53, CI: 5.49–16.05; site C = 31.89, CI: 14.52–47.14; site D = 24.83, CI: 11.92–98).
Figure 2.
Proportion of avian-derived Cx. pipiens blood meals at four study sites in CT, 2006 (n = 147). American robins comprised the greatest number of blood meals at all sites.
Mosquito abundance patterns as determined from light trap collections varied between sites. Abundance of Cx. pipiens peaked between late June and early August (electronic supplementary material, figure S2). The largest samples of Cx. pipiens were collected on 2 August (464) at site A, on 13 July (744) at site B, on 19 July (1276) at site C and on 26 June (419) at site D. The trap collections were used to parametrize the model. Further details on mosquito trapping results are provided in the electronic supplementary material.
(b). Model results
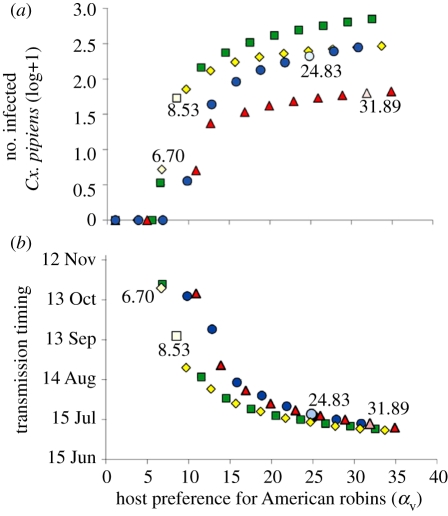
To address how strongly vector-feeding preferences influence WNV transmission relative to other biological mechanisms in the model, we varied the feeding index, αv, from baseline across a range of values (from 1 to 35). We observed its effect on two model outcomes; the intensity of transmission (measured as the peak number of WNV-infected Cx. pipiens at a site) and the timing of peak transmission (measured as the day of peak number of infected Cx. pipiens). Enzootic transmission of WNV was maintained at all sites in the model. Overall, sites with higher αv had more intense transmission occurring earlier in the season. Transmission peaked on day 295 (22 October) with 4.23 infected Cx. pipiens at site A, day 259 (16 September) with 52.72 infected Cx. pipiens at site B, day 193 (12 July) with 62.16 infected Cx. pipiens at site C, and day 200 (19 July) with 209.59 infected Cx. pipiens at site D.
We observed a consistent effect of αv on both transmission intensity and timing at all sites (figure 3). No transmission was maintained when αv was less than 6, and transmission intensity and timing was most sensitive to moderate αv values (7–15). For high αv (greater than 20), transmission intensity was approximately constant. This indicates that mosquito feeding preference has a large impact on transmission dynamics, and uncertainty in feeding preference estimates will most affect transmission intensity predictions when they are moderate.
Figure 3.
Model-predicted densities (log+1) of WNV-infected Cx. pipiens mosquitoes at four sites in CT in 2006. The influence of Cx. pipiens' preference for feeding on American robins (feeding index, αv) on peak abundance WNV-infected mosquitoes was compared. Site A (yellow diamonds) baseline αv was 6.70, site B (green squares) baseline αv was 8.53, site C (red triangles) baseline αv was 31.89 and site D (blue circles) baseline αv was 24.83. Transmission dynamics followed a consistent, nonlinear αv-dependent pattern across all sites.
These results are best illustrated when we simplify our model by assuming an equilibrium vector population of size (Nv). Under this assumption (see the electronic supplementary material) the basic reproductive number is given by:
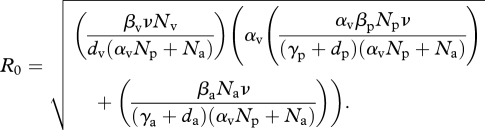 |
R0 must be larger than 1 for disease to invade the disease-free steady state, and depends heavily on the value αv.
(c). Sensitivity analysis
We explored the contributions of each of the model parameters on the prediction precision of the two transmission outcomes (intensity and timing) using LHS and PRCC's. The sensitivity analysis showed that αv was the most important parameter for both transmission intensity (electronic supplementary material, table S1) and timing (electronic supplementary material, table S2) at all four sites. A strong positive correlation was observed between αv and number of WNV-infected Cx. pipiens at all sites, and a strong negative correlation between αv and day of peak transmission at three sites. The positive correlation between αv and timing of peak transmission at the remaining site, site A, suggests a lack of transmission when αv values are very low and a late date onset when αv values are just high enough to maintain transmission.
Mosquito biting rate (v) was ranked second in the PRCC analysis. The biting rate was positively correlated with transmission intensity at all four sites and negatively correlated with timing, except again at site A, where it was positively correlated for the same reason as for αv.
The PRCCs also show that the least influential parameter was the rate of new susceptible birds entering a site (b), either through births or immigration.
(d). Model validation
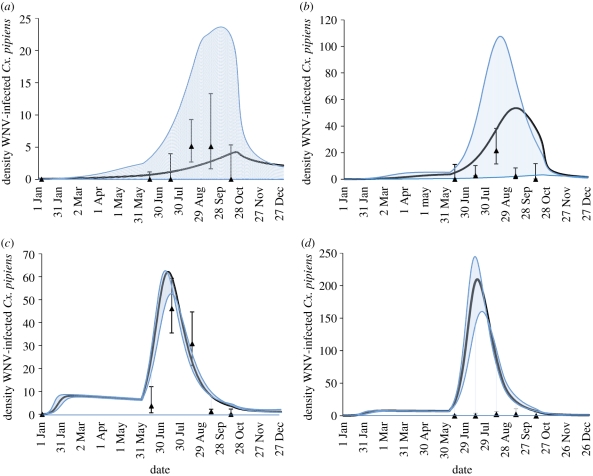
To determine whether model outcomes realistically represent enzootic transmission at the sites, we validated the model using a field-derived monthly mean Vector Index (VI) [38]. The VI is an estimate of the mean number of WNV-infected Cx. pipiens based on mosquito abundance and the MLE of the IR. A total of 16 676 adult female Cx. pipiens were collected and tested for WNV infection, of which 1267 individuals (53 pools) were from site A, 2823 (98 pools) from site B, 11 419 (273 pools) from site C, and 1167 (38 pools) from site D. The seasonal adjusted MLE (WNV-infected mosquitoes per 1000) for Cx. pipiens was 11.99 (13/53 pools tested positive for WNV), 4.93 (13/98), 10.91 (99/273) and 0.86 (1/38) at sites A–D, respectively. The first infected mosquitoes were identified on 29 June, the number of infected mosquitoes peaked in August or September and no positive mosquito pools were collected after 26 September at any of the sites.
We modelled the daily number of WNV-infected Cx. pipiens at each site when parameters were set to baseline. We show these results along with corresponding first and third quartiles from 1000 model runs (using LHS), and the monthly mean VI for validation in figure 4. The model predicts transmission outcomes falling within the confidence intervals of the monthly VI for three of the sites. Baseline model simulations matched the ranked order of the VI at three of the sites (low-level enzootic WNV transmission at site A, moderate at site B and higher at site C) but transmission was over-predicted at site D. The model was broadly able to corroborate outcomes, suggesting that our model realistically describes enzootic transmission at the study sites.
Figure 4.
Model results at baseline (black line), first (lower blue line) and third (upper blue line) quartiles (Q1 and Q3, respectively) from Latin hypercube sampling for the four sites in CT (A–D), along with 2006 Vector Indices (VI) represented as black triangles. (a) Site A, (b) site B, (c) site C, (d) site D. VI = monthly average maximum-likelihood estimate of infection prevalence × monthly average mosquito abundance. The lower (upper) error bar is derived from the lower (upper) limit of the MLE for infection prevalence multiplied by the monthly average mosquito abundance. Sites with low baseline αv had high variability, while sites with high baseline αv had less variability.
5. Discussion
Our model describes how vector–host contact heterogeneity owing to preferential mosquito feeding drives WNV enzootic transmission dynamics at four different locations in CT. Site-specific mosquito and bird abundances were represented in the model. From field data we identified host preferences for American robins by Cx. pipiens using the feeding index (αv) and with our model showed that this is the most influential parameter on both WNV transmission intensity and timing. This finding is consistent with host preferences by Cx. pipiens for American robins demonstrated by both field studies [7,8] and experimental trials [29]. As a consequence, it has been suggested that preference-induced contact heterogeneity [7,8,39] may influence pathogen transmission dynamics. Our model shows that strong host-feeding preference by Cx. pipiens for American robins is possibly a major driver for WNV transmission in CT.
Our model advances previous modelling efforts because it is both parametrized and validated using field data. We also used a parsimonious modelling approach: rather than modelling each site independently, we developed a single transmission model and subsequently incorporated site-specific field data from various locations of interest. This provides the advantage of a flexible model for exploring transmission dynamics both within and between multiple localities.
We identified a feeding preference threshold necessary for maintaining Cx. pipiens-mediated transmission of WNV (αv > 6). Increasing the degree of feeding on American robins resulted in more intense transmission occurring earlier in the season. The greatest influence was observed when preference was moderate (6 < αv <20); when αv > 20, transmission intensity and timing approached a plateau. This finding is consistent with how the threshold quantity Ro depends on αv (see the electronic supplementary material). Our model predicts maximum enzootic transmission potential at each site based on mosquito abundance and the relative abundance of preferred versus non-preferred hosts (figure 3). The level of host preference by Cx. pipiens, measured by αv, subsequently determines the timing and intensity of transmission and whether the plateau is reached. Inter-site differences in host preferences may be driven by landscape structure influencing the microhabitat availability of hosts to mosquitoes, availability of alternative hosts, etc; further research at a finer spatial scale may be warranted.
We used the VI to qualitatively validate that the model provides realistic estimates of infection in Cx. pipiens. The model agreed well with site-specific VI at sites A–C, predicting high densities of infected Cx. pipiens at site C, moderate densities at site B, and low densities at site A, but no agreement was observed between model and field data for site D. Lack of fit between model predictions and VI could be owing to simplified model assumptions, such as stable bird population densities, only two avian species represented, or similar infectivity and transmissibility for both avian hosts. On the other hand, the VI estimate is limited by field sampling biases and parameter estimation errors and low sample sizes, requiring using a monthly average of the VI. Unreliable IR estimates owing to small sample sizes for site D (38 pools tested, only 1 positive pool) may have led to the lack of fit with the model prediction.
Model results, along with IRs in Cx. pipiens in our study, further support the role of Cx. pipiens as the primary enzootic vector in this region. Culex pipiens has been established as an important enzootic vector by consistent isolations of WNV from mosquito trap collections [11,13,20], by its ornithophilic feeding behaviour [13,40,41], and associations between virus-infected mosquitoes and dead bird reports [42–46]. This species has also been incriminated as a bridge vector in Illinois [47]. However, in our study, 94.65 per cent (139 out of 147) of blood meals examined were avian-derived and no human-derived samples were obtained, indicating that other mosquito species also serve as bridge vectors during epidemics in CT. Models to determine how feeding behaviours of bridge vectors influence epidemic transmission patterns would be useful to determine the risk of spill-over of WNV from birds to humans.
Our model builds on previously published epidemiological models that use generalized parameters to understand WNV transmission dynamics [27,48,49]. Previous models were rarely field-validated, since many were not empirically informed. Field-derived parameters are difficult to measure precisely and labour-intensive to obtain, but are vital to understanding real-world transmission dynamics scenarios. In contrast to previous models [27,48–53], we used raw counts from mosquito traps to derive the rate of change in abundance. Other models have used differing assumptions to parametrize the vector population (reviewed in [54]). They may postulate some level of constant population growth [55], use a step-function [27], simulate growth over a season [51], or estimate production in a compartmental model consisting of egg, larvae and emerging susceptible adult stages [49,52]. By contrast, our method wholly captures the variation in the adult mosquito abundance data and does not require estimates for immature or other mosquito age classes.
WNV provides a suitable model for studying transmission dynamics of multi-host vector-borne pathogens because it is maintained in an enzootic cycle between mosquito vectors and multiple avian host species, with epizootic events in the United States every year. Given the strength of our findings, these results may be extended to other vector-borne diseases. A multitude of host-choice experiments provide convincing evidence that many malaria vectors, including Anopheles gambiae, prefer humans over cattle [56–63], for example. Culex nigripalpus, a major vector for Saint Louis encephalitis in the southern United States, preferred chickens to bobwhite quail in a host-choice experiment in Florida, USA [64]. Host preferences are not exclusive to mosquitoes: Triatoma bugs that transmit Chagas' disease demonstrated strong feeding preferences for dogs over both chickens and cats in a recent study [65]. At least one species of tsetse fly (Glossina morsitans morsitans), vector of human (sleeping sickness) and animal trypanosomiasis in Africa, show feeding preferences towards buffalo and oxen but avoid other hosts such as waterbuck (reviewed in [66]). Extension of this model to other systems could determine how host preferences influence transmission dynamics.
In this study, we identified vector host preferences as the most important transmission parameter and quantified the contribution of preference-induced contact heterogeneity to enzootic transmission. The ‘dilution effect’ [5], whereby the presence of less pathogen-competent hosts in a more diverse community results in ‘wasted’ transmission events and therefore lower overall pathogen prevalence, relies on the assumption that vectors are generalists. Extensive application of the dilution effect to Lyme disease, for example, has been based on the assumption of no or low host preferences by ticks ([67], but see [68] for evidence of host preferences by ticks). Our study indicates that the host community composition experienced by the pathogen may be quite different from the host community we measured; incorporating host preferences and community reservoir competence into transmission models may be essential for modelling transmission dynamics and predicting epizootics.
Acknowledgements
We would like to thank the 2006 Yale field crew, especially Nick Bonomo for conducting bird surveys and to Elyse LeeVan, Anna Milkowski and Lindsay Rollend for their assistance in mosquito collection. We are also grateful to the CT Ag Station, John Shepard and Mike Thomas for providing equipment, mosquito trapping and identification, and Phil Armstrong for virus testing. We thank Angela Luis for helping with the R scripts to fit smoothed splines to the raw mosquito data. We thank the organizers of the EEID workshop (DEB—0722115, ‘Training Workshops on the Ecology and Evolution of Infectious Diseases’) for organizing the modelling training workshop that facilitated collaboration for several coauthors. We also thank Durland Fish, Jamie Childs, Corrine Folsom-O'Keefe and Paul Cislo at Yale University for their valuable scientific feedback and support. Funding was provided by the Ruth L. Kirschstein NIH Training Grant, CDC Laboratory Capacity for Infectious Diseases Cooperative Agreement no. U50/CCU6808-01-1, USDA-ARDS Cooperative Agreement 58-6615-1-218, USDA-ARDS Cooperative Agreement 58-0790-5-068, USDA Hatch Funds CONH00768 to the CAES, Yale Institute for Biospheric Studies Faculty Support Endowment Fund.
References
- 1.Cunningham A. A. 2005. A walk on the wild side: emerging wildlife diseases. Br. Med. J. 331, 1214–1215 10.1136/bmj.331.7527.1214 (doi:10.1136/bmj.331.7527.1214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor L. H., Latham S. M., Woolhouse M. E. J. 2001. Risk factors for human disease emergence. Phil. Trans. R. Soc. Lond. B 356, 983–989 10.1098/rstb.2001.0888 (doi:10.1098/rstb.2001.0888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woolhouse M. E. J., Gowtage-Sequeria S. G. 2005. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 11, 1842–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woolhouse M. E. J. 1997. Heterogeneities in the transmission of infectious agents: implications for the design of control programs. Proc. Natl Acad. Sci. USA 94, 338–342 10.1073/pnas.94.1.338 (doi:10.1073/pnas.94.1.338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norman R., Bowers R. G., Begon M., Hudson P. J. 1999. Presistence of tick-borne virus in the presence of multiple host species: tick reservoirs and parasite mediated competition. J. Theor. Biol. 200, 111–118 10.1006/jtbi.1999.0982 (doi:10.1006/jtbi.1999.0982) [DOI] [PubMed] [Google Scholar]
- 6.Keesing F., Holt R. D., Ostfeld R. S. 2006. Effects of species diversity on disease risk. Ecol. Lett. 9, 485–498 10.1111/j.1461-0248.2006.00885.x (doi:10.1111/j.1461-0248.2006.00885.x) [DOI] [PubMed] [Google Scholar]
- 7.Kilpatrick A. M., Daszak P., Jones M. J., Marra P. P., Kramer L. D. 2006. Host heterogeneity dominates West Nile virus transmission. Proc. R. Soc. B 273, 2327–2333 10.1098/rspb.2006.3575 (doi:10.1098/rspb.2006.3575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamer G. L., Kitron U. D., Goldberg T. L., Brawn J. D., Loss S. R., Ruiz M. O., Hayes D. B., Walker E. D. 2009. Host selection by Culex pipiens mosquitoes and West Nile virus amplification. Am. J. Trop. Med. Hyg. 80, 268–278 [PubMed] [Google Scholar]
- 9.Kent R., Juliusson L., Weissmann M., Evans S., Komar N. 2009. Seasonal blood-feeding behavior of Culex tarsalis (Diptera: Culicidae) in Weld County, Colorado, 2007. J. Med. Entomol. 46, 380–390 10.1603/033.046.0226 (doi:10.1603/033.046.0226) [DOI] [PubMed] [Google Scholar]
- 10.Kay B. H., Boreham P. F. L., Edman J. D. 1979. Application of the feeding index concept to studies of mosquito host-feeding patterns. Mosq. News 39, 68–72 [Google Scholar]
- 11.Andreadis T. G., Anderson J. F., Vossbrinck C. R. 2001. Mosquito surveillance for West Nile virus in Connecticut, 2000; isolation from Culex pipiens, Cx. restuans, Cx. salinarius, and Culiseta melanura. Emerg. Infect. Dis. 7, 670–674 10.3201/eid0704.010413 (doi:10.3201/eid0704.010413) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apperson C. S., et al. 2004. Host feeding patterns of established and potential mosquito vectors of West Nile virus in the eastern United States. Vector Borne Zoonotic Dis. 4, 71–82 10.1089/153036604773083013 (doi:10.1089/153036604773083013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molaei G., Andreadis T. G., Armstrong P. M., Anderson J. F., Vossbrinck C. R. 2006. Host feeding patterns of Culex mosquitoes and West Nile virus transmission, northeastern United States. Emerg. Infect. Dis. 12, 468–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreadis T. G., Anderson J. F., Vossbrinck C. R., Main A. J. 2004. Epidemiology of West Nile virus in Connecticut, USA: a five year analysis of mosquito data 1999–2003. Vector Borne Zoonotic Dis. 4, 360–378 10.1089/vbz.2004.4.360 (doi:10.1089/vbz.2004.4.360) [DOI] [PubMed] [Google Scholar]
- 15.Bibby C. J., Burgess N. D., Hill D. A., Mustoe S. H. 2008. Bird census techniques, pp. 91–112, 2nd edn New York, NY: Academic Press [Google Scholar]
- 16.Andreadis T. G., Thomas M. C., Shepard J. J. 2005. Identification guide to the mosquitoes of Connecticut. Bull. Conn. Agr. Exp. Stat. 966, 1–173 [Google Scholar]
- 17.Molaei G., Andreadis T. G. 2006. Identification of avian- and mammalian-derived blood meals in Aedes vexans and Culiseta melanura (Diptera: Culicidae) and its implication for West Nile virus transmission in Connecticut, USA. J. Med. Entomol. 43, 1088–1093 10.1603/0022-2585(2006)43[1088:IOAAMB]2.0.CO;2 (doi:10.1603/0022-2585(2006)43[1088:IOAAMB]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 18.Molaei G., Andreadis T. G., Armstrong P. M., Diuk-Wasser M. 2008. Host-feeding patterns of potential mosquito vectors in Connecticut, USA: molecular analysis of blood meals from twenty-three species of Aedes, Anopheles, Culex, Coquillettidia, Psorophora, and Uranotaenia. J. Med. Entomol. 45, 1143–1151 10.1603/0022-2585(2008)45[1143:HPOPMV]2.0.CO;2 (doi:10.1603/0022-2585(2008)45[1143:HPOPMV]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 19.Lanciotti R. S., et al. 2000. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J. Clin. Microbiol. 38, 4066–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biggerstaff B. J. 2006. PooledInfRate, Version 3.0: a Microsoft Excel Add-in to compute prevalence estimates from pooled samples. Fort Collins, CO: Centers for disease Control and Prevention [Google Scholar]
- 21.Anderson R. M., May R. M. 1991. Infectious diseases of humans: dynamics and control. Oxford, UK: Oxford University Press [Google Scholar]
- 22.Morneau F., Lépine C., Décarie R., Villard M., DesGranges J. 1995. Reproduction of American robin (Turdus migratorius) in a suburban environment. Landscape Urban Plann. 32, 55–62 10.1016/0169-2046(94)00177-5 (doi:10.1016/0169-2046(94)00177-5) [DOI] [Google Scholar]
- 23.Farner D. S. 1949. Age groups and longevity in the American robin: comments, further discussion and certain revisions. Wilson Bull. 61, 68–81 [Google Scholar]
- 24.Coulson J. C. 1960. A study of the mortality of the starling based on ringing recoveries. J. Anim. Ecol. 29, 251–271 10.2307/2203 (doi:10.2307/2203) [DOI] [Google Scholar]
- 25.Botkin D. B., Miller R. S. 1974. Mortality rates and survival of birds. Am. Nat. 108, 181–192 10.1086/282898 (doi:10.1086/282898) [DOI] [Google Scholar]
- 26.Karr J. R., Nichols J. D., Klimkiewicz M. K., Brawn J. D. 1990. Survival rates of birds of tropical and temperate forests: will the dogma survive. Am. Nat. 136, 277–291 10.1086/285098 (doi:10.1086/285098) [DOI] [Google Scholar]
- 27.Wonham M. J., de-Camino-Beck T., Lewis M. A. 2004. An epidemiological model for West Nile virus: invasion analysis and control applications. Proc. R. Soc. Lond. B 271, 501–507 10.1098/rspb.2003.2608 (doi:10.1098/rspb.2003.2608) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vinogradova E. B. 2000. Culex pipiens pipiens mosquitoes: taxonomy, distribution, ecology, physiology, genetics, applied importance and control, pp. 46–67 Sofia, Bulgaria: Pensoft [Google Scholar]
- 29.Simpson J. E., Folsom-O'Keefe C. M., Childs J. E., Simons L. E., Andreadis T. G., Diuk-Wasser M. A. 2009. Avian host-selection by Culex pipiens in experimental trials. PLoS ONE 4, e7861. 10.1371/journal.pone.0007861 (doi:10.1371/journal.pone.0007861) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turell M. J., O'Guinn M., Oliver J. 2000. The potential for New York mosquitoes to transmit West Nile virus. Am. J. Trop. Med. Hyg. 62, 413–414 [DOI] [PubMed] [Google Scholar]
- 31.Tiawsirisup S. T., Platt K. B., Evans R. B., Rowley W. A. 2005. A comparison of West Nile virus transmission by Ochlerotatus trivittatus (COQ.), Culex pipiens (L.), and Aedes albopictus (Skuse). Vector Borne Zoonotic Dis. 5, 40–47 10.1089/vbz.2005.5.40 (doi:10.1089/vbz.2005.5.40) [DOI] [PubMed] [Google Scholar]
- 32.Komar N., Langevin S., Hinten S., Nemeth N., Edwards E., Hettler D., Davis B., Bowen R., Bunning M. 2003. Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerg. Infect. Dis. 9, 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tiawsirisup S. T., Platt K. B., Evans R. B., Roawley W. A. 2004. Susceptibility of Ochlerotatus trivittatus (Coq.), Aedes albopictus (Skuse), and Culex pipiens (L.) to West Nile virus infection. Vector Borne Zoonotic Dis. 4, 190–197 [DOI] [PubMed] [Google Scholar]
- 34.Blower S. M., Dowlatabadi H. 1994. Sensitivity and uncertainty analysis of complex models of disease transmission: an HIV model, as an example. Int. Stat. Rev. 62, 229–243 10.2307/1403510 (doi:10.2307/1403510) [DOI] [Google Scholar]
- 35.Iman R. L., Helton J. C. 1988. An investigation of uncertainty and sensitivity analysis techniques for computer models. Risk Anal. 8, 71–90 10.1111/j.1539-6924.1988.tb01155.x (doi:10.1111/j.1539-6924.1988.tb01155.x) [DOI] [Google Scholar]
- 36.McKay M. D., Beckman R. J., Conover W. J. 1979. A comparison of three methods for selecting values of input variables in the analysis of output from a computer code. Technometrics 21, 239–245 10.2307/1268522 (doi:10.2307/1268522) [DOI] [Google Scholar]
- 37.Hamby D. M. 1994. A review of techniques for parameter sensitivity analysis of environmental models. Environ. Monit. Assess. 32, 135–154 10.1007/BF00547132 (doi:10.1007/BF00547132) [DOI] [PubMed] [Google Scholar]
- 38.Gujral I. B., Zielinski-Gutierrez E. C., LeBailly A., Nasci R. 2007. Behavioral risks for West Nile virus disease, Northern Colorado, 2003. Emerg. Infect. Dis. 13, 419–425 10.3201/eid1303.060941 (doi:10.3201/eid1303.060941) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kilpatrick A. M., Kramer L. D., Jones M. J., Marra P. P., Daszak P. 2006. West Nile virus epidemics in North America are driven by shifts in mosquito feeding behavior. PLoS Biol. 4, 606–610 10.1371/journal.pbio.0040082 (doi:10.1371/journal.pbio.0040082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diuk-Wasser M. A., Molaei G., Simpson J. E., Folsom-O'Keefe C. M., Armstrong P. M., Andreadis T. G. 2010. Avian communal roosts as amplification foci for West Nile virus in urban areas in northeastern United States. Am. J. Trop. Med. Hyg. 82, 337–343 10.4269/ajtmh.2010.09-0506 (doi:10.4269/ajtmh.2010.09-0506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patrican L. A., Hackett L. E., Briggs J. E., McGowan J. W., Unnasch T. R., Lee J. H. 2007. Host-feeding patterns of Culex mosquitoes in relation to trap habitat. Emerg. Infect. Dis. 13, 1921–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernard K. A., et al. 2001. West Nile virus infection in birds and mosquitoes, New York State, 2000. Emerg. Infect. Dis. 7, 679–685 10.3201/eid0704.010415 (doi:10.3201/eid0704.010415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eidson M., Kramer L., Stone W., Hagiwara Y., Schmit K. & New York State West Nile Virus Avian Surveillance Team 2001. Dead bird surveillance as an early warning system for West Nile virus. Emerg. Infect. Dis. 7, 631–635 10.3201/eid0704.010405 (doi:10.3201/eid0704.010405) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nasci R. S., et al. 2002. Detection of West Nile virus-infected mosquitoes and seropositive juvenile birds in the vicinity of virus-positive dead birds. Am. J. Trop. Med. Hyg. 67, 492–496 [DOI] [PubMed] [Google Scholar]
- 45.Mostashari F., Kulldorff M., Hartman J. J., Miller J. R., Kulasekera V. 2003. Dead bird clusters as an early warning system for West Nile virus activity. Emerg. Infect. Dis. 9, 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nielsen C. F., Reisen W. K. 2007. West Nile virus-infected dead corvids increase the risk of infection in Culex mosquitoes (Diptera: Culicidae) in domestic landscapes. J. Med. Entomol. 44, 1067–1073 10.1603/0022-2585(2007)44[1067:WNVDCI]2.0.CO;2 (doi:10.1603/0022-2585(2007)44[1067:WNVDCI]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 47.Hamer G. L., Kitron U. D., Brawn J. D., Loss S. R., Ruiz M. O., Goldberg T. L., Walker E. D. 2008. Culex pipiens (Diptera: Culicidae): a bridge vector of West Nile virus to humans. J. Med. Entomol. 45, 125–128 10.1603/0022-2585(2008)45[125:CPDCAB]2.0.CO;2 (doi:10.1603/0022-2585(2008)45[125:CPDCAB]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 48.Wonham M. J., Lewis M. A. 2008. A comparative analysis of models for West Nile virus. In Mathematical epidemiology (eds Brauer F., Van den Driessche P., Wu J.), pp. 365–390 Berlin/Heidelberg, Germany: Springer [Google Scholar]
- 49.Rappole J. H., Compton B. W., Leimgruber P., Robertson J., King D. I., Renner S. C. 2006. Modeling movement of West Nile virus in the Western Hemisphere. Vector Borne Zoonotic Dis. 6, 128–139 10.1089/vbz.2006.6.128 (doi:10.1089/vbz.2006.6.128) [DOI] [PubMed] [Google Scholar]
- 50.Cruz-Pachedo G., Esteva L., Montaño-Hirose J. A., Vargas C. 2005. Modeling dynamics of West Nile virus. Bull. Math. Biol. 67, 1157–1172 10.1016/j.bulm.2004.11.008 (doi:10.1016/j.bulm.2004.11.008) [DOI] [PubMed] [Google Scholar]
- 51.Lord C. C., Day J. F. 2001. Simulation studies of St. Louis encephalitis in south Florida. Vector Borne Zoonotic Dis. 1, 299–316 10.1089/15303660160025921 (doi:10.1089/15303660160025921) [DOI] [PubMed] [Google Scholar]
- 52.Shaman J., Spiegelman M., Cane M., Steiglitz M. 2006. A hydrologically driven model of swamp water mosquito population dynamics. Ecol. Model. 194, 395–404 10.1016/j.ecolmodel.2005.10.037 (doi:10.1016/j.ecolmodel.2005.10.037) [DOI] [Google Scholar]
- 53.Unnasch R. S., Sprenger T., Katholi C. R., Cupp E. W., Hill G. E., Unnasch T. R. 2006. A dynamic transmission model of eastern equine encephalitis virus. Ecol. Model. 192, 425–440 10.1016/j.ecolmodel.2005.07.011 (doi:10.1016/j.ecolmodel.2005.07.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lord C. C. 2004. Seasonal population dynamics and behaviour of insects in models of vector-borne pathogens. Physiol. Entomol. 29, 214–222 10.1111/j.0307-6962.2004.00411.x (doi:10.1111/j.0307-6962.2004.00411.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chitnis N., Hyman J. M., Cushing J. M. 2008. Determining important parameters in the spread of malaria through the sensitivity analysis of a mathematical model. Bull. Math. Biol. 70, 1272–1296 10.1007/s11538-008-9299-0 (doi:10.1007/s11538-008-9299-0) [DOI] [PubMed] [Google Scholar]
- 56.Gillies M. T. 1964. Selection for host preference in Anopheles gambiae. Nature 203, 852–854 10.1038/203852a0 (doi:10.1038/203852a0) [DOI] [PubMed] [Google Scholar]
- 57.Knols B. G. J., de Jong R., Takken W. 1995. Differential attractiveness of isolated humans to mosquitoes in Tanzania. Trans. R. Soc. Trop. Med. Hyg. 89, 604–606 10.1016/0035-9203(95)90406-9 (doi:10.1016/0035-9203(95)90406-9) [DOI] [PubMed] [Google Scholar]
- 58.Dekker T., Takken W. 1998. Differential responses of mosquito sibling species Anopheles arabiensis and An. quadriannulatus to carbon dioxide, a man or a calf. Med. Vet. Entomol. 12, 136–140 10.1046/j.1365-2915.1998.00073.x (doi:10.1046/j.1365-2915.1998.00073.x) [DOI] [PubMed] [Google Scholar]
- 59.Diatta M., Spiegel A., Lochouarn L., Fontenille D. 1998. Similar feeding preferences of Anopheles gambiae and A. arabiensis in Senegal. Trans. R. Soc. Trop. Med. Hyg. 92, 270–272 10.1016/S0035-9203(98)91005-7 (doi:10.1016/S0035-9203(98)91005-7) [DOI] [PubMed] [Google Scholar]
- 60.Costantini C., Sagnon N., Torre A. D., Diallo M., Brady J., Gibson G., Coluzzi M. 1998. Odor mediated host preferences of West African mosquitoes, with particular reference to malaria vectors. Am. J. Trop. Med. Hyg. 58, 56–63 [DOI] [PubMed] [Google Scholar]
- 61.Duchemin J. B., Tsy J. M., Rabarison P., Roux J., Coluzzi M., Costantini C. 2001. Zoophily of Anopheles arabiensis and An. gambiae in Madagascar demonstrated by odour-baited entry traps. Med. Vet. Entomol. 15, 50–57 10.1046/j.1365-2915.2001.00276.x (doi:10.1046/j.1365-2915.2001.00276.x) [DOI] [PubMed] [Google Scholar]
- 62.Tirados I., Costantini C., Gibson G., Torr S. J. 2006. Blood-feeding behaviour of the malarial mosquito Anopheles arabiensis: implications for vector control. Med. Vet. Entomol. 20, 425–437 10.1111/j.1365-2915.2006.652.x (doi:10.1111/j.1365-2915.2006.652.x) [DOI] [PubMed] [Google Scholar]
- 63.Lefèvre T., Gouagna L. C., Dabire K. R., Elguero E., Fontenille D., Costantini C., Thomas F. 2009. Evolutionary lability of odour-mediated host preference by the malaria vector Anopheles gambiae sensu stricto. Trop. Med. Int. Health 14, 1–9 10.1111/j.1365-3156.2009.02206.x (doi:10.1111/j.1365-3156.2009.02206.x) [DOI] [PubMed] [Google Scholar]
- 64.Lord C. C., Day J. F. 2000. Attractiveness of chickens and bobwhite quail for Culex nigripalpus. J. Am. Mosq. Control Assoc. 16, 271–273 [PubMed] [Google Scholar]
- 65.Gürtler R. E., Ceballos L. A., Ordóñez-Krasnowski P., Lanati L. A., Stariolo R., Kitron U. 2009. Strong host-feeding preferences of the vector Triatoma infestans modified by vector density: implications for epidemiology of Chagas disease. PLoS Negl. Trop. Dis. 3, e447. 10.1371/journal.pntd.0000447 (doi:10.1371/journal.pntd.0000447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gikonyo N. K., Hassanali A., Njagi P. G., Saini R. K. 2003. Responses of Glossina morsitans morsitans to blends of electroantennographically active compounds in the odors of its preferred (buffalo and ox) and nonpreferred (waterbuck) hosts. J. Chem. Ecol. 29, 2331–2345 10.1023/A:1026230615877 (doi:10.1023/A:1026230615877) [DOI] [PubMed] [Google Scholar]
- 67.Ostfeld R. S., Keesing F. 2000. The function of biodiversity in the ecology of vector-borne zoonotic diseases. Can. J. Zool. 78, 2061–2078 10.1139/z00-172 (doi:10.1139/z00-172) [DOI] [Google Scholar]
- 68.Slowik T. J., Lane R. S. 2009. Feeding preferences of the immature stages of three Western North American Ixodid ticks (Acari) for avian, reptilian, or rodent hosts. J. Med. Ent. 46, 115–122 10.1603/033.046.0115 (doi:10.1603/033.046.0115) [DOI] [PubMed] [Google Scholar]