Abstract
Plant nuclear genome size (GS) varies over three orders of magnitude and is correlated with cell size and growth rate. We explore whether these relationships can be owing to geometrical scaling constraints. These would produce an isometric GS–cell volume relationship, with the GS–cell diameter relationship with the exponent of 1/3. In the GS–cell division relationship, duration of processes limited by membrane transport would scale at the 1/3 exponent, whereas those limited by metabolism would show no relationship. We tested these predictions by estimating scaling exponents from 11 published datasets on differentiated and meristematic cells in diploid herbaceous plants. We found scaling of GS–cell size to almost perfectly match the prediction. The scaling exponent of the relationship between GS and cell cycle duration did not match the prediction. However, this relationship consists of two components: (i) S phase duration, which depends on GS, and has the predicted 1/3 exponent, and (ii) a GS-independent threshold reflecting the duration of the G1 and G2 phases. The matches we found for the relationships between GS and both cell size and S phase duration are signatures of geometrical scaling. We propose that a similar approach can be used to examine GS effects at tissue and whole plant levels.
Keywords: scaling exponent, standard major axis regression regression, nuclear size, cell size, cell cycle phases, root meristem
1. Introduction
Sizes of plant nuclear genomes vary over three orders of magnitude from about 0.065 (Genlisea margaretae) to 152.23 pg/1C (Paris japonica) [1,2]. However, possible ecological implications have begun to be explored only recently [3,4], for a review see [5]. Although the full functional meaning of this variation is yet unknown, it is clear that genome size (GS) must represent certain phenotypic constraints. Cells must be larger to accommodate large genomes, and larger genomes typically take more time to multiply [6,7] (but see [8–10]), thus seemingly constraining the cell growth rate and other plant functional traits. These assumptions led to the formulation of the large genome constraint hypothesis (LGCH) [11]. According to the LGCH, species with large genomes are disadvantaged owing to the costs associated with accumulation and replication of non-coding repetitive DNA. The strongest support for this hypothesis is the correlation between GS and cell size [3,5,12,13] as well as between GS and cell cycle duration [6,14].
In addition to these basic relationships, there are numerous reports of correlations between GS and various plant traits, as well as environmental or evolutionary characteristics [5,11,12,14–20]. Angiosperms with larger genomes often have lower specific leaf areas and, consequently, lower maximum photosynthetic rate [5,11] and lower growth rate [14,15], possibly owing to the competition for phosphorus between DNA and RNA [16]. At the same time, GS in herbs is often associated with larger seed mass [12,17,18] and leaf lifespan [19]. Furthermore, herbaceous species with large genomes are not adapted to extreme environments, as they cannot have a large number of small stomata [12]. The same constraint also holds for woody species, which consequently have smaller genomes [12]. From an evolutionary viewpoint, plant lineages with large genomes also show reduced capacity for fast evolutionary differentiation [11], and are rare in species groups that show adaptive radiation on oceanic islands [20]. Genome size has thus been suggested to be one of the important attributes that constrain plant fitness [21].
In spite of extensive research on correlations of GS with various plant traits, we are still far from understanding specific processes that underlie these correlations. One of the possible yet little explored quantitative approaches is to characterize scaling relationships between GS and proximal variables such as cell size, tissue density and cell division rate. Scaling relationships arise owing to universal (often geometrical) processes or constraints that act over a large range of scales and take the form of scale-invariant power laws [22–26]. The signature of such a scaling relationship (namely, the value of the scaling exponent, typically measured as the slope in the log–log plot) can shed light on the type of the underlying process or constraint [27–31].
Although scaling relationships are common in the biological world and have been extensively studied [32–38], only scant attention has been paid to the scaling of GS with other plant traits. In an early study, Baetcke et al. [39] found that the slope of the interspecific relationship between GS and nuclear volume is approximately 1 (i.e. a linear relationship). Similarly, Gregory [40] explored different studies focusing on cell sizes of both plants and vertebrates. He showed that, in plants, nuclear volume scales linearly with cell volume; however, he did not use GS (dataset from Price et al. [41]). The only attempt to capture the scaling relationship between GS and cell cycle duration comes from Cavalier-Smith [42] who predicted that cell cycle duration should be proportional to the amount of material that a cell needs to grow and develop, which in turn is constrained by the rate of transport. Assuming that the rate of transport is proportional to the cell surface (which increases with the cell volume with the exponent 2/3), and the GS is an isometric function of cell volume, he inferred that the scaling exponent between GS and cell cycle duration should be 1/3. However, he did not test this prediction using statistical techniques.
All of these pioneering studies used ordinary least-squares (OLS) regression to estimate the scaling exponents. While OLS regression is a standard tool for analysing data where both predictors and dependent variables are clear, its use should be restricted to cases when errors on the x-axis are negligible relative to the errors on the y-axis, with this constraint particularly important when the relationship is not very strong (see [43]). Thus, this technique is not suited to analysing the relationship between GS and other cell properties, where the independent variable is unclear [44] and relationships are not always very strong [5,11]. In such cases, it is necessary to estimate the scaling exponent by more appropriate techniques, such as standard major axis regression (SMA, sometimes also called reduced major axis, RMA [45,46]).
In the present paper, we attempt to identify some possible relationships between GS and variables at the cell and tissue level based on simple geometrical assumptions and test them using empirical data on diploid herbaceous plant species. We first formulate several predictions on scaling relationships involving GS based on geometrical considerations. Next, we examine these predictions using datasets from a large number of plant species. We take advantage of the fact that the recent advent of fast cytometric techniques has made possible collection of extensive data on many plant species [1,21]. This enabled us to assemble empirical datasets on some of these relationships for quite large numbers of species in order to examine to what degree they conform to theoretical predictions. We have limited ourselves to considering diploid plants only. Although polyploidy does increase total DNA content, it introduces a number of different phenomena that do not occur when the base (monoploid) DNA amount increases [1,21]. This is supported by the evidence that the relationship between intraspecific cell size and GS differs between diploids and polyploids [47–50]. Moreover, it has been shown that increase in ploidy level does not increase cell cycle duration [8–10]. Analysing diploid and recent polyploid species together would thus confound the effects of polyploidy and monoploid DNA amount [21].
2. Predictions
We predict that GS, cell size and cell cycle length are constrained by geometrical relationships and thus should scale together. These geometrical relationships act both directly (as in the GS–nuclear size relationship), and indirectly, through limits for the transport of material to the cell and within the cell or through the cell metabolism. As all these constraints are possibly bidirectional, we explicitly do not assume any cause-and-effect chain and limit ourselves to symmetrical scaling relationships.
The simplest geometrical constraint concerns the relationship between nuclear volume and GS. As these are measured in volume units and mass units, they should correspond linearly to each other and their scaling exponent should consequently be 1 (prediction 1).
Further, cell size should scale with nuclear volume and hence with GS. The simplest possible relationship is a geometrical constraint as larger cells are needed to accommodate larger nuclei either physically, owing to cytoskeletal structure or owing to the necessary ratio between RNA synthesis and the rate of protein synthesis [51]. This scaling is between volume–volume units (or volume–mass units), meaning that the relationship should also be linear, i.e. with the scaling exponent of 1 (prediction 2a). Consequently, the scaling exponent for the cell area–GS relationship should correspond to 2/3 owing to the area : volume ratio (prediction 2b). Also, cell length should scale with GS according to the length : volume ratio, i.e. with scaling exponent 1/3 (prediction 2c).
When analysing data on cell lengths, it is necessary to take into account the fact that most empirical cell length data come from measurement of stomata guard cells. These cells are known to have rather constant sizes and typically do not endoreduplicate [48] (which makes them a favourite subject for comparative studies), but have a strongly constrained (curved) shape. The scaling relationship with other cell variables may be affected by the possible effect of this bend or curvature. We assume that the shape of this bend could be expressed by a power function y = axb, where x is the measured length, and a and b are positive numbers. This length of the bent shape (S) is always greater than x and scales nonlinearly with the measured length, with the scaling exponent depending on the shape of the cell (i.e. parameters a and b of the curve), but necessarily greater than 1 (derivation for the simplest case of a quadratic parabola is in the electronic supplementary material). If we denote this scaling exponent c, the measured length (x) will thus scale with GS with the exponent (1/3) into (1/c), which is always lower than 1/3 if c > 1 (prediction 2d).
Constraints on possible scaling relationships between GS and cell growth and division rates should be due either to matter transport to the cell [42] or to cell metabolism [52] as a function of GS and cell size. During cell growth, DNA synthesis and cell division, the cell would need amounts of material proportional to its volume, with the rate of transport limited by cell surface area. This area increases with a scaling of 2/3 in relationship with cell volume. If transport across the cell surface is the limiting factor, the cell cycle duration should thus be proportional to the amount of material that needs to pass into the cytoplasm, divided by the rate of transport [42]:
| 2.1 |
where T is duration of the cell cycle and V is the cell volume. Alternatively, cell cycle duration can also be driven by cell metabolism. Both respiration and photosynthesis take place on organelle membranes (owing to the location of ATP-synthesis complexes). Assuming that an increase in cell size leads to a proportional increase in the number of organelles [53], the rate of whole-cell metabolism should scale with GS with the scaling exponent 1. Therefore, mass-specific metabolic rate should be independent of GS (GS1/GS1). Assuming the duration of the cell cycle is inverse to the cell cycle rate, cell cycle duration should not depend on GS (scaling exponent 0). When considering both role of cell metabolism and the rate of transport as factors responsible for cell cycle duration, the scaling exponent between GS and cell cycle duration should vary between 1/3 (limitation by the surface : volume ratio only) and 0 (limitation by cell metabolism only; prediction 3).
This prediction is however problematic as the cell cycle consists of four phases and the duration of each phase may depend upon GS differently owing to the differences underlying or limiting each of these phases. We therefore make specific predictions for the relationship between GS and duration of each phase of the cycle. Because of the demands of the growth phases (G1 and G2), we expect that during these phases cell metabolic rate should be especially important. Thus, the duration of these two phases should not depend on GS; i.e. scaling exponents of relationships between G phase duration and GS should be 0. However, as metabolic rate might be constrained by the rate of transport through the cell membrane, we expect that the scaling coefficient between the duration of G phases and GS should vary between 1/3 and 0, as shown earlier (prediction 4a).
Limitation on the S phase is in principle owing to the number of replication origins and replication rate per replicon. However, the dataset of Hof & Bjerknes [7] shows that there is no significant relationship between GS and the fork rate (replication rate per replicon). We thus suggest that the main factor limiting duration of the S phase is the amount of DNA polymerase and other factors (e.g. CDC6 and CDT1, see [54]) important for the activation of replication origins. Given that the amount of the DNA polymerase as well as CDC6 and CDT1 needed for the replication scales linearly with GS, it should be limited by the rate of transport through the nuclear surface. S phase duration will then be constrained by the ratio between the total amount of material needed for replication (which scales at 1 with the GS) divided by the rate of transport (which scales at 2/3 with the nuclear volume). The predicted exponent for the relationship between S phase duration and GS should thus be 1/3 (prediction 4b). We hypothesize that during cell division (M phase), GS (and cell size) limits the development of nuclear membranes and cell walls. Therefore, duration of the M phase should scale linearly with the total surface area of nuclear membranes and cell walls, which in turn scales at 2/3 with the cell volume and thus at 2/3 with GS. We thus predict the scaling exponent between the duration of the M phase and GS to be 2/3 (prediction 4c).
3. Methods
(a). Datasets
We re-analysed 11 published datasets reporting nuclear volume, cell size, cell cycle duration and cell cycle phase duration (detailed description is given in table 1). Both differentiated and undifferentiated cells were used to obtain data on cell and nucleus sizes; for cell cycle duration, we used only root meristem data where potential constraints owing to GS would show up most clearly owing to their high cell division rates. To reduce noise in the data, we included data only on diploid herbaceous plants. We excluded woody plants, which usually have much smaller genomes because of strong selection on cambial and stomatal cell size [12].
Table 1.
Published datasets used in the study.
| reported variables | type of cells | species | families | reference |
|---|---|---|---|---|
| nuclear volume, GS | root and shoot meristem | 16 | 10 | [39] |
| nuclear volume, GS | epidermisa of young leaves | 5 | 4 | [49] |
| cell volume, nuclear volume | shoot meristem | 14 | 9 | [41] |
| pollen width, GS | pollen grains | 178 | 40 | [13] |
| cell area, guard cell length, GS | leaf epidermal and guard cells | 20 | 13 | [12] |
| pollen volume, GS | pollen grains | 10b | 1c | [55] |
| cell cycle duration, GS | root meristem | 62 | 16 | [6] |
| G1, S, G2, M phase duration | root meristem | 7 | 5 | [56–58] |
| G1, S, G2, M phase duration, GS | root meristem | 5 | 4 | [59] |
| S phase duration, GS | root meristem | 21 | 11 | [7] |
aWe used nuclear volume only for data of 2C nuclei reported in this study.
bWe excluded one outlier.
cPoaceae.
The data came from a total of 269 plant species from 46 families. Genome sizes (2C-value in pg) were taken from original publications and, if missing there, from the Angiosperm DNA C-value database [1]. Ploidy levels were also taken from original publications or from C-value database [1]; corrections of ploidy levels using other existing cytogenetic data were performed in cases of clear errors (J. Suda 2011, unpublished data). We were able to compile cell cycle phase duration data from different resources (table 1, rows 8 and 9) as they all were measured within the same temperature range (20–23°C) [7,56–59].
(b). Data analysis
We re-analysed each dataset using SMA regression (R language, package lmodel2; [60]). The scaling exponent for each relationship between GS and different cell parameters was calculated as the slope of the SMA regression from the power law relationship (logarithmic transformation of all variables). We calculated confidence intervals for all scaling exponents. The match of the estimated slopes with the theoretical predictions was carried out by assessing (i) simple numerical closeness of the theoretical and empirical values and (ii) statistical fit of the theoretical and empirical values using 95% confidence intervals of the empirical values.
As numerically high values of intercepts in untransformed data can seriously distort estimated values of scaling exponents in log–log transformed data [61], we checked all relationships for the intercept effect. We tested for nonlinearity of the relationship in the log–log transformed data by testing significance of the quadratic term in quadratic polynomial regression fitted by OLS. If the quadratic term was significant, we estimated the scaling exponent from nonlinear regression (SPSS, v. 10; SPSS Inc., Chicago, IL, USA). We used nonlinear regression to fit the equation y = Cxz + a, using the loss function proposed by Ebert & Russell [61] for model II regression (analogue to SMA) estimation of the y-axis intercept.
4. Results
(a). Nuclear volume and cell size
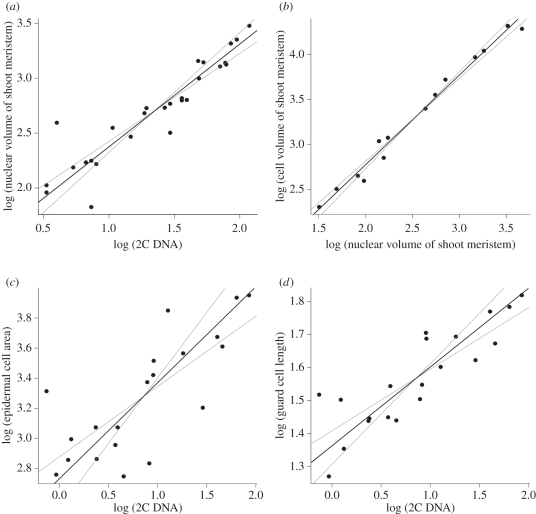
There was a strong linear relationship between GS and nuclear volume. The SMA scaling exponent between nuclear volume and GS was 0.96 for shoot meristem cells, 0.85 for root meristem cells and 1.01 for epidermal cells of young leaves (table 2 and figure 1a). Confidence intervals of all these relationships covered the predicted value of 1. Also, in accord with our prediction, GS (and nuclear volume) scaled linearly with the cell volume (table 2 and figure 1b–d). The strongest relationship occurred between the shoot meristem cells and nuclear volume (scaling exponent 1.01, r2 = 0.98; table 2 and figure 1b). A similar, but weaker relationship was found between epidermal cell area and GS (scaling exponent 0.64, close to the predicted value of 2/3; table 2 and figure 1c). Nevertheless, these results with epidermal cells may be partly affected by endoreduplication [48]. There was a very weak indication of a relationship between GS and pollen width having a scaling exponent of 0.34 (table 2). Although this was in agreement with our prediction (1/3), this relationship was not significant, as the p-value was 0.18 (0.08 for one-tailed test). In the case of guard cell length (table 2 and figure 1d), as the observed scaling exponent was 0.24, it was less than 1/3, and thus in accord with our prediction. The highest deviation from the predicted value was found in the relationship between pollen volume and GS (table 2), with the scaling exponent being 1.16 instead of the predicted 1.
Table 2.
Standard major axis power law regression between GS and nuclear volume, cell size, whole cell cycle duration and duration of cell cycle phases.
| power law relationship | prediction no. | predicted slope | estimated SMA slope | 95% confidence intervals | r2 | p | ref. |
|---|---|---|---|---|---|---|---|
| nuclear volume | |||||||
| nuclear volume–GS (shoot meristem) | 1 | 1 | 0.96 | (0.77;1.20) | 0.86 | <0.001 | [39] |
| nuclear volume–GS (root meristem) | 1 | 1 | 0.85 | (0.68;1.05) | 0.86 | <0.001 | [39] |
| nuclear volume (epidermal cells)–GS | 1 | 1 | 1.01 | (0.86;1.19) | 0.99 | <0.001 | [49] |
| cell size | |||||||
| cell volume–nuclear volume | 2a | 1 | 0.99 | (0.90;1.08) | 0.98 | <0.001 | [41] |
| pollen volume–GS | 2a | 1 | 1.16 | (0.96;1.41) | 0.89 | <0.001 | [55] |
| epidermal cell area–GS | 2b | 2/3 | 0.64 | (0.47;0.87) | 0.59 | <0.001 | [12] |
| pollen width–GS | 2c | 1/3 | 0.34 | (0.29;0.39) | 0.01 | 0.18a | [13] |
| guard cell length–GS | 2d | <1/3 | 0.24 | (0.19;0.30) | 0.75 | <0.001 | [12] |
| cell cycle duration | |||||||
| cell cycle duration–GS | 3 | 1/3–0 | 0.69b | — | — | — | [6] |
| G1 phase duration–GS | 4a | 1/3–0 | n.s. | — | 0.06 | 0.48 | [56–59] |
| S phase duration–GS | 4b | 1/3 | 0.36 | (0.21;0.53) | 0.50 | 0.02 | [56–59] |
| G2 phase duration–GS | 4a | 1/3–0 | n.s. | — | <0.01 | 0.87 | [56–59] |
| M phase duration–GS | 4c | 2/3 | 0.40 | (0.20;0.79) | 0.46 | 0.07 | [56–59] |
| S phase duration (synthesis of all data available) –GS | 4b | 1/3 | 0.36 | (0.27;0.48) | 0.64 | <0.001 | [56–59] |
ap = 0.08 for one-sided test.
bScaling exponent is estimated from the nonlinear regression.
Figure 1.
Relationship between genome size and nuclear and cell sizes: (a) relationship between genome size (GS) and nuclear volume of shoot meristem cells (slope = 0.96); (b) relationship between nuclear volume and cell volume of shoot meristem cells (slope = 0.99); (c) relationship between GS and epidermal cell area (slope = 0.64); and (d) relationship between GS and guard cell length. Black line represents SMA regression line (slope = 0.24); grey lines are SMA 95% confidence intervals. The regression results are listed in table 2, rows 1, 4, 6 and 8.
(b). Cell cycle duration
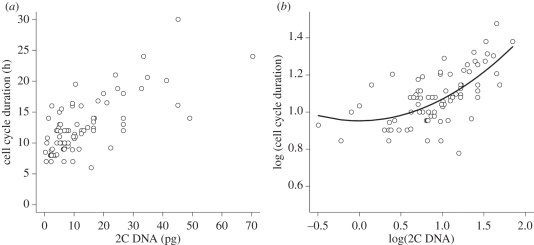
There was strong nonlinearity in the power-law function of the relationship of cell cycle duration to GS, as showed by the high significance of the GS as a quadratic term (p = 0.004). This was the only relationship with a significant quadratic term. This relationship also had a high value of the intercept in non-transformed data (figure 2a,b). The value of the intercept (6.14) corresponded to the minimum value of cell cycle duration. The scaling exponent estimated by nonlinear regression (0.69) did not match the value predicted by the surface : volume or metabolic limitation (table 2).
Figure 2.
The relationship between cell cycle duration and genome size: (a) untransformed, (b) with logarithmic transformation. The line shown in part (b) is the quadratic function. The model containing log(GS) both as linear and quadratic term has a significantly better fit than the linear-only model (p-value = 0.004, n = 83).
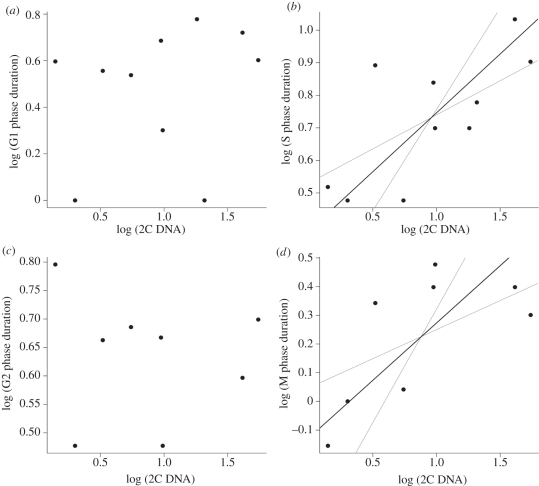
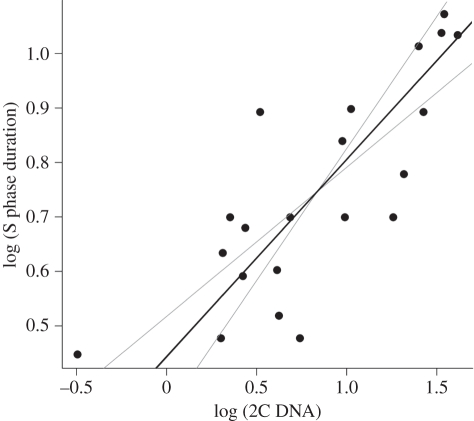
The relationships between GS and individual cell cycle phases varied widely. Although there was no relationship between G phases and GS (table 2 and figure 3a,c), the relationship between duration of the S phase and GS was significantly positive (table 2 and figure 3b) and the scaling exponent (0.36) corresponded to the predicted 1/3 (when the same dataset as for other phases was used). When we analysed the larger set of all available S phase data, the scaling exponent remained the same and the relationship was even tighter (table 2 and figure 4). Duration of the M phase increased with GS marginally significantly (table 2 and figure 3d). The estimated scaling exponent 0.40 did not match numerically the value we predict (0.67), although the difference was not significant owing to high variation and the low number of observations.
Figure 3.
Relationships between duration of individual cell cycle phases and genome size: (a) G1 phase, (b) S phase (slope = 0.36), (c) G2 phase and (d) M phase (slope = 0.40). Black line is the slope of SMA regression; grey lines are SMA 95% confidence intervals. Regression results are listed in table 2, rows 10–13.
Figure 4.
The relationship between S phase and genome size (slope = 0.36). Black line is the slope of SMA regression; grey lines are SMA 95% confidence intervals. Regression results are listed in table 2, row 14.
5. Discussion
We found strong support for the role of geometry in the relationship between GS, nuclear size and cell size. Scaling exponents of these relationships almost exactly matched expectations based on geometrical considerations. In the case of pollen volume, however, the scaling exponent (1.16) was above the predicted value of 1, although the confidence interval (0.96–1.41) did include this value. This was probably owing to variation in the shape of pollen grains and the fact that an individual pollen grain could contain several nuclei. In addition, their sizes can be affected by other factors, namely potential selection for the small sizes necessary for successful transport to female flowers. All these factors are likely to confound the relationship by adding unexplained variation. However, the fact that the size of pollen grain still scales with GS despite such a large amount of unexplained variability, indicates that GS still acts as an important constraint for the pollen grain size. Indeed, the difference from the predicted value of 1 was not significant when using the 95% confidence interval.
Most importantly, the relationships between GS and both cell size and nuclear size are of simple isometry with no nonlinear component. This indicates that size relationships within cells are not constrained by transport, as this process typically involves a mass or volume over plane relationship, which is not isometric. However, our data show that nucleus : genome in a cell is constrained by aspects of its cellular background, such as cytoskeletal structure or ratio between rates of RNA synthesis and of protein synthesis, that are simply proportional to its size. While it has been pointed out earlier by many authors that small cells cannot contain large genomes [40,55], the fact of simple isometry shows that the reverse is also true, i.e. large cells do not contain small genomes, and this relationship is likely owing to a geometrical constraint.
Although there was a strong relationship between GS and cell cycle duration, it did not match our prediction that was based on a single geometrical constraint. This is clearly owing to differences in correlations between GS and different cell cycle phases. Whereas there was almost no relationship between GS and the duration of growth phases (G1 and G2), M phase and particularly S phase scaled with GS rather strongly. Still none of the relationships involved simple isometry, indicating the strong role of transport or similar processes in determining cell cycle duration.
The fact that cell growth phases (G1 and G2) were independent of GS, and consequently, of cell size, indicates that the duration of phases of cell growth is not limited by any transport process involving the nucleus. These phases are probably driven by the rate of cell metabolism, which is possibly maximized by number of organelles increasing with GS [53]. In contrast with the G phases, duration of the S phase increased with GS relatively strongly, with the scaling exponent corresponding to the 1/3 predicted by the limitation of the DNA-polymerase transport rate into the nucleus. Duration of this part of the cell cycle thus seems to be limited by a geometrical constraint on DNA replication. The role of geometry in the M phase is unclear owing to the weak relationship, with the estimated value not only differing from the predicted value of 2/3 (limitation by membrane or cell wall construction), but also weakly differing from zero.
Taking into account the different results that we found for individual cell cycle phases, the relationship between GS and total cell cycle duration seems to consist of two components. The first is the duration of the G phases (which are virtually independent of the GS) in determining the cell cycle duration threshold. The second is the role of the S (and possibly M) phases in determining the slope of this relationship. GS thus plays a significant role in the whole plant growth rate by increasing the time needed for DNA polymerization. However, owing to the different proportions of the cell cycle constituted by particular cell cycle phases among different plants [59,62], the slope of the scaling relationship between GS and cell cycle duration is not universal. Its numerical value may, however, be indicative of the relative time that cells spend in the individual cell division phases.
It is known that a decrease in cell cycle duration is associated with an increase in relative growth rate [47,63], and thus indirectly with photosynthetic rate [64] despite the fact that the relationship need not be straightforward in a population of proliferating cells. These relationships scale up to other plant traits [5] and are fundamental for other biological rates. Cell size scaling has been much less studied in spite of its key role in controlling organ or tissue complexity and thus whole-organism metabolic rate [52]. The separate scaling relationships for cell cycle duration and cell size indicate that their actions are likely to be independent.
While the geometrical scaling strongly indicates intimate functional ties between GS, nuclear volume, cell size and cell cycle duration, it does not say anything about the causal direction of the relationships (see also [44]). In particular, it is not clear whether larger genomes result only from passive accumulation of junk DNA, as is commonly believed. In this scenario, the passive accumulation of junk DNA then constrains plant traits and life histories [11]. However, this is not the only possible basis for evolution of large genomes. For example, small cells may be economically disadvantaged because of their higher metabolic rates and consequently higher energetic demands [65,66]. Increase in the GS may thus be a possible option to diminish the energetic costs of the cell [67] as large cells cannot have small genomes. Moreover, organisms have been demonstrated to both increase and decrease the GS in their cells by accumulation and deletion mechanisms (see [68] for review) and to change the proportion of heterochromatin in response to GS (data from Nagl [69]), suggesting possible mechanisms to cope with excess DNA amounts.
In conclusion, the numerical fit between the predicted and empirical scaling exponents provides strong support for the concept of GS as a phenotypic trait (nucleotypic theory of Bennett [3], etc.). It has also made it possible to identify specific components of this limitation. In particular, size relationships and rate relationships are likely owing to different limitations. At a finer scale, absence of any relationship between GS and both G phases is a strong indication that the surface area : volume ratio is not limiting during cell growth, but does limit the S phase and mitosis. These findings, although tentative and concerning only undifferentiated meristematic cells, show the power of the scaling relationships at the cellular level in spite of the low number of data points available. We are convinced that even finer analysis of geometrical and mechanical constraints in the GS relationships will be possible as additional comparative data are collected. A similar approach might also be possible for examining GS effects at tissue and whole plant levels.
Acknowledgements
We thank Matyáš Fendrych, Lukáš Kratochvíl, Jonathan Rosenthal, Zuzana Starostová, David Storch, Jan Suda, Arnošt Šizling, Martin Weiser, Viktor Žárský, Hirokazu Tsukaya and three anonymous reviewers for discussions and/or comments on an earlier draft of this paper. The research reported here was supported by research grants MSM0021620845, MSM 0021620828, LC06073 from the Czech Ministry of Education and AV0Z60050516 from the Academy of Sciences of the Czech Republic.
References
- 1.Bennett M. D., Leitch I. J. 2010. Plant DNA C-values database (release 5.0, December 2010). See http://www.kew.org/cvalues/.
- 2.Pellicer J., Fay M., Leitch I. J. 2010. The largest eukaryotic genome of them all? Bot. J. Linn. Soc. 164, 10–15 10.1111/j.1095-8339.2010.01072.x (doi:10.1111/j.1095-8339.2010.01072.x) [DOI] [Google Scholar]
- 3.Bennett M. D. 1987. Variation in genomic form in plants and its ecological implications. New Phytol. 106, 177–200 10.1111/j.1469-8137.1987.tb04689.x (doi:10.1111/j.1469-8137.1987.tb04689.x) [DOI] [Google Scholar]
- 4.Thompson K. 1990. Genome size, seed size and germination temperature in herbaceous angiosperms. Evol. Trends Plants 4, 113–116 [Google Scholar]
- 5.Knight C., Beaulieu J. 2008. Genome size scaling through phenotype space. Ann. Bot. (Lond.) 101, 759–766 10.1093/aob/mcm321 (doi:10.1093/aob/mcm321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis D., Davies M., Barlow P. 2008. A strong nucleotypic effect on the cell cycle regardless of ploidy level. Ann. Bot. (Lond.) 101, 747–757 10.1093/aob/mcn038 (doi:10.1093/aob/mcn038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hof J. V., Bjerknes C. 1981. Similar replicon properties of higher-plant cells with different S periods and genome sizes. Exp. Cell Res. 136, 461–465 10.1016/0014-4827(81)90027-6 (doi:10.1016/0014-4827(81)90027-6) [DOI] [PubMed] [Google Scholar]
- 8.Bayliss M. W. 1976. Variation of cell cycle duration within suspension cultures of Daucus carota and its consequence for the induction of ploidy changes with colchicine. Protoplasma 88, 279–285 10.1007/BF01283252 (doi:10.1007/BF01283252) [DOI] [PubMed] [Google Scholar]
- 9.Gong J., Traganos F., Darzynkiewicz Z. 1993. Simultaneous analysis of cell cycle kinetics at two different DNA ploidy levels based on DNA content and cyclin B measurements. Cancer Res. 53, 5096–5099 [PubMed] [Google Scholar]
- 10.Storchová Z., Breneman A., Cande J., Dunn J., Burbank K., O'Toole E., Pellman D. 2006. Genome-wide genetic analysis of polyploidy in yeast. Nature 443, 541–547 10.1038/nature05178 (doi:10.1038/nature05178) [DOI] [PubMed] [Google Scholar]
- 11.Knight C., Molinari N., Petrov D. 2005. The large genome constraint hypothesis: evolution, ecology and phenotype. Ann. Bot. (Lond.) 95, 177–190 10.1093/aob/rnci011 (doi:10.1093/aob/rnci011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaulieu J., Leitch I., Patel S., Pendharkar A., Knight C. 2008. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytol. 179, 975–986 10.1111/j.1469-8137.2008.02528.x (doi:10.1111/j.1469-8137.2008.02528.x) [DOI] [PubMed] [Google Scholar]
- 13.Knight C. A., Clancy R. B., Götzenberger L., Dann L., Beaulieu J. M. 2010. On the relationship between pollen size and genome size. J. Bot. 2010, 7. 10.1155/2010/612017 (doi:10.1155/2010/612017) [DOI] [Google Scholar]
- 14.Gruner A., Hoverter N., Smith T., Knight C. A. 2010. Genome size is a strong predictor of root meristem growth rate. J. Bot. 2010, 4. (doi:10.1155/2010/390414) [Google Scholar]
- 15.Leishman M. 1999. How well do plant traits correlate with establishment ability? Evidence from a study of 16 calcareous grassland species. New Phytol. 141, 487–496 10.1046/j.1469-8137.1999.00354.x (doi:10.1046/j.1469-8137.1999.00354.x) [DOI] [Google Scholar]
- 16.Hessen D., Jeyasingh P., Neiman M., Weider L. 2010. Genome streamlining and the elemental costs of growth. Trends Ecol. Evol. 25, 75–80 10.1016/j.tree.2009.08.004 (doi:10.1016/j.tree.2009.08.004) [DOI] [PubMed] [Google Scholar]
- 17.Maranon T., Grubb P. J. 1993. Physiological basis and ecological significance of the seed size and relative growth rate relationship in Mediterranean annuals. Funct. Ecol. 7, 591–599 10.2307/2390136 (doi:10.2307/2390136) [DOI] [Google Scholar]
- 18.Knight C., Ackerly D. 2002. Variation in nuclear DNA content across environmental gradients: a quantile regression analysis. Ecol. Lett. 5, 66–76 10.1046/j.1461-0248.2002.00283.x (doi:10.1046/j.1461-0248.2002.00283.x) [DOI] [Google Scholar]
- 19.Morgan H., Westoby M. 2005. The relationship between nuclear DNA content and leaf strategy in seed plants. Ann. Bot. (Lond.) 96, 1321–1330 10.1093/aob/mci284 (doi:10.1093/aob/mci284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suda J., Kyncl T., Freiova R. 2003. Nuclear DNA amounts in Macaronesian angiosperms. Ann. Bot. (Lond.) 92, 153–164 10.1093/aob/mcg104 (doi:10.1093/aob/mcg104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leitch I. J., Bennett M. D. 2007. Genome size and its uses: the impact of flow cytometry. In Flow cytometry with plant cells (eds Dolezel J., Greilhuber J., Suda J.), pp. 153–176 Weinheim, Germany: Wiley-VCH [Google Scholar]
- 22.Peters R. H. 1986. The ecological implications of body size. Cambridge, UK: Cambridge University Press [Google Scholar]
- 23.Connor E., McCoy E. 1979. Statistics and biology of the species-area relationship. Am. Nat. 113, 791–833 10.1086/283438 (doi:10.1086/283438) [DOI] [Google Scholar]
- 24.Martinez N. 1992. Constant connectance in community food webs. Am. Nat. 139, 1208–1218 10.1086/285382 (doi:10.1086/285382) [DOI] [Google Scholar]
- 25.Reich P., Oleksyn J., Wright I., Niklas K., Hedin L., Elser J. 2010. Evidence of a general 2/3-power law of scaling leaf nitrogen to phosphorus among major plant groups and biomes. Proc. R. Soc. B 277, 877–883 10.1098/rspb.2009.1818 (doi:10.1098/rspb.2009.1818) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tilman D. 1999. The ecological consequences of changes in biodiversity: a search for general principles. Ecology 80, 1455–1474 [Google Scholar]
- 27.Niklas K. 1994. The scaling of plant and animal body-mass, length and diameter. Evolution 48, 44–54 10.2307/2410002 (doi:10.2307/2410002) [DOI] [PubMed] [Google Scholar]
- 28.West G., Brown J., Enquist B. 1997. A general model for the origin of allometric scaling laws in biology. Science 276, 122–126 10.1126/science.276.5309.122 (doi:10.1126/science.276.5309.122) [DOI] [PubMed] [Google Scholar]
- 29.Moses M., Brown J. 2003. Allometry of human fertility and energy use. Ecol. Lett. 6, 295–300 10.1046/j.1461-0248.2003.00446.x (doi:10.1046/j.1461-0248.2003.00446.x) [DOI] [Google Scholar]
- 30.Savage V., Gillooly J., Woodruff W., West G., Allen A., Enquist B., Brown J. 2004. The predominance of quarter-power scaling in biology. Funct. Ecol. 18, 257–282 10.1111/j.0269-8463.2004.00856.x (doi:10.1111/j.0269-8463.2004.00856.x) [DOI] [Google Scholar]
- 31.Reich P., Tjoelker M., Machado J., Oleksyn J. 2006. Universal scaling of respiratory metabolism, size and nitrogen in plants. Nature 439, 457–461 10.1038/nature04282 (doi:10.1038/nature04282) [DOI] [PubMed] [Google Scholar]
- 32.Brown J., West G. 2001. Scaling in biology. Oxford, UK: Oxford University Press [Google Scholar]
- 33.Price C., Enquist B. 2007. Scaling mass and morphology in leaves: an extension of the WBE model. Ecology 88, 1132–1141 10.1890/06-1158 (doi:10.1890/06-1158) [DOI] [PubMed] [Google Scholar]
- 34.Enquist B., Kerkhoff A., Stark S., Swenson N., McCarthy M., Price C. 2007. A general integrative model for scaling plant growth, carbon flux and functional trait spectra. Nature 449, 218–222 10.1038/nature06061 (doi:10.1038/nature06061) [DOI] [PubMed] [Google Scholar]
- 35.Milla R., Reich P. 2007. The scaling of leaf area and mass: the cost of light interception increases with leaf size. Proc. R. Soc. B 274, 2109–2114 10.1098/rspb.2007.0417 (doi:10.1098/rspb.2007.0417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niklas K., Cobb E., Niinemets U., Reich P., Sellin A., Shipley B., Wright I. 2007. ‘Diminishing returns’ in the scaling of functional leaf traits across and within species groups. Proc. Natl Acad. Sci. USA 104, 8891–8896 10.1073/pnas.0701135104 (doi:10.1073/pnas.0701135104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.West G., Enquist B., Brown J. 2009. A general quantitative theory of forest structure and dynamics. Proc. Natl Acad. Sci. USA 106, 7040–7045 10.1073/pnas.0812294106 (doi:10.1073/pnas.0812294106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori S., et al. 2010. Mixed-power scaling of whole-plant respiration from seedlings to giant trees. Proc. Natl Acad. Sci. USA 107, 1447–1451 10.1073/pnas.0902554107 (doi:10.1073/pnas.0902554107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baetcke K., Sparrow A., Nauman C., Schwemme S. S. 1967. Relationship of DNA content to nuclear and chromosome volumes and to radiosensitivity (LD50). Proc. Natl Acad. Sci. USA 58, 533–540 10.1073/pnas.58.2.533 (doi:10.1073/pnas.58.2.533) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregory T. 2001. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol. Rev. 76, 65–101 10.1017/S1464793100005595 (doi:10.1017/S1464793100005595) [DOI] [PubMed] [Google Scholar]
- 41.Price H., Sparrow A., Nauman A. 1973. Correlations between nuclear volume, cell volume and DNA content in meristematic cells of herbaceous angiosperms. Experientia 29, 1028–1029 10.1007/BF01930444 (doi:10.1007/BF01930444) [DOI] [Google Scholar]
- 42.Cavalier-Smith T. 1978. Nuclear volume control by nucleoskeletal DNA, selection for cell volume and cell growth rate, and the solution of the DNA C-value paradox. J. Cell Sci. 34, 247. [DOI] [PubMed] [Google Scholar]
- 43.Smith R. 2009. Use and misuse of the reduced major axis for line-fitting. Am. J. Phys. Anthropol. 140, 476–486 10.1002/ajpa.21090 (doi:10.1002/ajpa.21090) [DOI] [PubMed] [Google Scholar]
- 44.Hodgson J., et al. 2010. Stomatal vs. genome size in angiosperms: the somatic tail wagging the genomic dog? Ann. Bot. (Lond.) 105, 573–584 10.1093/aob/mcq011 (doi:10.1093/aob/mcq011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jolicoeur P. 1975. Linear regressions in fishery research: come comments. J. Fish. Res. Board Can. 32, 1491–1494 10.1139/f75-171 (doi:10.1139/f75-171) [DOI] [Google Scholar]
- 46.Legendre P., Legendre L. 1998. Numerical ecology. Amsterdam, The Netherlands: Elsevier Science [Google Scholar]
- 47.Beemster G., Vusser K., De Tavernier E., De Bock K., De Inze D. 2002. Variation in growth rate between Arabidopsis ecotypes is correlated with cell division and A-type cyclin-dependent kinase activity. Plant Physiol. 129, 854–864 10.1104/pp.002923 (doi:10.1104/pp.002923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melaragno J. E., Mehrotra B., Coleman A. W. 1993. Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5, 1661–1668 10.1105/tpc.5.11.1661 (doi:10.1105/tpc.5.11.1661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jovtchev G., Schubert V., Meister A., Barow M., Schubert I. 2006. Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Genome Res. 114, 77–82 10.1159/000091932 (doi:10.1159/000091932) [DOI] [PubMed] [Google Scholar]
- 50.Gray J., Kolesik P., Hoj P., Coombe B. 1999. Confocal measurement of the three-dimensional size and shape of plant parenchyma cells in a developing fruit tissue. Plant J. 19, 229–236 10.1046/j.1365-313X.1999.00512.x (doi:10.1046/j.1365-313X.1999.00512.x) [DOI] [PubMed] [Google Scholar]
- 51.Cavalier-Smith T. 2005. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann. Bot. (Lond.) 95, 147–175 10.1093/aob/mci010 (doi:10.1093/aob/mci010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kozlowski J., Konarzewski M., Gawelczyk A. 2003. Cell size as a link between noncoding DNA and metabolic rate scaling. Proc. Natl Acad. Sci. USA 100, 14 080–14 085 10.1073/pnas.2334605100 (doi:10.1073/pnas.2334605100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeLong J., Okie J., Moses M., Sibly R., Brown J. 2010. Shifts in metabolic scaling, production and efficiency across major evolutionary transitions of life. Proc. Natl Acad. Sci. USA 107, 12 941–12 945 10.1073/pnas.1007783107 (doi:10.1073/pnas.1007783107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Méchali M. 2010. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat. Rev. Mol. Cell. Biol. 11, 728–738 10.1038/nrm2976 (doi:10.1038/nrm2976) [DOI] [PubMed] [Google Scholar]
- 55.Bennett M. D. 1972. Nuclear DNA content and minimum generation time in herbaceous plants. Proc. R. Soc. Lond. B 181, 109–135 10.1098/rspb.1972.0042 (doi:10.1098/rspb.1972.0042) [DOI] [PubMed] [Google Scholar]
- 56.Kuroki Y., Tanaka R. 1973. Duration of mitotic cell cycle in root tip cells of male Rumex acetosa. Jpn. J. Genet. 48, 19–26 10.1266/jjg.48.19 (doi:10.1266/jjg.48.19) [DOI] [Google Scholar]
- 57.Powell M., Davies M., Francis D. 1986. The influence of zinc on the cell cycle in the root meristem of a zinc-tolerant and zoinc-nontolerant cultivar of Festuca rubra L. New Phytol. 102, 419–428 10.1111/j.1469-8137.1986.tb00819.x (doi:10.1111/j.1469-8137.1986.tb00819.x) [DOI] [PubMed] [Google Scholar]
- 58.King P. 1980. Plant tissue culture and the cell cycle. Adv. Biomed. Eng. 18, 1–38 [Google Scholar]
- 59.Barlow P. 1973. Mitotic cycles in root meristems. In The cell cycle in development and differentiation (eds Balls M., Billet F. S.), pp. 133–165 Cambridge, UK: British Society for Developmental Biology Symposium [Google Scholar]
- 60.R Development Core Team 2010. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org/. [Google Scholar]
- 61.Ebert T., Russell M. 1994. Allometry and model II nonlinear regression. J. Theor. Biol. 168, 367–372 10.1006/jtbi.1994.1116 (doi:10.1006/jtbi.1994.1116) [DOI] [Google Scholar]
- 62.Francis D., Barlow P. 1988. Temperature and the cell cycle. In Plants and temperature (eds Long S. P., Woodward F. I.), pp. 181–201 Cambridge, U: Company of Biologists; [PubMed] [Google Scholar]
- 63.Ivanov V., Dubrovsky J. 1997. Estimation of the cell-cycle duration in the root apical meristem: a model of linkage between cell-cycle duration, rate of cell production, and rate of root growth. Int. J. Plant Sci. 158, 757–763 10.1086/297487 (doi:10.1086/297487) [DOI] [Google Scholar]
- 64.Garnier E. 1991. Resource capture, biomass allocation and growth in herbaceous plants. Trends Ecol. Evol. 6, 126–131 10.1016/0169-5347(91)90091-B (doi:10.1016/0169-5347(91)90091-B) [DOI] [PubMed] [Google Scholar]
- 65.Vinogradov A. 1995. Nucleotypic effect in homeotherms: body-mass-corrected basal metabolic rate of mammals is related to genome size. Evolution 49, 1249–1259 10.2307/2410449 (doi:10.2307/2410449) [DOI] [PubMed] [Google Scholar]
- 66.Starostova Z., Kratochvil L., Frynta D. 2005. Dwarf and giant geckos from the cellular perspective: the bigger the animal, the bigger its erythrocytes? Funct. Ecol. 19, 744–749 10.1111/j.1365-2435.2005.01020.x (doi:10.1111/j.1365-2435.2005.01020.x) [DOI] [Google Scholar]
- 67.Vinogradov A., Anatskaya O., Kudryavtsev B. 2001. Relationship of hepatocyte ploidy levels with body size and growth rate in mammals. Genome 44, 350–360 10.1139/g01-015 (doi:10.1139/g01-015) [DOI] [PubMed] [Google Scholar]
- 68.Gregory T. 2005. The evolution of the genome. San Diego, CA: Elsevier [Google Scholar]
- 69.Nagl W. 1974. Mitotic cycle time in perennial and annual plants with various amounts of DNA and heterochromatin. Dev. Biol. 39, 342–346 [DOI] [PubMed] [Google Scholar]