Abstract
Statistical models are helping palaeontologists to elucidate the history of biodiversity. Sampling standardization has been extensively applied to remedy the effects of uneven sampling in large datasets of fossil invertebrates. However, many vertebrate datasets are smaller, and the issue of uneven sampling has commonly been ignored, or approached using pairwise comparisons with a numerical proxy for sampling effort. Although most authors find a strong correlation between palaeodiversity and sampling proxies, weak correlation is recorded in some datasets. This has led several authors to conclude that uneven sampling does not influence our view of vertebrate macroevolution. We demonstrate that multi-variate regression models incorporating a model of underlying biological diversification, as well as a sampling proxy, fit observed sauropodomorph dinosaur palaeodiversity best. This bivariate model is a better fit than separate univariate models, and illustrates that observed palaeodiversity is a composite pattern, representing a biological signal overprinted by variation in sampling effort. Multi-variate models and other approaches that consider sampling as an essential component of palaeodiversity are central to gaining a more complete understanding of deep time vertebrate diversification.
Keywords: multi-variate models, palaeodiversity, sauropodomorpha
1. Introduction
Understanding how biodiversity has fluctuated over extended intervals of deep time, and how it responds to extinction events, is central to understanding the significance of modern biodiversity loss and responses to climate change. Fossils provide the only data on these issues, and attempts to understand the diversification of life using fossil data have a long pedigree (e.g. [1–3]). Initial taxonomic compilations (e.g. [4–6]) formed the basis of ‘palaeodiversity curves’, which were commonly interpreted as a literal reading of ancient biodiversity [2,3]. However, it is possible that much of the variation in observed ‘global’ palaeodiversity is driven by temporal and spatial variation in the amount of available fossiliferous rock, or by disparities in collection effort by palaeontologists (e.g. [7,8]). To remedy this, recent palaeodiversity databases, exemplified by The Palaeobiology Database (PBDB; http://www.paleodb.org/), incorporate subsampling mechanisms that allow for the simulation of even fossil sampling through time. This has resulted in substantial revisions to palaeodiversity curves, and to our understanding of the history of life on Earth [9,10].
Large-scale patterns across the entire Phanerozoic (542 million years ago (Mya)–present) have been the focus of palaeodiversity research (e.g. [2,3,9,10]). The Phanerozoic record primarily comprises shallow marine invertebrates inhabiting the submerged continental shelves, where most rock deposition occurs. Terrestrial and open ocean palaeodiversity have received comparatively little attention, and studies of vertebrate palaeodiversity were relatively rare until recently. Vertebrate fossils are less abundant. For some taxonomic groups, facies and time intervals, many collecting localities have yielded only a single specimen or taxon, instead of a more complete faunal sample (PBDB, accessed on 27 May 2011). These factors restrict the utility of subsampling approaches. Thus, many vertebrate palaeodiversity studies have relied on modelling approaches to ‘correct’ data for uneven sampling (e.g. [11–15]). However, perspectives on the importance of sampling to our understanding of vertebrate macroevolution are polarized. Some workers have stated that macroevolutionary patterns contain a strong biological signal, and can be interpreted at face value [16–18], whereas others suggest that these signals are distorted by sampling biases, and emphasize caution (e.g. [12–14,19,20]).
Because of this lack of consensus, some high-profile macroevolutionary studies of vertebrates recently have ignored sampling biases altogether [18,21]. This runs counter to scientific intuition, and the fact that several studies detected a significant correlation between observed vertebrate palaeodiversity and proxies representing sampling effort (e.g. [12–14,20]). Several authors have argued that the apparent absence of this correlation for some datasets indicates that vertebrate palaeodiversity signals are not influenced by sampling bias [17,18,22]. However, this may not be correct: only if biodiversity was constant through time would we expect to see a perfect correlation between sampling and observed palaeodiversity. In cases where underlying biodiversity exhibits high levels of variation, this correlation should become weaker. However, this would not be a special case of an ‘unbiased’ fossil record, because the strong, ‘genuine’ biodiversity signal should still be overprinted by variation in sampling effort.
We illustrate this principle using multi-variate regression models for sauropodomorph dinosaur palaeodiversity. These show that including even a simple numerical representation of biological diversification in a multi-variate model can improve the fit of a sampling proxy beyond that obtained by a univariate, sampling-only, model. Furthermore, some representations of biodiversity, though justified by palaeontological observations, may show poor fit to observed palaeodiversity unless sampling is explicitly considered by following a multi-variate approach.
2. Material and methods
Sauropodomorpha is a dinosaurian clade of primarily long-necked, large-bodied, herbivorous taxa. It includes more than 200 known taxa (e.g. Diplodocus, Brachiosaurus), first appearing in the Late Triassic (228 Mya) and surviving until the end of the Cretaceous (65.5 Mya) [14,23]. Sauropodomorphs have been the focus of several palaeodiversity studies [14,24]. Compared with other dinosaurian clades, observed sauropodomorph palaeodiversity shows only a weak correlation with sampling proxies [12]. The significance of this is disputed; it may indicate that sampling does not influence sauropodomorph palaeodiversity (e.g. [18], see the electronic supplementary material) or that sauropodomorph biodiversity showed genuinely greater fluctuations, obscuring the relationship between sampling and palaeodiversity in univariate comparisons [14]. Notably, sauropodomorphs suffered a substantial extinction event at the end of the Jurassic (approx. 145 Mya), resulting in the disappearance of broad-toothed forms and their replacement by ornithischians in Cretaceous ecosystems [14,24,25]. If this is correct then sauropodomorph biodiversity may be approximated by a simple model (‘TJK’ model) in which Triassic–Jurassic (TJ) biodiversity is assigned one value (‘0’) and Cretaceous (K) biodiversity is assigned a different value (‘1’).
To test between interpretations of sauropodomorph palaeodiversity, we analysed a comprehensive species-level dataset of Late Triassic–Cretaceous sauropodomorphs [14,23] parsed into 26 stage-level time bins (figure 1a; electronic supplementary material, appendix S1). We compared this with our simple model of sauropodomorph biodiversity, and to two sampling proxies (counts of geological formations (dinosaur-bearing formations, DBFs) and collections (dinosaur-bearing collections, DBCs) yielding dinosaur body fossils; figure 1a) [14,23], using generalized least-squares regression. This approach incorporates an autoregressive model to account for potential non-independence of successive data points in a time series. Variables were log-e transformed prior to analysis. Residuals from the regression models were normally distributed and homoskedastic. Three types of regression model were compared: univariate models comprising (i) a sampling proxy, (ii) our simple biodiversity model, and (iii) a bivariate model comprising a sampling proxy and our simple biodiversity model. All analyses were implemented in R v. 2.10.1 [26], following the approach of Hunt et al. [27] and Marx & Uhen [28] (electronic supplementary material, appendix S2).
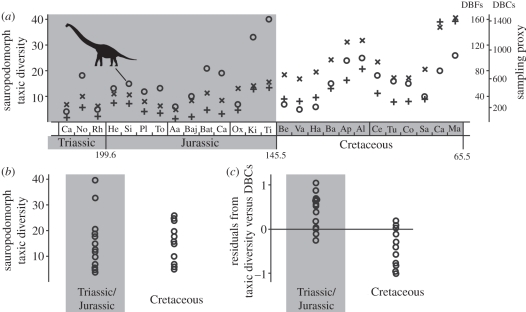
Figure 1.
(a) Sauropodomorph taxic diversity (circles) and counts of dinosaur-bearing collections (DBCs, plus symbols) and formations (DBFs, crosses) through 26 Mesozoic stages (names abbreviated), shading indicates division into two phases of standing diversity in the TJK model; (b) clumped taxic diversity data displayed for Triassic–Jurassic and Cretaceous stages showing no clear difference in observed palaeodiversity despite documented end-Jurassic decimation of sauropodomorph ecotypes (t-test statistics: t = −0.044, d.f. = 23.269, p = 0.9652); (c) residual taxic after regression against DBCs shows a clear difference in diversity between the Triassic–Jurassic and Cretaceous intervals (t = 4.933, d.f. = 22.501, p > = 0.0001).
3. Results
All models including a sampling proxy fit significantly better than the null model in which palaeodiversity is constant and variation is subsumed by an error term. However, substantially higher R2 (proportion of variance explained) and lower Akaike information criterion (AICc) scores [29,30] indicate that the bivariate model is the best explanation of sauropodomorph palaeodiversity (table 1). The univariate model including only our simple biodiversity model is a poorer fit than the null model (figure 1b and table 1), indicating that it is significantly different from the impression of sauropodomorph diversity obtained by inspecting ‘raw’ palaeodiversity data. Importantly, both sampling proxies show a greater t-value and stronger statistical significance within the bivariate models than they do in the univariate models (table 1). Thus, including an estimate of sauropodomorph biodiversity in the regression model actually improves the fit of the sampling proxy. This is not consistent with the suggestion that sauropodomorph palaeodiversity is independent of sampling bias [18]. These results are independent of the effects of the variable duration of geological stages (electronic supplementary material, appendix S2).
Table 1.
Summary of regression model fits to observed palaeodiversity. ‘TJK’ is the simple biodiversity model; ‘DBC’ is dinosaur-bearing collections; ‘DBF’ is dinosaur-bearing formations. N = 26 stage-level time bins. Rows represent combinations of explanatory variables (‘models’, listed on the left), columns represent the coefficients of variables within the models (columns 2–7) and model fit parameters (columns 8–12). AICc is the version of the Akaike information criterion for small sample sizes [29]; R2 is the generalized coefficient of determination ([30]; negative values are incurred when the model explains less variance than the null model).
| sampling (DBC or DBF) |
biodiversity model (TJK) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| slope | t-value | p-value | slope | t-value | p-value | R2 | log-likelihood | AICc | AICc (weight) | |
| null model | — | — | — | — | — | — | — | −25.2 | 54.8 | <0.001 |
| TJK | — | — | — | 0.08 | 0.34 | 0.7360 | −0.033 | −25.6 | 58.2 | <0.001 |
| DBC | 0.761 | 5.21 | >0.0001 | — | — | — | 0.422 | −18.0 | 45.2 | 0.029 |
| DBC + TJK | 0.860 | 6.91 | >0.0001 | −1.07 | −3.54 | 0.0018 | 0.603 | −13.2 | 38.2 | 0.927 |
| DBF | 0.488 | 2.52 | 0.0186 | — | — | — | 0.156 | −22.9 | 53.0 | <0.001 |
| DBF + TJK | 1.370 | 4.97 | 0.0001 | −1.22 | −3.85 | 0.0008 | 0.457 | −17.2 | 44.3 | 0.044 |
4. Discussion
Our results illustrate a general principle relevant to all palaeodiversity studies. Poor fit of univariate sampling models (e.g. pairwise statistical tests of correlation) may not always result from an ‘unbiased’ palaeodiversity curve. Instead, all palaeodiversity curves represent a composite signal, comprising both sampling and genuine biological diversity. Our sauropodomorph biodiversity model is coarse, postulating two phases of standing biodiversity, a Late Triassic–Jurassic interval of relatively high diversity followed by a Cretaceous interval of lower diversity. Nonetheless, when considered alongside uneven sampling, this model fits the observed palaeodiversity data extremely well. In principle, more complex biodiversity models could be applied. For example, logistic dynamics, which incorporate an initial phase of increasing diversity followed by the attainment of ‘carrying capacity’, describe sampling-standardized palaeodiversity curves for most invertebrate clades [10,31].
Multi-variate models (e.g. [32,33]), and other approaches, which isolate biological and sampling signals (e.g. subsampling [9,10,14,19,31] and the ‘residuals’ method of Smith & McGowan [11–15]), are key to understanding palaeodiversity. This knowledge is central to correctly interpreting macroevolutionary patterns, and we strongly urge that all palaeodiversity studies employ appropriate methods to account for sampling biases to elucidate the biological significance of the fossil record.
References
- 1.Phillips J. 1860. Life on the Earth: its origin and succession. Cambridge, UK: Macmillan [Google Scholar]
- 2.Sepkoski J. J., Bambach R. K., Raup D. M., Valentine J. W. 1981. Phanerozoic marine diversity and the fossil record. Nature 293, 435–437 10.1038/293435a0 (doi:10.1038/293435a0) [DOI] [Google Scholar]
- 3.Benton M. J. 1995. Diversification and extinction in the history of life. Science 268, 52–58 10.1126/science.7701342 (doi:10.1126/science.7701342) [DOI] [PubMed] [Google Scholar]
- 4.Sepkoski J. J. 1982. A compendium of fossil marine families. Milwaukee Public Mus. Contrib. Biol. Geol. 51, 1–125 [PubMed] [Google Scholar]
- 5.Sepkoski J. J. 2002. A compendium of fossil marine animal genera. Bull. Am. Paleontol. 363, 1–560 [Google Scholar]
- 6.Benton M. J. 1993. (ed.) The fossil record 2. London, UK: Chapman & Hall [Google Scholar]
- 7.Raup D. M. 1972. Taxonomic diversity during the Phanerozoic. Science 177, 1065–1071 10.1126/science.177.4054.1065 (doi:10.1126/science.177.4054.1065) [DOI] [PubMed] [Google Scholar]
- 8.Smith A. B. 2001. Large-scale heterogeneity of the fossil record: implications for Phanerozoic biodiversity studies. Phil. Trans. R. Soc. Lond. B 356, 351–367 10.1098/rstb.2000.0768 (doi:10.1098/rstb.2000.0768) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alroy J., et al. 2008. Phanerozoic trends in the global diversity of marine invertebrates. Science 321, 97–100 10.1126/science.1156963 (doi:10.1126/science.1156963) [DOI] [PubMed] [Google Scholar]
- 10.Alroy J. 2010. The shifting balance of diversity among major marine animal groups.. Science 329, 1191–1194 10.1126/science.1189910 (doi:10.1126/science.1189910) [DOI] [PubMed] [Google Scholar]
- 11.Smith A. B., McGowan A. J. 2007. The shape of the Phanerozoic marine palaeodiversity curve: how much can be predicted from the sedimentary rock record of western Europe? Palaeontology 50, 765–774 10.1111/j.1475-4983.2007.00693.x (doi:10.1111/j.1475-4983.2007.00693.x) [DOI] [Google Scholar]
- 12.Barrett P. M., McGowan A. J., Page V. 2009. Dinosaur diversity and the rock record. Proc. R. Soc. B 276, 2667–2674 10.1098/rspb.2009.0352 (doi:10.1098/rspb.2009.0352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson R. J. B., Butler R. J., Lindgren J., Smith A. S. 2010. Mesozoic marine tetrapod diversity: mass extinctions and temporal heterogeneity in geological megabiases affecting vertebrates. Proc. R. Soc. B 277, 829–834 10.1098/rspb.2009.1845 (doi:10.1098/rspb.2009.1845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mannion P. D., Upchurch P., Carrano M. T., Barrett P. M. 2011. Testing the effect of the rock record on diversity: a multidisciplinary approach to elucidating the generic richness of sauropodomorph dinosaurs through time. Biol. Rev. 86, 157–181 10.1111/j.1469-185X.2010.00139.x (doi:10.1111/j.1469-185X.2010.00139.x) [DOI] [PubMed] [Google Scholar]
- 15.Lloyd G. T. 2012. A refined modelling approach to assess the influence of sampling on palaeobiodiversity curves: new support for declining Cretaceous dinosaur richness. Biol. Lett. 8, 123–126 (doi:10.1098/rsbl.2011.0210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benton M. J., Emerson B. C. 2007. How did life become so diverse? The dynamics of diversification according to the fossil record and molecular phylogenetics. Palaeontology 50, 23–40 10.1111/j.1475-4983.2006.00612.x (doi:10.1111/j.1475-4983.2006.00612.x) [DOI] [Google Scholar]
- 17.Marx F. G. 2009. Marine mammals through time: when less is more in studying palaeodiversity. Proc. R. Soc. B 276, 887–892 10.1098/rspb.2008.1473 (doi:10.1098/rspb.2008.1473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahney S., Benton M. J., Ferry P. A. 2010. Links between global taxonomic diversity, ecological diversity and the expansion of vertebrates on land. Biol. Lett. 6, 540–543 10.1098/rsbl.2009.1024 (doi:10.1098/rsbl.2009.1024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alroy J. 2000. New methods for quantifying macroevolutionary patterns and processes. Paleobiology 26, 707–733 (doi:10.1666/0094-8373(2000)026<0707:NMFQMP>2.0.CO;2) [DOI] [Google Scholar]
- 20.Fröbisch J. 2008. Global taxonomic diversity of anomodonts (Tetrapoda, Therapsida) and the terrestrial rock record across the Permian–Triassic boundary. PLoS ONE 3, e3733. 10.1371/journal.pone.0003733 (doi:10.1371/journal.pone.0003733) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahney S., Benton M. J., Falcon Lang H. J. 2010. Rainforest collapse triggered Pennsylvanian tetrapod diversification. Geology 38, 1079–1082 10.1130/G31182.1 (doi:10.1130/G31182.1) [DOI] [Google Scholar]
- 22.Uhen M. D., Pyenson N. D. 2007. Diversity estimates, biases, and historiographic effects: resolving cetacean diversity in the Tertiary. Palaeo. Electron. 10, 11A [Google Scholar]
- 23.Butler R. J., Benson R. B. J., Carrano M. T., Mannion P. D., Upchurch P. 2011. Sea-level, dinosaur diversity, and sampling biases: investigating the ‘common cause’ hypothesis in the terrestrial realm. Proc. R. Soc. B 278, 1165–1170 10.1098/rspb.2010.1754 (doi:10.1098/rspb.2010.1754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Upchurch P., Barrett P. M. 2005. A phylogenetic perspective on sauropod diversity. In The Sauropods: evolution and paleobiology (eds Curry-Rogers K. A., Wilson J. A.), pp. 104–124 Berkeley, CA: University of California Press [Google Scholar]
- 25.Barrett P. M., Upchurch P. 2005. Sauropod diversity through time: possible macroevolutionary and palaeoecological implications. In The Sauropods: evolution and paleobiology (eds Curry-Rogers K. A., Wilson J. A.), pp. 125–156 Berkeley, CA: University of California Press [Google Scholar]
- 26.R Development Core Team 2010. R: a language and environment for statistical computing. See http://www.R-project.org
- 27.Hunt G., Cronin T. M., Roy K. 2005. Species-energy relationships in the deep sea: a test using the Quaternary fossil record. Ecol. Lett. 8, 739–747 10.1111/j.1461-0248.2005.00778.x (doi:10.1111/j.1461-0248.2005.00778.x) [DOI] [Google Scholar]
- 28.Marx F. G., Uhen M. D. 2010. Climate, critters, and cetaceans: Cenozoic drivers of the evolution of modern whales. Science 327, 993–996 10.1126/science.1185581 (doi:10.1126/science.1185581) [DOI] [PubMed] [Google Scholar]
- 29.Sugiura N. 1978. Further analysis of the data by Akaike's Information Criterion and the Finite Corrections. Commun. Stat. Theory Methods A 7, 13–26 10.1080/03610927808827599 (doi:10.1080/03610927808827599) [DOI] [Google Scholar]
- 30.Nagelkerke N. J. D. 1991. A note on a general definition of the coefficient of determination. Biometrika 78, 691–692 10.1093/biomet/78.3.691 (doi:10.1093/biomet/78.3.691) [DOI] [Google Scholar]
- 31.Alroy J. 2010. Geographical, environmental and intrinsic biotic controls on Phanerozoic marine diversification. Palaeontology 53, 1211–1235 10.1111/j.1475-4983.2010.01011.x (doi:10.1111/j.1475-4983.2010.01011.x) [DOI] [Google Scholar]
- 32.Jablonski D. 2008. Extinction and the spatial dynamics of biodiversity. Proc. Natl Acad. Sci. USA 105, 11 528–11 535 10.1073/pnas.0801919105 (doi:10.1073/pnas.0801919105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedman M. 2009. Ecomorphological selectivity among marine teleost fishes during the end-Cretaceous extinction. Proc. Natl Acad. Sci. USA 106, 5218–5223 10.1073/pnas.0808468106 (doi:10.1073/pnas.0808468106) [DOI] [PMC free article] [PubMed] [Google Scholar]