Abstract
The distribution of species among genera and higher taxa has largely untapped potential to reveal among-clade variation in rates of origination and extinction. The probability distribution of the number of species within a genus is modelled with a stochastic, time-homogeneous birth–death model having two parameters: the rate of species extinction, μ, and the rate of genus origination, γ, each scaled as a multiple of the rate of within-genus speciation, λ. The distribution is more sensitive to γ than to μ, although μ affects the size of the largest genera. The species : genus ratio depends strongly on both γ and μ, and so is not a good diagnostic of evolutionary dynamics. The proportion of monotypic genera, however, depends mainly on γ, and so may provide an index of the genus origination rate. Application to living marine molluscs of New Zealand shows that bivalves have a higher relative rate of genus origination than gastropods. This is supported by the analysis of palaeontological data. This concordance suggests that analysis of living taxonomic distributions may allow inference of macroevolutionary dynamics even without a fossil record.
Keywords: evolutionary rates, hollow curve, mathematical modelling, New Zealand, Mollusca
1. Introduction
Documenting rates of lineage origination and extinction is central in understanding macroevolutionary dynamics. Biologists have focused attention on differences in diversification between clades [1] and on temporal changes in diversification within clades [2]. Taxonomic structure—how species within a clade are distributed among genera or higher taxonomic units—has not been as widely studied [3,4], yet it has real potential to reveal the dynamics of diversification [3]. Here, I will outline one way to infer rates of species extinction and genus origination, based on the proportion of living genera with one species, two species, etc., i.e. the size–frequency distribution of genera. This approach provides a complement to palaeontological analyses and phylogenetic methods.
2. Material and methods
(a). Evolutionary model
The model is a continuous time-variant of the discrete time, hierarchical branching model of Patzkowsky [5] and is similar to that of Reed & Hughes [6] except in the treatment of genus origination (electronic supplementary material). Let μ be the per capita extinction rate per lineage million years (Lmy), λ be the within-genus speciation rate per Lmy and γ be the rate of origination of new genera per Lmy. Thus, a proportion λ/(λ + γ) of speciation events found new species within existing genera, and a proportion γ/(λ + γ) found new, monotypic genera. Because the diversification rate within the entire clade equals (λ + γ − μ), a clade may diversify even if μ > λ (see bivalve example below). I will assume that rates are time-homogeneous and uniform within a clade, although these assumptions can be relaxed (electronic supplementary material).
Both speciation and genus origination are assumed to be cladogenetic, although associated anagenesis is generally required to recognize new species and genera. The events that found new genera could be larger evolutionary steps, the accumulated change from a series of smaller steps (in which case genus origination would presumably take longer than speciation) or even arbitrary taxonomic decisions. This will affect the interpretation of parameters, but not the resulting size–frequency distribution, and differences between clades will be of interest regardless of how the rate of genus origination is interpreted.
(b). Living molluscs of New Zealand
Data were taken from the New Zealand Inventory of Biodiversity [7]. I elevated subgenera to genus rank, and I omitted unpublished species, species questionably assigned to genera, species prefixed with ‘aff.’ or ‘cf.’ and introduced species. Collectively, the data consist of 454 bivalve species in 237 genera, and 1780 gastropod species in 713 genera (electronic supplementary material, table S1) giving species: genus ratios of 1.9 and 2.5, respectively.
The growth of molecular phylogenetics has led to intensified efforts to infer evolutionary rates and other aspects of macroevolution from living taxa alone [2,8–10]. As a complement to these approaches, it is worth exploring whether evolutionary rates can be inferred from the size–frequency distribution of living genera [3].
If fn is the probability that a genus has n species, given (λ,γ,μ) (electronic supplementary material, equation S3), and Nn is the number of observed genera with n species, then the support (log likelihood) is given by
Support was maximized via the optim() function in R [11], using option ‘L-BFGS-B’.
(c). Cenozoic fossil molluscs of New Zealand
I used a compilation of times of first and last appearances of species, the synoptic dataset [12]. This has the advantages of being comprehensive in scope and largely standardized taxonomically. I vetted the data following the protocols of Crampton et al. [12].
I tabulated the number of within-genus speciation events as the difference between the total number of new species and the total number of new genera. This allows the ratios between parameters to be estimated as
and
where NF and NL are the number of first and last appearances, the subscripts s and g refer to species- and genus-level data and the sum is taken over all time intervals.
3. Results
(a). Modelling
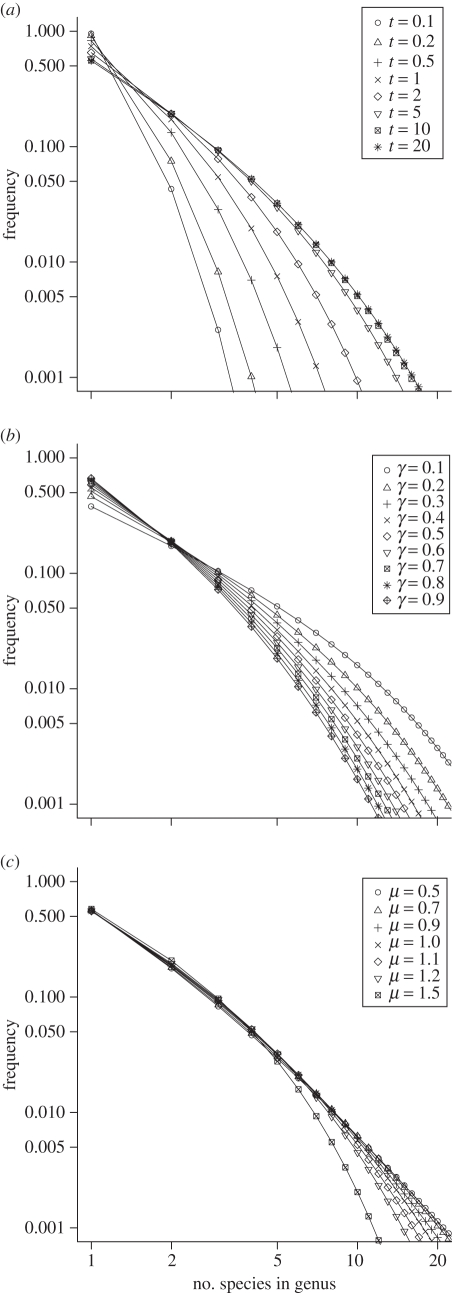
Figure 1a shows the effects of increasing the elapsed time with evolutionary parameters held fixed. The size–frequency distribution stabilizes [5,6], barely changing after about 10 times the average species duration (i.e. after t = 10/μ). I will therefore assume an asymptotic distribution, which allows the evolutionary process to be based on two parameters, the ratios γ* = γ/λ and μ* = μ/λ.
Figure 1.
Modelled genus size–frequency distributions. (a) Variation in elapsed time with parameters held fixed at λ = 1, μ = 1.1 and γ = 0.4. (b) Variation in γ* with μ* = 1.1 and assuming the asymptotic distribution (in which elapsed time is effectively infinite and μ* and γ* are expressed in terms of multiples of λ). Convergence of curves at a genus size of two species is a consequence of these particular parameters and is not a general feature of the model. (c) Variation in μ* with γ* = 0.4 and assuming the asymptotic distribution.
When γ* increases, new, monotypic genera are produced at a higher rate, so the size–frequency distribution becomes progressively dominated by smaller genera (figure 1b). Varying extinction rate has a smaller effect on the distribution (figure 1c), but it is important in determining the size of the few largest genera. These results suggest that better empirical constraints may be expected on γ* than on μ*. Perhaps the most commonly used metric of taxonomic structure is the species : genus ratio, but this depends significantly on both γ* and μ* (electronic supplementary material). The proportion of monotypic genera, however, is primarily sensitive to γ* and so may be a reasonable index of the relative genus origination rate, for example, when data are too sparse for parameter estimation.
(b). Living molluscs
The size–frequency distributions for New Zealand molluscs are depicted in figure 2a, along with the best-fitting curves. The best-fitting parameters are depicted in figure 2b,c. Consistent with expectations developed from figure 1b,c, the spacing of contours in figure 2 shows that support in the neighbourhood of the parameter estimates changes more rapidly with respect to γ* than μ*.
Figure 2.
Analysis of size–frequency distributions of living New Zealand molluscs. (a) Observed distributions (points) with best model fits (solid line with filled squares, bivalves; dashed line open circles, gastropods). (b) Support contours for bivalves, with best-fitting parameter pair shown by the point. (c) The same for gastropods. Data from Spencer et al. [7].
A greater proportion of speciation events in bivalves found new genera than is the case for gastropods (γ* = 0.53 versus γ* = 0.40). Bivalves also have a higher relative extinction rate (μ* = 1.29 versus μ* = 0.90) but this may not be a robust result (see below).
Table 1 compares a model in which bivalves and gastropods are governed by different rates to one in which they have the same rates. The mean rate estimates, weighted by the number of genera in each group, are also presented; these are within 10 per cent of the estimates for the combined data. The two-group model is substantially better supported, with an Akaike weight [13] of 93 per cent, suggesting that it is reasonable to treat bivalves and gastropods as having distinct taxonomic structures.
Table 1.
Parameter estimates and model comparison for living marine molluscs of New Zealand. AICc is the sample size-corrected Akaike Information Criterion [13]; w is the Akaike weight [13]. Sample sizes (number of genera) are 237 for bivalves and 713 for gastropods (electronic supplementary material, table S1).
| model | bivalves |
gastropods |
combined |
support | AICc | w | |||
|---|---|---|---|---|---|---|---|---|---|
| γ* | μ* | γ* | μ* | γ* | μ* | ||||
| two groups | 0.53 | 1.29 | 0.40 | 0.90 | — | — | −1418.2 | 2844.4 | 0.93 |
| one group | — | — | — | — | 0.45 | 0.92 | −1422.8 | 2849.6 | 0.07 |
| weighted mean | — | — | — | — | 0.43 | 1.00 | — | — | |
(c). Fossil molluscs
The number of first and last appearances in the synoptic data (table 2) imply relative genus origination rates of γ* = 0.54 ± 0.017 for bivalves and γ* = 0.38 ± 0.01 for gastropods. The relative extinction rates are μ* = 1.13 ± 0.028 for bivalves and μ* = 1.12 ± 0.014 for gastropods. The maximum-likelihood values of γ* estimated from the living size–frequency distribution are consistent with the fossil estimates. Because bivalves are better sampled and improved sampling, all else being equal, decreases the apparent value of γ*, the difference is unlikely to be a sampling artefact (electronic supplementary material). In contrast to the living species, the fossil data suggest no appreciable difference in relative extinction rate.
Table 2.
Summary statistics on molluscan species and genera from the Cenozoic of New Zealand. Rate estimates ±1 s.e. based on bootstrap resampling of stratigraphic ranges.
| higher taxon | NFs | NLs | NFg | γ* | μ* |
|---|---|---|---|---|---|
| Bivalvia | 589 | 434 | 206 | 0.54 ± 0.017 | 1.13 ± 0.028 |
| Gastropoda | 1502 | 1087 | 415 | 0.38 ± 0.010 | 1.12 ± 0.014 |
4. Discussion
The model herein makes no explicit assumptions regarding taxonomic practice. Implicitly, however, the distribution of species among genera is assumed not to depend on the size of the clade as a whole. The modelling could be made more realistic, but potentially unwieldy, by overlaying the evolutionary results with a model of taxonomic practice. Another promising direction would be to determine empirically how taxonomic structure changes as new systematic approaches are adopted.
The analysis of living molluscs tacitly assumed that differences in sampling between bivalves and gastropods do not dominate the difference in taxonomic structure. Incomplete sampling and how it might be circumvented are considered in the electronic supplementary material, where it is suggested that reconstructing size–frequency distributions is possible if species sampling probabilities can be estimated. The question of how taxonomic practice changes as sampling of species improves deserves further attention.
I have treated New Zealand as a closed system. This simplification is not entirely unwarranted, given the high degree of species endemism [7,12]; 80 per cent of bivalve species and 77 per cent of gastropod species used herein are endemic, although the figures for genera are only 16 and 15 per cent. In addition to high species endemism, there are at least two reasons to think that the bivalve–gastropod difference is not simply owing to migration. First, the rates of endemism do not differ appreciably between the two groups. Second, the difference in taxonomic structure is also present within the strictly endemic genera. Although the data are too sparse to estimate γ* and μ*, the endemic bivalve genera have a higher proportion of monotypics than do endemic gastropods (61 versus 48 per cent) and a lower species : genus ratio (1.6 versus 2.7).
An obvious caveat is that there is uncertainty as to what the origin of a genus means. At one extreme, it could represent a larger evolutionary transition than a within-genus speciation event. At the other extreme, it could represent a mere result of taxonomic practice. Either way, the results raise a compelling question. Why do gastropods produce more species that are minor variations on their congeners? Or why are students of bivalves quicker to erect new genera for newly described species? Differences in maturity of practice or intensity of effort do not seem a likely explanation, as there is little difference in the year of publication of bivalve and gastropod species, and as similar genus origination rates are obtained with an older compendium of New Zealand Mollusca (electronic supplementary material).
If we accept that the rate of genus origination is biologically meaningful, it is encouraging that the difference between bivalves and gastropods inferred from living species agrees with that inferred from the fossil record. Such an agreement should be tested with other groups that have a rich fossil record and comprehensive taxonomic treatment, for example, planktonic foraminifera [14] and mammals [15,16]. If it is consistently supported, this raises the possibility of inferring from living species alone a parameter that should be of general evolutionary interest, namely, the rate of origin of supraspecific taxa [17]. Differences in taxonomic structure have also been documented among biogeographic provinces [3,4], and the approach taken here could help to understand the evolutionary basis for those differences. Interprovincial differences will be complicated by immigration and emigration, however, and these processes would initially have to be subsumed within the origination and extinction parameters for the sake of analysis, with the distinction between speciation and immigration and between extinction and emigration constrained independently. The methods used here could also be helpful in determining whether new genera preferentially originate in certain environments or biogeographic areas [18,19] simply because there are more species to serve as a source for new genera, or because those species also have a higher per capita rate of genus production (γ).
Acknowledgements
For advice and discussion, I thank D. S. Abbot, A. G. Beu, J. S. Crampton, L. J. Harmon, P. G. Harnik, G. Hunt, A. Z. Krug and B. A. Marshall. I am grateful to A. G. Beu for providing the synoptic data and to J. S. Crampton for providing data from Spencer et al. [7]. P. G. Harnik and A. I. Miller made helpful suggestions on the manuscript. G. Hunt, A. Phillimore, A. Purvis and another referee provided thoughtful reviews. Supported by NASA Exobiology (NNX10AQ44G).
References
- 1.Magallón S., Sanderson M. J. 2001. Absolute diversification rates in angiosperm clades. Evolution 55, 1762–1780 10.1111/j.0014-3820.2001.tb00826.x (doi:10.1111/j.0014-3820.2001.tb00826.x) [DOI] [PubMed] [Google Scholar]
- 2.Rabosky D. L. 2010. Primary controls on species richness in higher taxa. Syst. Biol. 59, 634–645 10.1093/sysbio/syq060 (doi:10.1093/sysbio/syq060) [DOI] [PubMed] [Google Scholar]
- 3.Harnik P. G., Jablonski D., Krug A. Z., Valentine J. W. 2010. Genus age, provincial area and the taxonomic structure of marine faunas. Proc. R. Soc. B 277, 3427–3435 10.1098/rspb.2010.0628 (doi:10.1098/rspb.2010.0628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krug A. Z., Jablonski D., Valentine J. W. 2008. Species-genus ratios reflect a global history of diversification and range expansion in marine bivalves. Proc. R. Soc. B 275, 1117–1123 10.1098/rspb.2007.1729 (doi:10.1098/rspb.2007.1729) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patzkowsky M. E. 1995. A hierarchical branching model of evolutionary radiations. Paleobiology 21, 440–460 [Google Scholar]
- 6.Reed W. J., Hughes B. D. 2002. On the size distribution of live genera. J. Theor. Biol. 217, 125–135 10.1006/yjtbi.3009 (doi:10.1006/yjtbi.3009) [DOI] [PubMed] [Google Scholar]
- 7.Spencer H. G., et al. 2009. Phylum Mollusca: chitons, clams, tusk shells, snails, squids, and kin. In New Zealand inventory of biodiversity, volume one. Kingdom Animalia: Radiata, Lophotrochozoa, Deuterostomia (ed. Gordon D. P.), pp. 161–209 Christchurch: Canterbury University Press [Google Scholar]
- 8.Hey J. 1992. Using phylogenetic trees to study speciation and extinction. Evolution 46, 627–640 10.2307/2409633 (doi:10.2307/2409633) [DOI] [PubMed] [Google Scholar]
- 9.Nee S., May R. M., Harvey P. H. 1994. The reconstructed evolutionary process. Phil. Trans. R. Soc. Lond. B 344, 305–311 10.1098/rstb.1994.0068 (doi:10.1098/rstb.1994.0068) [DOI] [PubMed] [Google Scholar]
- 10.Harmon L. J., et al. 2010. Early bursts of body size and shape evolution are rare in comparative data. Evolution 64, 2385–2396 10.1111/j.1558-5646.2010.01025.x (doi:10.1111/j.1558-5646.2010.01025.x) [DOI] [PubMed] [Google Scholar]
- 11.R Development Core Team 2010. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; See http://www.R-project.org/ [Google Scholar]
- 12.Crampton J. S., Foote M., Beu A. G., Maxwell P. A., Cooper R. A., Matcham I., Marshall B. A., Jones C. M. 2006. The ark was full! Constant to declining Cenozoic shallow marine biodiversity on an isolated midlatitude continent. Paleobiology 32, 509–532 10.1666/06014.1 (doi:10.1666/06014.1) [DOI] [Google Scholar]
- 13.Burnham K. P., Anderson D. R. 2002. Model selection and multimodel inference: a practical information-theoretic approach. New York, NY: Springer [Google Scholar]
- 14.Aze T., Ezard T. H. G., Purvis A., Coxall H. K., Steward D. R. M., Wade B. S., Pearson P. N. 2011. A phylogeny of Cenozoic macroperforate planktonic foraminifera from fossil data. Biol. Rev. 26 10.1111/j.1469-185X.2011.00178.x (doi:10.1111/j.1469-185X.2011.00178.x) [DOI] [PubMed] [Google Scholar]
- 15.Wilson D. E., Reeder D. M. (). 2005. Mammal species of the world: a taxonomic and geographic reference, 3rd edn. Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 16.Bininda-Emonds O. R. P., et al. 2007. The delayed rise of present-day mammals. Nature 446, 507–512 10.1038/nature05634 (doi:10.1038/nature05634) [DOI] [PubMed] [Google Scholar]
- 17.Valentine J. W. 1969. Patterns of taxonomic and ecological structure of the shelf benthos during Phanerozoic time. Palaeontology 12, 684–709 [Google Scholar]
- 18.Jablonski D., Roy K., Valentine J. W. 2006. Out of the tropics: evolutionary dynamics of the latitudinal diversity gradient. Science 314, 102–106 10.1126/science.1130880 (doi:10.1126/science.1130880) [DOI] [PubMed] [Google Scholar]
- 19.Kiessling W., Simpson C., Foote M. 2010. Reefs as cradles of evolution and sources of biodiversity in the Phanerozoic. Science 327, 196–198 10.1126/science.1182241 (doi:10.1126/science.1182241) [DOI] [PubMed] [Google Scholar]