Abstract
We have used a polymorphism dataset on introns and coding sequences of X-linked loci in Drosophila americana to estimate the strength of selection on codon usage and/or biased gene conversion (BGC), taking into account a recent population expansion detected by a maximum-likelihood method. Drosophila americana was previously thought to have a stable demographic history, so that this evidence for a recent population expansion means that previous estimates of selection need revision. There was evidence for natural selection or BGC favouring GC over AT variants in introns, which is stronger for GC-rich than GC-poor introns. By comparing introns and coding sequences, we found evidence for selection on codon usage bias, which is much stronger than the forces acting on GC versus AT basepairs in introns.
Keywords: Drosophila americana, codon usage, biased gene conversion, population expansion
1. Introduction
In bacteria, yeast, Drosophila and plants, there is evidence for selection on codon usage at synonymous coding sites, probably because of selection on translational efficiency and/or accuracy [1]. Several population genetic studies of Drosophila have used polymorphism data to estimate the intensity of selection on codon usage [2–7]. In addition, genome evolution is affected by the process of biased gene conversion (BGC), which tends to favour GC over AT basepairs in the meiotic products of GC/AT heterozygotes, and acts in a similar way to directional selection [8]. Its effects and strength can be inferred from polymorphism data on non-coding sequences [9,10].
Here, we present results on the nature and intensity of selection and/or BGC on non-coding and synonymous sites, using polymorphism data on X-linked loci of Drosophila americana, a close relative of Drosophila virilis. The virilis group diverged from the Drosophila melanogaster group about 62 Ma [11] and has somewhat different patterns of codon usage and base composition [12,13] making it of special interest for studies of these genomic features. Drosophila americana has been used in evolutionary genetic studies for several decades [14–18]. It has a well-defined ecology, independent of human activity [14], and might thus be expected to have a relatively stable demographic history, which is advantageous for estimating the parameters of natural selection from polymorphism data [3].
This paper presents, to our knowledge, the first analysis of a species in the virilis group to detect both selection on codon usage and BGC from polymorphism data, using a population genetic method that allows for a recent population size change [19], whereas a previous study of selection on codon usage assumed demographic equilibrium [3]. We provide evidence for a recent population expansion, and for selection on codon usage at synonymous sites, as well as selection or BGC favouring GC over AT in GC-rich introns.
2. Material and methods
For DNA extractions, we used males from 14 D. americana isofemale lines from the HI99 population on the south bank of the Missouri River (http://www.biology.uiowa.edu/mcallister/HI.html), provided by Bryant McAllister. About 85 per cent of genomes from this population have a fusion between the X and chromosome 4 [15,16]. Because genes located near the fusion region or in inversions may suffer from hitchhiking effects of the rearrangements, regions affected by the X/4 fusion or known segregating inversions were excluded.
Details of DNA extraction, amplification, sequencing and alignment of sequences are provided in the electronic supplementary material. The resulting dataset contains sequences for 32 introns sampled from 18 loci, including 12 short introns and 20 long introns (electronic supplementary material, figure S1). We also obtained the coding sequences of 15 X-linked genes, and retrieved four additional X-linked coding sequences from Maside & Charlesworth [17], in order to compare synonymous sites and introns. Sequences were deposited in GenBank (accession numbers JN246676–JN246926).
Using the codon preference table for D. virilis from Betancourt et al. [20], we assigned preferred (P) and unpreferred (U) alternatives to each synonymous site in both species, and then used parsimony to determine whether the synonymous site change within D. americana was P > P, U > U, P > U or U > P. Similarly, we obtained the counts and frequencies of AT > TA, GC > CG, GC > AT and AT > GC polymorphic changes for each intron in the D. americana intron dataset to test for selection or BGC favouring GC over AT basepairs [9,10].
We used the maximum-likelihood (ML) method of Zeng & Charlesworth [4], as modified by Haddrill et al. [6], for fitting the observed frequencies of variants to models of selection and demography, to estimate the strength of selection/BGC on U > P synonymous polymorphisms or GC > AT basepairs and the extent of mutational bias in favour of GC > AT versus GC > AT changes, allowing for the possibility of a recent population size change in D. americana. Details are given in the electronic supplementary material.
3. Results
Our major findings are presented below; other results are described in the electronic supplementary material. The mean values of various summary statistics are shown in table 1. The mean diversity and divergence values are broadly consistent with those reported previously, even after excluding the four coding sequences in common with Maside & Charlesworth [17]. There are no significant differences in mean Tajima's D values between the different classes of sites, or in variation and divergence values among intronic versus synonymous sites. The consistently negative Tajima's D values suggest a recent population expansion [21], as confirmed by the analysis below.
Table 1.
Summary statistics of polymorphism and divergence for the different classes of sites. (S is the number of segregating sites; π and θW are the standard measures of the nucleotide diversity based on the mean pairwise divergence per nucleotide site between alleles and the number of segregating sites, respectively; D is the number of fixed differences between D. virilis and D. americana; KJC is the mean Jukes–Cantor-corrected divergence from D. virilis; and DT is Tajima's D statistic.)
| category | S | π (%) (s.e.) | θW (%) (s.e.) | D | KJC (%) (s.e.) | DT (s.e.) |
|---|---|---|---|---|---|---|
| introns | 803 | 2.17 (0.23) | 2.32 (0.22) | 609 | 9.98 (0.92) | −0.75 (0.09) |
| synonymous | 173 | 1.96 (0.56) | 1.77 (0.32) | 245 | 9.84 (1.01) | −0.73 (0.13) |
| non-synonymous | 29 | 0.09 (0.03) | 0.11 (0.03) | 68 | 0.78 (0.22) | −0.93 (0.20) |
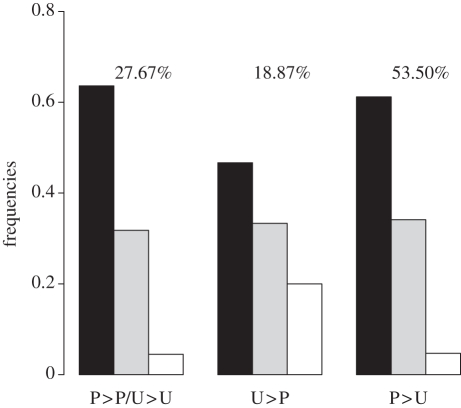
We first examined selection on variants affecting codon usage, using data on 19 X-linked coding sequences. There are four classes of mutations: P > P and U > U (expected to be selectively nearly neutral), P > U (potentially deleterious), and U > P mutations (potentially advantageous) [2,22]. Selection favouring P versus U variants is usually expected to yield an excess of P > U over U > P variants [2–4]. Consistent with this, we found nearly three times as many P > U variants as U > P variants (162 versus 56). In addition, P > U variants are disproportionately present at low frequencies compared with U > P variants (figure 1); the mean frequency of U > P mutations over the segregating sites in the sample was significantly higher than that of both P > U changes (Wilcoxon's W = 916.5, p = 0.022) and the pooled P > P and U > U changes (W = 856, p = 0.030).
Figure 1.
The frequency classes of polymorphisms for different types of synonymous site changes (see text for explanation of P and U). The numbers above each type of synonymous site change indicate the percentages of the total number of synonymous polymorphisms contributed by each type. Black bars, less than 0.2; grey bars, greater than or equal to 0.2 and less than or equal to 0.8; unfilled bars, greater than 0.8.
We also explored the possible effect of BGC on intronic base composition, which is expected to favour GC over AT variants [8]. The total numbers of GC > AT and AT > GC variants over the set of 32 introns are similar (248 versus 242), whereas the mean frequency of AT > GC variants is higher than that of GC > AT variants (0.28 versus 0.19) (W = 197.5, p = 0.002).
We also analysed these datasets by the method of Zeng & Charlesworth [4,6]. The ML estimates of mutational bias under all models examined indicate higher rates of mutations towards P > U and GC > AT variants compared with the reverse mutations, as found in previous Drosophila studies [4]. The contrasts between the model with no expansion, but with all other parameters fitted (L0), and the other models (L1) indicate a recent 4.2-fold increase in population size (table 2), with an ML estimate of the time since the event of τ = 0.11, where τ is the number of generations since the expansion divided by twice the current effective population size.
Table 2.
Estimates of the mutation, selection and demographic parameters for introns and synonymous sites. (Na and Nb are the effective population sizes after and before the population expansion; g = Na/Nb; τ is the time since the expansion (in units of 2Na generations); κ is the mutational bias; γ is the equivalent of the selection coefficient in favour of heterozygotes at a site, multiplied by 4Na. L0 is a model with selection on GC versus AT basepairs at intronic sites and P versus U codons at synonymous sites, but no population expansion. The full L1 model is the same as model L0 with population expansion; the other L1 models all have one parameter different from L1; the last L1 model has two estimates for γint—the smaller value is for introns with low GC content and the larger value is for introns with high GC content. The p-values correspond to the likelihood-ratio test of each alternative model against the full L1 model, and the ln L rank gives the rank of the log-likelihood among the eight models considered, where 1 indicates the most likely model.)
| model | g(Na/Nb) | τ(t/2Na) | γcod | κcod P > U | γint | κint GC > AT | ln L | p-value | ln L rank |
|---|---|---|---|---|---|---|---|---|---|
| L0 | — | — | 1.89 | 5.27 | 0.42 | 2.43 | −12770.70 | <0.0001 | 8 |
| L1 (γcod = 0) | 4.49 | 0.10 | 0 | 0.85 | 0.36 | 2.31 | −12749.21 | <0.0001 | 7 |
| L1 (κcod = 1) | 4.59 | 0.09 | 0.19 | 1 | 0.36 | 2.31 | −12 745.88 | <0.0001 | 6 |
| L1 (κint = 1) | 18.40 | 0.49 | 3.02 | 20.42 | −0.46 | 1 | −12743.26 | <0.0001 | 5 |
| L1 (γcod = γint) | 4.40 | 0.10 | 0.55 | 1.45 | 0.55 | 2.77 | −12741.58 | <0.0001 | 4 |
| L1 (γint = 0) | 4.25 | 0.11 | 1.55 | 3.87 | 0 | 1.65 | −12738.40 | 0.004 | 3 |
| full L1 | 4.21 | 0.11 | 1.56 | 3.87 | 0.36 | 2.31 | −12734.27 | — | 2 |
| L1 (γint low GC, γint high GC) | 4.20 | 0.11 | 1.55 | 3.87 | 0.27, 0.45 | 2.32 | −12724.84 | <0.0001 | 1 |
To test for selection on codon usage, we compared the full L1 model with the reduced version with γcod = 0, where γcod is the estimate of the strength of selection/BGC at a synonymous site, scaled by four times the effective population size before the expansion. The full model has strong statistical support (χ21 = 29.9, p < 0.0001), with γcod = 1.6, implying selection in favour of preferred codons, consistent with the patterns of P > U versus U > P variants described above. To test for selection/BGC on intronic variants, we compared the full L1 model with γint = 0 (χ12 = 8.27, p = 0.004). Selection or BGC in favour of GC intronic basepairs is thus implied, with γint = 0.36. We tested whether γcod is significantly larger than γint, by comparing a model with a single γ for both categories: the full L1 model is significantly more likely than that with γcod = γint (χ21 = 14.6, p < 0.0001). We similarly found that the γint estimates are significantly different for introns with high and low GC content (χ21 = 18.9, p < 0.0001).
4. Discussion
Our analysis provides evidence for a fairly large, recent increase in population size in D. americana, within a time-span of approximately 0.11 × 2Ne generations. This is consistent with the results for another widespread North American species, Drosophila pseudoobscura [6]. Given the mean silent site diversity values of about 2 per cent (table 1), using the standard formula for equilibrium neutral diversity (4Neμ) together with the D. melanogaster mutation rate estimate of 3.5 × 10−9 [23], we estimate that the current Ne of D. americana is about 1.4 million, implying that the expansion took place about 308 000 generations ago. Assuming five generations per year for this slowly breeding species [14], this corresponds to 61 600 years, although there is considerable uncertainty about the exact value.
The results in table 2 show that both synonymous sites and intron sequences in D. americana are influenced by selection and/or BGC, even after the recent population expansion was taken into account. The γ estimate of about 1.6 for selection favouring preferred over unpreferred codons is in line with values for other Drosophila species [6,19], but is lower than the value of 2.6 found previously in D. americana [3], suggesting that population expansion caused the strength of selection to be overestimated, as expected theoretically [4]. Consistent with other evidence from Drosophila for selection or BGC favouring GC over AT base pairs in non-coding sequences [10,19], we found evidence for natural selection or BGC favouring GC over AT basepairs. As in Haddrill & Charlesworth [10], selection/BGC appears to be significantly stronger in GC-rich compared with GC-poor introns, consistent with the idea that the intensity of BGC shapes the GC content of genomes [8].
As preferred codons are mostly GC-ending, selection for codon usage largely works in the same direction as BGC. The difference in γ between the synonymous sites and introns almost certainly reflects the action of selection on codon usage bias at synonymous sites, possibly in addition to the effects of BGC, whereas the apparent selection on intron sites may result from BGC alone [8]. This difference could also be owing to a higher rate of recombination in exons than in introns, resulting in a higher rate of BGC in exons [3], although we did not find any evidence for this (see electronic supplementary material).
Acknowledgements
This work formed part of the GENACT Project, funded by a Marie Curie Host Fellowship for Early Stage Training awarded to S.M.P., as part of the Framework 6 Programme of the European Commission. K.Z. was supported by a Biomedical Personal Research Fellowship, awarded by the Royal Society of Edinburgh and the Caledonian Research Foundation. A.J.B. was supported by a research grant from the Biotechnology and Biological Sciences Research Council. We thank Penelope Haddrill and three anonymous reviewers for helpful comments on the manuscript.
References
- 1.Hershberg R., Petrov D. A. 2008. Selection on codon bias. Annu. Rev. Genet. 42, 287–299 10.1146/Annurev.Genet.42.110807.091442 (doi:10.1146/Annurev.Genet.42.110807.091442) [DOI] [PubMed] [Google Scholar]
- 2.Akashi H. 1995. Inferring weak selection from patterns of polymorphism and divergence at silent sites in Drosophila DNA. Genetics 139, 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maside X. L., Lee A. W. S., Charlesworth B. 2004. Selection on codon usage in Drosophila americana. Curr. Biol. 14, 150–154 10.1016/J.Cub.2003.12.055 (doi:10.1016/J.Cub.2003.12.055) [DOI] [PubMed] [Google Scholar]
- 4.Zeng K., Charlesworth B. 2009. Estimating selection intensity on synonymous codon usage in a nonequilibrium population. Genetics 183, 651–662 10.1534/Genetics.109.101782 (doi:10.1534/Genetics.109.101782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comeron J. M., Guthrie T. B. 2005. Intragenic Hill–Robertson interference influences selection intensity on synonymous mutations in Drosophila. Mol. Biol. Evol. 22, 2519–2530 10.1093/molbev/msi246 (doi:10.1093/molbev/msi246) [DOI] [PubMed] [Google Scholar]
- 6.Haddrill P. R., Zeng K., Charlesworth B. 2011. Determinants of synonymous and nonsynonymous variability in three species of Drosophila. Mol. Biol. Evol. 28, 1731–1743 10.1093/molbev/msq354 (doi:10.1093/molbev/msq354) [DOI] [PubMed] [Google Scholar]
- 7.dos Reis M., Wernisch L. 2009. Estimating translational selection in eukaryotic genomes. Mol. Biol. Evol. 26, 451–461 10.1093/Molbev/Msn272 (doi:10.1093/Molbev/Msn272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marais G. 2003. Biased gene conversion: implications for genome and sex evolution. Trends Genet. 19, 330–338 10.1016/S0168-9525(03)00116-1 (doi:10.1016/S0168-9525(03)00116-1) [DOI] [PubMed] [Google Scholar]
- 9.Galtier N., Bazin E., Bierne N. 2006. GC-biased segregation of noncoding polymorphisms in Drosophila. Genetics 172, 221–228 10.1534/genetics.105.046524 (doi:10.1534/genetics.105.046524) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haddrill P. R., Charlesworth B. 2008. Non-neutral processes drive the nucleotide composition of non-coding sequences in Drosophila. Biol. Lett. 4, 438–441 10.1098/Rsbl.2008.0174 (doi:10.1098/Rsbl.2008.0174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura K., Subramanian S., Kumar S. 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21, 36–44 10.1093/molbev/msg236 (doi:10.1093/molbev/msg236) [DOI] [PubMed] [Google Scholar]
- 12.McVean G. A., Vieira J. 2001. Inferring parameters of mutation, selection and demography from patterns of synonymous site evolution in Drosophila. Genetics 157, 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heger A., Ponting C. P. 2007. Variable strength of translational selection among 12 Drosophila species. Genetics 177, 1337–1348 10.1534/genetics.107.070466 (doi:10.1534/genetics.107.070466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Throckmorton L. H. 1982. The virilis species group. In The genetics and biology of Drosophila (eds Ahsburner M., Carson H. L., Thompson J. N.), pp. 227–296 New York, NY: Academic Press [Google Scholar]
- 15.Vieira J., McAllister B. F., Charlesworth B. 2001. Evidence for selection at the fused1 locus of Drosophila americana. Genetics 158, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McAllister B. F. 2002. Chromosomal and allelic variation in Drosophila americana: selective maintenance of a chromosomal cline. Genome 45, 13–21 10.1139/g01-112 (doi:10.1139/g01-112) [DOI] [PubMed] [Google Scholar]
- 17.Maside X., Charlesworth B. 2007. Patterns of molecular variation and evolution in Drosophila americana and its relatives. Genetics 176, 2293–2305 10.1534/Genetics.107.071191 (doi:10.1534/Genetics.107.071191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vieira C. P., Almeida A., Dias J. D., Vieira J. 2006. On the location of the gene(s) harbouring the advantageous variant that maintains the X4 fusion of Drosophila americana. Genet. Res. 87, 163–174 10.1017/S0016672306008147 (doi:10.1017/S0016672306008147) [DOI] [PubMed] [Google Scholar]
- 19.Zeng K., Charlesworth B. 2010. Studying patterns of recent evolution at synonymous sites and intronic sites in Drosophila melanogaster. J. Mol. Evol. 70, 116–128 10.1007/S00239-009-9314-6 (doi:10.1007/S00239-009-9314-6) [DOI] [PubMed] [Google Scholar]
- 20.Betancourt A. J., Welch J. J., Charlesworth B. 2009. Reduced effectiveness of selection caused by a lack of recombination. Curr. Biol. 19, 655–660 10.1016/J.Cub.2009.02.039 (doi:10.1016/J.Cub.2009.02.039) [DOI] [PubMed] [Google Scholar]
- 21.Tajima F. 1989. The effect of change in population size on DNA polymorphism. Genetics 123, 597–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haddrill P. R., Bachtrog D., Andolfatto P. 2008. Positive and negative selection on noncoding DNA in Drosophila simulans. Mol. Biol. Evol. 25, 1825–1834 10.1093/Molbev/Msn125 (doi:10.1093/Molbev/Msn125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keightley P. D., Trivedi U., Thomson M., Oliver F., Kumar S., Blaxter M. L. 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19, 1195–1201 10.1101/Gr.091231.109 (doi:10.1101/Gr.091231.109) [DOI] [PMC free article] [PubMed] [Google Scholar]