Abstract
Males and females usually invest asymmetrically in offspring. In species lacking parental care, females influence offspring in many ways, while males only contribute genetic material via their sperm. For this reason, maternal effects have long been considered an important source of phenotypic variation, while paternal effects have been presumed to be absent or negligible. The recent surge of studies showing trans-generational epigenetic effects questions this assumption, and indicates that paternal effects may be far more important than previously appreciated. Here, we test for sex-linked paternal effects in Drosophila melanogaster on a life-history trait, and find substantial support for both X- and Y-linked effects.
Keywords: Drosophila melanogaster, paternal effects, X-chromosome, Y-chromosome, trans-generational epigenetic effects, life history
1. Introduction
Understanding the full set of causative components of genetic variation that contribute to phenotypic variation is of fundamental importance in the study of evolution and the aetiology of disease [1,2]. Traditionally, paternal effects have been ignored in studies of quantitative genetics, as they have been assumed to be negligible in the majority of species and because most crossing designs do not estimate this variance component (e.g. [3,4]). Despite the fact that sperm is stripped of most of its cytoplasm, paternal effects can potentially be mediated in several ways, including paternal care (reviewed in [5]), male courtship (e.g. [6]), seminal fluid products, genomic imprinting (reviewed in [7]) and other trans-generational epigenetic effects (reviewed in [8]). Sex chromosomes offer a unique opportunity to test for genetic paternal effects, because each is only transmitted to one sex of a sire's offspring. Here, we report a proof-of-principle study testing for paternal effects in Drosophila melanogaster, and find evidence for both X- and Y-linked effects.
2. Material and methods
We backcrossed intact X- and Y-chromosomes from each of four strains of D. melanogaster into the same genetic background (LHM) [9], to produce four iso-Xi/Y lines and four iso-X/Yi lines (see the electronic supplementary material, figure S1 for the crossing protocol). The four strains (i = 1–4) came from Zimbabwe, Africa (Z-53), Congo, Africa (Congo), north-central North America (Canton-S, hereafter CS) and north-eastern North America (Massachusetts-1, hereafter Mass-1).
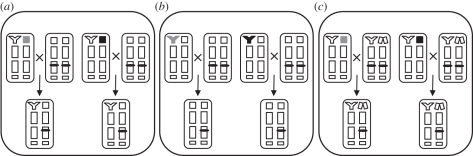
To test for an egg-to-adult survival-effect of the four paternal Xi chromosomes on the sex of offspring that does not carry them (males), we began by mating eight groups of 50 LHM-Sxl-eGFP dams (a replica of LHM homozygous for an autosomal construct with an eGFP reporter expressed only in female embryos [10]) to 50 Xi/Y sires, with two groups randomly assigned to each type of Xi/Y sire (figure 1a). Sons from these crosses had an identical genotype and cytotype, and only differed with respect to their father's X-chromosome. After 24 h, females were allowed to oviposit for 1 h on a Petri dish filled with food medium. Six hours later, the embryos were sorted by sex under a fluorescence microscope. Fifty non-fluorescing male embryos were placed in a vial with 300 competitor embryos of the same age, from a replica of LHM homozygous for the recessive mutation w (causing flies to have white eyes). The eye-colour of the competitor flies permitted eclosing target flies to be distinguished from competitor flies. This procedure was repeated over 4 days with the same parents (continuously housed together for the duration of the experiment). The number of target flies that emerged was scored daily until day 20 post egg deposition, and the sum of offspring from each set of the parents made one independent data point. The entire protocol was repeated three times, each with an independent set of parents, to produce three independent blocks of data and a total of 24 data points. The same protocol was used to test for Yi-linked paternal effects by substituting X/Yi males for Xi/Y males and measuring the survival of non-carrier daughters (figure 1c). A third experiment was done to test for X-linked paternal effects on daughters. The protocol was identical to that used to test for paternal Xi-effects on sons except that attached-X/Y dams (C(1)DX y,f homozygous for the autosomal eGFP reporter) replaced the X/X dams (figure 1b). In this experiment, four independent sets of parents per Xi/Y crossing were used, repeated over two blocks, for a total of 32 data points.
Figure 1.
Experimental design. Fathers carrying an intact X (upper rectangle) or Y (letter) chromosome from one of four geographically isolated populations (black or grey colour) and a standardized genetic background (depicted by white letters or rectangles) were crossed to females with a standardized genetic background, homozygous for an eGFP reporter (black bar on chromosome 3). These crosses produced non-carrier offspring of identical sex, genotype and cytotype. (a) Test for X-linked paternal effects on sons. (b) Test for Y-linked paternal effects on daughters. (c) Test for X-linked paternal effects on daughters (in this cross, males were crossed to females carrying a Y-chromosome and an attached-X [C(1)DX,y,f], depicted as two connected rectangles). Chromosomes are arranged with sex chromosomes in the top tier and the three autosomes in the lower tiers.
To test for a possible mechanism by which a paternal effect might be produced (i.e. egg provisioning), we also measured the length (L) and width (W) of eggs produced by Xi/Y sires mated with X/X females. Egg volume was calculated using the formula πW2L/6. Two replicates, each based on 45–50 eggs, were measured for each of the four types of Xi/Y sires.
Egg-to-adult survival was analysed using generalized linear models (GLM) of the JMP v. 8.0 software package with binomial, or beta-binomial, error terms and the logit linker function. The factors source of the paternal X- or Y-chromosome and block was included as fixed effects in the models. The interaction between paternal X- or Y-chromosome and block was non-significant in all models and therefore excluded from the analyses. Variation in egg size was analysed with ANOVA, with replicate nested within X-chromosome. Pairwise comparisons were carried out using the contrast function in the GLM routine of JMP v. 8.0.
3. Results
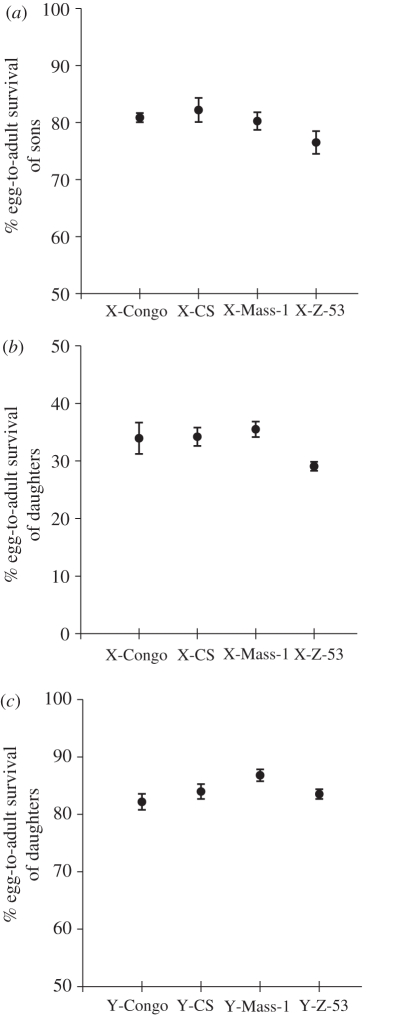
We found significant X-linked paternal effects on egg-to-adult survival in non-carrier sons (Chromosome: X2 = 16.16, d.f. = 3, p = 0.001; Block: X2 = 12.39, d.f. = 2, p = 0.002; figure 2a). Contrast analysis indicated that X-Z-53 differed from all other X-chromosomes (false discovery rate (FDR), q-values ≤ 0.05). A similar, but weaker pattern of X-linked paternal effects was found for egg-to-adult survival in non-carrier daughters (Chromosome: X2 = 8.02, d.f. = 3, p = 0.046; Block: X2 = 0.80, d.f. = 1, p = 0.37; figure 2b). Contrast analysis indicated that X-Z-53 differed from the X-Mass (FDR, q-value ≤ 0.05) and approached significance with X-Congo and X-CS (FDR, q-values ≤ 0.09). We also found significant Y-linked paternal effects influencing non-carrier daughters' egg-to-adult survival (Chromosome: X2 = 10.73, d.f. = 3, p = 0.0133; Block: X2 = 8.95, d.f. = 2, p = 0.0114; figure 2c). Contrast analysis indicated that X-Mass differed from the X-Congo (FDR, q-value ≤ 0.01) and approached significance with Z-53 (FDR, q-values ≤ 0.06) and X-CS (FDR, q-value ≤ 0.1). We did not find a significant paternal X-linked effect on egg volume (Chromosome: F3,3.99 = 1.44, p = 0.35).
Figure 2.
Per cent egg-to-adult survival in offspring, in response to the paternal X- and Y-chromosome they did not carry. (a) X-linked paternal effects on sons from X/X dams. (b) X-linked paternal effects on daughters from attached-X dams. (c) Y-linked paternal effects on daughters from X/X dams. Note that the egg-to-adult survival of daughters from attached-X mothers (b) is reduced by about a factor of 2 compared with the survival from the other crosses, because half of the female offspring are triploid for the X and die prior to eclosion.
4. Discussion
Despite the fact that our screen only surveyed X- and Y-linked genes (approx. 20% of the genome), we uncovered substantial variation in egg-to-adult survival owing to paternal effects. Although egg-to-adult survival is an inclusive measure of total juvenile fitness, paternal effects on adult-stage phenotypes of offspring are also possible and will need to be addressed in future studies. The X- and Y-linked effects we detected could have been mediated through any one of many potential mechanisms. For example, harmful male–female behavioural interactions are known to occur during courtship [6] and X- and Y-coded variation in male phenotypes could cause males to harm their mates to different degrees. Such variation in a sire's harm to his dams could feasibly lead to paternal effects on offspring survival by indirectly influencing the quality of eggs produced. Alternatively, X- and Y-coded seminal fluid phenotypes could directly induce females to alter their investment in eggs or induce different degrees of harm in terms of female survival [11] and/or fecundity [12].
Our failure to detect a sire effect on egg size rules out the simplest mechanism by which paternal effects might be mediated by courtship or seminal-fluid, although these traits could influence other maternal factors such as the quality/ratio of nutrients placed in eggs. Also, in the case of Y- and X-coded paternal effects on daughters, variation in sire fertility could not contribute to the observed paternal influence on offspring survival, since only living (fluorescing) embryos were screened. Sire fertility potentially contributed to our observed X-coded paternal effect on the survival of ‘sons’ (non-fluorescing, and therefore potentially unfertilized). However, the strong similarity of the pattern of the paternal effects of this chromosome on daughters (figure 2a,b) indicates that fertility was not the major contributor to the paternal X's effect on non-carrier sons.
We suggest that trans-generational epigenetic effects are a feasible candidate for the paternal effects on egg-to-adult survival found here. This may seem unlikely, especially in the case of the Y-chromosome, where only about a dozen structural genes have been found. Recent studies of Drosophila have, however, shown that the Y-chromosome influences expression of many hundreds of genes in males [13]. The Y-chromosome also has a phenotypic effect similar to that of the autosomes on a range of behavioural and physiological traits in mice [14]. Recent evidence in mice indicates that a father's Y-chromosome strongly influences quantitative traits in his daughters, apparently through trans-generational epigenetic effects [15]. Finally, there is convincing evidence showing that the Y is imprinted in D. melanogaster [16,17]. Collectively, these new findings indicate that the Y-, as well as the X-chromosome may have the potential to code for trans-generational epigenetic effects, e.g. by altering the chromatin structure on other chromosomes.
Our results have important implications for the field of quantitative genetics. Most quantitative genetic models designed to decompose genetic variation into its causative components ignore paternal effects despite their inclusion of maternal effects. One commonly employed method in quantitative genetics is the paternal half-sib design. This design is primarily used to test for additive genetic variation while controlling for maternal effects. Given that paternal effects are present, this and other similar designs will inflate estimates of additive genetic variance and thus the heritability of traits. Our results add to a growing list studies indicating that paternal effects may be an important component of total phenotypic variation (reviewed in [18]).
As a final point, sex-linked paternal effects are of special relevance to the theory of sexually antagonistic-zygotic drive (SA-zygotic drive). This theory predicts that sib-competition and/or harmful sib-mating cause the X and Y to evolve paternal effects that harm the sex of offspring that do not carry them [19,20]. The finding that the X-coded paternal effect influences both sons and daughters does not appear to support the operation of SA-zygotic drive. However, this finding would be consistent with SA-zygotic drive, if the X-coded paternal effect was accomplished by imprinting the Y in a manner that harmed any carrier, irrespective of its sex (since the Y is male-limited in nature).
This work demonstrates the ability of non-transmitted paternal chromosomes to influence fitness of offspring; however, it remains to be seen how variable such effects are within and between species.
Acknowledgements
We thank Michael Dickinson for providing a fluorescence dissecting microscope. This study was supported by grants to W.R.R. (National Science Foundation (DEB-0128780 and DEB-0111613) and National Institutes of Health (1R01HD057974-01)). U.F. was founded by a scholarship from the Wenner-Gren Foundations.
References
- 1.Bonduriansky R., Day T. 2009. Nongenetic inheritance and its evolutionary implications. Annu. Rev. Ecol. Syst. 40, 103–125 10.1146/annurev.ecolsys.39.110707.173441 (doi:10.1146/annurev.ecolsys.39.110707.173441) [DOI] [Google Scholar]
- 2.Uller T. 2008. Developmental plasticity and the evolution of parental effects. Trends Ecol. Evol. 23, 432–438 10.1016/j.tree.2008.04.005 (doi:10.1016/j.tree.2008.04.005) [DOI] [PubMed] [Google Scholar]
- 3.Falconer D. S., Mackay T. F. C. 1996. Introduction to quantitative genetics. New York, NY: Longman Group [Google Scholar]
- 4.Lynch M., Walsh B. 1998. Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer [Google Scholar]
- 5.Ridley M. 1978. Paternal care. Anim. Behav. 26, 904–932 10.1016/0003-3472(78)90156-2 (doi:10.1016/0003-3472(78)90156-2) [DOI] [Google Scholar]
- 6.Partridge L., Fowler K. 1990. Nonmating costs of exposure to males in female Drosophila melanogaster. J. Insect Physiol. 36, 419–425 10.1016/0022-1910(90)90059-O (doi:10.1016/0022-1910(90)90059-O) [DOI] [Google Scholar]
- 7.Wilkins J. F., Haig D. 2003. Inbreeding, maternal care and genomic imprinting. J. Theor. Biol. 221, 559–564 10.1006/jtbi.2003.3206 (doi:10.1006/jtbi.2003.3206) [DOI] [PubMed] [Google Scholar]
- 8.Lange U. C., Schneider R. 2010. What an epigenome remembers. Bioessays 32, 659–668 10.1002/bies.201000030 (doi:10.1002/bies.201000030) [DOI] [PubMed] [Google Scholar]
- 9.Rice W. R., Linder J. E., Friberg U., Lew T. A., Morrow E. H., Stewart A. D. 2005. Inter-locus antagonistic coevolution as an engine of speciation: assessment with hemiclonal analysis. Proc. Natl Acad. Sci. USA 102(Suppl. 1), 6527–6534 10.1073/pnas.0501889102 (doi:10.1073/pnas.0501889102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hempel L. U., Oliver B. 2007. Sex-specific DoublesexM expression in subsets of Drosophila somatic gonad cells. BMC Dev. Biol. 7, 113. 10.1186/1471-213X-7-113 (doi:10.1186/1471-213X-7-113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman T., Liddle L. F., Kalb J. M., Wolfner M. F., Partridge L. 1995. Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature 373, 241–244 10.1038/373241a0 (doi:10.1038/373241a0) [DOI] [PubMed] [Google Scholar]
- 12.Friberg U., Lew T. A., Byrne P. G., Rice W. R. 2005. Assessing the potential for an ongoing arms race within and between the sexes: selection and heritable variation. Evol. Int. J. Org. Evol. 59, 1540–1551 [PubMed] [Google Scholar]
- 13.Lemos B., Araripe L. O., Hartl D. L. 2008. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science 319, 91–93 10.1126/science.1148861 (doi:10.1126/science.1148861) [DOI] [PubMed] [Google Scholar]
- 14.Singer J. B., et al. 2004. Genetic dissection of complex traits with chromosome substitution strains of mice. Science 304, 445–448 10.1126/science.1093139 (doi:10.1126/science.1093139) [DOI] [PubMed] [Google Scholar]
- 15.Nelson V. R., Spiezio S. H., Nadeau J. H. 2010. Transgenerational genetic effects of the paternal Y chromosomes on daughters' phenotypes. Epigenomics 2, 513–521 10.2217/epi.10.26 (doi:10.2217/epi.10.26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menon D. U., Meller V. H. 2009. Imprinting of the Y chromosome influences dosage compensation in roX1 roX2 Drosophila melanogaster. Genetics 183, 811–820 10.1534/genetics.109.107219 (doi:10.1534/genetics.109.107219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golic K. G., Golic M. M., Pimpinelli S. 1998. Imprinted control of gene activity in Drosophila. Curr. Biol. 8, 1273–1276 10.1016/S0960-9822(07)00537-4 (doi:10.1016/S0960-9822(07)00537-4) [DOI] [PubMed] [Google Scholar]
- 18.Jablonka E., Raz G. 2009. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84, 131–176 10.1086/598822 (doi:10.1086/598822) [DOI] [PubMed] [Google Scholar]
- 19.Rice W. R., Gavrilets S., Friberg U. 2008. Sexually antagonistic ‘zygotic drive’ of the sex chromosomes. PLoS Genet. 4, e1000313. 10.1371/journal.pgen.1000313 (doi:10.1371/journal.pgen.1000313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice W. R., Gavrilets S., Friberg U. 2009. Sexually antagonistic chromosomal cuckoos. Biol. Lett. 5, 686–688 10.1098/rsbl.2009.0061 (doi:10.1098/rsbl.2009.0061) [DOI] [PMC free article] [PubMed] [Google Scholar]