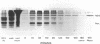Abstract
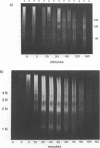
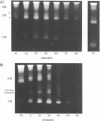
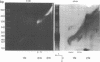
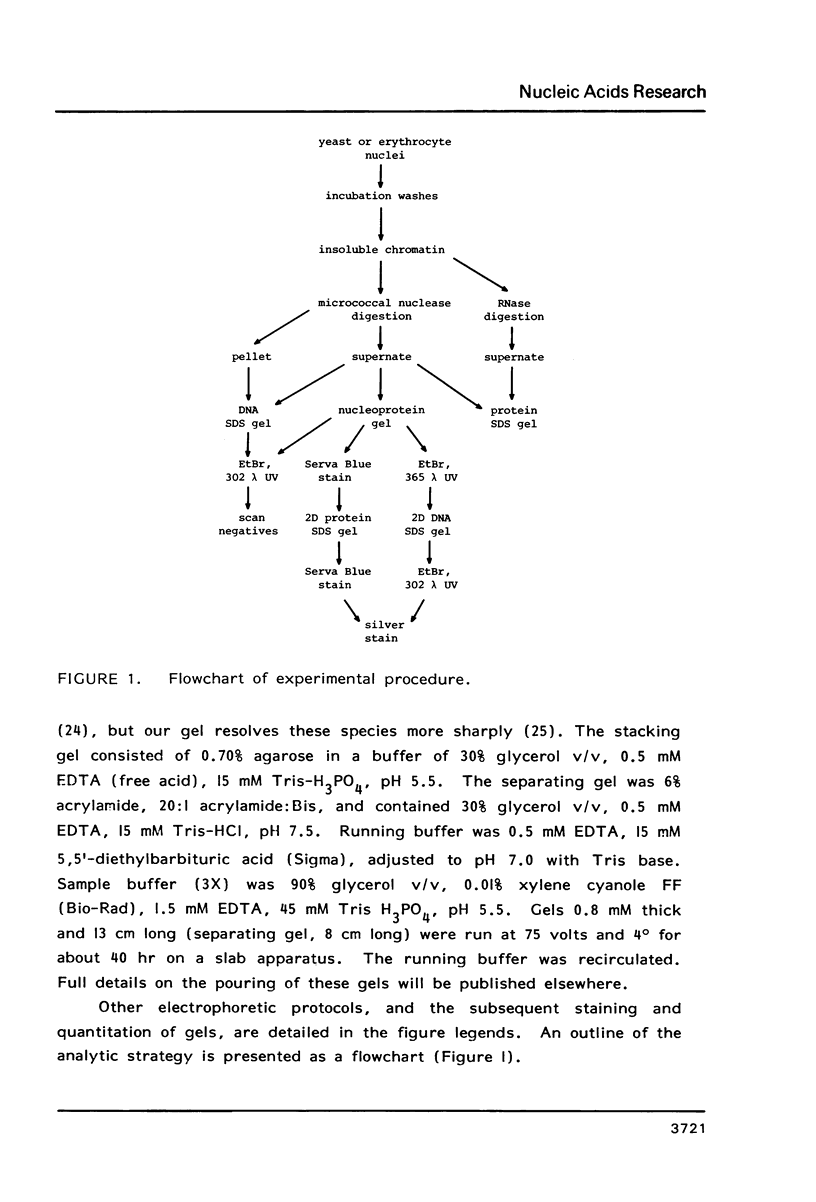
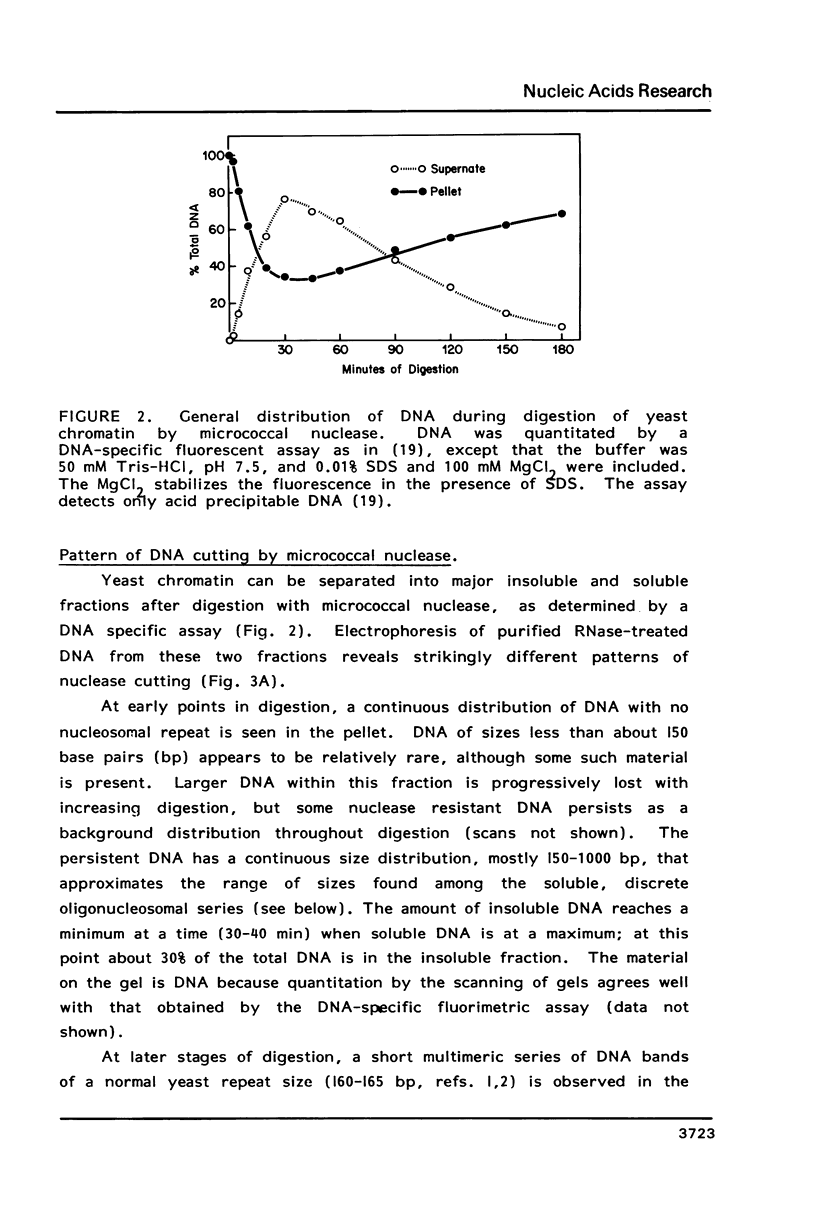
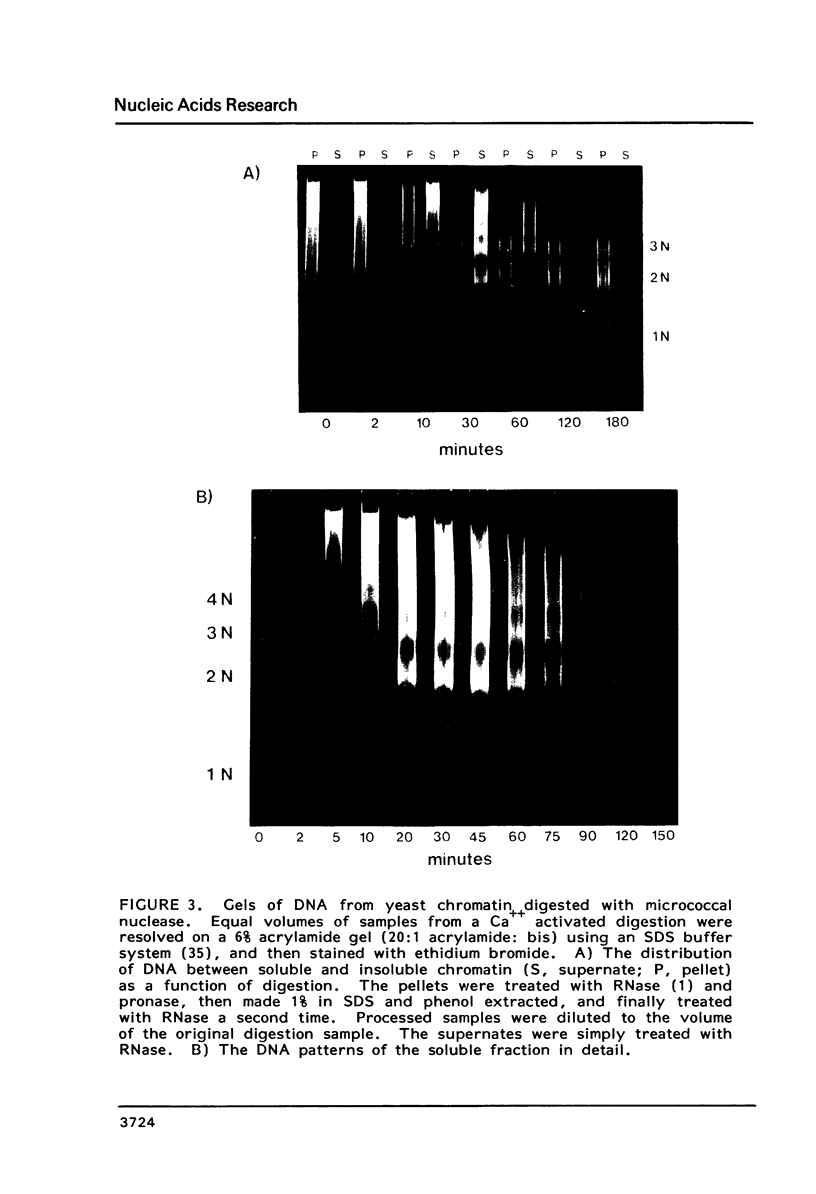
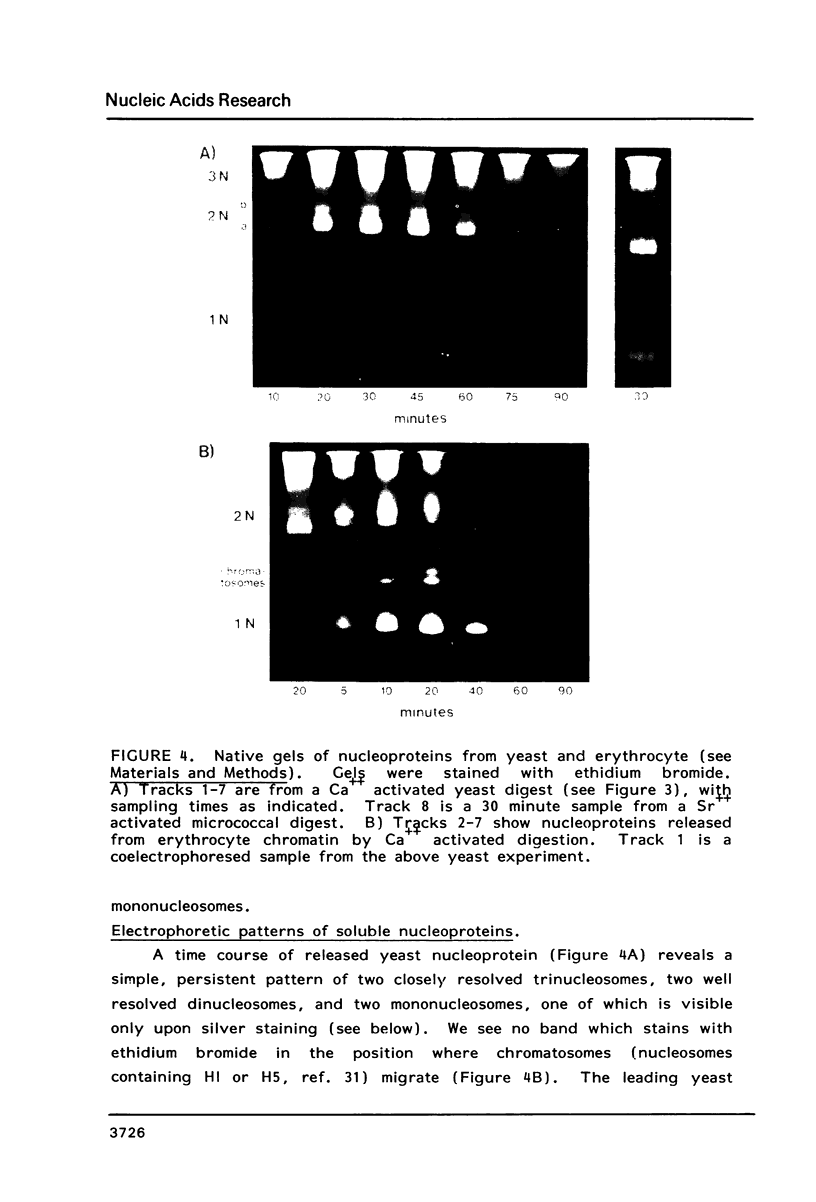
We have developed a method of preparing yeast chromatin that facilitates the analysis of nucleoprotein organization. Yeast chromatin, isolated as an insoluble complex, is digested with micrococcal nuclease and fractionated into major insoluble and soluble fractions. No nucleosomal repeat is seen early in digestion for the insoluble fraction. Nucleosomal complexes of the soluble fraction are excised by nuclease in a distinctive non-random pattern; they are markedly depleted in mononucleosomes. When we analyze the soluble material by high resolution native electrophoresis, we find that the nucleoproteins resolve into two bands for each DNA multimer of the nucleosomal repeat. Our results suggest that there are structural similarities between bulk yeast chromatin and chromatin configurations found in transcribing genes of complex eukaryotes.
Full text
PDF





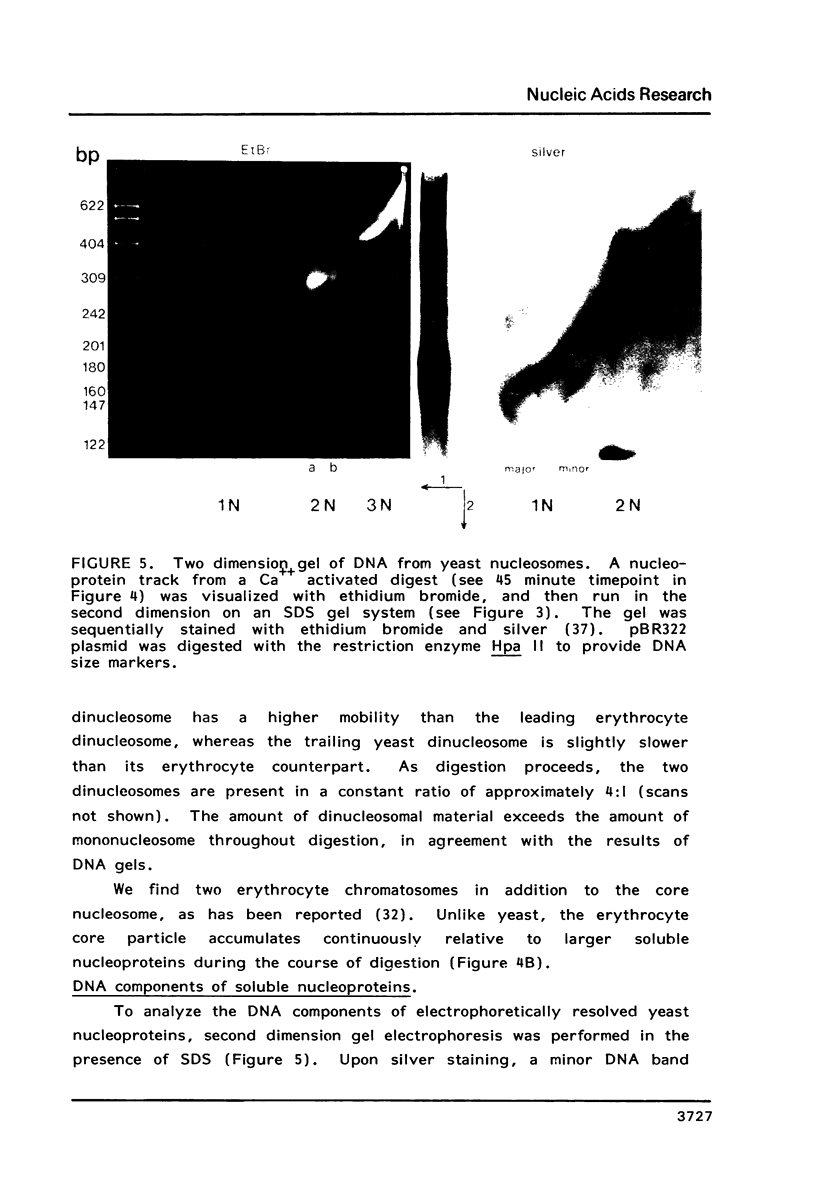
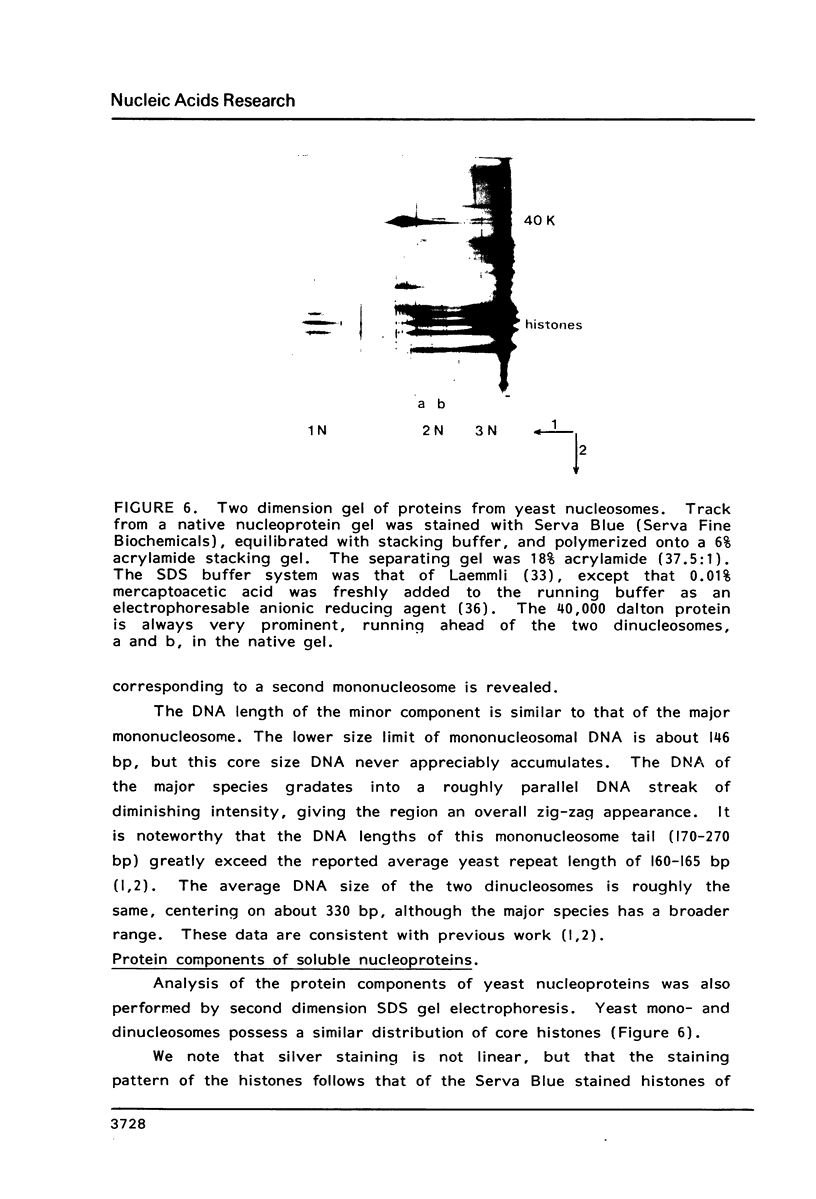








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bakayeva T. G., Bakayev V. V. Separation of nucleosomes containing histones H1 and H5. Mol Biol Rep. 1978 Oct 16;4(3):185–189. doi: 10.1007/BF00777522. [DOI] [PubMed] [Google Scholar]
- Basu S. Superstructures of wet inactive chromatin and the chromosome surface. J Supramol Struct. 1979;10(4):377–395. doi: 10.1002/jss.400100402. [DOI] [PubMed] [Google Scholar]
- Bellard M., Dretzen G., Bellard F., Oudet P., Chambon P. Disruption of the typical chromatin structure in a 2500 base-pair region at the 5' end of the actively transcribed ovalbumin gene. EMBO J. 1982;1(2):223–230. doi: 10.1002/j.1460-2075.1982.tb01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer J. M. Artifact produced in disc electrophoresis by ammonium persulfate. Science. 1967 Apr 14;156(3772):256–257. doi: 10.1126/science.156.3772.256. [DOI] [PubMed] [Google Scholar]
- Ciejek E. M., Nordstrom J. L., Tsai M. J., O'Malley B. W. Ribonucleic acid precursors are associated with the chick oviduct nuclear matrix. Biochemistry. 1982 Sep 28;21(20):4945–4953. doi: 10.1021/bi00263a018. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Lang J., Hayday A., Lania L., Fried M., Chiswell D. J., Wyke J. A. Active viral genes in transformed cells lie close to the nuclear cage. EMBO J. 1982;1(4):447–452. doi: 10.1002/j.1460-2075.1982.tb01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuatrecasas P., Fuchs S., Anfinsen C. B. Catalytic properties and specificity of the extracellular nuclease of Staphylococcus aureus. J Biol Chem. 1967 Apr 10;242(7):1541–1547. [PubMed] [Google Scholar]
- Davie J. R., Saunders C. A., Walsh J. M., Weber S. C. Histone modifications in the yeast S. Cerevisiae. Nucleic Acids Res. 1981 Jul 10;9(13):3205–3216. doi: 10.1093/nar/9.13.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffus J. H. The isolation of yeast nuclei and methods to study their properties. Methods Cell Biol. 1975;12:77–97. doi: 10.1016/s0091-679x(08)60953-x. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring E. S., Grossman L. I., Krupnick D., Cryer D. R., Marmur J. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J Mol Biol. 1970 Sep 14;52(2):323–335. doi: 10.1016/0022-2836(70)90033-1. [DOI] [PubMed] [Google Scholar]
- Hall R. M., Nagley P., Linnane A. W. Biogenesis of mitochondria. XLII. Genetic analysis of the control of cellular mitochondrial DNA levels in Saccharomyces cerevisiae. Mol Gen Genet. 1976 May 7;145(2):169–175. doi: 10.1007/BF00269590. [DOI] [PubMed] [Google Scholar]
- Hereford L. M., Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977 Mar;10(3):453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- Karpov V. L., Bavykin S. G., Preobrazhenskaya O. V., Belyavsky A. V., Mirzabekov A. D. Alignment of nucleosomes along DNA and organization of spacer DNA in Drosophila chromatin. Nucleic Acids Res. 1982 Jul 24;10(14):4321–4337. doi: 10.1093/nar/10.14.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarca C., Paigen K. A simple, rapid, and sensitive DNA assay procedure. Anal Biochem. 1980 Mar 1;102(2):344–352. doi: 10.1016/0003-2697(80)90165-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee K. P., Baxter H. J., Guillemette J. G., Lawford H. G., Lewis P. N. Structural studies on yeast nucleosomes. Can J Biochem. 1982 Mar;60(3):379–388. doi: 10.1139/o82-045. [DOI] [PubMed] [Google Scholar]
- Levy A., Noll M. Chromatin fine structure of active and repressed genes. Nature. 1981 Jan 15;289(5794):198–203. doi: 10.1038/289198a0. [DOI] [PubMed] [Google Scholar]
- Libertini L. J., Small E. W. Salt induced transitions of chromatin core particles studied by tyrosine fluorescence anisotropy. Nucleic Acids Res. 1980 Aug 25;8(16):3517–3534. doi: 10.1093/nar/8.16.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D. E. Detailed analysis of the nucleosomal organization of transcribed DNA in yeast chromatin. Biochemistry. 1981 Oct 13;20(21):5966–5972. doi: 10.1021/bi00524a007. [DOI] [PubMed] [Google Scholar]
- Lohr D. Chromatin structure differs between coding and upstream flanking sequences of the yeast 35S ribosomal genes. Biochemistry. 1983 Feb 15;22(4):927–934. doi: 10.1021/bi00273a034. [DOI] [PubMed] [Google Scholar]
- Lohr D., Kovacic R. T., Van Holde K. E. Quantitative analysis of the digestion of yeast chromatin by staphylococcal nuclease. Biochemistry. 1977 Feb 8;16(3):463–471. doi: 10.1021/bi00622a020. [DOI] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Organization of spacer DNA in chromatin. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6326–6330. doi: 10.1073/pnas.76.12.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardian J. K., Isenberg I. Yeast inner histones and the evolutionary conservation of histone-histone interactions. Biochemistry. 1978 Sep 5;17(18):3825–3833. doi: 10.1021/bi00611a023. [DOI] [PubMed] [Google Scholar]
- May R. Isolationsbedingungen für Zelkerne aus Hefeprotoplasten. Z Allg Mikrobiol. 1971;11(2):131–142. doi: 10.1002/jobm.3630110208. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Rau D. C., Charney E., Felsenfeld G. Orientation of the nucleosome within the higher order structure of chromatin. Cell. 1980 Nov;22(1 Pt 1):87–96. doi: 10.1016/0092-8674(80)90157-9. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982 Sep;30(2):567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- Nelson D. A., Beltz W. R., Rill R. L. Chromatin subunits from baker's yeast: isolation and partial characterization. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1343–1347. doi: 10.1073/pnas.74.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. A. Histone acetylation in baker's yeast. Maintenance of the hyperacetylated configuration in log phase protoplasts. J Biol Chem. 1982 Feb 25;257(4):1565–1568. [PubMed] [Google Scholar]
- Rattner J. B., Saunders C., Davie J. R., Hamkalo B. A. Ultrastructural organization of yeast chromatin. J Cell Biol. 1982 Apr;93(1):217–222. doi: 10.1083/jcb.93.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz M., Nehls P., Hozier J. Involvement of histone H1 in the organization of the chromosome fiber. Proc Natl Acad Sci U S A. 1977 May;74(5):1879–1883. doi: 10.1073/pnas.74.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S. I., Nelkin B. D., Vogelstein B. The ovalbumin gene is associated with the nuclear matrix of chicken oviduct cells. Cell. 1982 Jan;28(1):99–106. doi: 10.1016/0092-8674(82)90379-8. [DOI] [PubMed] [Google Scholar]
- Sajdel-Sulkowska E. M., Bhargava M. M., Arnaud M. V., Halvorson H. O. An improved method for the isolation of yeast nuclei active in RNA synthesis in vitro. Biochem Biophys Res Commun. 1974 Jan 23;56(2):496–502. doi: 10.1016/0006-291x(74)90870-5. [DOI] [PubMed] [Google Scholar]
- Samal B., Worcel A., Louis C., Schedl P. Chromatin structure of the histone genes of D. melanogaster. Cell. 1981 Feb;23(2):401–409. doi: 10.1016/0092-8674(81)90135-5. [DOI] [PubMed] [Google Scholar]
- Scheer U., Zentgraf H., Sauer H. W. Different chromatin structures in Physarum polycephalum: a special form of transcriptionally active chromatin devoid of nucleosomal particles. Chromosoma. 1981;84(2):279–290. doi: 10.1007/BF00399138. [DOI] [PubMed] [Google Scholar]
- Scott J. H., Schekman R. Lyticase: endoglucanase and protease activities that act together in yeast cell lysis. J Bacteriol. 1980 May;142(2):414–423. doi: 10.1128/jb.142.2.414-423.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Sledziewski A., Young E. T. Chromatin conformational changes accompany transcriptional activation of a glucose-repressed gene in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1982 Jan;79(2):253–256. doi: 10.1073/pnas.79.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Furber V. Yeast chromatin structure. FEBS Lett. 1976 Jul 15;66(2):274–280. doi: 10.1016/0014-5793(76)80521-2. [DOI] [PubMed] [Google Scholar]
- Todd R. D., Garrard W. T. Overall pathway of mononucleosome production. J Biol Chem. 1979 Apr 25;254(8):3074–3083. [PubMed] [Google Scholar]
- WILLIAMS D. E., REISFELD R. A. DISC ELECTROPHORESIS IN POLYACRYLAMIDE GELS: EXTENSION TO NEW CONDITIONS OF PH AND BUFFER. Ann N Y Acad Sci. 1964 Dec 28;121:373–381. doi: 10.1111/j.1749-6632.1964.tb14210.x. [DOI] [PubMed] [Google Scholar]
- Weischet W. O., Allen J. R., Riedel G., Van Holde K. E. The effects of salt concentration and H-1 depletion on the digestion of calf thymus chromatin by micrococcal nuclease. Nucleic Acids Res. 1979;6(5):1843–1862. doi: 10.1093/nar/6.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Strogatz S., Riley D. Structure of chromatin and the linking number of DNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1461–1465. doi: 10.1073/pnas.78.3.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- Zenke M., Sauer G. Spliced and unspliced virus specific RNA sequences are associated with purified simian virus 40 chromatin. Nucleic Acids Res. 1982 Aug 11;10(15):4543–4550. doi: 10.1093/nar/10.15.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]