Abstract
Calibration is a critical step in every molecular clock analysis but it has been the least considered. Bayesian approaches to divergence time estimation make it possible to incorporate the uncertainty in the degree to which fossil evidence approximates the true time of divergence. We explored the impact of different approaches in expressing this relationship, using arthropod phylogeny as an example for which we established novel calibrations. We demonstrate that the parameters distinguishing calibration densities have a major impact upon the prior and posterior of the divergence times, and it is critically important that users evaluate the joint prior distribution of divergence times used by their dating programmes. We illustrate a procedure for deriving calibration densities in Bayesian divergence dating through the use of soft maximum constraints.
Keywords: molecular clock, calibration, fossil record, evolution, BEAST, MCMCTree
1. Introduction
The fossil record was long perceived to be the only basis for a timescale for evolutionary history but this role has been usurped by the molecular clock. The relationship between molecular clocks and the fossil record has not always been easy, but fossil data remain integral in establishing absolute divergence times using molecular clock methodology. After a long period of methodological development to accommodate rate variation, attention is now focused on accurately implementing calibrations, particularly, since widely adopted Bayesian methods afford a means of accommodating the degree to which fossil evidence approximates the divergence date that they constrain [1,2]. Fossil evidence can be used directly to derive a minimum constraint only. However, divergence time estimation is not possible with minimum constraints alone, as the substitution rate is not known; at least one point calibration or maximum constraint is required in order to calculate the substitution rate and absolute divergence times.
The two most popular approaches that have been adopted to establish a maximum constraint are: (i) to employ mathematical functions of probability densities to describe the degree to which a minimum constraint approximates a divergence date [3–8], and (ii) to establish explicitly justified fossil-based ‘soft maximum’ constraints [2,3]. Both approaches provide a means of constraining node ages in a way that is compatible with the imperfect nature of the evidence. However, little is known about how they perform, or the effect of parameters specifying the shape of probability densities. Indeed, a survey of Bayesian molecular clock studies (2010–2011; see electronic supplementary material, table S1) reveals that, where they are employed, justification is rarely provided for calibration densities or their parameters, despite precautionary examples [7,8]. To explore the performance of alternative approaches to accommodating calibration uncertainty, we established a suite of minimum and maximum constraints on the divergences of arthropod species whose genomes have been, or are being, sequenced. We conducted a series of molecular clock analyses employing widely used calibration densities to highlight the impact of parameter choices in those densities on the posterior time estimation, and to illustrate a procedure of implementing such densities, in order to reflect uncertainties in fossil calibrations.
2. Material and methods
We selected 15 arthropod species that have reached draft assembly stage on the NCBI Entrez Genome Project database (April 2011) and constructed a matrix based on the amino acid sequences of seven housekeeping genes (electronic supplementary material). We fixed the topology throughout our analyses (electronic supplementary material). We used or revised established calibrations [5], supplemented by new calibrations for the remaining nodes in our topology (electronic supplementary material).
To investigate the impact of calibration densities on the Bayesian estimation of divergence times, we conducted three sets of analysis using the programs BEAST v. 1.6.1 [1] and MCMCTree [9]. First, we employed the minimum and maximum constraints for each node. All constraints were hard in BEAST, but in MCMCTree minimum constraints were hard and maximum constraints had a 2.5 per cent soft tail. Uniform distributions spanned the minimum and maximum constraints.
In the second set of analyses, we considered different combinations of arbitrary parameter choices for the lognormal distribution used in BEAST [1], and for the truncated Cauchy distribution used in MCMCTree [2,8]. In BEAST, we analysed the data with three different values for the standard deviation s (0.5, 1 and 2) and four different values for the mean in log space m (1, 1.5, 2 and 2.5), where the offset of the lognormal distribution corresponded to our minimum constraints. In MCMCTree, we permuted the location parameter p (0.1 and 0.5) and the scale parameter c (0.2, 0.5, 1 and 2). In both programs, we constrained the age at the root to be between 636.1 and 515 Ma, based on the fossil record (electronic supplementary material). As before, the minimum bound is hard in both programs, whereas the maximum bound is hard in BEAST and soft in MCMCTree. MCMCTree requires an upper constraint and we found that if this was not implemented in BEAST, divergence time estimates became unreasonably ancient. These analyses mimic current common practice in molecular clock dating (electronic supplementary material, table S1).
In a third set of analyses, we compared the use of non-uniform distributions between the fossil-based minimum and maximum constraints, using soft maximum bounds to guide our parameter choices in specifying the fossil calibration densities. We considered the lognormal distribution in BEAST and the skew-t in MCMCTree, and in each case implemented hard minima and attempted to match the maximum bound for each node with the 97.5% percentile of the distribution. In both programs, we retained a uniform distribution at the root of the tree as above.
For further details and Markov chain Monte Carlo settings, see the electronic supplementary material. All molecular clock analyses were carried out using the computational facilities of the Advanced Computing Research Centre, University of Bristol.
3. Results
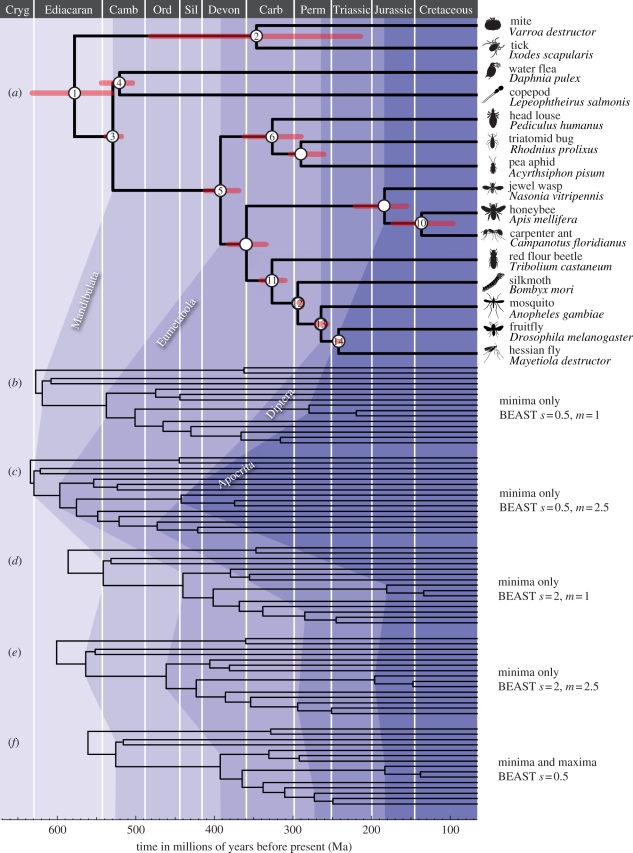
Analytical results are presented in the electronic supplementary material, table S2. Figure 1 illustrates the effects of changing parameters in the calibration densities. The divergence time estimates calculated in both BEAST and MCMCTree using minimum and maximum constraints on every node are highly congruent, although the BEAST estimates are consistently slightly younger because hard maximum constraints were employed.
Figure 1.
(a) Posterior time estimates when uniform and (f) lognormal priors are used to constrain node ages with fossil-based minima and maxima in BEAST. (b–e) Posterior mean estimates showing the impact of changing the parameters of the lognormal distribution (m = mean and s = standard deviation) relative to fossil minima.
When non-uniform distributions, such as lognormal or truncated Cauchy, are employed to describe the calibration density relative to the minimum constraint alone, the results show that divergence estimates are extremely sensitive to parameter choice. For instance, changing the mean or the standard deviation of the lognormal distribution can cause the mean divergence estimates, as well as lower and upper 95% posterior intervals, to differ by hundreds of millions of years (figure 1b–d; electronic supplementary material, table S2). The effect of increasing the mean of the lognormal distribution is more apparent with smaller values of the standard deviation (s). As the mean increases, the upper range of probable ages is truncated by the inferred ages of the deepest nodes within the topology and, ultimately, the maximum constraint on the root, which permits estimates from becoming unreasonably ancient. The effect of increasing the standard deviation of the lognormal distribution is that the mode of the distribution is shifted towards the minimum bound and exhibits increased kurtosis; younger ages and narrower intervals result.
Similar outcomes are achieved when permuting the parameters of the truncated Cauchy distribution, applied to the minimum bounds in MCMCTree [8]. When the location parameter (p) is increased, the peak in the prior is shifted away from the minimum constraint towards older ages, and when the scale parameter (c) is increased, the prior distribution becomes flatter and the 95% limit of the soft maximum constraint becomes older. Increasing both parameters tends to produce older and more diffuse estimates until they are constrained by the prior at the root. Evidently, the results are sensitive to the maximum constraint placed at the root. In representing the oldest possible divergence time in the tree, this constraint dictates the maximum envelope of time during which all speciation events can occur. When increasingly diffuse arbitrary densities are applied, divergence times can only become as ancient as this constraint will allow.
When the prior density is constrained by both minimum and maximum constraints, rather than just minimum constraints, analyses that employ a lognormal distribution in BEAST, a skew-t distribution in MCMCTree or a uniform distribution in either program, calculate posterior estimates and 95% highest posterior density intervals that are closely comparable (figure 1a,f; electronic supplementary material, table S2).
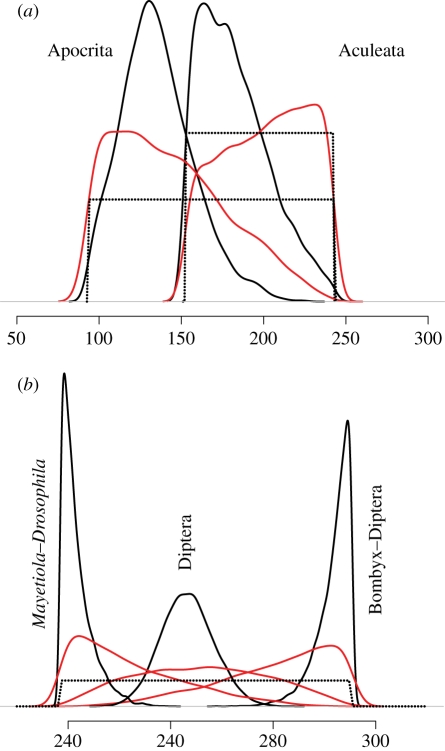
Analyses without molecular data show that, regardless of which prior density function is employed, the specified calibration densities are not those implemented in the estimation of divergence times. For example, the initial uniform densities specified for Apocrita (node 9) and Aculeata (node 10) are transformed into non-uniform effective priors (figure 2a) while the common uniform initial prior for Lepidoptera–Diptera (node 12), Diptera (node 13) and Drosophila–Mayetiola (node 14) is transformed into three distinct non-uniform effective priors (figure 2b).
Figure 2.
The effect of truncation in the establishment of the joint time prior. The dashed line represents the user-specified uniform prior, the red line represents the effective priors, and the solid black lines represent the marginal posteriors. (a) Interaction of the overlapping priors used to constrain the age of nodes 9 and 10 of figure 1. (b) Interaction between the priors used to constrain nodes 12–14 of figure 1, all of which share the same fossil-based minimum and maximum constraints.
4. Discussion
It will surprise some to discover that user-specified (initial) prior probabilities on the timing of clade divergence are not the effective priors implemented in calculating the posterior time estimates [2,7–9]. This occurs because programs like BEAST and MCMCTree truncate the initial calibration densities so that the joint (effective) prior of divergence times satisfies the biological requirement that ancestral nodes should be at least as old as descendent nodes. In BEAST, such discrepancies between the specified and effective priors can occur even when a single calibration is used because the program generates the (effective) joint time prior using a heuristic procedure of multiplicative construction, multiplying directly the calibration densities with the density of times specified by the Yule process [10]. MCMCTree instead uses the conditional construction and does not suffer from this problem [2]. However, effective and specified divergence time priors will always differ when specified priors on nested clades overlap temporally. At the least, it is imperative that users follow the guidance of software manuals, to evaluate the effective priors (by running analyses without sequence data) to ensure that they are in reasonable accord with the palaeontological evidence used to establish the initial priors. If heed to the effective priors is not taken, any effort expended in debate over the use of one fossil or another, or one probability density versus another, will have been squandered.
A variety of mathematical distributions have been proposed to describe the errors in calibration when, for instance, using fossil or biogeographic evidence [1,3–7,11]. These have been advocated based on generalized perceptions of the degree to which the constraints provided by these classes of evidence approximate the timing of divergence. The results of our analyses show that, where these prior probability densities reflect minimum constraints alone—as is commonplace—the divergence time estimates can be extremely sensitive to the arbitrary choice of prior density and of parameters in the density. However, the impact on divergence time estimates of different prior densities is minimized when both minimum and maximum constraints are used. Thus, we advocate the use of minimum and soft maximum constraints where the choice of prior probability distribution is not evidence-based.
However, it should be noted that this is only a pragmatic solution in place of fully justified divergence time priors. In this endeavour, fossil occurrence data have the potential to be of greater use than merely establishing minimum and maximum constraints on node age. The stratigraphic distribution of fossil occurrences is distinctly non-random, and the biases that control their stratigraphic distribution are a topic of active research. There have been few attempts to model divergence time priors based on fossil occurrence data [12], perhaps because of the challenge of deriving a global composite database. This represents a critical area for future research, and requires the expertise of molecular biologists and palaeontologists. Improved accuracy and precision of divergence time estimation is more likely to result from improvement of divergence time priors to better summarize the information in the fossil record than from accumulation of molecular sequence data [13].
Acknowledgements
We acknowledge with thanks the help of Allison Daley, Greg Edgecombe, Jim Jago, Xiaoya Ma, John Paterson and Omar Rota-Stabelli in deriving the calibration constraints. This work was funded by a studentship from the Natural Environment Research Council to R.C.M.W., and grants from NERC to P.C.J.D., and from the Biotechnology and Biological Sciences Research Council to Z.Y. and P.C.J.D.
References
- 1.Drummond A. J., Ho S. Y. W., Phillips M. J., Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88. 10.1371/journal.pbio.0040088 (doi:10.1371/journal.pbio.0040088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Z., Rannala B. 2006. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol. Biol. Evol. 23, 212–226 10.1093/molbev/msj024 (doi:10.1093/molbev/msj024) [DOI] [PubMed] [Google Scholar]
- 3.Benton M. J., Donoghue P. C. J. 2007. Paleontological evidence to date the Tree of Life. Mol. Biol. Evol. 24, 26–53 10.1093/molbev/msl150 (doi:10.1093/molbev/msl150) [DOI] [PubMed] [Google Scholar]
- 4.Donoghue P. C. J., Benton M. J. 2007. Rocks and clocks: calibrating the Tree of Life using fossils and molecules. Trends Ecol. Evol. 22, 424–431 10.1016/j.tree.2007.05.005 (doi:10.1016/j.tree.2007.05.005) [DOI] [PubMed] [Google Scholar]
- 5.Benton M. J., Donoghue P. C. J., Asher R. J. 2009. Calibrating and constraining molecular clocks. In The timetree of life (eds Hedges S. B., Kumar S.), pp. 35–86 Oxford, UK: Oxford University Press [Google Scholar]
- 6.Ho S. Y. W. 2007. Calibrating molecular estimates of substitution rates and divergence times in birds. J. Avian Biol. 38, 409–414 10.1111/j.2007.0908-8857.04168.x (doi:10.1111/j.2007.0908-8857.04168.x) [DOI] [Google Scholar]
- 7.Ho S. Y. W., Phillips M. J. 2009. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. System. Biol. 58, 367–380 10.1093/sysbio/syp035 (doi:10.1093/sysbio/syp035) [DOI] [PubMed] [Google Scholar]
- 8.Inoue J. G., Donoghue P. C. J., Yang Z. 2010. The impact of the representation of fossil calibrations on Bayesian estimation of species divergence times. System. Biol. 59, 74–89 10.1093/sysbio/syp078 (doi:10.1093/sysbio/syp078) [DOI] [PubMed] [Google Scholar]
- 9.Yang Z. 2007. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 10.1093/molbev/msm088 (doi:10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 10.Heled J., Drummond A. J. In press. Calibrated tree priors for relaxed phylogenetics and divergence time estimation. System. Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedges S. B., Kumar S. 2004. Precision of molecular time estimates. Trends Genet. 20, 242–247 10.1016/j.tig.2004.03.004 (doi:10.1016/j.tig.2004.03.004) [DOI] [PubMed] [Google Scholar]
- 12.Wilkinson R. D., Steiper M. E., Soligo C., Martin R. D., Yang Z., Tavaré S. 2011. Dating primate divergences through an integrated analysis of palaeontological and molecular data. System. Biol. 60, 16–31 10.1093/sysbio/syq054 (doi:10.1093/sysbio/syq054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rannala B., Yang Z. H. 2007. Inferring speciation times under an episodic molecular clock. System. Biol. 56, 453–466 10.1080/10635150701420643 (doi:10.1080/10635150701420643) [DOI] [PubMed] [Google Scholar]