Abstract
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide, however, genetic-environmental interactions and mechanisms associated with the development of HCC remains largely unclear. Our recent work described novel inactivating mutations of ARID2 (AT-rich interactive domain 2) in four major subtypes of HCC through exomic sequencing of ten HCV-associated HCCs and subsequent evaluation of the tumors from additional affected individuals. Here, we summarize the current knowledge about the relevance of ARID2 in HCC and the implication in future patient care.
Keywords: Hepatocellular Carcinoma, Hepatitis C Virus, ARID2, Tumor Suppressor, Interferon
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most frequent malignant diseases worldwide. With an estimated 748,000 newly diagnosed cases per year and a low five-year survival rate, HCC is the third leading cause of cancer deaths [1, 2]. Epidemiologic studies have conclusively linked viruses and chemicals to the development of HCC [3]. Among the viruses, hepatitis B (HBV) and C (HCV) viruses attribute to HCC development in more than 80% of the HCC cases [4]. The non-viral risk factors, including dietary aflatoxin B1 (AFB1) exposure, cigarette smoking and heavy alcohol consumption, can have synergistic effects [5]. Besides these risk factors, several disorders such as cirrhosis alone and hemochromatosis are associated with an increased risk of HCC [6].
In recent years, the mechanisms and genetic-environmental interactions associated with the development of HCC have been elucidated [7, 8]. Genomic and gene expression analyses have identified key dysregulated signal transduction pathways involved in liver carcinogenesis [9]. The genomic structural changes including recurrent allelic deletions and regional losses and gains have been found on several chromosomes [10-14]. Epigenetic changes in genomic DNA appear to act by directly suppressing gene expression as well as indirectly by creating conditions that increase the chance of generating hepatocyte populations containing critical combinations of structurally and functionally aberrant genes [10-12]. Several critical genes including oncogenes such as c-myc and N-ras [15] and tumor suppressor genes such as TP53, Rb1, CDKN2A, Axin1 are located in the chromosome regions of genomic and epigenetic changes [9-11, 15, 16].
MUTATIONS IDENTIFIED IN HCC
To gain additional insights into the genetic basis of HCC, we performed exomic sequencing for ~18,000 protein-coding genes in the cancers and normal tissues of ten individuals with HCV-associated HCC [17]. Four hundred and twenty-nine non-synonymous somatic mutations in 411 genes were identified. Five genes, which were somatically mutated in more than one tumor, were further analyzed. Among these, CTNNB1 was mutated in four tumors, TP53 was mutated in three tumors, and ARID2, DMXL1 and NLRP1 were each mutated in two tumors. The former two genes, CTNNB1 and TP53, have been previously observed in HCC as tumor suppressor genes, but the other three have not been reported in any tumor type to our knowledge. The ARID2 mutations, which seemed enriched in HCV-associated HCC in the US and European patient populations (14%, 6 out of 43 tumors) compared with the overall mutation frequency (6.5%, 9 out of 139 tumors), attracted much interest.
Human ARID2 and its structural characteristics
ARID2 (AT-rich interactive domain 2) was initially identified in the Polybromo-associated BRG1-associated factor (PBAF) complex, a SWI/SNF chromatin-remodeling complex involved in ligand-dependent transcriptional activation by nuclear receptors [18-20]. Human ARID superfamily includes fifteen members which are classified into seven subfamilies named ARID1 through ARID5, JARID1 and JARID2 [21, 22]. ARID1 consists of two members, ARID1a and ARID1b. ARID1a, ARID1b, and ARID2, also known as BAF250a, BAF250b and BAF200, respectively, are all subunits of the SWI/SNF complexes.
The human ARID2 gene is located on chromosome 12q and consists of 21 exons (Figure 1a). Its orthologs have been found in mouse, rat, cattle, chicken, and mosquito. The ARID2 protein contains a conservative N-terminal AT-rich DNA interaction domain (ARID), a RFX-type winged-helix, a proline- and glutamine-rich region, and two conservative C-terminal C2H2 Zn-fingers motifs (Figure 1b) [23]. ARID-containing proteins are involved in a variety of biological processes including embryonic development, cell lineage gene regulation, and cell cycle control [24]. Besides the N-terminal domain, the RFX domain is another DNA-binding domain and was named after Regulatory Factor X, a protein that binds to the X-box of the MHC class II genes [25]. The two C2H2 Zn-fingers form the tandem CWCH2 (tCWCH2) motif that is the most popular DNA-binding motif among putative eukaryotic transcription factors [26]. Recent studies regarding the binding capability of Zinc-finger domains revealed that zinc fingers can bind not only to DNA but also to RNA and protein. Therefore, it is plausible that the double Zinc-finger of ARID2 has the potential to interact with DNA, RNA, and/or proteins [27, 28].
Figure 1. Somatic ARID2alterations identified in HCC.
a, Somatic alterations identified in the ARID2 gene. *, nonsense mutations; #, frame-shifting indels; &, splice site mutation. b, ARID2 protein and the inactivating alterations (red arrows) Truncating mutations are indicated by red arrows. ARID, AT-rich interaction domain; RFX, RFX-like DNA binding motif; GLN, Proline-and glutamine-rich region; ZnF, C2H2 Zinc Fingers.
ARID2: A NEW TUMOR SUPPRESSOR GENE IN HCC
The three types of ARID2 mutations identified in the HCV-associated HCCs are frame-shifting deletion, nonsense mutation and splice site alteration [17] (Figure 1a). These alterations are predicted to truncate and inactivate the ARID2 protein (Figure 1b). Interestingly, some of the mutations disrupt the Zn-finger motifs only, suggesting the importance of these motifs in the biological activity of ARID2.
In addition to genetic evidence, functional studies have shown that ARID2 was the only subunit in PBAF with short transcript half-life and suppression of ARID2 by small interfering RNA reduced the protein levels of other subunits in the PBAF complex [20]. Thus, ARID2 is essential for the stability of the PBAF complex. Interestingly, a recent study employing exomic sequencing has identified high-frequency (92/227, 41%) truncating mutations in the PBRM1 gene in renal clear cell carcinomas [29]. PBRM1 encodes the BAF180 protein, another chromatin targeting subunit of the PBAF complex. A third PBAF protein likely involved in tumor genesis is BRD7. The BRD7 gene is frequently deleted in the breast cancers with wild-type p53 [30]. Mutations in subunits of BAF, another SWI/SNF complex, have also been found in human cancers. Inactivating mutations in ARID1A (BAF250a) were identified in approximately 50% of ovarian clear-cell carcinomas [31], 30% of endometrioid carcinomas [32], 83% of gastric cancers with microsatellite instability [33], 10% of colorectal cancers [34], and 19% of transitional cell carcinoma of the bladder[35]. In addition, biallelic inactivating alterations in the hSNF5/INI1 gene which encodes a subunit shared by the PBAF and BAF complexes were found in almost all malignant rhabdoid tumors [36-38]. These observations strongly suggest that the SWI/SNF complexes have tumor-suppressing activities and ARID2 is a tumor suppressor gene.
ARID2 AND IFN SIGNAL TRANSDUCTION
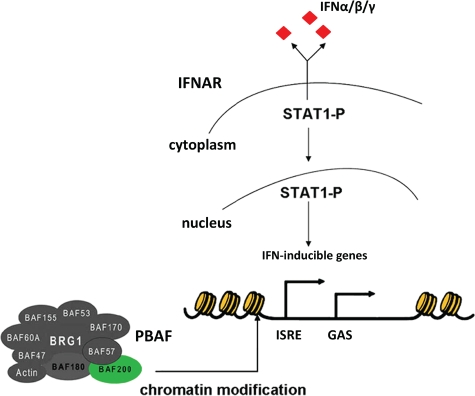
Increasing evidence suggests that the SWI/SNF complexes mediate cellular antiviral activities by binding to the IFN-inducible promoters to facilitate chromatin remodeling in response to IFN signaling [39-41] (Figure 2). A recent study has identified ARID2 as a specificity subunit in PBAF [20]. Functional analysis showed that suppression of ARID2 by small interfering RNAs specifically abolished transcription of the interferon-α-induced IFITM1 (interferon-induced transmembrane protein 1) gene, but not the others examined. Interestingly, IFITM1 is required for the IFN-induced anti-proliferative activity in hepatocellular carcinoma cells and non-malignant hepatocytes [42]. Thus, ARID2 seems to play an important role in regulating the expression of a subset of the interferon responsive genes and in mediating the anti-proliferative activity. It is conceivable that the ARID2 mutations abrogate the induced expression of these genes upon IFN signal transduction, which sets a stage for HCV virus and the infected host cells to escape from the IFN anti-proliferative activities. In addition, the infected host cells harboring ARID2 mutations may have lost the ability to express higher levels of the class I MHC molecules in response to IFN signaling, making them less visible to the cytotoxic T lymphocytes [43]. These cells will then proliferate in an uncontrolled fashion and have the opportunity to acquire more genetic alterations and clonally expand into full-blown cancers.
Figure 2. Schematic representation of the PBAF complex containing ARID2 (BAF200) which is involved in the transcriptional initiation of the human IFN-α/β/γ genes.
IFN, Interferon; IFNAR, Interferon-α/β/γ receptor; PBAF, Polybromo-associated BRG1-associated factor; ISRE, Interferon-sensitive response element; GAS, Interferon-γ-activated site; STAT1, Signal transducers and activators of transcription 1.
FUTURE STUDIES
Strong genetic and functional data have been provided to support the notion that ARID2 is an important tumor suppressor gene in HCC. The future work should capitalize on this discovery by focusing on the following research efforts that could lead to significant improvement in patient care.
Clinical studies should be conducted to investigate whether ARID2 mutations impact the prognosis of HCC patients in general and the prognosis of patients who have undergone treatment (e.g. curative therapy, IFN therapy, etc.). Adjuvant IFN therapy following curative HCC treatment has shown encouraging results, but more clinical studies are needed before it can be accepted as the standard of care for HCC patients [44-47]. Based on the observation that ARID2 was involved in IFN signaling, it is tempting to hypothesize that the ARID2 mutations might impact the outcome of the adjuvant IFN therapy. Thus, in future studies for adjuvant IFN therapy, the resected tumors should be genotyped for ARID2 mutations. The information obtained from these studies could be used to redirect specific health care resources to the patient populations that benefit the most.
We have observed an enrichment of the ARID2 mutations in HCV-associated HCC in the US and European patient populations (6/43 tumors, 14%), compared with HBV-associated HCC in the Chinese population (1/50 tumors, 2%, P = 0.046) [16]. However, a larger study including more Chinese and Western patients with HCV-associated HCC is required to confirm the result and rule out the possibility that ethnic or environmental factors contribute to this difference. It should be noted that HCV has six major genotypes which are clustered based on geographic regions [48, 49]. For example, type 1a and 1b of HCV are dominant among the US patients, whereas type 1b and 2a are more common in China. Thus, HCV genotyping should also be performed for Chinese and Western patients to investigate whether the ARID2 mutations are correlated with specific HCV subtypes.
The PBAF chromatin-remodeling complex seems to be a preferred target of tumorigenesis in both HCC and renal clear cell carcinoma. Furthermore, its specificity subunit ARID2 is required for the expression of a subset of the IFN-inducible proteins, including IFITM1 that mediates the IFN anti-proliferative activity. Studies to further characterize the biochemical and biological activities of the PBAF complex and its subunits, as well as the signal transduction pathways it regulates will provide insights that could help design novel therapeutic approaches.
HCC remains a leading cause of cancer deaths, despite worldwide efforts to develop more effective therapeutic approaches. Personalized therapy brings new hope for HCC patient care. Stratification of patients based on the genetic defects identified in the tumors could help redirect precious patient care resources to those who would benefit most and thus, reduce the health care cost. More importantly, treatment plans tailored to individual patients could result in better therapeutic response. The identification of ARID2 as an important tumor suppressor gene in HCC provides another target that can be exploited for personalized medicine.
REFERENCES
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Society AC. Cancer Facts and Figures. 2009 [Google Scholar]
- 3.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–87. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 4.Wang XW, Hussain SP, Huo TI, Wu CG, Forgues M, Hofseth LJ, Brechot C, Harris CC. Molecular pathogenesis of human hepatocellular carcinoma. Toxicology. 2002;181-182:43–7. doi: 10.1016/s0300-483x(02)00253-6. [DOI] [PubMed] [Google Scholar]
- 5.Yu MC, Yuan JM. Environmental factors and risk for hepatocellular carcinoma. Gastroenterology. 2004;127:S72–8. doi: 10.1016/j.gastro.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 6.Kowdley KV. Iron, hemochromatosis, and hepatocellular carcinoma. Gastroenterology. 2004;127:S79–86. doi: 10.1016/j.gastro.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 7.Brechot C. Molecular mechanisms of hepatitis B and C viruses related to liver carcinogenesis. Hepatogastroenterology. 1998;45(Suppl 3):1189–96. [PubMed] [Google Scholar]
- 8.Smela ME, Currier SS, Bailey EA, Essigmann JM. The chemistry and biology of aflatoxin B(1): from mutational spectrometry to carcinogenesis. Carcinogenesis. 2001;22:535–45. doi: 10.1093/carcin/22.4.535. [DOI] [PubMed] [Google Scholar]
- 9.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–46. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 10.Buendia MA. Genetics of hepatocellular carcinoma. Semin Cancer Biol. 2000;10:185–200. doi: 10.1006/scbi.2000.0319. [DOI] [PubMed] [Google Scholar]
- 11.Chen YJ, Yeh SH, Chen JT, Wu CC, Hsu MT, Tsai SF, Chen PJ, Lin CH. Chromosomal changes and clonality relationship between primary and recurrent hepatocellular carcinoma. Gastroenterology. 2000;119:431–40. doi: 10.1053/gast.2000.9373. [DOI] [PubMed] [Google Scholar]
- 12.Okabe H, Ikai I, Matsuo K, Satoh S, Momoi H, Kamikawa T, Katsura N, Nishitai R, Takeyama O, Fukumoto M, Yamaoka Y. Comprehensive allelotype study of hepatocellular carcinoma: potential differences in pathways to hepatocellular carcinoma between hepatitis B virus-positive and -negative tumors. Hepatology. 2000;31:1073–9. doi: 10.1053/he.2000.6409. [DOI] [PubMed] [Google Scholar]
- 13.Marchio A, Pineau P, Meddeb M, Terris B, Tiollais P, Bernheim A, Dejean A. Distinct chromosomal abnormality pattern in primary liver cancer of non-B, non-C patients. Oncogene. 2000;19:3733–8. doi: 10.1038/sj.onc.1203713. [DOI] [PubMed] [Google Scholar]
- 14.Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis--A comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32:970–9. doi: 10.1053/jhep.2000.19797. [DOI] [PubMed] [Google Scholar]
- 15.Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26:2166–76. doi: 10.1038/sj.onc.1210279. [DOI] [PubMed] [Google Scholar]
- 16.Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T, Sasaki Y, Imaoka S, Murata M, Shimano T, Yamaoka Y, Nakamura Y. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. 2000;24:245–50. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 17.Li M, Zhao H, Zhang X, Wood LD, Anders RA, Choti MA, Pawlik TM, Daniel HD, Kannangai R, Offerhaus GJ, Velculescu VE, Wang L, Zhou S, Vogelstein B, Hruban RH, Papadopoulos N, Cai J, Torbenson MS, Kinzler KW. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43:828–9. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, Workman JL, Crabtree GR. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–82. [PMC free article] [PubMed] [Google Scholar]
- 19.Xue Y, Canman JC, Lee CS, Nie Z, Yang D, Moreno GT, Young MK, Salmon ED, Wang W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci U S A. 2000;97:13015–20. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K, Wang W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–7. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patsialou A, Wilsker D, Moran E. DNA-binding properties of ARID family proteins. Nucleic Acids Res. 2005;33:66–80. doi: 10.1093/nar/gki145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilsker D, Probst L, Wain HM, Maltais L, Tucker PW, Moran E. Nomenclature of the ARID family of DNA-binding proteins. Genomics. 2005;86:242–51. doi: 10.1016/j.ygeno.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Mohrmann L, Langenberg K, Krijgsveld J, Kal AJ, Heck AJ, Verrijzer CP. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol Cell Biol. 2004;24:3077–88. doi: 10.1128/MCB.24.8.3077-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilsker D, Patsialou A, Dallas PB, Moran E. ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. 2002;13:95–106. [PubMed] [Google Scholar]
- 25.Steimle V, Mach B. Complementation cloning of mammalian transcriptional regulators: the example of MHC class II gene regulators. Curr Opin Genet Dev. 1995;5:646–51. doi: 10.1016/0959-437x(95)80034-4. [DOI] [PubMed] [Google Scholar]
- 26.Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409:832–3. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- 27.Hatayama M, Aruga J. Characterization of the tandem CWCH2 sequence motif: a hallmark of inter-zinc finger interactions. BMC Evol Biol. 2010;10:53. doi: 10.1186/1471-2148-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iuchi S. Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci. 2001;58:625–35. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, Bignell G, Butler A, Cho J, Dalgliesh GL, Galappaththige D, Greenman C, Hardy C, Jia M, Latimer C, Lau KW, Marshall J, McLaren S, Menzies A, Mudie L, Stebbings L, Largaespada DA, Wessels LF, Richard S, Kahnoski RJ, Anema J, Tuveson DA, Perez-Mancera PA, Mustonen V, Fischer A, Adams DJ, Rust A, Chan-on W, Subimerb C, Dykema K, Furge K, Campbell PJ, Teh BT, Stratton MR, Futreal PA. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–42. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drost J, Mantovani F, Tocco F, Elkon R, Comel A, Holstege H, Kerkhoven R, Jonkers J, Voorhoeve PM, Agami R, Del SG. BRD7 is a candidate tumour suppressor gene required for p53 function. Nat Cell Biol. 2010;12:380–9. doi: 10.1038/ncb2038. [DOI] [PubMed] [Google Scholar]
- 31.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, Heravi-Moussavi A, Giuliany R, Chow C, Fee J, Zayed A, Prentice L, Melnyk N, Turashvili G, Delaney AD, Madore J, Yip S, McPherson AW, Ha G, Bell L, Fereday S, Tam A, Galletta L, Tonin PN, Provencher D, Miller D, Jones SJ, Moore RA, Morin GB, Oloumi A, Boyd N, Aparicio SA, IeM S, Mes-Masson AM, Bowtell DD, Hirst M, Gilks B, Marra MA, Huntsman DG. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–43. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones S, Wang TL, IeM S, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Jr, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, Chan TL, Kan Z, Chan AS, Tsui WY, Lee SP, Ho SL, Chan AK, Cheng GH, Roberts PC, Rejto PA, Gibson NW, Pocalyko DJ, Mao M, Xu J, Leung SY. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011 doi: 10.1038/ng.982. LID - 10.1038/ng.982 [doi]. [DOI] [PubMed] [Google Scholar]
- 34.Jones1 S, Li M, Parsons DW, Zhang X, Wesseling J, Kristel P, Schmidt MK, Markowitz S, Yan H, Bigner D, Hruban RH, Eshleman JR, Iacobuzio-Donahue CA, Goggins M, Maitra A, Malek SN, Powell S, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2011 doi: 10.1002/humu.21633. LID - 10.1002/humu.21633 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, He M, Li Z, Sun X, Jia W, Chen J, Yang S, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–8. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–6. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 37.Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–9. [PubMed] [Google Scholar]
- 38.Jackson EM, Sievert AJ, Gai X, Hakonarson H, Judkins AR, Tooke L, Perin JC, Xie H, Shaikh TH, Biegel JA. Genomic analysis using high-density single nucleotide polymorphism-based oligonucleotide arrays and multiplex ligation-dependent probe amplification provides a comprehensive analysis of INI1/SMARCB1 in malignant rhabdoid tumors. Clin Cancer Res. 2009;15:1923–30. doi: 10.1158/1078-0432.CCR-08-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–78. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 40.Liu H, Kang H, Liu R, Chen X, Zhao K. Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex. Mol Cell Biol. 2002;22:6471–9. doi: 10.1128/MCB.22.18.6471-6479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui K, Tailor P, Liu H, Chen X, Ozato K, Zhao K. The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming. Mol Cell Biol. 2004;24:4476–86. doi: 10.1128/MCB.24.10.4476-4486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang G, Xu Y, Chen X, Hu G. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene. 2007;26:594–603. doi: 10.1038/sj.onc.1209807. [DOI] [PubMed] [Google Scholar]
- 43.Zhou F. Molecular mechanisms of IFN-gamma to up-regulate MHC class I antigen processing and presentation. Int Rev Immunol. 2009;28:239–60. doi: 10.1080/08830180902978120. [DOI] [PubMed] [Google Scholar]
- 44.Shen YC, Hsu C, Chen LT, Cheng CC, Hu FC, Cheng AL. Adjuvant interferon therapy after curative therapy for hepatocellular carcinoma (HCC): a meta-regression approach. J Hepatol. 2010;52:889–94. doi: 10.1016/j.jhep.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 45.Ikeda K, Arase Y, Saitoh S, Kobayashi M, Suzuki Y, Suzuki F, Tsubota A, Chayama K, Murashima N, Kumada H. Interferon beta prevents recurrence of hepatocellular carcinoma after complete resection or ablation of the primary tumor-A prospective randomized study of hepatitis C virus-related liver cancer. Hepatology. 2000;32:228–32. doi: 10.1053/jhep.2000.9409. [DOI] [PubMed] [Google Scholar]
- 46.Miyaguchi S, Watanabe T, Takahashi H, Nakamura M, Saito H, Ishii H. Interferon therapy for hepatocellular carcinoma patients with low HCV-RNA levels. Hepatogastroenterology. 2002;49:724–9. [PubMed] [Google Scholar]
- 47.Llovet JM, Di BAM, Bruix J, Kramer BS, Lencioni R, Zhu AX, Sherman M, Schwartz M, Lotze M, Talwalkar J, Gores GJ. Design and endpoints of clinical trials in hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:698–711. doi: 10.1093/jnci/djn134. [DOI] [PubMed] [Google Scholar]
- 48.Nainan OV, Alter MJ, Kruszon-Moran D, Gao FX, Xia G, McQuillan G, Margolis HS. Hepatitis C virus genotypes and viral concentrations in participants of a general population survey in the United States. Gastroenterology. 2006;131:478–84. doi: 10.1053/j.gastro.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 49.Lu L, Nakano T, He Y, Fu Y, Hagedorn CH, Robertson BH. Hepatitis C virus genotype distribution in China: predominance of closely related subtype 1b isolates and existence of new genotype 6 variants. J Med Virol. 2005;75:538–49. doi: 10.1002/jmv.20307. [DOI] [PubMed] [Google Scholar]