Abstract
Hepatitis C virus (HCV) is a considerable global health problem for which new classes of therapeutics are needed. We developed a high-throughput assay to identify compounds that selectively block translation initiation from the HCV internal ribosome entry site (HCV IRES). Rabbit reticulocyte lysate conditions were optimized to faithfully report on authentic HCV IRES-dependent translation relative to a 5′ capped mRNA control. We screened a library of ~430,000 small molecules for IRES inhibition, leading to ~1,700 initial hits. After secondary counter screening the vast majority of hits proved to be luciferase and general translation inhibitors. Despite well-optimized in vitro translation conditions, in the end we found no selective HCV IRES inhibitors but did discover a new scaffold of general translation inhibitor. The analysis of these molecules, and the finding that a large fraction of false positives resulted from off-target effects, highlights the challenges inherent in screens for RNA-specific inhibitors.
Keywords: Hepatitis C virus (HCV), IRES, luciferase, high-throughput screen, rabbit reticulocyte lysate
Introduction
Hepatitis C virus (HCV) is a significant global health problem with ~180 million people infected worldwide.1,2 Most patients (75–85%) develop chronic disease, leading to cirrhosis and hepatocellular carcinoma in approximately 20% of these cases.1 The current treatment for HCV is the combination of pegylated interferon-α and ribavirin, which has limited efficacy and causes significant side effects. New classes of anti-HCV compounds with varied mechanisms of action are needed to improve the current standard of care.3
The 5′-untranslated region (5′-UTR) of the HCV genome, a positive-sense, single-stranded RNA, is attractive as an anti-viral target due to its importance in the viral life cycle. In particular, nucleotides (nts) 40–372 comprise an internal ribosome entry site (IRES) by forming a specific secondary and tertiary RNA structure essential for viral protein synthesis.4,5 Both the structure and mechanism of the IRES have been studied extensively, revealing some of the molecular events that lead to translation initiation.6 The IRES RNA binds directly to the human 40S ribosomal subunit and to eukaryotic initiation factor 3 (eIF3).7,8 The IRES thereby bypasses the need for most initiation factors involved in typical cellular translation initiation, such as the cap-binding eIF4F complex and scanning-associated factors eIFs 1, 1A, and 4A.7 Specific steps in this pathway can be blocked by mutation or deletion of particular regions of the IRES RNA.9–11
The IRES is one of the most conserved regions of the HCV genome because it must function both in ribosome recruitment to the 5′ end of the plus-strand RNA and in viral replication from the 3′ end of the minus-strand.2 Despite its functional importance, the IRES is a challenging drug target, as it has no enzymatic activity or explicit active site, but instead uses an extended surface to interact with both the 40S ribosomal subunit and eIF3.12,13 While inhibition of HCV IRES translation with an antisense oligonucleotide to disrupt the structure of a key region of the IRES has been preliminarily successful,14,15 direct inhibition of IRES binding to 40S subunits or to eIF3 may be difficult to achieve with a small molecule.
Importantly, however, the IRES is more than a molecular scaffold, since mutations that do not affect its affinity for the host translation machinery can nonetheless dramatically reduce translation efficiency.8,11 For example, conformational flexibility of the domain II hairpin is required to induce a conformational change in the 40S subunit,13 a necessary precursor to formation of active 80S ribosomes. An attractive possibility is that small molecules can be found that specifically disrupt local structures or conformational dynamics of the RNA required for IRES function.
In an effort to discover such small molecule inhibitors of the HCV IRES, we developed a high-throughput assay based on IRES-dependent in vitro translation in rabbit reticulocyte lysate. Conditions for this assay were designed to report on authentic HCV IRES translation relative to a 5′ capped mRNA control. We screened a library of 426,736 small molecules for IRES inhibitors, leading to ~1,700 initial hits of which 59 appeared promising after secondary assays. That a large number of these molecules proved to be off-target luciferase inhibitors underscores the challenges in screens for compounds that recognize RNA targets and in using enzymes as read-outs for activity.
Materials and Methods
Reagents
Restriction endonucleases, calf intestinal alkaline phosphatase and T4 DNA ligase were purchased from New England Biolabs (Beverly, MA). [35S]methionine (>1000 Ci/mmol) was obtained from GE Life Sciences (Boston, MA). Plasmid DNA preparation, restriction enzyme digestion, agarose gel electrophoresis of DNA and RNA, DNA ligation and bacterial transformations were carried out using standard methods.16
Plasmid Construction and RNA Transcription
A DNA fragment encoding the HCV IRES sequence was generated by PCR using a genotype 1a HCV IRES construct (H77 strain,17 nts 40–372) as the template. The resulting DNA fragment was ligated into the EcoRI and BamHI restriction sites of pUC19 to form the parent plasmid for all subsequent constructs. Derivative plasmids encoding the IRES with domain II deleted (ΔdomII, nts 40–119 deleted) were generated by using QuikChange mutagenesis (Stratagene). The firefly (FF) and Renilla (RN) luciferase reporter genes were amplified from pGL3 and pRL-TK, respectively (Promega), and cloned between the BamHI and HindIII restriction sites of pUC19. Capped messages had a template-encoded poly(A) tail of 62 nt. All constructs were verified by DNA sequencing. IRES and 5′ capped RNAs were prepared by in vitro transcription (Megascript and mMESSAGE T7 system, respectively, Ambion) according to the manufacturer’s instructions. The 5′ capped-messages were generated with a GTP:m7GpppGTP ratio of 1:10 to ensure >90% capping efficiency.
In vitro translation
In vitro translations were performed in rabbit reticulocyte lysate and translation activity was measured using either a luciferase reporter assay or radiolabeling with [35 S]methionine, according to the manufacturers’ instructions (Promega and Life Technologies). Unless otherwise indicated, in vitro translations were carried out in 15 μl reactions, containing 2 μL of nuclease-treated RRL (0.5 μL Ambion + 1.5 μL Promega; the composition of RRL was optimized for translation efficiency and accuracy, data not shown), 1 ng/μl and 3 ng/μL of IRES-RN and 5′ cap-FF reporter mRNAs, respectively, and 5 ng/μL polyC RNA as a carrier. Translation reactions were adjusted to final concentrations of 2.6 mM Mg(OAc)2, 45 mM KCl and 90 mM KOAc, and also contained 1 mM amino acids, 1% dimethyl sulfoxide (DMSO) and 0.1% complete protease inhibitor (Roche). Translation reactions were incubated at 30°C for 90 min and terminated by the addition of puromycin to a final concentration of 20 μM.
For high-throughput screening (HTS), reactions were scaled proportionally to 5 μL total volume and the positive controls for IRES-RN and 5′ cap-FF translation inhibition were a DNA oligonucleotide complementary to the IIId loop of the IRES (10 μM) and puromycin (20 μM), respectively. The IIId oligo sequence used was 5′-ACCCAACACTACTCGGC-3′.14 A combination of DualGlo, BrightGlo, and Dual Luciferase Assay System kits (Promega) was used in preliminary assays, as noted in figure legends, to measure firefly and Renilla luciferase activities from the same well. The DualGlo luciferase assay kit (Promega) was selected for HTS due to its long half-life of luminescence, and was used according to the manufacturer’s instructions with minor adjustments. Luminescence was measured by TopCount (PerkinElmer) for 96-well plates and by Envision for 384-well plates (PerkinElmer). For the secondary enzyme interference screen, in vitro translation reactions were performed as above but in the absence of compounds. After quenching translation reactions with puromycin, compounds were added and incubated with the translated enzymes for 30 min at room temperature before measuring luciferase activity.
For [35S]methionine incorporation experiments, translation reactions were performed as above, except that methionine was omitted from the amino acid mixture, and 10μCi of [35S]methionine was added. Aliquots from translation mixtures were mixed with SDS sample buffer, boiled for 2 min and resolved on a 10% SDS polyacrylamide gel. Gels were fixed in 10% methanol, 7.5% acetic acid, and treated with ENHANCE (New England Nuclear). The level of translation of FF luciferase (61 kDa) and RN luciferase (36 kDa) was quantified by phosphorimaging and densitometry using ImageQuant TL (Molecular Dynamics).
Compound library and liquid handling
The chemical library for high-throughput screening consisted of 426,736 compounds (Gilead Sciences, Inc). Liquid handling protocols were optimized for distribution of (a) the compound library (Bravo, Velocity11), (b) translation mix to minimize formation of air bubbles (Deerac, Tecan), (c) RNA templates to ensure precise dispensing at small volumes (Deerac, Labcyte), and (d) luciferase substrates (microFill, Biotek).
Data analysis
Data analysis was carried out by Graphpad Prizm5.1 and Spotfire. The Z-factor was calculated as previously described.18 The normalized percentage of inhibition values (NPI, %) were calculated for each HTS plate by setting the average signal of the negative control (1% DMSO) as 0% inhibition and the average signal of the positive control (IIId oligo or puromycin) as 100% inhibition.
Results
Translation inhibition screen design
A robust functional assay was established to screen for small molecule inhibitors that block HCV IRES-mediated translation. To enable independent assessment of 5′ cap-dependent and cap-independent translation initiation from the same well, we designed two monocistronic mRNAs encoding distinct luciferase enzymes, one driven by a 5′ cap and the other by the HCV IRES (Fig. 1A; genotype 1a, H77 strain, nts 40–372). The IRES primary sequence and tertiary structure are highly conserved among all 6 genotypes,19,20 making it likely that inhibitors found against this genotype would have pan-genotypic activity. For the translation of the reporter constructs, we considered both HeLa cell extract and rabbit reticulocyte lysate (RRL). Although HeLa translation extracts are reported to faithfully recapitulate the translation behavior observed in cells,21 low activity prevented obtaining sufficient signal for the statistical analysis required for a high-throughput screen. In contrast, unmodified RRL has limited 5′ cap-dependence and low sensitivity to mutations in the HCV IRES,22,23,this study but produces high levels of translation. These observations led us to focus on optimization of RRL as the system in which to conduct inhibitor screens.
FIG. 1.
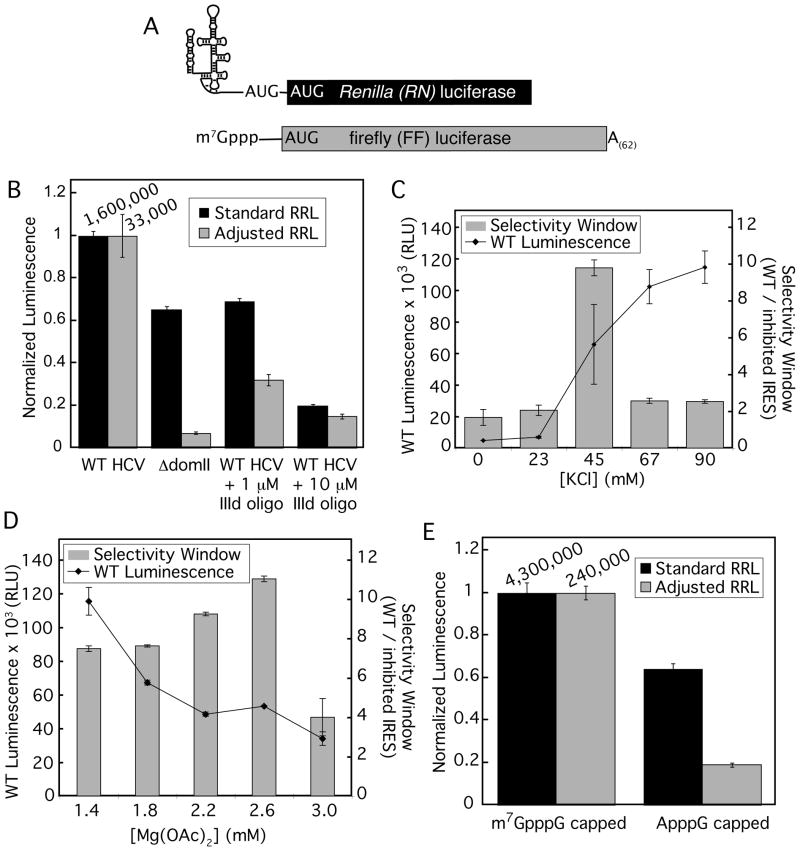
Optimization of rabbit reticulocyte lysate for authentic translation of HCV IRES with a firefly luciferase reporter. (A) Schematic showing IRES-RN and 5′ cap-FF reporter mRNAs used in this study. (B) In vitro translation of HCV IRES-FF mRNA in standard or adjusted RRL and the effect of deleting domain II (ΔdomII), or addition of a DNA oligo that hybridizes to the IIId domain of the IRES (IIId oligo). Effects of (C) KCl or (D) Mg(OAc)2 titration on translation efficiency of WT IRES and the ΔdomII mutant IRES. The absolute luminescence signal of WT IRES-containing mRNA is shown in relative light units (RLU) and selectivity window is defined as the ratio of signal from WT over the ΔdomII IRES. (E) Comparison of translation of m7GpppG- and ApppG-capped FF RNAs in standard and adjusted RRL. Luciferase activity was measured using Luciferase Assay System. Note that all of the above in vitro translation reactions were carried out in pre-HTS conditions (8.5 μL RRL/15 μL reaction, in the absence of polyC RNA and DMSO, with 100% Promega RRL).
In light of previous results showing that the salt concentrations in RRL could be adjusted to stimulate faithful 5′ cap-dependent scanning,24 we examined the effect of RRL salt concentration on the fidelity of HCV IRES-mediated translation. To distinguish IRES-dependent from promiscuous translation, we utilized two established methods for inhibiting HCV IRES function: (a) deletion of domain II (nts 40–119, referred to as ΔdomII9) and (b) inhibition with a DNA oligonucleotide complementary to the IIId loop of the IRES, a region required for recruitment of the 40S ribosomal subunit (referred to as ‘IIId oligo’).14
Under standard RRL conditions (79 mM KOAc, 0.5 mM MgCl2, Promega), a reporter mRNA containing the ΔdomII IRES was translated with 65% of the activity of an mRNA containing the wild type IRES (WT; Fig. 1B, black columns). By comparison, the ΔdomII mRNA has been shown to have only 3%, 2%, and 20% activity in mice, HeLa cells and HeLa S10 extracts, respectively.11,21 Notably, the inhibitory activity of the IIId oligo against IRES-mediated translation was also limited under these conditions (Fig. 1B). When the salt concentration of the RRL reactions was adjusted to a previously identified condition (2.2 mM Mg2+, 45 mM KCl, 90 mM KOAc),24 the ΔdomII-containing mRNA showed a significant reduction in translation activity (7% of the wild type IRES-containing mRNA). Furthermore, the IIId oligo was more effective at inhibiting IRES-driven translation in the adjusted RRL when compared to standard RRL (Fig. 1B). The ratio of luciferase activities from translation of WT IRES-containing mRNA versus either ΔdomII IRES or WT IRES in the presence of the IIId oligo defined the “selectivity window” of the assay. This selectivity window can be considered to report both on the authenticity of IRES translationn and our ability to detect inhibition of this translation. Further optimization of KCl (Fig. 1C) and Mg(OAc)2 (Fig. 1D) showed that the largest selectivity window of 11-fold occurred at 45 mM KCl and 2.6 mM Mg(OAc)2.
Under these adjusted RRL conditions, a 5′ cap-containing mRNA was translated ~5-fold better than the same mRNA containing a non-physiological cap (Fig. 1E). Thus, the adjusted RRL displays increased fidelity for both IRES- and 5′ cap-dependent translation. Although the overall activity of the salt-adjusted RRL was decreased relative to unaltered conditions (nearly 20-fold reduction for 5′ capped mRNA and ~50-fold for IRES-containing mRNA), these activity levels were significantly higher than those observed with HeLa extracts (which showed at least 20-fold lower activity in the presence of greater than 100-fold more mRNA; data not shown).
Reporter mRNA choice and assay optimization
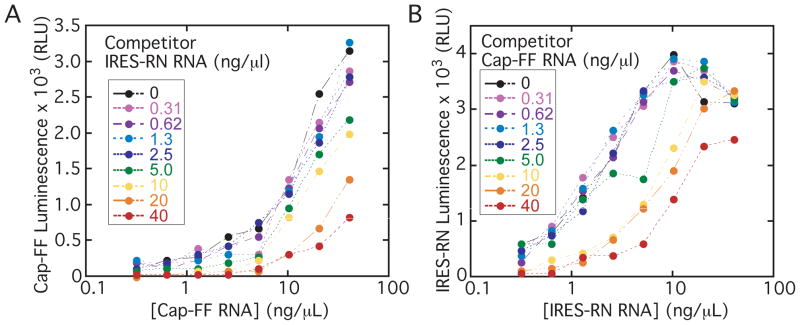
To identify IRES-specific inhibitors, we designed a high-throughput screen using two reporter mRNAs encoding distinct luciferase enzymes, Renilla (RN) and firefly (FF). To maximize the signal:noise ratio in the primary screen, we wanted each well to contain as much of the two monocistronic mRNAs as possible without introducing competition between them, which could result from one reporter preferentially monopolizing ribosomes and other translation factors in the extract. Titrations of 5′ capped-FF and IRES-RN mRNAs were performed using the optimized salt conditions established above, measuring the signal from one reporter at a time. As IRES-RN mRNA concentration was increased to moderate to high concentrations, the signal from 5′ capped-FF translation dropped (Fig. 2A, note the vertical spread), which is indicative of interference between the two messages. Similarly, the highest concentrations of 5′ capped-FF mRNA interfered with the signal from IRES-RN translation (Fig. 2B), although competition in this direction does not appear to be as strong. Significant interference at high concentrations of either reporter mRNA was also demonstrated for the converse reporter pair (IRES-FF and 5′ capped-RN; see Supplementary Fig. 1 at http://jbx.sagepub.com/supplemental). Based on the results of these two-dimensional titrations of reporter mRNAs, we chose roughly 1 ng/μL IRES-RN RNA and 3 ng/μL 5′ capped-FF RNA for the primary screen; the concentration of templates used was adjusted, within 3-fold, for each new batch of in vitro transcribed RNA. Different concentrations of each monocistronic mRNA were ideal to maximize signal while avoiding competition between the mRNAs, underscoring an important advantage of using two monocistronic messages as opposed to a single bicistronic message where the relative concentrations cannot be varied.
FIG. 2.
Two-dimensional titrations of IRES-RN and 5′ cap-FF mRNAs in adjusted RRL. The concentration of each of the two mRNAs was varied between 0 ng/μl and 40 ng/μl and the absolute luminescence activities of (A) 5′ cap-FF and (B) IRES-RN mRNA are plotted. Luciferase activities were measured using the DualGlo system.
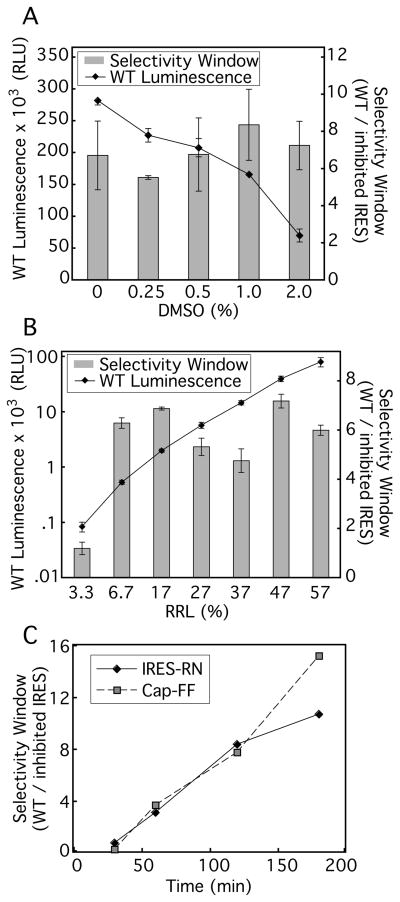
The RRL-based translation assay conditions were further adjusted to be suitable for high-throughput screening (HTS) of potential small molecule inhibitors. Because the small molecules to be tested were dissolved in DMSO, we examined the effect of this organic solvent on the activity and selectivity window of the translation reactions. Although the signal from IRES-FF mRNA steadily decreased as DMSO concentration increased,(Fig. 3A), the selectivity window between WT IRES- andΔdomII IRES-containing mRNAs, representing the faithfulness of the extract, remained greater than 6.5 in the presence of up to 2% DMSO. These data show that the final DMSO concentration of 1% was compatible with this assay.
FIG. 3.
Optimization of RRL translation assay for high-throughput screening. The absolute luminescence signal of WT IRES-FF mRNA is shown in relative light units (RLU). (A) Effect of DMSO titration on efficiency (luminescence) and authenticity of IRES translation (WT signal over ΔdomII signal defined the selectivity window). These in vitro translation reactions were conducted at the pre-HTS RRL concentration (8.5 μL RRL/15 μL reaction, with 100% Promega RRL). (B) Effect of RRL concentration on efficiency and authenticity of IRES translation. The selectivity window was measured using the ΔdomII IRES for RRL concentrations greater than and equal to 17% (vol/vol), and addition of the 10 μM IIId oligo for RRL concentrations less than 17% (vol/vol). (C) Time course of in vitro translation assay. 3 ng/μl 5′ cap-FF and 1 ng/μl IRES-RN mRNA were translated in the optimized RRL conditions for 60–180 minutes. The selectivity window for 5′ cap-FF translation was calculated as the ratio of the signal in the absence of puromycin over the signal at 20 μM puromycin. The selectivity window for IRES-RN was calculated as the ratio of the signal in the absence of IIId oligo over the signal at 10 μM IIId oligo. Luciferase activity in all panels was measured using DualGlo system.
It was also possible to significantly reduce the amount of RRL used in the assay for implementation in the high-throughput screen. The selectivity window and the signal from WT IRES-containing mRNA were measured as RRL concentration was varied from 3.3–57% (vol/vol), while the overall salt concentration was kept constant. The luminescence signal decreased by ~1000-fold over the range of titration, but the selectivity window remained relatively stable from ~57% down to 6.7% (Fig. 3B). Lastly, it was important to ensure the linearity of reaction kinetics. At 13% RRL, the condition chosen for HTS, the 5′ capped-FF signal remained linear for at least three hours while the IRES-RN signal began to plateau at ~120 minutes (Fig. 3C). Thus, an incubation time of 90 minutes was chosen for the high-throughput screen, as it gave substantial signal and was within the linear range. In the final format for the primary screen, 5 μL translation reactions containing 13% RRL (Ambion:Promega = 1:3) were incubated at 30 °C for 90 min.
Primary Screen
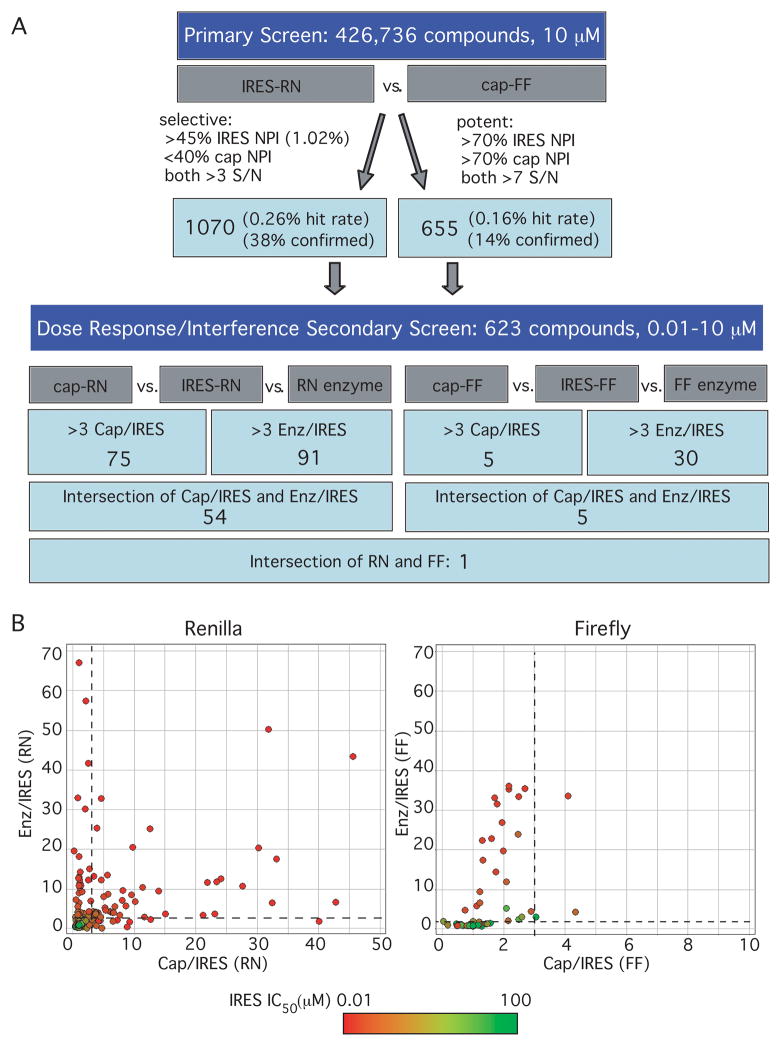
Using the assay conditions outlined above, a total of 426,736 compounds (average MW ~ 550) were tested in a primary screen. The Z-factor was determined for each 384-well plate. The average Z-factors for 5′ cap-FF and IRES-RN reporters were 0.71 and 0.62, respectively (ranging between 0.5 and 0.86 (see Supplementary Fig. 2 at http://jbx.sagepub.com/supplemental). The results from plates with Z-factors which did not meet the cut-off of 0.5 were discarded and the assay plates rerun. Three known translation-inhibiting antibiotics were present in the library and inhibited both the IRES-RN and 5′ cap-FF message in the primary screen (see Supplementary Table 1 at http://jbx.sagepub.com/supplemental), confirming the robustness of screen. Approximately 1% of the total compounds tested reduced IRES-RN luciferase signal by at least 45% (reported as normalized percentage of inhibition (NPI)) (Fig. 4A). Within this group, ~25% were IRES-RN selective as the 5′ cap-FF signal had an NPI less than 40%. In addition to these IRES-RN selective hits, ~650 compounds were identified as highly potent inhibitors of both IRES-RN and 5′ capped-FF reporters (>70% NPI). These inhibitors were included with the selective inhibitors to be tested in a secondary screen since selectivity for the IRES over a 5′ cap might not be observed for a potent inhibitor at the single concentration of 10 μM tested in the primary assay.
FIG. 4.
Results of high-throughput screen. (A) Screening cascade for primary and secondary screens, showing the criteria for hits and the observed hit rates. Normalized percentage of inhibition (NPI) was defined as the relative translation inhibition compared to positive controls (puromycin for 5′ cap-mRNA or IIId oligo for IRES-mRNA). The ~1,700 hits from the primary screen were tested in an identical confirmation assay in triplicates (at least two out of three repeated hits were counted as positive) and the confirmation rates are shown. These confirmation rates are similar to what has been observed for other high-throughput screens (observations at Gilead Sciences, Inc). (B) Selectivity index (SI) plots. Enz/IRES SI was defined as the ratio of the IC50 of a compound against the luciferase enzyme vs. IRES-driven translation of the enzyme, and the Cap/IRES SI was the ratio of a compound’s IC50 against 5′ cap-mRNA vs. IC50 against IRES-mRNA translation. Data points are shown for the 623 compounds analyzed in the secondary screen, and are colored by their IC50 against the IRES reporter, with red being potent inhibitors and green showing no inhibition of the IRES reporter up to 100 μM. Dashed lines indicate the SI>3 cut-off of the secondary assays.
Secondary Screen
Of these ~1,700 primary hit compounds, a representative group of 623 compounds was purchased for follow-up dose-response assays. These compounds were chosen based on commercial availability and their drug-like nature, determined by an automated in silico triage based on molecular weight (<600 Daltons), lack of reactive groups (e.g. methylating groups), and solubility (calculated partition coefficient, log P < 5). The goals of these assays were (a) to define the selectivity window between IRES- and 5′ cap-dependent inhibition, even for potent hits, and (b) to distinguish authentic IRES inhibitors from compounds that inhibit Renilla luciferase preferentially over firefly luciferase. Two formats of secondary screens were used. One was essentially identical to the primary screen except it consisted of the opposite pair of reporters: IRES-FF and 5′ capped-RN mRNA. The second test was a luciferase enzyme interference assay in which the translation of IRES-RN and 5′ capped-FF was performed as in the primary screen but in the absence of any compounds. Once the translation reaction was quenched with puromycin, compounds were added and incubated with the translated enzymes before measuring luciferase activity. In total, these assays yielded six sets of data: IC50 values for each compound against IRES-RN, 5′ capped-RN, IRES-FF, 5′ capped-FF (compounds present during translation reactions), and IC50 values against Renilla enzyme and firefly enzyme (compounds added after the translation of enzyme). Data for these dose-response follow up assays were plotted as the selectivity index (SI; ratio of IC50) calculated for the 5′ cap/IRES and the enzyme (Enz)/IRES comparisons made for each luciferase (Fig. 4B).
The vast majority of compounds in both the Renilla and the firefly assays showed a selectivity index of less than 3 for both Enz/IRES and 5′ cap/IRES, falling into the lower left-hand quadrant in Figure 4B. Some of these compounds, colored in green, had very high IC50s against the IRES-containing message, suggesting that they were simply false positives from the primary screen. Many other compounds, colored in red or orange, had low IC50s against the IRES-containing message, indicating that they were correctly identified by the primary screen as inhibitors of the IRES-containing message. These compounds are therefore most likely luciferase inhibitors that equally inhibit the luciferase signal, whether it is driven by the IRES, 5′ cap, or comes directly from the enzyme.
There were also a large number of compounds present in the upper left-hand quadrants of the plots in Figure 4B, showing selectivity for IRES-driven translation over the enzyme alone, but not over 5′ cap-dependent translation. Many of these molecules are likely to be general translation inhibitors capable of blocking translation driven either by the 5′ cap or the IRES. Consistent with this inhibition acting at the level of general translation, these compounds behaved similarly against both RN and FF reporters: of the 21 compounds that were found in this upper left-hand quadrant for FF and the 34 compounds in this quadrant for RN, 18 overlap. It is also possible that some of these compounds are general luciferase inhibitors that were not hits in the enzyme alone assays due to unavoidable differences in the setup of this assay (see Discussion).
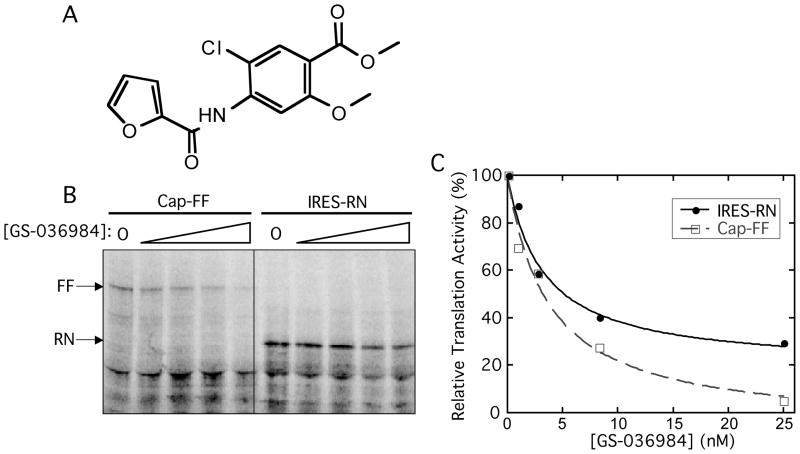
We chose compounds that had a selectivity index greater than 3 for both Enz/IRES and 5′ cap/IRES as potential IRES-selective inhibitors. These compounds fell into the upper right quadrant of either plot in Figure 4B. As the screening cascade shows (Fig. 4A), many more compounds met this criterion with the Renilla reporter than the firefly reporter. Of these compounds, only one appeared to be a selective inhibitor with both reporter enzymes (GS-036984, Fig. 5A).
FIG. 5.
Non-selective inhibition of GS-036984 against IRES- and 5′ cap-dependent translation. (A) Structure of GS-036984. (B,C) [35S]methionine incorporation translation assay of IRES-RN and 5′ cap-FF reporter in the presence of increasing concentrations of GS-036984 from 0 to 25 μM. Translation activity was measured by phosphorimaging of the fixed and dried SDS PAGE gel (B), and normalized to levels in the absence of the compound. (C) The percentage of inhibition was quantified by phosphorimaging and densitometry as the ratio between the intensity of the band in the presence of absence of compounds, after subtracting the background. Data were fit in GraphPad Prism, yielding IC50 values of 3.6 ±1.0 against 5′ cap-FF and 3.2±0.8 against IRES-RN.
Validation Assay
While GS-036984 appeared to be an IRES-selective inhibitor after the secondary screens using both FF and RN reporters, the selectivity index of 5′ cap/IRES (RN) for this compound was 3.9, barely meeting the cut-off of 3. Therefore we wanted to additionally validate this compound’s activity in an assay completely independent of luciferase activity. [35S]methionine incorporation followed by SDS PAGE analysis was used to observe translation of the IRES-RN and 5′ capped-FF messages in the presence of GS-036984 (Fig. 5B). Contrary to expectations from the screen, this compound was found to inhibit translation from both 5′ cap-FF mRNA and IRES-RN reporters and thus is in fact a general translation inhibitor (Fig. 5C; see discussion).
We also examined the activity of the 59 compounds that were identified as selective IRES inhibitors with either FF or RN reporters, but not both, using the [35S]methionine-incorporation translation assay. One additional compound related to the scaffold above also inhibited both 5′ cap- and IRES-driven translation. To the best of our knowledge, these compounds represent a new scaffold of general translation inhibitor. The other compounds tested, however, failed to inhibit translation of either message (data not shown).
Discussion
The HCV IRES RNA is an attractive drug target due to its central role in regulating viral protein synthesis and pan-genotypic sequence conservation. In this study, we screened a small molecule library for specific inhibitors of HCV IRES function. The results of this work highlight both successes in high-throughput assay development and challenges inherent in identifying compounds that recognize RNA targets.
Previous work established that direct IRES binding by an antisense oligonucleotide can block IRES-mediated protein synthesis.14 Although this nucleic acid based candidate (ISIS 14803) reached Phase I clinical trials, limited on-target activity and aminotransferase flares led to a halt of clinical development.15 Additionally, there have been other attempts to identify IRES-specific inhibitors by high-throughput screens with smaller libraries. Fragment-based screening using mass spectrometry has identified small molecule binders to domain II of the IRES which are inhibitors of an HCV replicon assay.25 A dual luciferase-based HTS assay in Krebs extracts with a bicistronic reporter did not reveal any IRES-specific compounds.26 We sought to identify new chemical scaffolds that directly target IRES function and could be further optimized to yield potent oral antiviral agents.
A screening assay was developed using RRL supplemented with additional salts, which improved the fidelity of both cap-dependent and HCV IRES-mediated translation. These results are analogous to what has previously been seen with the encephalomyocarditis virus (EMCV) IRES (although the EMCV IRES requires a different set of translation initiation factors from the HCV IRES).27 The use of two monocistronic reporters (one IRES-driven and one 5′ cap-driven) enabled each reporter’s signal to be maximized independently, while avoiding competition between the two reporter templates, which cannot be achieved using a single bicistronic mRNA. We noted that in general, many more compounds in the secondary screen inhibited the RN enzyme than the FF enzyme, and no compounds inhibited both. This result suggests that the primary screen was effective at excluding general luciferase inhibitors, such as competitive inhibitors of the ATP substrate or protein aggregants. However, ~3% of the primary hits appeared to be general translation inhibitors (in the upper left-hand quadrant of Fig. 4B) which would have ideally been eliminated in the primary screen.
Due to issues with luciferase interference, an [35S]methionine-incorporation validation assay was essential to verify the inhibition activity of the fifty-nine compounds which appeared to be selective IRES inhibitors in the secondary screen with either the firefly or Renilla reporter. Two of these compounds were found to be general translation inhibitors and, to the best of our knowledge, represent a new chemical scaffold of general translation inhibitors. The compound GS-036984 was selected from the primary screen as a potent hit, consistent with it inhibiting both 5′ cap- and IRES-dependent translation. The selectivity indices determined in the secondary screen for 5′ cap/IRES were barely above the cut-off of 3.0 (3.9 with the Renilla reporter and 3.3 with the firefly reporter). While this low selectivity suggested a potential preference for inhibiting the IRES, the direct visualization of translation products without interference of luciferase reporters clearly showed that this compound inhibits both 5′ cap-dependent and IRES-dependent translation (Figs. 5B and C).
In the [35S]methionine-incorporation validation assay, 57 out of 59 of the compounds identified in the secondary luciferase screen showed no inhibition against either IRES- or 5′ cap-dependent translation. This observation suggests that these compounds are likely to be direct inhibitors of Renilla luciferase enzyme. Several scaffolds emerged from the compounds showing selective Renilla luciferase inhibition, such as para-amino-sulfonamides, sulfanyltriazoles, and dihydropyrrolones. The ten most potent Renilla-specific inhibitors identified in the screen are shown in Supplementary Table 2 at http://jbx.sagepub.com/supplemental. These Renilla luciferase inhibitors may not have been eliminated in the secondary screen due to differences in the assay conditions, such as the temperature of incubation and the higher final luciferase protein levels in the enzymatic interference assay due to the lack of DMSO during translation. A subset of compounds that showed no inhibition at the HTS conditions for the enzymatic interference assay (incubation at room temperature for 30 minutes) demonstrated enhanced inhibitory activity against the luciferase enzymes when the incubation conditions were changed to more closely match the primary assay (30°C for 90 minutes). Additionally, when the interference assay was performed using recombinant Renilla luciferase enzyme rather than in vitro translated enzyme, an even larger subset of compounds (~80% of the 54 hits) showed inhibition when incubated at 30°C for 90 minutes.
The relative abundance of luciferase inhibitors identified in this study reflects the fact that it is much simpler chemically for a compound to block an enzyme active site than to bind to a macromolecular surface and inhibit a conformational change or an intermolecular interaction (note that a large fraction (~40%) of current drug targets are enzymes and many other drug targets also naturally bind to small molecules).28 Our secondary counter screens eliminated most, but not all, of the facile and prevalent direct inhibitors of Renilla luciferase. The high level of Renilla luciferase inhibitors selected in this screen suggests there is an inherent problem in using an enzymatic reporter assay as a means to screen for compounds that interact with a particularly challenging target, such as a structured RNA, as the reporter enzyme is itself a classical target of small molecules.
In order to expand our ability to devise new and diverse drugs, the scope of small molecule activities must be expanded to non-traditional targets, such as influencing macromolecular interactions and conformations, and efforts are underway to expand the diversity of small molecules in screening libraries.29 Based on our experience with enzyme inhibition masking the desired RNA-based inhibition, assays for new functions would do well to avoid traditional small molecule targets as read-outs for activity. Fluorescence, as opposed to luminescence, may be an alternative approach to couple translation activity to a spectroscopic signal. For example, members of the green fluorescence protein family do not have a defined active site for binding small molecules and may therefore be less subject to direct reporter inhibition. A dual reporter screen could be conducted with a pair of red and green fluorescent proteins, though spectroscopic interference from heterocyclic compounds in the library would need to be considered.30 Additionally, in an effort to reduce enzyme interference, the active site of firefly luciferase has been redesigned and evolved to discourage small molecules binding to the luciferin and ATP pockets. This new enzyme, ‘Ultra-Glo’, shows a marked loss of inhibition from several scaffolds that inhibited the original firefly luciferase.31
Other strategies for identifying RNA-binding small molecules as inhibitors that do not require enzymatic read-outs, such as fragment-based or in silico screening, could also be considered as alterative approaches.25,32,33 A FRET-based screen for small molecule binders to RNA has previously been used to find compounds that bind to and stabilize the HCV IRES IIId loop,34 but whether these compounds are effective inhibitors of HCV IRES translation has yet to be determined. As a complement to such small molecule binding screens, it is certainly attractive to envision direct functional screens to look for inhibitors of IRES-mediated translation. However, the present study shows that such assays are not as straightforward as they may first appear.
In this study we validated the use of rabbit reticulocyte lysate with increased salt concentrations as a system to study the effects of mutations and inhibitors of the HCV IRES. The current screen has provided insights into how deleterious direct enzyme inhibition can be to an otherwise robust high-throughput assay for a difficult target, such as the HCV IRES. Additionally, a new scaffold has been identified of a general eukaryotic translation inhibitor, the mechanism of which will be interesting to investigate in the future.
Supplementary Material
Acknowledgments
We thank members of the Doudna laboratory for helpful discussions and comments on the manuscript, S. Coyle, A. Law and B. Reid for assistance with data collection, and Gilead IRES team members, M. Desai, M. McGrath, R. Sakowicz, S. Swaminathan and J. Ward for insightful discussion and comments on project implementation. This work was supported by a Program project grant from the National Institutes of Health and a research gift generously provided by Gilead Inc. [to J.A.D].
References
- 1.Webster DP, Klenerman P, Collier J, Jeffery KJ. Development of novel treatments for hepatitis C. Lancet Infect Dis. 2009;9:108–117. doi: 10.1016/S1473-3099(09)70020-9. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki T, Aizaki H, Murakami K, Shoji I, Wakita T. Molecular biology of hepatitis C virus. J Gastroenterol. 2007;42:411–423. doi: 10.1007/s00535-007-2030-3. [DOI] [PubMed] [Google Scholar]
- 3.Nelson DR. Hepatitis C drug development at a crossroads. Hepatology. 2009;50:997–9. doi: 10.1002/hep.23208. [DOI] [PubMed] [Google Scholar]
- 4.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513–29. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 6.Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 7.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci USA. 2004;101:16990–95. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–80. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 12.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–5. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 13.Spahn CM, Kieft JS, Grassucci RA, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 14.Tallet-Lopez B, Aldaz-Carroll L, Chabas S, Dausse E, Staedel C, Toulme J. Antisense oligonucleotides targeted to the domain IIId of the hepatitis C virus IRES compete with 40S ribosomal subunit binding and prevent in vitro translation. Nucleic Acids Res. 2003;31:734–742. doi: 10.1093/nar/gkg139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McHutchinson JG, Patel K, Pockros P, et al. A phase I trial of an antisense inhibitor of hepatitis C virus (ISIS 14803), administered to chronic hepatitis C patients. J Hepatol. 2006;44:88–96. doi: 10.1016/j.jhep.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 17.Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci U S A. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 19.Han JH, Shyamala V, Richman KH, et al. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at the 3′ end. Proc Natl Acad Sci U S A. 1991;88:1711–5. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bukh J, Purcell RH, Miller RH. Sequence analysis of the 5′ noncoding region of hepatitis C virus. Proc Natl Acad Sci U S A. 1992;89:4942–6. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCaffrey AP, Ohashi K, Meuse L, et al. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol Ther. 2002;5:676–84. doi: 10.1006/mthe.2002.0600. [DOI] [PubMed] [Google Scholar]
- 22.Soto Rifo R, Ricci EP, Decimo D, Moncorge O, Ohlmann T. Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res. 2007;35:e121. doi: 10.1093/nar/gkm682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dasso MC, Jackson RJ. On the fidelity of mRNA translation in the nuclease-treated rabbit reticulocyte lysate system. Nucleic Acids Res. 1989;17:3129–44. doi: 10.1093/nar/17.8.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozak M. Evaluation of the fidelity of initiation of translation in reticulocyte lysates from commercial sources. Nucleic Acids Res. 1990;18:2828. doi: 10.1093/nar/18.9.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seth PP, Miyaji A, Jefferson EA, et al. SAR by MS. discovery of a new class of RNA-binding small molecules for the hepatitis C virus: internal ribosome entry site IIA subdomain. J Med Chem. 2005;48:7099–102. doi: 10.1021/jm050815o. [DOI] [PubMed] [Google Scholar]
- 26.Novac O, Guenier AS, Pelletier J. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 2004;32:902–15. doi: 10.1093/nar/gkh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson RJ. Potassium salts influence the fidelity of mRNA translation initiation in rabbit reticulocyte lysates: unique features of encephalomyocarditis virus RNA translation. Biochem Biophys Acta. 1991;1088:345–48. doi: 10.1016/0167-4781(91)90124-5. [DOI] [PubMed] [Google Scholar]
- 28.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–6. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen TE, Schreiber SL. Towards the optimal screening collection: a synthesis strategy. Angew Chem Int Ed Engl. 2008;47:48–56. doi: 10.1002/anie.200703073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inglese J, Johnson RL, Simeonov A, et al. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–79. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 31.Auld DS, Zhang YQ, Southall NT, et al. A Basis for Reduced Chemical Library Inhibition of Firefly Luciferase Obtained from Directed Evolution. J Med Chem. 2009;52:1450–58. doi: 10.1021/jm8014525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hermann T, Westhof E. Rational drug design and high-throughput techniques for RNA targets. Comb Chem High Throughput Screen. 2000;3:219–34. doi: 10.2174/1386207003331652. [DOI] [PubMed] [Google Scholar]
- 33.Bodoor K, Boyapati V, Gopu V, et al. Design and implementation of an ribonucleic acid (RNA) directed fragment library. J Med Chem. 2009;52:3753–61. doi: 10.1021/jm9000659. [DOI] [PubMed] [Google Scholar]
- 34.Baugh C, Wang S, Li B, Appleman JR, Thompson PA. SCAN--a high- throughput assay for detecting small molecule binding to RNA targets. J Biomol Screen. 2009;14:219–29. doi: 10.1177/1087057108330111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.