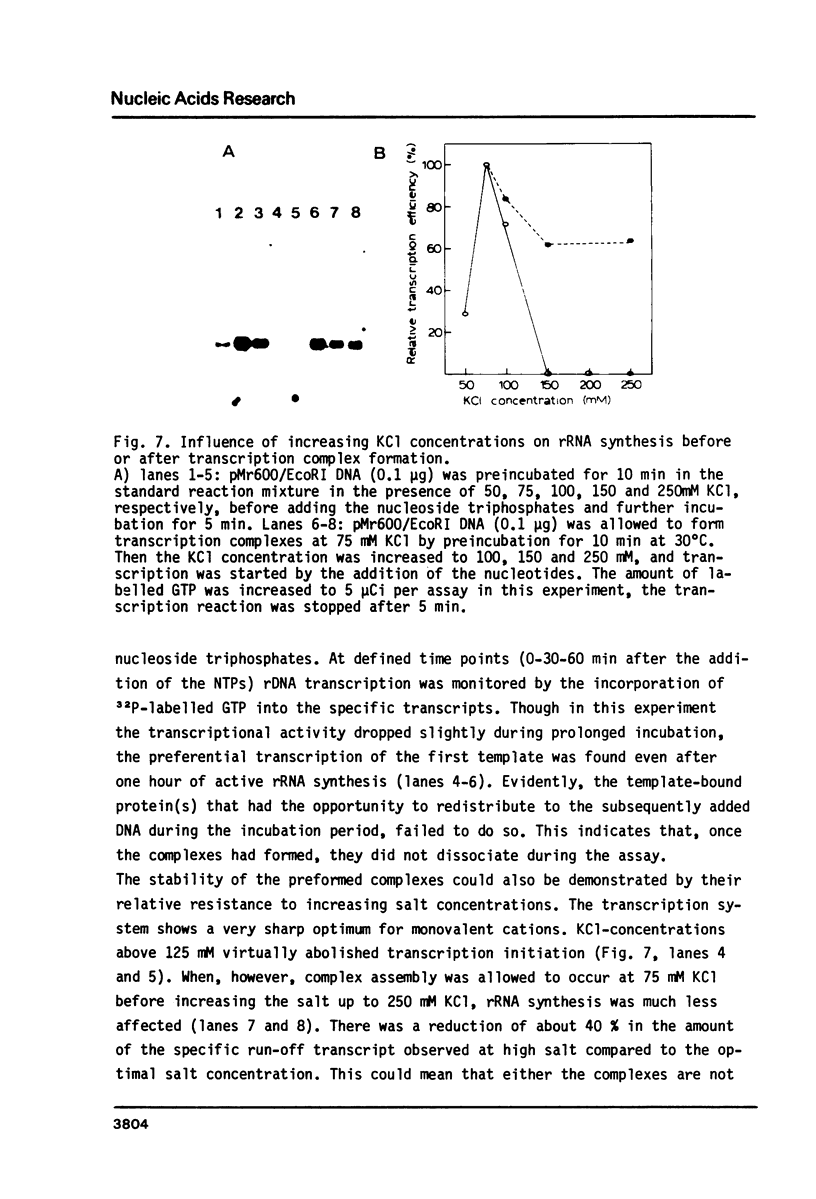
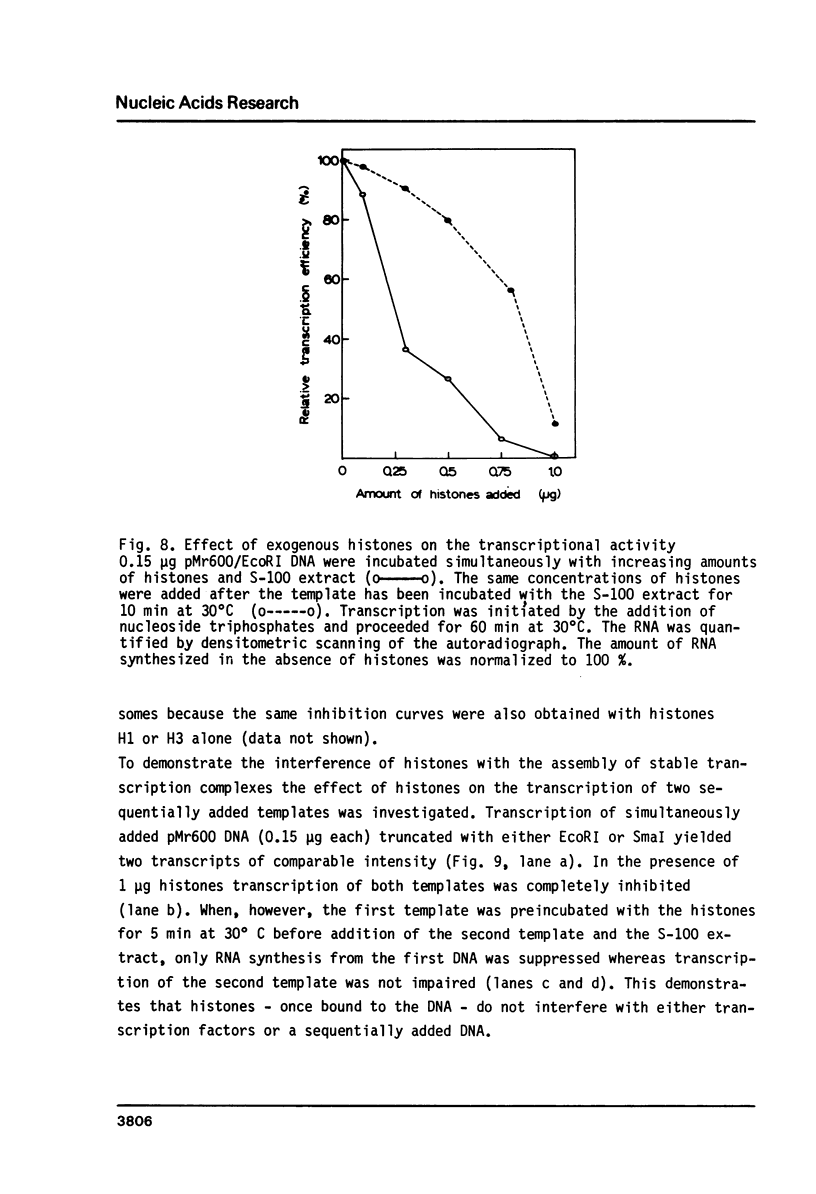
Abstract
Cytoplasmic extracts from cultured mouse cells contain the factor(s) required for specific transcription initiation of rDNA by RNA polymerase I. Prior to transcription the essential proteins bind to the ribosomal gene and remain bound to the template for several rounds of transcription. The assembly of these preinitiation complexes in vitro has been demonstrated by kinetic analysis of the transcription reaction and by competition experiments. Complex formation involves an initial, rapid binding of transcription factor(s) to rDNA sequences followed by additional events which arrange the DNA-protein complex into a transcriptionally active state. Once the complexes have formed they persist for at least 2 hours in vitro and are resistant to elevated salt concentrations. The assembly of the complexes was inhibited when the template DNA was incubated with histones prior to the addition of S-100 extract. If, however, preinitiation complex formation was allowed to occur before the addition of histones, the interference of histones with specific transcription was much less pronounced.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Davison B. L., Egly J. M., Mulvihill E. R., Chambon P. Formation of stable preinitiation complexes between eukaryotic class B transcription factors and promoter sequences. Nature. 1983 Feb 24;301(5902):680–686. doi: 10.1038/301680a0. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Fowlkes D. M., Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980 Nov;22(2 Pt 2):405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- Grummt I. Specific transcription of mouse ribosomal DNA in a cell-free system that mimics control in vivo. Proc Natl Acad Sci U S A. 1981 Feb;78(2):727–731. doi: 10.1073/pnas.78.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R., Brown D. D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Weil P. A., Segall J., Harris B., Ng S. Y., Roeder R. G. Faithful transcription of eukaryotic genes by RNA polymerase III in systems reconstituted with purified DNA templates. J Biol Chem. 1979 Jul 10;254(13):6163–6173. [PubMed] [Google Scholar]