Abstract
Myelination of peripheral nerves by Schwann cells depends upon a gene regulatory network controlled by Egr2/Krox20, which is specifically required for Schwann cells to initiate and maintain myelination. To elucidate the mechanism by which Egr2 regulates gene expression during myelination, we have performed chromatin immunoprecipitation analysis on myelinating rat sciatic nerve in vivo. The resulting samples were applied to a tiled microarray consisting of a broad spectrum of genes that are activated or repressed in Egr2-deficient mice. The results show extensive binding within myelin-associated genes, as well as some genes that become repressed in myelinating Schwann cells. Many of the Egr2 peaks coincide with regions of open chromatin, which is a marker of enhancer regions. In addition, further analysis showed that there is substantial colocalization of Egr2 binding with Sox10, a transcription factor required for Schwann cell specification and other stages of Schwann cell development. Finally, we have found that Egr2 binds to promoters of several lipid biosynthetic genes, which is consistent with their dramatic upregulation during the formation of lipid-rich myelin. Overall, this analysis provides a locus-wide profile of Egr2 binding patterns in major myelin-associated genes using myelinating peripheral nerve.
Keywords: Chromatin, Krox20, Schwann, myelination, ChIP
Introduction
During the postnatal development of the PNS, Schwann cells establish a one-on-one relationship with neurons and initiate the formation of the multilayered membrane structure around large diameter axons known as the myelin sheath (reviewed in Jessen and Mirsky 2005). The integrity of myelin is required for rapid, saltatory nerve conduction of action potentials. Defective formation and maintenance of myelin by Schwann cells is a major cause of human peripheral neuropathies, such as Charcot-Marie-Tooth and Dejerine-Sottas disease (reviewed in Suter and Scherer 2003; Wrabetz et al. 2004). Profiling studies have provided extensive characterization of gene expression changes during the myelination process, including induction of major myelin genes such as Myelin Protein Zero and Connexin 32, as well as genes involved in lipid synthesis to support the synthesis of myelin membrane by Schwann cells (Nagarajan et al. 2002; Verheijen et al. 2003).
To coordinate these gene induction events, peripheral nerve myelination entails intricate networks of transcription factors, including Egr2/Krox20, Oct6/Scip/Pou3f1, Brn2/Pou3f2, Sox10, SREBP factors, NFATc3/c4 and NF-κB (Topilko et al. 1994; Bermingham et al. 1996; Jaegle et al. 1996; Britsch et al. 2001; Jaegle et al. 2003; Nickols et al. 2003; Kao et al. 2009). Although many of these transcription factors are expressed in several cell types, it is thought that combinatorial actions of these transcriptional regulators contribute to tissue-specificity of the transcriptome in myelinating Schwann cells (reviewed in Svaren and Meijer 2008). At least some of these combinatorial interactions involve Egr2/Krox20 (hereafter referred to as Egr2), which is significantly induced at the onset of myelination. Consistent with this, Egr2 null and hypomorphic mice display a severe hypomyelination phenotype in peripheral nerve (Topilko et al. 1994; Le et al. 2005a; Decker et al. 2006), and target gene analysis indicates that Egr2 induces many of the major myelin genes (Nagarajan et al. 2001; Parkinson et al. 2004; Le et al. 2005a). Egr2 is also required for high levels of expression of several lipid biosynthetic genes—including HMG CoA reductase and Stearoyl CoA desaturase (Nagarajan et al. 2001; Le et al. 2005a; LeBlanc et al. 2005). Mutations of the EGR2 gene itself are associated with a spectrum of human peripheral neuropathies (Warner et al. 1998; Bellone et al. 1999; Timmerman et al. 1999; Pareyson et al. 2000; Mikesova et al. 2005). While a large number of genes are altered in Egr2-deficient mice, it is unclear if most of them are directly or indirectly regulated by Egr2.
Several lines of evidence suggest that Egr2 also represses other genes as myelination proceeds. Successful myelination depends upon interaction of Egr2 with the Nab1/2 transcriptional corepressors (Russo et al. 1995; Svaren et al. 1996). Targeted disruption of Nab1/2 causes myelination defects in peripheral nerve (Le et al. 2005b), and similar results have recently been obtained using knockins of Nab-resistant alleles of Egr2 (Desmazieres et al. 2008; Baloh et al. 2009). Accordingly, Egr2 and Nab proteins are required to downregulate genes that become repressed as myelination proceeds, such as Pou3f1/Oct6/Scip, Sox2, and Id2/Id4 (Zorick et al. 1999; Le et al. 2005a; Le et al. 2005b; Mager et al. 2008).
Myelination is coordinated by induction and modulation of specific transcription factors, which bind to cognate elements within dynamically regulated myelin genes. Therefore, such elements constitute the sites where positive and negative effects are integrated to establish the correct expression level of myelin genes. Another critical component of transcriptional regulation in Schwann cells is Sox10, a high mobility group (HMG) box-containing transcription factor required for Schwann cell specification and later stages of Schwann cell development (Kuhlbrodt et al. 1998; Britsch et al. 2001; Schreiner et al. 2007; Finzsch et al. 2010). Binding sites for Egr2 and Sox10 have been identified in a composite configuration in several myelin genes such as Connexin32, Mbp, Mpz, and Mag (Bondurand et al. 2001; Denarier et al. 2005; LeBlanc et al. 2007; Jang and Svaren 2009). However, it remains poorly understood how and to what extent Egr2 regulates expression of its target genes in conjunction with Sox10 during PNS myelination.
To identify Egr2 binding sites, we have developed a method for in vivo chromatin immunoprecipitation (ChIP) on rat sciatic nerve during the peak of PNS myelination (postnatal day 10–15) (Jang et al. 2006). It should be noted that the success of these ChIP assays depends in part on the large proportion of myelinating Schwann cells in sciatic nerve, which is the only cell type in peripheral nerve that expresses Egr2 (Zorick et al. 1996; Topilko et al. 1997). Since standard ChIP assays targeted at selected sites is impractical for efficient discovery of novel sites, we performed a more unbiased screen for Egr2 binding sites by coupling in vivo ChIP on myelinating rat sciatic nerve with a tiled microarray (hereafter referred to as ChIP-chip). The array represents gene sets that are induced or repressed in Egr2-deficient peripheral nerve, many of which have yet to be analyzed for the mechanism of their regulation. The analysis allowed us to test if Egr2-regulated genes are generally associated with Egr2 binding in upstream, intragenic, and downstream regions. Finally, the locus-wide analysis of Egr2 binding sites provided the basis to test if Egr2 is generally colocalized with Sox10 binding in genes that are regulated during myelination.
Materials and Methods
In vivo chromatin immunoprecipitation
In vivo ChIP assays on rat myelinating sciatic nerve were performed on Sprague-Dawley rat pups at postnatal day 15 (P15) as previously described (Jang et al. 2006; Jang and Svaren 2009), except with the omission of herring sperm DNA in the blocking procedure. All experiments on rats were performed in strict accordance with experimental protocols approved by the Institutional Animal Care and Use Committee, University of Wisconsin, School of Veterinary Medicine. The antibodies used in this study include Egr2 antibodies (Abcam cat. no.43020 and Covance cat no. PRB-236P, designated antibodies 1 and 2, respectively), Sox10 (Santa Cruz Biotechnology cat# sc-17342X), and control IgG (normal rabbit IgG: Millipore 12–370 and normal goat IgG: Santa Cruz Biotechnology cat# sc-2028). Following ChIP, quantitative PCR was performed in duplicate to calculate the fold recovery of a given segment relative to the non-specific IgG, using the Comparative Ct method (Livak and Schmittgen 2001). Primer sequences used in this study are available upon request.
ChIP analysis using tiled microarrays
To combine ChIP with microarray analysis, amplicons were first generated from ChIP products by whole genome amplification (Sigma). Labeling of the samples with Cy5 (experimental-Egr2) or Cy3 (control-total input) followed by microarray hybridization was performed as described (Jang and Svaren 2009) by Nimblegen, using a custom microarray designed with isothermic probes staggered by 17 bp representing the selected gene loci listed in Table 1. Complete listing and coordinates of tiled regions in the rn4 genome build are provided in Supplemental Table S1. Gaps in the tiling represent repetitive DNA regions for which unique, optimal probes could not be designed. The enrichment ratio of Cy5 to Cy3 was plotted on a log2 scale and further processed to display a moving average using a window size of 5 probes. Peak finding was performed using the CMARRT (Correlation, Moving Average, Robust and Rapid method on Tiling array) algorithm, which identifies peaks in ChIP-chip data based on a moving average model that incorporates the correlation structure of tiled microarrays (Kuan et al. 2008), with a false discovery rate of 0.05. Displayed peaks are those that are common to the two independent replicates with each antibody. All raw data sets for the custom tiled array are available from the NCBI Gene Expression Omnibus website: accession number: GSE23648.
Table 1.
Egr2-regulated genes present on custom tiled array
| Myelin Genes |
| Connexin 32, Gjb1* |
| N-myc downstream regulated gene, Ndrg1* |
| Periaxin, Prx* |
| Peripheral Myelin Protein 22, Pmp22* |
| Peripheral Myelin Protein 2, Pmp2 |
| Myelin Protein Zero, Mpz* |
| Myelin Basic Protein, Mbp |
| Myelin protein 11 kd, Mp11 |
| Myelin-associated glycoprotein, Mag |
| Mal, myelin and lymphocyte protein |
| Plasmolipin, Pllp |
| Transcription Factors |
| NGFI-A binding protein 1, Nab1 |
| NGFI-A binding protein 2, Nab2 |
| Pou3f1/Scip |
| Pou3f2/Brn2 |
| Inhibitor of differentiation 1, Id1 |
| Inhibitor of differentiation 2, Id2 |
| Inhibitor of differentiation 3, Id3 |
| Inhibitor of differentiation 4, Id4 |
| Sox2 |
| T-box protein 2, Tbx2 |
| c-myc |
| Early Growth Response, Egr1 |
| c-jun |
| Genes mutated in peripheral neuropathies |
| Neurofilament light, Nefl* |
| Sox10* |
| Myotubularin-related protein 2, Mtmr2* |
| Early Growth Response 2, Egr2* |
| Set-binding factor 2, Sbf2/Mtmr13* |
| lipopolysaccharide-ind. TNF factor, Litaf* |
| Signaling |
| Nerve growth factor receptor, Ngfr |
| Mapk8 interacting protein/jip1, Mapk8ip |
| Eph receptor B6, EphB6 |
| Membrane metalloendopeptidase, Mme |
| Desert Hedgehog, Dhh |
| Ras homolog in diabetes, Rad |
| L1 cell adhesion molecule, L1cam |
| Lipid Synthesis/Transport |
| HMG CoA Synthase, Hmgcs1 |
| HMG CoA Reductase, Hmgcr |
| Phosophomevalonate kinase, Pmvk |
| Mevalonate decarboxylase, Mvd |
| Isopentenyl diphosphate isomerase, Idi1 |
| Farnesyl diphosphate synthase, Fdps |
| Farnesyl diphosphate farnesyl trans., Fdft1 |
| Squalene epoxidase, Sqle |
| Lanosterol synthase, LSS |
| Lanosterol demethylase (Cyp51) |
| NADP-dependent steroid dehyd., NSDHL |
| Sterol C5 desaturase, Sc5d |
| Stearoyl CoA desaturase, Scd2 |
| Lipoprotein lipase, Lpl |
| Spot14, Thrsp |
| Malic Enzyme, Me1 |
| LDL receptor, Ldlr |
| UDP glycosyl transferase, Ugt8 |
| Cell Cycle |
| Cdk inhibitor p21, Cdkn1a |
| Cdk inhibitor p57, Cdkn1c |
| Cyclin D1, Ccnd1 |
Genes in bold text are induced in Egr2-deficient peripheral nerve (Le et al. 2005a; Le et al. 2005b), and those in normal text are reduced (with the exception of Nab2). Genes that are mutated in human peripheral neuropathies are marked with an asterisk. Additional genes in this category are listed in the lower left column. A complete listing of tiled genes and the chromosomal coordinates are included in Supplemental Table S1.
In vivo FAIRE
The in vivo FAIRE assay on rat myelinating sciatic nerve was performed at P14 using pooled nerves from 4 rat pups. Freshly dissected sciatic nerves were minced in phosphate-buffered saline (PBS) containing 1% formaldehyde for 5 minutes at room temperature (22°C). The nerves were washed with cold PBS, pooled, and frozen at −8°C. The nerves were thawed and homogenized in buffer (150 mM NaCl, 10% glycerol, 50 mM Tris pH 8.0) containing protease inhibitor cocktail (Sigma, St Louis, MO, USA; 5μl of cocktail per ml of buffer). The cells were then lysed using incubation in three buffers, L1, L2 and L3 as previously described (Giresi and Lieb 2009). After cell lysis, cells were sonicated using the Bioruptor (Diagenode), and extracted with phenol:chloroform as previously described (Giresi and Lieb 2009). The protocol only differed in that the cells were sonicated in 2 ml of Lysis Buffer 3 on high with 30 seconds on followed by 30 seconds off, all on ice. 20% of the crosslinked chromatin was used as an input control, and the samples were reverse crosslinked at 65°C overnight.
Transfection Assays
The B16 (mouse melanoma) cell line was grown and transfected as described (Jones et al. 2007; LeBlanc et al. 2007). The reporter constructs contain the following coordinates from the rat genome (Rn4 build in UCSC Genome Browser), cloned upstream of the pGL4 luciferase reporter. The Ndrg1and Mag constructs also contain a minimal promoter.
Ndrg1 +9.3 kb: chr7:104335109-104335666; Ndrg1 +20.6 kb: chr7:104324019-104324460 c-jun: chr5:115361190-115362027; Id4: chr17:22528145-22528659; mouse Mag: mm9 chr7:31697999-31698377
Sequence analysis
Sequences obtained from rat genomic regions of Egr2 enrichment called as peaks in the ChIP-chip analysis were evaluated by comparison with the previously defined Egr2 binding sites (Nardelli et al. 1992; Swirnoff and Milbrandt 1995), using Transfac matrix M00246, or Egr1 position weight matrices defined recently (Badis et al. 2009). Analysis for composite elements containing Sox10 and Egr2 sites was performed as described using a constructed SOX dimer binding (Jones et al. 2007), allowing for 3–6 bp spacing between inverted repeats of the Sox10 monomeric site.
Results
Design of genomic tiling array with candidate Egr2 target genes
In an attempt to obtain an unbiased survey of Egr2 binding patterns in genes regulated during myelination, we combined in vivo Egr2-ChIP with a high-resolution custom microarray representing a broad representation of genes that are induced or repressed in peripheral nerve of Egr2-deficient mice (Le et al. 2005a; Le et al. 2005b). Overall, these genes are categorized into five functional groups in Table 1. In addition, the array included additional rat orthologues of genes that are mutated in human peripheral neuropathies (e.g. Egr2, Litaf). The custom microarray incorporated tiled probes representing ~50–100 kb windows surrounding the genes of interest in the rat genome. A complete listing of tiled genes and the chromosomal coordinates are included in Supplemental Table S1.
Immunoprecipitations were performed using two independent Egr2 antibodies with sciatic nerve samples obtained from pooled littermate samples (~10–12) at P15. The immunoprecipitations were performed on two independent litters for each antibody. The ChIP samples were labeled and co-hybridized to the tiled microarray along with corresponding input DNA as the control. These data were analyzed by the CMARRT algorithm, which identifies peaks in ChIP-chip data based on a moving average model that incorporates the correlation structure of tiled microarrays (Kuan et al. 2008). All previously identified Egr2 binding sites in Mag, Mbp, Mpz, Nab1, Nab2, Dhh, and Prx genes (Denarier et al. 2005; Jang et al. 2006; Jones et al. 2007; Srinivasan et al. 2007; Jang and Svaren 2009) were identified as peaks in at least 3 out of 4 ChIP-chip analyses. Overall, the degree of overlap between the two Egr2 antibodies was fairly high as 65.5% of the Egr2 Ab.2 peaks overlapped with an Egr2 Ab.1 peak (Table 2). In general, the signal to noise ratio for Ab. 2 was greater than that of Ab. 1, resulting in a larger Ab.2 peak set identified by the CMARRT analysis. Western analysis shows that both antibodies detect Egr2, but Ab. 2 is somewhat more specific (Supplementary Figure S1).
Table 2.
Peak overlap analysis in sciatic nerve ChIP-chip data
| 1st ↓ 2nd → | Egr2-Ab.1 | Egr2-Ab.2 | Sox10 |
|---|---|---|---|
| Egr2-Ab.1 | -- | 65.51% | 32.35% |
| Egr2-Ab.2 | 47.98% | -- | 25.56% |
| Sox10 | 64.39% | 72.68% | -- |
Each of the numbers describes the percentages of peaks within a given dataset listed in the first column that overlap with peaks in an independent data set listed in the first row. For example, 64.39% of the Sox10 peaks were found to overlap with peaks in the Egr2 Ab.1 data set. Each of the peak sets are defined as those peaks that were positive in two independent replicate experiments for that antibody. The minimum required overlap is 1 bp.
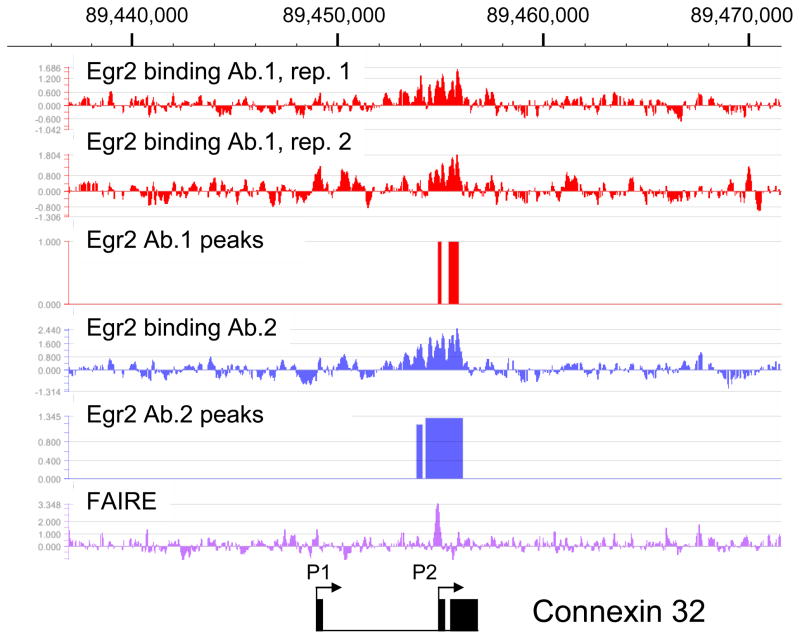
Egr2 binding to the Connexin 32 promoter
Mutations of the human Connexin 32 gene (GJB1, gap junction protein, beta1) cause an X-linked form of Charcot-Marie-Tooth Disease (CMT1X), which is the 2nd most common form of inherited peripheral myelinopathy. Connexin 32 expression is induced during myelination, and its expression in mouse peripheral nerve is dependent upon Egr2 (Le et al. 2005a; Le et al. 2005b). Previous studies of the Connexin 32 gene had identified two alternate promoters, but the P2 promoter is used exclusively in peripheral nerve (Neuhaus et al. 1995). The P2 promoter has binding sites for both Egr2 and Sox10, and this promoter has been shown to respond to these factors in transient transfection assays (Bondurand et al. 2001; Musso et al. 2003). Analysis of the Egr2 ChIP-chip data showed one enriched region of Egr2 binding over the P2 promoter region (Figure 1). In contrast, there does not appear to be any significant binding in the P1 promoter region of the Connexin 32 locus. An independent ChIP-chip assay was performed with a second antibody (Egr2 Ab.2) exhibited essentially the same major peak, although there were some minor differences perhaps reflecting some differences in non-specific binding.
Figure 1. Egr2 binding in the Connexin 32 (Gjb1) locus.
Diagram shows peak data for binding of Egr2 within the rat Gjb1 locus on chromosome X. In vivo ChIP assays were performed on rat sciatic nerve at postnatal day 15 (P15) using two independent antibodies to Egr2. The ChIP samples were labeled with Cy5 (Egr2) or Cy3 (total input DNA) for hybridization to the genomic tiling array. The enrichment ratio of Cy5 to Cy3 was plotted on a log2 scale and further processed to display a 5 point moving average. The top two tracks show replicates of Egr2 antibody 1, and the 4th track shows an independent assay with Egr2 antibody 2. Peak analysis was performed as described in the Methods section at a false discovery rate of <0.05. The bottom track shows analysis of sciatic nerve chromatin using the FAIRE technique to identify disruptions in nucleosomal structure. The two alternate promoters in the Gjb1 locus are shown, along with the 3 exons (filled boxes). The P2 promoter is active in peripheral nerve.
A general characteristic of regulatory elements is that they lie within nucleosome-free regions. To map regions of open chromatin we used FAIRE (formaldehyde-assisted identification of regulatory elements), which identifies regions of DNA that are not efficiently crosslinked to histones. Data from the ENCODE project has shown that FAIRE sites largely colocalize with DNaseI hypersensitive sites (Giresi et al. 2007; Giresi and Lieb 2009). Interestingly, FAIRE analysis of the Connexin 32 locus identified a single major peak that coincides with the previously identified binding sites for Egr2 and Sox10 (Bondurand et al. 2001; Musso et al. 2003), confirming previous reports that this site is a functional enhancer. The FAIRE peak is more narrowly focused over the reported Egr2 binding sites than the ChIP-chip analysis of Egr2 itself. Moreover, this analysis did not identify any additional regulatory elements outside of the P2 promoter region in the 50 kb tiled window surrounding the Connexin 32 gene.
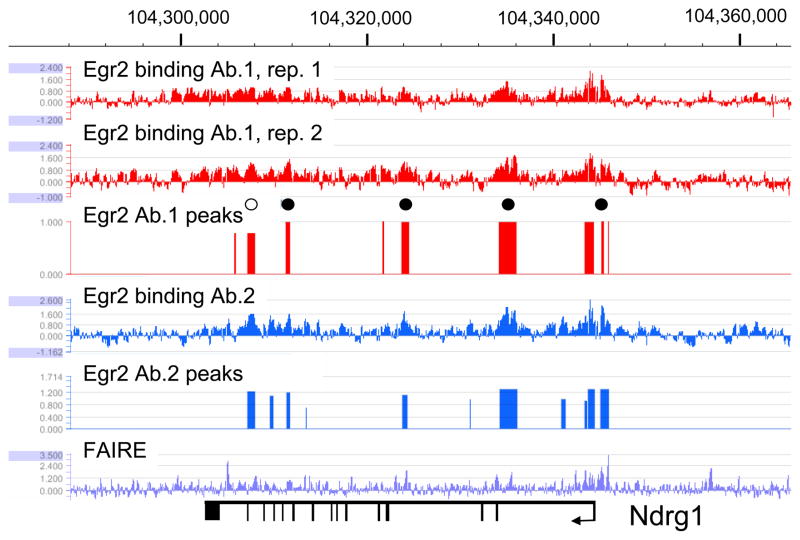
Egr2 regulation of Ndrg1
Figure 2 shows Egr2 binding patterns in the Ndrg1 gene (N-myc downstream regulated gene), which is mutated in a recessive peripheral neuropathy classified as Charcot-Marie-Tooth disease type 4D (Kalaydjieva et al. 1996; Kalaydjieva et al. 1998; Kalaydjieva et al. 2000; Berger et al. 2002). Targeted disruption of this gene leads to progressive demyelination in peripheral nerves, and its expression is dynamically regulated during peripheral myelination (Nagarajan et al. 2002; Verheijen et al. 2003; Okuda et al. 2004). Although Ndrg1 is downregulated in Egr2-deficient mice, the sites of regulation have not yet been identified. The ChIP-chip assay identified several regions of Egr2 enrichment called as peaks throughout the Ndrg1 locus (Fig. 2). One major peak was found in the promoter region, which corresponds to a previously identified Egr1 site (Zhang et al. 2007), and other peaks were found in intron-associated sites. Several peaks correlate with open chromatin as defined by FAIRE analysis (i.e. promoter region, and intronic sites at ~104,324,000 and ~104,335,000). These data provide the first evidence for direct regulation of Ndrg1 by Egr2 during peripheral nerve myelination and are consistent with the observed downregulation of Ndrg1 in Egr2-deficient mice (Le et al. 2005a).
Figure 2. Egr2 is associated with the proximal promoter and intragenic region of the Ndrg1 locus.
In vivo ChIP-chip data for binding of Egr2 to the Ndrg1 gene in rat sciatic nerve at postnatal day 15 (P15) was provided by the experiment described for Figure 1. Chromosome 7 numbering is from the Rn4 genome build, and exons are indicated by vertical lines in the diagram at the bottom. Circles in the diagram indicate selected peaks at which QPCR reactions on independent ChIP samples were used to validate binding (data not shown). Filled circles represent confirmed sites, and open circles indicate sites at which binding above background was not confirmed. Selected peaks were identified in both replicates for each Egr2 antibody.
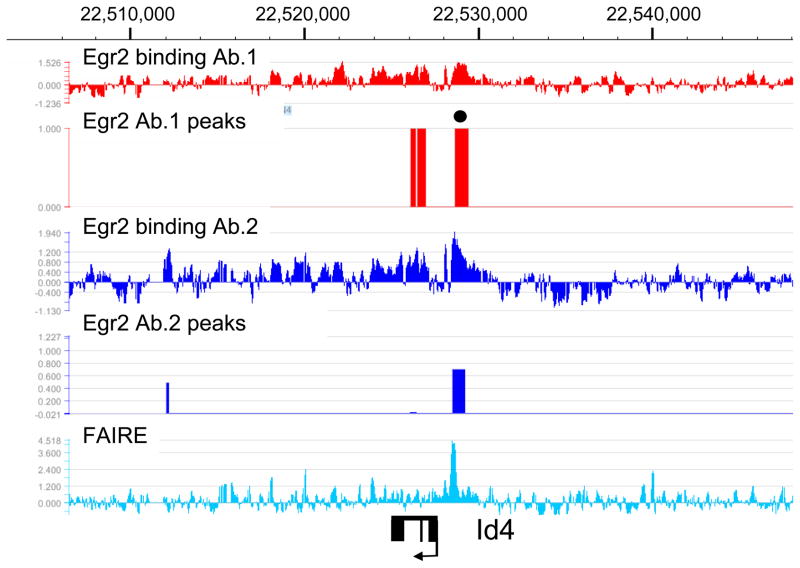
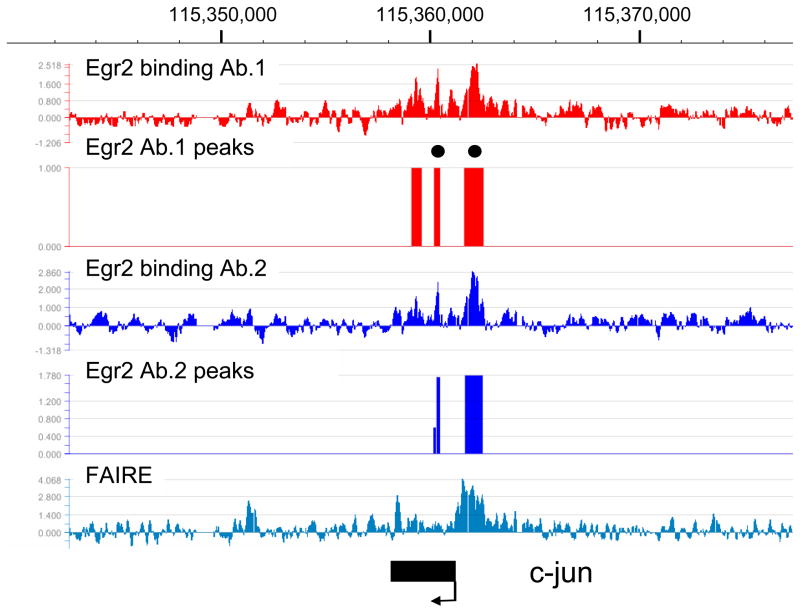
Egr2 binding to genes repressed during myelination
There is an extensive program of gene repression that occurs during the myelination process (reviewed in Jessen and Mirsky 2008). Many of these genes have been shown to antagonize the myelination process, such as Sox2 and c-jun (Parkinson et al. 2004; Le et al. 2005a). Interestingly, many such genes are normally repressed during myelination and become induced in peripheral nerve of mice that are deficient in Egr2/Krox20 (marked as bold text in Table 1), leading to the proposal that Egr2 is directly involved in gene repression (Zorick et al. 1999; Le et al. 2005a; Le et al. 2005b; Decker et al. 2006; Desmazieres et al. 2008; Mager et al. 2008). Development of our ChIP-chip array allowed us to examine locus-wide Egr2 binding patterns in several such genes. The examples shown are from Id4 and c-jun (Figures 3 and 4). Interestingly, binding of Egr2 is observed as a major peak in the promoter regions of both genes, with another notable peak peak within the c-jun coding region. The major binding of Egr2 in the promoters of both genes is observed within the context of open chromatin as determined by the FAIRE analysis. These data are consistent with direct repression of these genes by Egr2 in myelinating Schwann cells.
Figure 3. Egr2 binds to the repressed Id4 gene.
Binding of Egr2 is shown in the Id4 gene on chromosome 17, which is activated in Egr2-deficient mice. Analysis and peak finding were performed as described in Figure 1. One peak (filled circle) was selected for validation by the ChIP-qPCR assay (data not shown). The FAIRE data show one major peak of open chromatin in the Id4 promoter. Exons for Id4 are indicated by filled rectangles.
Figure 4. Egr2 binding in the c-jun locus.
ChIP-chip analysis shows Egr2 binding patterns in the c-jun locus on chromosome 5, which is repressed by Egr2. Analysis and peak finding were performed as described in Figure 1. Two peaks (filled circles) were selected for validated by the ChIP-qPCR assay (data not shown). The diagram at the bottom indicates the position of the single exon constituting the c-jun gene.
Distribution of Egr2 binding within target genes
Of all the Egr2-regulated genes listed in Table 1, all but two of the genes (Nefl and Lpl) had at least one Egr2 peak identified in the tiled region surrounding the gene in at least 3 out of the 4 datasets by CMARRT analysis. We analyzed the distribution of Egr2 binding sites in target gene loci with respect to the transcription start site (Figure 5A). The Egr2 peaks do not exhibit a significant preference for upstream regions with a high proportion of its binding sites found in intragenic regions. This is consistent with our previous analysis of selected genes (Jang et al. 2006; LeBlanc et al. 2006; Jones et al. 2007), in which functional Egr2 binding sites have been found within introns.
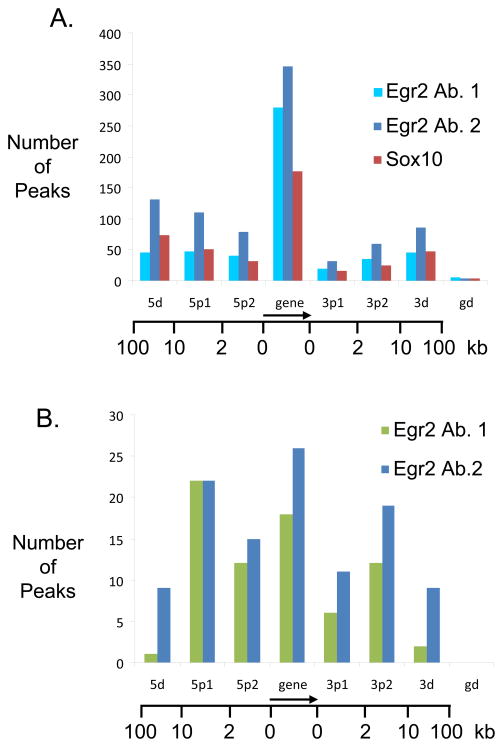
Figure 5. Location profile of Egr2 binding sites.
A. The graph describes location of sites associated with Egr2 and Sox10 with respect to transcription start sites of genes present within the tiled regions of the custom array. Binding sites are classified as 5d (10kb–100kb upstream), 5p1 (2kb–10kb upstream), 5p2 (less than 2kb upstream), gene (any exon or intron), 3p1 (less than 2kb downstream), 3p2 (2kb–10kb downstream), 3d (10kb–100kb downstream), and gd (greater than 100kb).
B. The tiled array contains 15 genes that are elevated in the Egr2-deficient peripheral nerve (bold text in Table 1) and therefore apparently repressed by Egr2 activity. A similar location analysis was performed on this subset of genes as described in panel A.
In the two repressed genes presented in Figures 3 and 4, there was a predominant peak in the promoter region. Overall, Egr2 binding was observed in virtually all of the repressed genes included on the custom microarray. A similar location analysis was therefore performed with the 15 genes on the array that are elevated in Egr2-deficient nerves (Le et al. 2005a; Le et al. 2005b; Mager et al. 2008), and are therefore potentially repressed by Egr2. In contrast to the total set, there seems to be more significant shift to binding to the promoter and upstream regions (Figure 5B), suggesting that repression of genes by Egr2 during myelination may involve more promoter-proximal mechanisms of interaction. However, it remains to be seen if this correlation observed in 15 genes will also be observed in a more genome-wide analysis.
Colocalization of Egr2 and Sox10 in regulated genes
Cooperative regulation by Egr2 and Sox10 has been suggested by protein interaction studies and analysis of myelin gene regulatory elements in which their respective binding sites are juxtaposed (Bondurand et al. 2001; Denarier et al. 2005; Wissmuller et al. 2006; Jones et al. 2007; LeBlanc et al. 2007). In an attempt to construct a more comprehensive integrated map of Egr2 and Sox10 interplay, we performed a similar ChIP-chip analysis of Sox10 binding in P15 rat sciatic nerve. Peak analysis for Sox10 was performed using the CMARRT algorithm as described above, and overlapping peaks of Egr2 and Sox10 binding (found in both replicates) were identified by comparison with the Egr2 data sets. The results of this overlap analysis (requiring overlap of ≥1 bp) are summarized in Table 2. Approximately 65% and 73% of the Sox10 peaks overlapped with the Egr2 Ab.1 and Egr2 Ab.2 peak sets, respectively. The increased percentage overlap with Egr2 Ab. 2 reflects the larger number of peaks found in both replicates using this antibody. Using the somewhat larger Egr2 peak sets as a starting point, approximately 32% (Ab. 1) and 26% (Ab. 2) of the Egr2 peaks overlapped with a Sox10 peak. To assess the statistical significance of the overlap, randomly sampled regions (with matched chromosome and peak width) were analyzed for similar levels of overlap. After 3000 iterations, none of the randomly sampled peak sets attained a similar degree of overlap (i.e. >26%). Overall, these data suggest that the relatively high degree of binding overlap between Egr2 and Sox10 binding is significantly greater than would be expected by chance.
We have recently proposed that many myelin gene regulatory elements contain a conserved composite module of Egr2 and Sox10 binding sites, and that this module may have predictive value in identifying myelin gene regulatory elements (Jones et al. 2007). In particular, this module incorporated an inverted pair of Sox10 binding sites with a 3–6 bp spacing (Jones et al. 2007), based on previous work showing binding of Sox10 dimers to such inverted pairs of sites (Peirano and Wegner 2000). We performed binding site analysis on a high confidence peak set, consisting of 97 overlap regions that were positive in both replicates for Egr2 Ab.1, Egr2 Ab.2, and Sox10 (see Supplemental Table S2 for complete listing). Approximately 88% of these peaks had at least one binding site that conformed (p<0.001) to the defined Egr2 binding site (Swirnoff et al. 1998), and 80% had at least one match to a recently defined matrix for the closely related Egr1 factor (Badis et al. 2009). More than half of the peaks had at least one match to the inverted dimeric Sox10 binding site (p<0.0001). There did not seem to be any apparent preference for a particular spacing between the Sox10 monomeric sites in this peak set. At a lower stringency (p<0.001), all but one of the peaks in this set had at least one match to the Sox10 site, but many of these could be Sox10 monomeric sites, which also are functionally active in myelin gene regulatory elements (Peirano and Wegner 2000). Overall, this analysis identified a number of novel composite elements containing Egr2 and dimeric Sox10 binding sites, some of which are listed in Table 3.
Table 3.
Predicted Composite Binding Sites for Egr2 and Sox10
| Gene | Predicted Sox10 sites | Predicted Egr2 sites |
|---|---|---|
| Ndrg1 (+9.5 kb) | AACAAAGtcagTTTTATA CACAAAGgccaCAGTGAG |
GCGTAGGCG |
| Jun promoter | GAAAATGaagTGTTGTG | GAGGGGGAG |
| Id4 promoter | AACAAGA |
GCGCGGGCG GCGAGGGCG |
| Mapk8ip (+5kb) | AACAAAAggctcTTATGAA | ATGGGGGCA |
Predicted binding sites for Sox10 were identified in comparison to a matrix of inverted dimeric sites (Jones et al. 2007), based on previous characterization of Sox10 binding sites (Peirano and Wegner 2000). Egr2 sites in bold text have been shown previously to bind Egr2 in vitro (Nardelli et al. 1992; Swirnoff and Milbrandt 1995). Predicted binding sites for all overlapping peaks are presented in Supplementary Table 5.
Transfection analysis of Egr2 binding sites
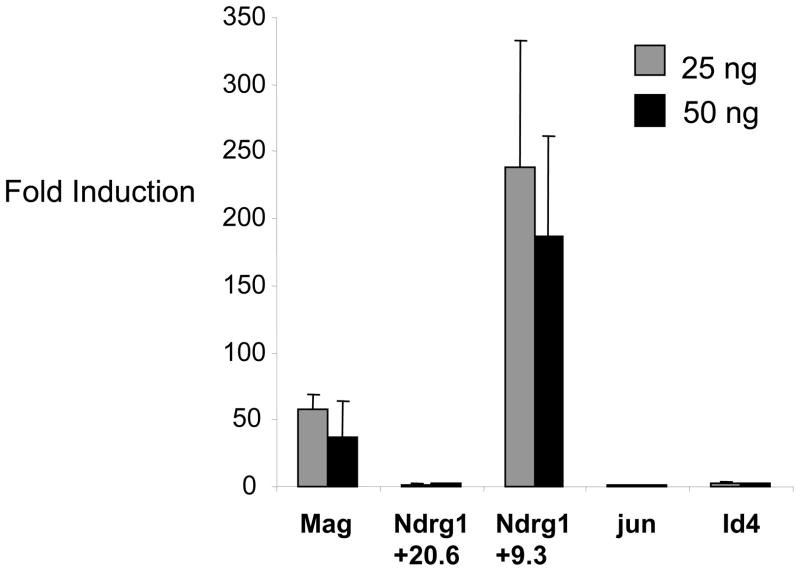
Transient transfection assays have shown that Egr2 can activate the Connexin 32 promoter (Bondurand et al. 2001; Musso et al. 2003), and that the Ndrg1 proximal promoter region is responsive to the closely related Egr1 (Zhang et al. 2007). To explore the function of some of the novel binding sites identified in our analysis, we cloned selected sites upstream of a luciferase reporter gene, and tested their ability to respond to Egr2 expression in transfection assays using the B16 melanoma cell line, which expresses Sox10 (Slutsky et al. 2003). An intronic Egr2 binding site in the Mag gene—first identified in a previous study (Jang et al. 2006)—was strongly activated by Egr2 (Figure 6). Similarly, an intronic binding site in Ndrg1 (+9.3 kb) was activated to a high level, whereas another binding region at +20.6 kb did not respond in this assay. This correlates with an apparently stronger ChIP signal at +9.3 kb as measured by qPCR validation (data not shown). We also tested the two repressed promoters (c-jun and Id4), but we observed only a ~2.8-fold activation of the Id4 promoter by Egr2 in a transfection assay. The c-jun promoter was not activated, but its basal activity was extremely high, indicating that endogenous activators may preclude activation by Egr2 in this assay.
Figure 6. Transfection analysis of novel Egr2 binding sites.
The indicated Egr2 binding sites were cloned upstream of a luciferase reporter gene and were transfected into B16/F10 cells along with increasing amounts (25 or 50 ng) of Egr2. Fold induction was calculated relative to the reporter activity in the absence of Egr2 for each construct. Id4 was activated 2.85- and 2.5-fold, respectively. Means and standard deviations for 6 independent assays are shown (for Mag, n=4).
Egr2 regulation of lipid biosynthesis genes
The peripheral nerve myelin membrane is extremely lipid-rich (70–80% of total dry mass), and the lipid content is unusually rich in cholesterol (composing 20–30% of total lipid, reviewed in Garbay et al. 2000). Interestingly, most of the cholesterol used for myelination is internally synthesized rather than being externally supplied from the circulation (Jurevics and Morell 1994). Egr2 is a convergent point of this coordination as several lipid synthesis genes are downregulated in Egr2-deficient mice (Nagarajan et al. 2001; Le et al. 2005a; LeBlanc et al. 2005). The ChIP-chip analysis revealed Egr2 binding sites in many of the lipid biosynthetic genes, implying that Egr2 directly regulates these genes. We confirmed binding of Egr2 to several of these promoters—HMG CoA Reductase, HMG CoA Synthase, Lanosterol Demethylase, Squalene Epoxidase, and Stearoyl CoA Desaturase—by quantitative PCR analysis of independent ChIP assays (Figure S2). Of the genes involved in lipid synthesis included in our array, 16 out of 17 had at least one Egr2 peak within 1 kb of the transcription start site.
Discussion
Profound peripheral nerve defects in knockout mice (Topilko et al. 1994; Le et al. 2005a; Decker et al. 2006) and the genetic mutations associated with human peripheral neuropathies (Warner et al. 1998; Bellone et al. 1999; Timmerman et al. 1999) have implicated Egr2 as a master regulator of the myelination program in the PNS (reviewed in Svaren and Meijer 2008). Expression profiling studies have yielded an array of genes that are dynamically regulated during myelination by Schwann cells and also sensitive to Egr2 expression (Nagarajan et al. 2001; Nagarajan et al. 2002; Verheijen et al. 2003; Le et al. 2005a; D’Antonio et al. 2006). However, for most of these genes, it is not known if they are directly or indirectly regulated by Egr2. In this study, application of in vivo ChIP to microarray analysis revealed sites of Egr2 binding in a wide range of target genes identified by previous analysis of Egr2-deficient mice.
FAIRE analysis showed that many peaks of Egr2 binding lie within areas of open chromatin, which generally correlates with regulatory regions (Giresi et al. 2007; Giresi and Lieb 2009). Interestingly, many of the peaks of Egr2 binding revealed by ChIP-chip analysis extended over several hundred basepairs (e.g. Figure 1), which may reflect some bridging interactions (through various cofactors) to adjacent DNA segments. In contrast, the FAIRE analysis often provided a more finely focused peak that could be used to more accurately pinpoint Egr2 sites.
There seems to be considerable heterogeneity in the location of Egr2 binding sites. The simplest case is that of the Connexin 32 gene, in which both Egr2 and Sox10 bind to the proximal promoter region of the regulated P2 promoter. Interestingly, Connexin 32 is the only myelin gene in which noncoding promoter mutations have been identified associated with human peripheral neuropathy (Bondurand et al. 2001; Houlden et al. 2004). In contrast, activation of other myelin genes such as Ndrg1 (Figure 2) and Mpz (Jang and Svaren 2009) is associated with multiple distributed Egr2 binding sites, including intragenic regions (often in conserved sites within introns). Based on similar analyses of other transcription factors, it is expected that not all sites are functionally required for regulation of specific genes in Schwann cells. It has been proposed that at least some binding sites may simply reflect the substantial affinity of transcription factors for DNA in general, but that functional sites are bound at quantitatively higher levels (Li et al. 2008). Accordingly, a strong site in the Ndrg1 gene was more highly activated in our tranfection assay compared to another intragenic site with a lower signal in our quantitative PCR validation. However, it is possible that weaker binding sites may still contribute to gene activation in the context of the whole locus.
The ChIP-chip analysis showed that Egr2 binds to the promoters of many of the lipid biosynthesis genes included on this array. Previous studies have indicated that the major regulators for lipid biosynthetic genes—sterol regulatory element binding proteins (SREBPs)—are required for efficient myelination (Verheijen et al. 2003; Verheijen et al. 2009). Since SREBPs generally bind to proximal promoter regions of their target genes (Seo et al. 2009), this may indicate that Egr2 interacts with SREBP factors in these promoters. Consistent with this, some lipid biosynthetic genes are synergistically activated by Egr2 and SREBPs in transfection experiments (LeBlanc et al. 2005). Egr2 has a binding specificity that overlaps to some extent with that of Sp1, which has been reported to bind adjacent to SREBPs on the promoters of many genes involved in lipid metabolism (reviewed in Shimano 2001). Therefore, it is possible that Egr2 utilizes some previously defined Sp1 binding sites to augment the activation of lipid biosynthetic genes to the high levels required for myelination.
Tissue-specific expression of myelin genes in Schwann cells depends upon combinatorial interactions between transcription factors. For example, both Egr2 and Sox10 are present in tissues that do not express myelin genes (e.g. T cells and melanocytes, respectively). Therefore, expression of either of these factors alone is not sufficient for high level expression of myelin genes. We had proposed that a cluster of conserved Egr2 and Sox10 dimeric sites may be a common theme in regulatory elements of developmentally regulated myelin genes in Schwann cells (Jones et al. 2007). The ChIP-chip analysis revealed extensive colocalization of Egr2 with Sox10, and binding site analysis identified putative binding sites for Sox10 in most Egr2 binding peaks. The extensive colocalization of Egr2 with Sox10 at various sites may reflect direct interaction of the two factors (Wissmuller et al. 2006; Jones et al. 2007; LeBlanc et al. 2007).
Many Egr2 target genes are dynamically induced during myelination since they are required for the formation of myelin structure. On the other hand, some genes are shown to be downregulated as myelination proceeds, including Sox2, Id2, Id4, and c-jun (Stewart 1995; Shy et al. 1996; Verheijen et al. 2003; Le et al. 2005a). Based on the elevated levels of these genes in peripheral nerve of Egr2- and NAB-deficient mice, it has been proposed that Egr2 and associated NAB corepressors downregulate their expression (Zorick et al. 1999; Le et al. 2005a; Le et al. 2005b; Decker et al. 2006; Mager et al. 2008). Our previous work showed that Egr2 and its corepressors, Nab1 and Nab2, bind to the Id2 gene in a developmentally-regulated manner as it is being repressed in peripheral nerve (Mager et al. 2008). The ChIP-chip analysis extends these findings by showing Egr2 binds to the promoters of virtually all of the genes (included on our array) that decline as myelination proceeds. It has not been determined how Egr2/Nab complexes repress some genes, while Egr2 activates myelin genes. Interestingly, Sox10 binding was also observed in repressed genes. Sox10 has been shown to be sumoylated on several conserved lysines, which is generally correlated with gene repression (Taylor and Labonne 2005; Girard and Goossens 2006), although further analysis will be required to determine if this plays a role in gene repression during myelination.
Overall, genome-wide mapping of Egr2 binding using ChIP-chip (or ChIP-Seq) in a physiologically relevant system will help elucidate the cognate elements that mediate gene regulation by Egr2 during PNS myelination. Moreover, it is expected that combining such studies with analyses of open chromatin (by DNaseI or FAIRE), cross-species sequence conservation, and colocalization with other transcription factors will accelerate discovery of the regulatory elements required for the developmental regulation of myelination.
Supplementary Material
Acknowledgments
We thank Stephen Johnson, Aseem Ansari, and Nolan Gokey for assistance with ChIP-chip data analysis, Jennifer Abraham for technical assistance, and Richard Quarles for providing the S16 cell line. This work was supported by grants from the National Institutes of Health: HD41590 and its ARRA supplement to JS, HG03747 to SK, and a core grant to the Waisman Center from the National Institute of Child Health and Human Development (P30 HD03352). EJ was supported by the Wayne and Jean Roper Distinguished Fellowship. The authors declare that there is no financial conflict of interest.
Abbreviations
- Egr2
early growth response 2
- FAIRE
formaldehyde-assisted identification of regulatory elements
- ChIP
chromatin immunoprecipitation
- Ndrg1
N-myc downstream regulated gene 1
- Nab
NGFI-A/Egr binding protein
References
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, Kuznetsov H, Wang CF, Coburn D, Newburger DE, Morris Q, Hughes TR, Bulyk ML. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloh RH, Strickland A, Ryu E, Le N, Fahrner T, Yang M, Nagarajan R, Milbrandt J. Congenital hypomyelinating neuropathy with lethal conduction failure in mice carrying the Egr2 I268N mutation. J Neurosci. 2009;29:2312–2321. doi: 10.1523/JNEUROSCI.2168-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellone E, Di Maria E, Soriani S, Varese A, Doria LL, Ajmar F, Mandich P. A novel mutation (D305V) in the early growth response 2 gene is associated with severe Charcot-Marie-Tooth type 1 disease. Hum Mutat. 1999;14:353–354. doi: 10.1002/(SICI)1098-1004(199910)14:4<353::AID-HUMU17>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Berger P, Young P, Suter U. Molecular cell biology of Charcot-Marie-Tooth disease. Neurogenetics. 2002;4:1–15. doi: 10.1007/s10048-002-0130-z. [DOI] [PubMed] [Google Scholar]
- Bermingham JR, Jr, Scherer SS, O’Connell S, Arroyo E, Kalla KA, Powell FL, Rosenfeld MG. Tst-1/Oct-6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes Dev. 1996;10:1751–1762. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- Bondurand N, Girard M, Pingault V, Lemort N, Dubourg O, Goossens M. Human Connexin 32, a gap junction protein altered in the X-linked form of Charcot-Marie-Tooth disease, is directly regulated by the transcription factor SOX10. Hum Mol Genet. 2001;10:2783–2795. doi: 10.1093/hmg/10.24.2783. [DOI] [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Antonio M, Michalovich D, Paterson M, Droggiti A, Woodhoo A, Mirsky R, Jessen KR. Gene profiling and bioinformatic analysis of Schwann cell embryonic development and myelination. Glia. 2006;53:501–515. doi: 10.1002/glia.20309. [DOI] [PubMed] [Google Scholar]
- Decker L, Desmarquet-Trin-Dinh C, Taillebourg E, Ghislain J, Vallat JM, Charnay P. Peripheral myelin maintenance is a dynamic process requiring constant Krox20 expression. J Neurosci. 2006;26:9771–9779. doi: 10.1523/JNEUROSCI.0716-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denarier E, Forghani R, Farhadi HF, Dib S, Dionne N, Friedman HC, Lepage P, Hudson TJ, Drouin R, Peterson A. Functional organization of a Schwann cell enhancer. J Neurosci. 2005;25:11210–11217. doi: 10.1523/JNEUROSCI.2596-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmazieres A, Decker L, Vallat JM, Charnay P, Gilardi-Hebenstreit P. Disruption of Krox20-Nab interaction in the mouse leads to peripheral neuropathy with biphasic evolution. J Neurosci. 2008;28:5891–5900. doi: 10.1523/JNEUROSCI.5187-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzsch M, Schreiner S, Kichko T, Reeh P, Tamm ER, Bosl MR, Meijer D, Wegner M. Sox10 is required for Schwann cell identity and progression beyond the immature Schwann cell stage. J Cell Biol. 2010;189:701–712. doi: 10.1083/jcb.200912142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbay B, Heape AM, Sargueil F, Cassagne C. Myelin synthesis in the peripheral nervous system. Prog Neurobiol. 2000;61:267–304. doi: 10.1016/s0301-0082(99)00049-0. [DOI] [PubMed] [Google Scholar]
- Girard M, Goossens M. Sumoylation of the SOX10 transcription factor regulates its transcriptional activity. FEBS Lett. 2006;580:1635–1641. doi: 10.1016/j.febslet.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlden H, Girard M, Cockerell C, Ingram D, Wood NW, Goossens M, Walker RW, Reilly MM. Connexin 32 promoter P2 mutations: a mechanism of peripheral nerve dysfunction. Ann Neurol. 2004;56:730–734. doi: 10.1002/ana.20267. [DOI] [PubMed] [Google Scholar]
- Jaegle M, Mandemakers W, Broos L, Zwart R, Karis A, Visser P, Grosveld F, Meijer D. The POU factor Oct-6 and Schwann cell differentiation. Science. 1996;273:507–510. doi: 10.1126/science.273.5274.507. [DOI] [PubMed] [Google Scholar]
- Jaegle M, Ghazvini M, Mandemakers W, Piirsoo M, Driegen S, Levavasseur F, Raghoenath S, Grosveld F, Meijer D. The POU proteins Brn-2 and Oct-6 share important functions in Schwann cell development. Genes Dev. 2003;17:1380–1391. doi: 10.1101/gad.258203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SW, Svaren J. Induction of myelin protein zero by early growth response 2 through upstream and intragenic elements. J Biol Chem. 2009;284:20111–20120. doi: 10.1074/jbc.M109.022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SW, LeBlanc SE, Roopra A, Wrabetz L, Svaren J. In vivo detection of Egr2 binding to target genes during peripheral nerve myelination. Journal of Neurochemistry. 2006;98:1678–1687. doi: 10.1111/j.1471-4159.2006.04069.x. [DOI] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci. 2005;6:671–682. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. Negative regulation of myelination: relevance for development, injury, and demyelinating disease. Glia. 2008;56:1552–1565. doi: 10.1002/glia.20761. [DOI] [PubMed] [Google Scholar]
- Jones EA, Jang SW, Mager GM, Chang L-W, Srinivasan R, Gokey NG, Ward RM, Nagarajan R, Svaren J. Interactions of Sox10 and Egr2 in myelin gene regulation. Neuron Glia Biology. 2007;3:377–387. doi: 10.1017/S1740925X08000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurevics HA, Morell P. Sources of cholesterol for kidney and nerve during development. J Lipid Res. 1994;35:112–120. [PubMed] [Google Scholar]
- Kalaydjieva L, Gresham D, Gooding R, Heather L, Baas F, de Jonge R, Blechschmidt K, Angelicheva D, Chandler D, Worsley P, Rosenthal A, King RH, Thomas PK. N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy-Lom. Am J Hum Genet. 2000;67:47–58. doi: 10.1086/302978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaydjieva L, Nikolova A, Turnev I, Petrova J, Hristova A, Ishpekova B, Petkova I, Shmarov A, Stancheva S, Middleton L, Merlini L, Trogu A, Muddle JR, King RH, Thomas PK. Hereditary motor and sensory neuropathy--Lom, a novel demyelinating neuropathy associated with deafness in gypsies. Clinical, electrophysiological and nerve biopsy findings. Brain. 1998;121(Pt 3):399–408. doi: 10.1093/brain/121.3.399. [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, Hallmayer J, Chandler D, Savov A, Nikolova A, Angelicheva D, King RH, Ishpekova B, Honeyman K, Calafell F, Shmarov A, Petrova J, Turnev I, Hristova A, Moskov M, Stancheva S, Petkova I, Bittles AH, Georgieva V, Middleton L, Thomas PK. Gene mapping in Gypsies identifies a novel demyelinating neuropathy on chromosome 8q24. Nat Genet. 1996;14:214–217. doi: 10.1038/ng1096-214. [DOI] [PubMed] [Google Scholar]
- Kao SC, Wu H, Xie J, Chang CP, Ranish JA, Graef IA, Crabtree GR. Calcineurin/NFAT signaling is required for neuregulin-regulated Schwann cell differentiation. Science. 2009;323:651–654. doi: 10.1126/science.1166562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan PF, Chun H, Keles S. CMARRT: a tool for the analysis of ChIP-chip data from tiling arrays by incorporating the correlation structure. Pac Symp Biocomput. 2008:515–526. [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le N, Nagarajan R, Wang JY, Araki T, Schmidt RE, Milbrandt J. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc Natl Acad Sci U S A. 2005a;102:2596–2601. doi: 10.1073/pnas.0407836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le N, Nagarajan R, Wang JY, Svaren J, LaPash C, Araki T, Schmidt RE, Milbrandt J. Nab proteins are essential for peripheral nervous system myelination. Nat Neurosci. 2005b;8:932–940. doi: 10.1038/nn1490. [DOI] [PubMed] [Google Scholar]
- LeBlanc SE, Ward RM, Svaren J. Neuropathy-associated Egr2 mutants disrupt cooperative activation of myelin protein zero by Egr2 and Sox10. Mol Cell Biol. 2007;27:3521–3529. doi: 10.1128/MCB.01689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc SE, Jang SW, Ward RM, Wrabetz L, Svaren J. Direct Regulation of Myelin Protein Zero Expression by the Egr2 Transactivator. J Biol Chem. 2006;281:5453–5460. doi: 10.1074/jbc.M512159200. [DOI] [PubMed] [Google Scholar]
- LeBlanc SE, Srinivasan R, Ferri C, Mager GM, Gillian-Daniel AL, Wrabetz L, Svaren J. Regulation of cholesterol/lipid biosynthetic genes by Egr2/Krox20 during peripheral nerve myelination. J Neurochem. 2005;93:737–748. doi: 10.1111/j.1471-4159.2005.03056.x. [DOI] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, Chu HC, Ogawa N, Inwood W, Sementchenko V, Beaton A, Weiszmann R, Celniker SE, Knowles DW, Gingeras T, Speed TP, Eisen MB, Biggin MD. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real- time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mager GM, Ward RM, Srinivasan R, Jang SW, Wrabetz L, Svaren J. Active gene repression by the EGR2/NAB complex during peripheral nerve myelination. J Biol Chem. 2008;283:18187–18197. doi: 10.1074/jbc.M803330200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikesova E, Huhne K, Rautenstrauss B, Mazanec R, Barankova L, Vyhnalek M, Horacek O, Seeman P. Novel EGR2 mutation R359Q is associated with CMT type 1 and progressive scoliosis. Neuromuscul Disord. 2005;15:764–767. doi: 10.1016/j.nmd.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Musso M, Balestra P, Taroni F, Bellone E, Mandich P. Different consequences of EGR2 mutants on the transactivation of human Cx32 promoter. Neurobiol Dis. 2003;12:89–95. doi: 10.1016/s0969-9961(02)00018-9. [DOI] [PubMed] [Google Scholar]
- Nagarajan R, Le N, Mahoney H, Araki T, Milbrandt J. Deciphering peripheral nerve myelination by using Schwann cell expression profiling. Proc Natl Acad Sci U S A. 2002;99:8998–9003. doi: 10.1073/pnas.132080999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R, Svaren J, Le N, Araki T, Watson M, Milbrandt J. EGR2 mutations in inherited neuropathies dominant-negatively inhibit myelin gene expression. Neuron. 2001;30:355–368. doi: 10.1016/s0896-6273(01)00282-3. [DOI] [PubMed] [Google Scholar]
- Nardelli J, Gibson T, Charnay P. Zinc finger-DNA recognition: analysis of base specificity by site-directed mutagenesis. Nucleic Acids Res. 1992;20:4137–4144. doi: 10.1093/nar/20.16.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus IM, Dahl G, Werner R. Use of alternate promoters for tissue-specific expression of the gene coding for connexin32. Gene. 1995;158:257–262. doi: 10.1016/0378-1119(94)00899-4. [DOI] [PubMed] [Google Scholar]
- Nickols JC, Valentine W, Kanwal S, Carter BD. Activation of the transcription factor NF-kappaB in Schwann cells is required for peripheral myelin formation. Nat Neurosci. 2003;6:161–167. doi: 10.1038/nn995. [DOI] [PubMed] [Google Scholar]
- Okuda T, Higashi Y, Kokame K, Tanaka C, Kondoh H, Miyata T. Ndrg1-deficient mice exhibit a progressive demyelinating disorder of peripheral nerves. Mol Cell Biol. 2004;24:3949–3956. doi: 10.1128/MCB.24.9.3949-3956.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareyson D, Taroni F, Botti S, Morbin M, Baratta S, Lauria G, Ciano C, Sghirlanzoni A. Cranial nerve involvement in CMT disease type 1 due to early growth response 2 gene mutation. Neurology. 2000;54:1696–1698. doi: 10.1212/wnl.54.8.1696. [DOI] [PubMed] [Google Scholar]
- Parkinson DB, Bhaskaran A, Droggiti A, Dickinson S, D’Antonio M, Mirsky R, Jessen KR. Krox-20 inhibits Jun-NH2-terminal kinase/c-Jun to control Schwann cell proliferation and death. J Cell Biol. 2004;164:385–394. doi: 10.1083/jcb.200307132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirano RI, Wegner M. The glial transcription factor Sox10 binds to DNA both as monomer and dimer with different functional consequences. Nucleic Acids Res. 2000;28:3047–3055. doi: 10.1093/nar/28.16.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo MW, Sevetson BR, Milbrandt J. Identification of NAB1, a repressor of NGFI-A and Krox20 mediated transcription. Proc Natl Acad Sci(USA) 1995;92:6873–6877. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner S, Cossais F, Fischer K, Scholz S, Bosl MR, Holtmann B, Sendtner M, Wegner M. Hypomorphic Sox10 alleles reveal novel protein functions and unravel developmental differences in glial lineages. Development. 2007;134:3271–3281. doi: 10.1242/dev.003350. [DOI] [PubMed] [Google Scholar]
- Seo YK, Chong HK, Infante AM, Im SS, Xie X, Osborne TF. Genome-wide analysis of SREBP-1 binding in mouse liver chromatin reveals a preference for promoter proximal binding to a new motif. Proc Natl Acad Sci U S A. 2009;106:13765–13769. doi: 10.1073/pnas.0904246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H. Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog Lipid Res. 2001;40:439–452. doi: 10.1016/s0163-7827(01)00010-8. [DOI] [PubMed] [Google Scholar]
- Shy ME, Shi Y, Wrabetz L, Kamholz J, Scherer SS. Axon-Schwann cell interactions regulate the expression of c-jun in Schwann cells. J Neurosci Res. 1996;43:511–525. doi: 10.1002/(SICI)1097-4547(19960301)43:5<511::AID-JNR1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Slutsky SG, Kamaraju AK, Levy AM, Chebath J, Revel M. Activation of myelin genes during transdifferentiation from melanoma to glial cell phenotype. J Biol Chem. 2003;278:8960–8968. doi: 10.1074/jbc.m210569200. [DOI] [PubMed] [Google Scholar]
- Srinivasan R, Jang SW, Ward RM, Sachdev S, Ezashi T, Svaren J. Differential regulation of NAB corepressor genes in Schwann cells. BMC Mol Biol. 2007;8:117. doi: 10.1186/1471-2199-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart HJ. Expression of c-Jun, Jun B, Jun D and cAMP response element binding protein by Schwann cells and their precursors in vivo and in vitro. Eur J Neurosci. 1995;7:1366–1375. doi: 10.1111/j.1460-9568.1995.tb01128.x. [DOI] [PubMed] [Google Scholar]
- Suter U, Scherer SS. Disease mechanisms in inherited neuropathies. Nat Rev Neurosci. 2003;4:714–726. doi: 10.1038/nrn1196. [DOI] [PubMed] [Google Scholar]
- Svaren J, Meijer D. The molecular machinery of myelin gene transcription in Schwann cells. Glia. 2008;56:1541–1551. doi: 10.1002/glia.20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J, Sevetson BR, Apel ED, Zimonjic DB, Popescu NC, Milbrandt J. NAB2, a Corepressor of NGFI-A (Egr-1) and Krox20, Is Induced by Proliferative and Differentiative Stimuli. Molecular and Cellular Biology. 1996;16:3545–3553. doi: 10.1128/mcb.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swirnoff AH, Milbrandt J. DNA-binding specificity of NGFI-A and related zinc finger transcription factors. Mol Cell Biol. 1995;15:2275–2287. doi: 10.1128/mcb.15.4.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swirnoff AH, Apel ED, Svaren J, Sevetson BR, Zimonjic DB, Popescu NC, Milbrandt J. Nab1, a corepressor of NGFI-A (Egr-1), contains an active transcriptional repression domain. Mol Cell Biol. 1998;18:512–524. doi: 10.1128/mcb.18.1.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KM, Labonne C. SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev Cell. 2005;9:593–603. doi: 10.1016/j.devcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Timmerman V, De Jonghe P, Ceuterick C, De Vriendt E, Lofgren A, Nelis E, Warner LE, Lupski JR, Martin JJ, Van Broeckhoven C. Novel missense mutation in the early growth response 2 gene associated with Dejerine-Sottas syndrome phenotype. Neurology. 1999;52:1827–1832. doi: 10.1212/wnl.52.9.1827. [DOI] [PubMed] [Google Scholar]
- Topilko P, Levi G, Merlo G, Mantero S, Desmarquet C, Mancardi G, Charnay P. Differential regulation of the zinc finger genes Krox-20 and Krox-24 (Egr-1) suggests antagonistic roles in Schwann cells. J Neurosci Res. 1997;50:702–712. doi: 10.1002/(SICI)1097-4547(19971201)50:5<702::AID-JNR7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Topilko P, Schneider-Maunoury S, Levi G, Baron-Van Evercooren A, Chennoufi AB, Seitanidou T, Babinet C, Charnay P. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- Verheijen MH, Chrast R, Burrola P, Lemke G. Local regulation of fat metabolism in peripheral nerves. Genes Dev. 2003;17:2450–2464. doi: 10.1101/gad.1116203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheijen MH, Camargo N, Verdier V, Nadra K, de Preux Charles AS, Medard JJ, Luoma A, Crowther M, Inouye H, Shimano H, Chen S, Brouwers JF, Helms JB, Feltri ML, Wrabetz L, Kirschner D, Chrast R, Smit AB. SCAP is required for timely and proper myelin membrane synthesis. Proc Natl Acad Sci U S A. 2009;106:21383–21388. doi: 10.1073/pnas.0905633106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner LE, Mancias P, Butler IJ, McDonald CM, Keppen L, Koob KG, Lupski JR. Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat Genet. 1998;18:382–384. doi: 10.1038/ng0498-382. [DOI] [PubMed] [Google Scholar]
- Wissmuller S, Kosian T, Wolf M, Finzsch M, Wegner M. The high-mobility-group domain of Sox proteins interacts with DNA-binding domains of many transcription factors. Nucleic Acids Res. 2006;34:1735–1744. doi: 10.1093/nar/gkl105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrabetz L, Feltri ML, Kleopa KA, Scherer SS. Inherited Neuropathies: Clinical, Genetic and Biological Features. In: Lazzarini RA, editor. Myelin Biology and Disorders. Elsevier Academic Press; San Diego: 2004. pp. 905–951. [Google Scholar]
- Zhang P, Tchou-Wong KM, Costa M. Egr-1 mediates hypoxia-inducible transcription of the NDRG1 gene through an overlapping Egr-1/Sp1 binding site in the promoter. Cancer Res. 2007;67:9125–9133. doi: 10.1158/0008-5472.CAN-07-1525. [DOI] [PubMed] [Google Scholar]
- Zorick TS, Syroid DE, Arroyo E, Scherer SS, Lemke G. The Transcription Factors SCIP and Krox-20 Mark Distinct Stages and Cell Fates in Schwann Cell Differentiation. Mol Cell Neurosci. 1996;8:129–145. doi: 10.1006/mcne.1996.0052. [DOI] [PubMed] [Google Scholar]
- Zorick TS, Syroid DE, Brown A, Gridley T, Lemke G. Krox-20 controls SCIP expression, cell cycle exit and susceptibility to apoptosis in developing myelinating Schwann cells. Development. 1999;126:1397–1406. doi: 10.1242/dev.126.7.1397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.